Abstract
This study was conducted to determine the species distribution, antimicrobial resistance pheno- and genotypes and virulence traits of mannitol-positive methicillin-resistant staphylococci (MRS) isolated from pigs in Nsukka agricultural zone, Nigeria. Twenty mannitol-positive methicillin-resistant coagulase-negative staphylococcal (MRCoNS) strains harboring the mecA gene were detected among the 64 Staphylococcus isolates from 291 pigs. A total of 4 species were identified among the MRCoNS isolates, namely, Staphylococcus sciuri (10 strains), Staphylococcus lentus (6 strains), Staphylococcus cohnii (3 strains) and Staphylococcus haemolyticus (one strain). All MRCoNS isolates were multidrug-resistant. In addition to β-lactams, the strains were resistant to fusidic acid (85%), tetracycline (75%), streptomycin (65%), ciprofloxacin (65%), and trimethoprim/sulphamethoxazole (60%). In addition to the mecA and blaZ genes, other antimicrobial resistance genes detected were tet(K), tet(M), tet(L), erm(B), erm(C), aacA-aphD, aphA3, str, dfrK, dfrG, cat pC221, and cat pC223. Thirteen isolates were found to be ciprofloxacin-resistant, and all harbored a Ser84Leu mutation within the QRDR of the GyrA protein, with 3 isolates showing 2 extra substitutions, Ser98Ile and Arg100Lys (one strain) and Glu88Asp and Asp96Thr (2 strains). A phylogenetic tree of the QRDR nucleotide sequences in the gyrA gene revealed a high nucleotide diversity, with several major clusters not associated with the bacterial species. Our study highlights the possibility of transfer of mecA and other antimicrobial resistance genes from MRCoNS to pathogenic bacteria, which is a serious public health and veterinary concern.
Keywords: methicillin-resistant, mannitol-fermenting, staphylococci, quinolone resistance, pigs
Introduction
Staphylococci are Gram-positive bacteria, and they are classified into two groups, coagulase-positive (CoPS) and coagulase-negative (CoNS), based on their ability to produce the enzyme coagulase (Bergeron et al., 2011). Staphylococcus aureus and Staphylococcus pseudintermedius are the most important species in the CoPS group as they are major pathogens for both humans and animals, especially S. aureus. Although CoNS are saprophytic and rarely pathogenic (Kloos and Bannerman, 1994), multidrug-resistant (MDR) strains have been associated with severe cases of difficult to treat infections, especially in immunocompromised individuals (Zell et al., 2008).
Methicillin-resistant staphylococci (MRS) are among the most important bacteria in both human and veterinary medicine and of major clinical, public health and economic concern (Kolar et al., 2010). The problem is aggravated by the fact that MRS, in addition to β-lactam antibiotics, are commonly resistant to other classes of antimicrobial agents including aminoglycosides, macrolides, phenicols, tetracyclines and fluoroquinolones (Lee, 2003; Khanna et al., 2008). Methicillin resistance is conferred by the mecA gene which encodes an altered penicillin-binding protein (PBP2a or PBP2’) with a low affinity for β-lactam antimicrobials (Weese, 2010).
Methicillin-resistant S. aureus (MRSA) colonization in pigs was first reported in the Netherlands in 2005 (Voss et al., 2005). Since then, MRSA, particularly strains of lineage ST398, have been isolated from livestock, especially pigs, in several countries in Europe, America and Asia (Khanna et al., 2008; Denis et al., 2009; Wagenaar et al., 2009; Baba et al., 2010; Golding et al., 2010; Gomez-Sanz et al., 2010; Graveland et al., 2011; Lin et al., 2011; Larsen et al., 2012; Oppliger et al., 2012). Recently, Chah et al. (2014) characterized methicillin-resistant CoNS from dogs in Nsukka, Nigeria. There are, however, no documented reports on the characterization of methicillin-resistant staphylococci from pigs in Nigeria. Thus, this study was conducted to gain insight into the species distribution, antimicrobial resistance pheno- and genotypes and virulence traits of mannitol-positive MRS from intensively reared pigs in the Nsukka agricultural zone, Nigeria.
Materials and Methods
Bacterial strains
Sixty-four staphylococcal strains isolated from nasal and ear swabs of 291 pigs from 16 farms in the Nsukka agricultural zone, Enugu State, Nigeria, were used in the study. Informed consent of each pig owner was obtained prior to sample collection. Stocked cultures of the isolates were sub-cultured on nutrient agar and confirmed as staphylococci based on their microscopic and biochemical characteristics (Gram-positive cocci in bunches and catalase-positive). Each confirmed Staphylococcus isolate was cultured on oxacillin resistance screening agar base (ORSAB) (Oxoid) supplemented with 2 μg/mL oxacillin (Oxoid) and incubated at 37 °C for 24–48 hours. Blue colonies (mannitol-positive isolates) were picked from ORSAB and sub-cultured on brain heart infusion (BHI) agar (Oxoid). Cultures on BHI agar were tested for coagulase and DNAse activity. Oxacillin/methicillin-resistant mannitol-positive isolates were identified to the species level by PCR amplification and sequencing of the sodA and 16S rRNA genes (Mellmann et al., 2006). Detection of the mecA gene was carried out by PCR using specific primers (Al-Talib et al., 2009).
Pheno- and genotype of the mannitol-positive methicillin-resistant staphylococci
Mannitol-positive methicillin-resistant isolates were tested for susceptibility to 16 antimicrobial agents using the disk diffusion method, and the interpretive criteria were as specified by the Clinical and Laboratory Standards Institute (CLSI, 2014) guidelines. The antimicrobial agents tested were penicillin (P), oxacillin (OX), cefoxitin (FOX), erythromycin (E), clindamycin (CC), gentamicin (GM), kanamycin (K), streptomycin (S), tobramycin (NN), tetracycline (T), trimethoprim/sulphamethoxazole (SXT), chloramphenicol (C), ciprofloxacin (CIP), mupirocin (MUP), fusidic acid (FUS), and linezolid (LIN). The breakpoints for fusidic acid/mupirocin and streptomycin were as recommended by EUCAST (http://www.eucast.org) and the Societé Francaise de Microbiologie (www.sfm.asso.fr), respectively. A double-disk diffusion test (D-test) was performed on the isolates to detect inducible clindamycin resistance. Detection of antimicrobial resistance genes (with the exception of that for fusidic acid) was performed by specific PCRs (Gomez-Sanz et al., 2010). Positive and negative controls from the collection of the University of La Rioja were used in each reaction.
Molecular basis for quinolone resistance
Ciprofloxacin-resistant isolates were evaluated by amplification and sequencing of the quinolone resistance-determining region (QRDR) of the DNA gyrase A subunit gene gyrA between positions equivalent to Ala-68 and Gln-107 (Takahata et al., 1997), using primers gyrA_157-fw (5′-TTAAATGAACAAGGTATGAC-3′) and gyrA_539-fw (5′-GCCATACCTACCGCGATACC-3′) (Takahata et al., 1997). The phylogenetic relationships between the QRDR nucleotide sequences (nt 202–321 of gyrA) of all ciprofloxacin-resistant isolates were investigated by constructing a maximum likelihood phylogenetic tree using the SeaView program version 4.4.0. The QRDR nucleotide sequences of representative strains of quinolone-susceptible S. aureus (Genbank accession no. CP000253), S. pseudintermedius (Genbank accession no. AM262968), Staphylococcus epidermidis (Genbank accession no. AF127634), and Staphylococcus haemolyticus (Genbank accession no. AY341071) were included in the analysis for comparison purposes. The sequences were aligned using MUSCLE, and the trees were constructed with PhyML using the general time reversible (GTR) model.
Detection of virulence genes
The presence of the eta, etb, etc, and etdgenes encoding for exfoliative toxins was assessed by PCR (Chen et al., 2007). Genes involved in biofilm formation, icaA, icaB, icaC, icaD, icaR and IS256, as well as bap, were also investigated by specific PCRs as previously described (Ziebuhr et al., 1999; Cucarella et al., 2001).
Results
Methicillin-resistant Staphylococcus species
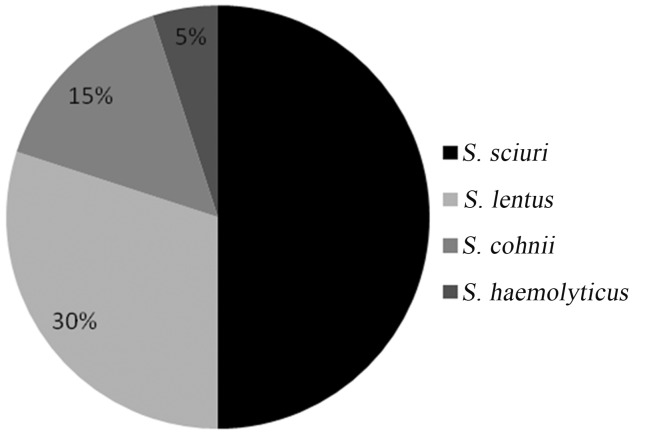
Out of the 64 Staphylococcus strains, 20 were able to grow on ORSAB supplemented with oxacillin and formed blue colonies, thus indicating mannitol fermentation. All 20 strains were negative in the coagulase and DNAse tests and harbored the mecA gene. Thus, the strains were all methicillin-resistant coagulase-negative staphylococci (MRCoNS). These mannitol-positive MRCoNS belonged to 4 species, namely, S. sciuri (10 strains), S. lentus (6 strains), S. cohnii (3 strains) and S. haemolyticus(one strain) (Figure 1).
Figure 1. Percentage distribution of mannitol-positive MRCoNS species from pigs in Nsukka agricultural zone, Nigeria.
Antimicrobial resistance phenotypes of the MRCoNS strains
Seventeen (85%) of the 20 MRCoNS strains were resistant to fusidic acid, while resistance to tetracycline, streptomycin and ciprofloxacin was shown by 15 (75%), 13 (65%) and 13 (65%) strains, respectively (Table 1). Five (25%) of the isolates expressed inducible clindamycin resistance as revealed by the typical D-shaped halo around a clindamycin disc, while the rest exhibited constitutive resistance to clindamycin. None of the isolates was resistant to mupirocin or linezolid. Sixteen different resistance patterns were found among the 20 MRCoNS strains, with P-OX-FOX-E-CCind-S-T-SXT-CIP-FUS being the predominant pattern (4/20). All the isolates were MDR as they displayed resistance to at least 3 classes of antimicrobial agents (Table 2).
Table 1. Antimicrobial resistance profile of mannitol-positive MRCoNS from pigs in Nsukka Agricultural Zone, Nigeria (n = 20).
| Antimicrobial agent (potency) | No. (%) of resistant isolates |
|---|---|
| Oxacillin (1 μg) | 20 (100) |
| Cefoxitin (30 μg) | 20 (100) |
| Penicillin (10 units) | 20 (100) |
| Fusidic acid (10 μg) | 17 (85) |
| Tetracycline (30 μg) | 15 (75) |
| Streptomycin (10 μg) | 13 (65) |
| Ciprofloxacin (5 μg) | 13 (65) |
| Trimethoprim/Sulphamethoxazole (25 μg) | 12 (60) |
| Clindamycin (2 μg) | 10 (50) |
| Erythromycin (15 μg) | 10 (50) |
| Gentamicin (10 μg) | 9 (45) |
| Kanamycin (30 μg) | 9 (45) |
| Tobramycin (10 μg) | 4 (20) |
| Chloramphenicol (30 μg) | 3 (12.5) |
| Mupirocin (200 μg) | 0 (0.0) |
| Linezolid (30 μg) | 0 (0.0) |
Table 2. Species and antimicrobial resistant pheno- and genotypes of mannitol-positive methicillin-resistant coagulase-negative staphylococci (MRCoNS) isolated from pigs in Nsukka, Nigeria.
| Strain No. | Staphylococcusspecies | Resistance phenotypea | Resistance genes detected |
|---|---|---|---|
| C4019 | S. sciuri | P-OX-FOX-E-CCind b-S-T-SXT-CIP-FUS | mecA, erm(C), str, tet(M), dfrG |
| C4017 | S. lentus | P-OX-FOX-S-T-SXT | mecA, str, dfrG |
| C4020 | S. sciuri | P-OX-FOX-SXT-CCi-C-FUS | mecA, dfr(G), cat pC221 |
| C3985 | S. sciuri | P-OX-FOX-E-CC-GM-K-S-T-SXT-FUS | mecA, erm(C), aacA/aphD, str, tet(K), tet(M) dfrG |
| C3986 | S. lentus | P-OX-FOX-S-T-SXT-FUS | mecA, str, dfrG |
| C4011 | S. sciuri | P-OX-FOX-E-CCind-S-T-SXT-CIP-FUS | mecA, erm(C), str, tet(M), dfrG |
| C4012 | S. cohnii | P-OX-FOX-E-CCind-GM-K-S-T-FUS | mecA, blaZ, erm(C), aacA/aphD, aphA3, str, tet(K) |
| C3988 | S. lentus | P-OX-FOX-E-CC-GM-NN-K-S-T-SXT-CIP-FUS | mecA, erm(B), aacA/aphD, str, tet(K), tet(M), [tet(L)-dfrK]c |
| C3989 | S. lentus | P-OX-FOX-E-CC-GM-NN-K-S-T-SXT-CIP-FUS | mecA, erm(B), aacA/aphD, str, tet(K), tet(M), [tet(L)-dfrK]c |
| C3991 | S. cohnii | P-OX-FOX-CCind-CIP-FUS | mecA |
| C3992 | S. sciuri | P-OX-FOX-GM-NN-K-CIP-FUS | mecA, blaZ, aacA/aphD |
| C4015 | S. cohnii | P-OX-FOX-E-CC-GM-K-S-T-SXT-CIP-FUS | mecA, erm(B), aacA/aphD, str, tet(K), tet(M), [tet(L)-dfrK]c |
| C4024 | S. sciuri | P-OX-FOX-T-FUS | mecA, tet(K) |
| C4001 | S. sciuri | P-OX-FOX-E-CCind-S-T-SXT-CIP-FUS | mecA, erm(C), str, tet(M), dfrG |
| C4002 | S. haemolyticus | P-OX-FOX-E-CCind-S-T-SXT-CIP-FUS | mecA, erm(C), str, tet(M), dfrG |
| C4005 | S. lentus | P-OX-FOX-GM-K-C-CIP-SXT | mecA, blaZ, aacA/aphD, aphA3, dfrG, cat pC223 |
| C3997 | S. sciuri | P-OX-FOX-GM-K-C-CIP | mecA, blaZ, aacA/aphD, aphA3, catpC223 |
| C4022 | S. lentus | P-OX-FOX-S-T-CIP-FUS | mecA, str, tet(K), tet(M) |
| C3998 | S. sciuri | P-OX-FOX-T-FUS | mecA, tet(K) |
| C4028 | S. sciuri | P-OX-FOX-E-CC-GM-NN-K-S-T-CIP-FUS | mecA, erm(B), aacA/aphD, aphA3, str, tet(K), tet(M) |
OX, oxacillin; FOX, cefoxitin; P, penicillin; FUS, fusidic acid; T, tetracycline; S, streptomycin; CC, clindamycin; CIP, ciprofloxacin; SXT, sulphamethoxazole-trimethoprin; E, erythromycin; K, kanamycin; GM, gentamycin; NN, tobramycin; C, chloramphenicol; VAN, vancomycin; LIN, linezolid; MUP, mupirocin.
CCind, Inducible clindamycin resistance.
Genes proved to be physically link.
Antimicrobial resistance and virulence genes detected
The antimicrobial resistance genes detected in the MRCoNS strains are presented in Table 2. The mecAand blaZ genes mediating β-lactam resistance were detected in all 20 and 4 MRCoNS strains, respectively. Tetracycline resistance was found to be mediated mainly by tet(K) alone (3 strains) or in combination with tet(M), or tet(M) and tet(L) (3 strains each). Two tetracycline-resistant strains lacked any of the genes encoding tetracycline resistance. Various combinations of the aacA-aphD, aphA3 and str genes were detected among the aminoglycoside-resistant MRCoNS strains. The cat p221 or cat p223genes were detected in the chloramphenicol-resistant strains. Of the 12 sulphamethoxazole/trimethoprim-resistant strains, 9 were positive for dfrG while the remaining 3 harbored the dfrK gene. These latter strains (2 strains of S. lentus and one strain of S. cohnii) also carried the tet(L) gene linked to dfrK, as determined by specific PCR using primers tet(L)-fw (5′-CATTTGGTCTTATTGGATCG-3′) and dfrK-rv (5′-CAAGAAGCTTTTCGCTCATAAA-3′) and by sequencing of a 1,732-bp fragment. All the MRCoNS strains were negative for the genes mediating exfoliative toxin and biofilm formation.
Analysis of the QRDR of quinolone-resistant strains
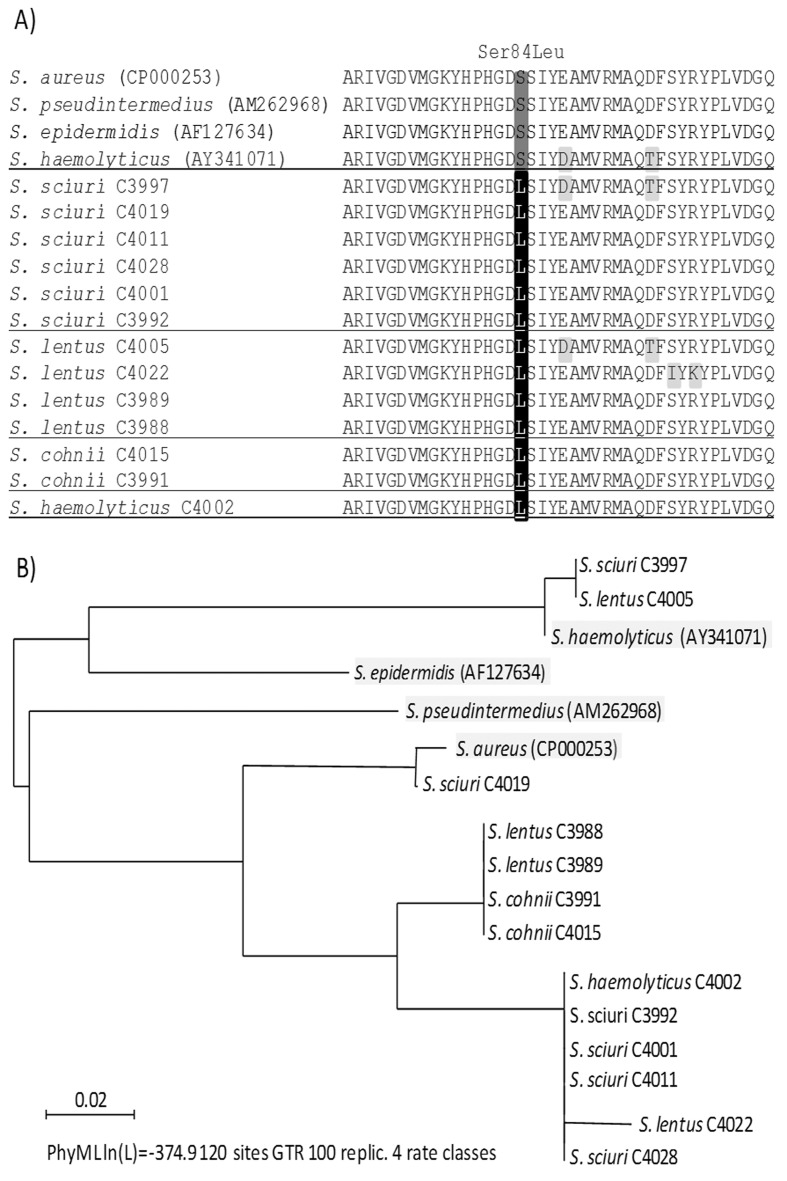
All 13 ciprofloxacin-resistant strains (6 strains of S. sciuri, 4 strains of S. lentus, 2 strains of S. cohniiand one strain of S. haemolyticus) harbored a Ser84Leu mutation within the QRDR of the GyrA protein at the position identical to that involved in quinolone resistance in S. aureus, S. pseudintermedius, S. epidermidis and S. haemolyticus (Sreedharan et al., 1991; Yonezawa et al., 1996; Gómez-Sanz et al., 2013) (Figure 2A). In addition to this, 3 isolates showed two extra substitutions, Ser98Ile and Arg100Lys in S. lentus strain C4022 and Glu88Asp and Asp96Thr in S. sciuri C3997 and S. lentus C4005, with the latter also present in the representative quinolone-susceptible S. haemolyticus strain (Figure 2A). The phylogenetic tree of the QRDR nucleotide sequences in the gyrA gene revealed a high nucleotide diversity, with several major clusters not associated with the bacterial species. The quinolone-susceptible S. pseudintermedius representative strain showed the most divergent QRDR (Figure 2B).
Figure 2. Molecular evaluation of the quinolone resistance determining region (QRDR) of 13 quinolone- and methicillin-resistant staphylococci isolated from healthy swine in Nsukka, Nigeria, in addition to the QRDR of representative quinolone-susceptible S. aureus (Genbank accession no. CP000253), S. pseudintermedius(AM262968), S. epidermidis (AF127634) and S. haemolyticus (AY341071). A) Amino acid sequence comparison of the QRDR of GyrA protein (aa 68–107). B) Phylogenetic relationships among the QRDR nucleotide sequences in gyrA gene (nt 202–321).
Discussion
In this study, the species distribution, antimicrobial resistance pheno- and genotypes and virulence traits of mannitol-fermenting MRS isolated from intensively reared pigs in the Nsukka agricultural zone, Nigeria, were investigated. The study provides the first report on MRS in pigs in Nigeria. MRCoPS, particularly MRSA, have been reported from pigs in several European, American and Asian countries (Khanna et al., 2008; Denis et al., 2009; Wagenaar et al., 2009; Baba et al., 2010; Golding et al., 2010; Gomez-Sanz et al., 2010; Graveland et al., 2011; Lin et al., 2011; Larsen et al., 2012; Oppliger et al., 2012). Surprisingly, none of the 20 strains in the present study was MRSA, despite the fact that they formed blue colonies on ORSAB. Oxacillin resistance screening agar base is a modification of mannitol-salt agar supplemented with 2 μg/mL of oxacillin, on which mannitol-fermenting isolates turn intensely blue due to the presence of aniline. Although ORSAB is intended for detection of MRSA (Simor et al., 2001), the results of the present study as well as those of previous work (Becker et al., 2002; Dzen et al., 2007) have shown that colonies of mannitol-positive CoNS on this medium are similar to those of MRSA. These findings, therefore, emphasize the need for accurate identification of mannitol-fermenting methicillin-resistant staphylococci.
Three of the four Staphylococcus species reported in the present study have recently been reported in dogs in the same study area (Chah et al., 2014). S. sciuri was the predominant MRCoNS species found among the pig Staphylococcus isolates in this study. In Switzerland, S. sciuri has been identified as the major MRCoNS species isolated from various animal sources, including pigs (Huber et al., 2011). S. sciuri was also reported as the major MRCoNS species in the bovine population in Belgium (Vanderhaeghen et al., 2013). Although the degree of importance of CoNS in veterinary medicine is not clearly understood, they are increasingly found to be implicated in cases of bovine mastitis (Inegol and Turkyilmaz, 2012; Kaynarca and Turkyilmaz, 2010) and canine pyoderma (Lima et al., 2012). S. sciuri has been reported to cause fatal exudative epidermitis in piglets in China (Chen et al., 2007). This species has also been associated with endocarditis (Wallet et al., 2000), urinary tract infections (Stepanovic et al., 2003) and wound infections (Shiyuu et al., 2004; Coimbra et al., 2011) in humans.
In addition to resistance to β-lactam antibiotics, the MRCoNS in this study also demonstrated high rates of resistance to other classes of antimicrobial agents, particularly tetracycline, aminoglycosides, fluoroquinolones and macrolides. Similar findings (except fluoroquinolones) have been reported for CoNS from dogs in Nsukka (Chah et al., 2014). As has been pointed out by Aslantas et al. (2013), MRCoNS could pose a major therapeutic challenge for veterinarians due to a limited choice of antimicrobial agents in the case of infection. A high diversity of antimicrobial resistance profiles was observed in this study, for the 20 MRCoNS strains exhibited 16 resistance patterns. This finding suggests the involvement of antimicrobial selective pressure in the pig farms.
Antimicrobial resistance in these MDR MRCoNS was mediated by multiple antimicrobial resistance genes. This observation is similar to that reported for MRCoNS isolated from dogs in Nsukka, Nigeria (Chah et al., 2014) and Hatay, Turkey (Aslantas et al., 2013). Although the degree of importance of MDR MRCoNS in veterinary medicine is not clearly understood, they may represent a reservoir of resistance genes which can be spread to pathogenic bacteria within and across species and genera. These MDR staphylococci, therefore, represent a public health risk as they may transfer their resistance genes to human pathogens such as S. aureus. The presence of these MDR bacteria in pigs may be due to the indiscriminate use of antimicrobial agents in pig production in Nigeria. The fact that the MRS strains harbor several resistance genes against the same antimicrobial agent indicates that the strains are capable of acquiring and maintaining multiple resistance genes at the same time. A possible adaptive advantage of carrying these redundant genes is unknown yet.
Detection of the resistance gene cluster tet(L)-dfrK in S. lentus and S. cohnii is a novel observation. A first description of the trimethoprim resistance gene dfrK located next to the tetracycline resistance gene tet(L) in plasmid pKKS2187 from a MRSA ST398 isolate of pig origin was reported in Germany (Kadlec and Schwarz, 2009). The presence of this gene cluster in MRCoNS isolates from swine in Nigeria indicates a broad geographic distribution of both resistance determinants. The use of tetracyclines and trimethoprim in pig farming in Nigeria may favor the spread and maintenance of their resistance genes.
All 13 quinolone-resistant MRCoNS strains harbored the Ser84Leu substitution in the GyrA protein. To our knowledge, this is first report of this mutation in S. sciuri, S. lentus and S. cohnii, suggesting that quinolone resistance in these bacterial species is commonly associated with this mutation. Until now, this type of mutation has been most frequently found in S. aureus, S. pseudintermedius, S. epidermidis and S. haemolyticus (Sreedharan et al., 1991; Yonezawa et al., 1996; Takahata et al., 1997; Gómez-Sanz et al., 2013). The majority of the strains carried a single amino acid substitution in the same region in the representative quinolone-susceptible S. aureus, S. pseudintermedius and S. epidermidis strains, while the representative S. haemolyticus strain exhibited two additional substitutions. Interestingly, analysis of the QRDR nucleotide sequences revealed relatively high intra-species divergence in this region, with S. pseudintermedius clustered farthest away from the others. These results broaden the current knowledge on the quinolone resistance mechanisms among different CoNS species.
This study demonstrated the presence of mannitol-positive MRCoNS in intensively reared pigs in the Nsukka agricultural zone, Nigeria, with S sciuribeing the predominant species. This is the first report on MRS in pigs in Nigeria. The study highlighted the fact that the CoNS strains were MDR and, therefore, could serve as a pool of resistance genes for pathogenic bacteria. Methicillin-resistant S. aureus which is frequently reported in pigs in other countries was not detected in this study. Our study provides a new insight into bacterial resistance mechanisms in MRCoNS from pigs, with special attention to quinolone resistance.
Acknowledgments
The molecular analysis was financially supported by the grant awarded to Ugwu, Chidozie Clifford by Universidad de La Rioja, Logroño, Spain. This work was partially supported by Project SAF2012-35474 from the Ministerio de Economía y Competitividad of Spain and the Fondo Europeo de Desarrollo Regional (FEDER).
References
- Al-Talib H, Yean CY, Al-Khateeb CY, et al. A pentaplex PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus and Panton-Valentine Leucocidin. BMC Microbiol. 2009;9:113. doi: 10.1186/1471-2180-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslantas O, Turkyilmaz S, Yilmaz MA, et al. Prevalence of methicillin-resistant staphylococci in dogs. Kafkas Univ Vet Fak Derg. 2013;19:37–42. [Google Scholar]
- Baba K, Ishihara K, Ozawa M, et al. Isolation of methicillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int J Antimicrob Agents. 2010;36:352–354. doi: 10.1016/j.ijantimicag.2010.06.040. [DOI] [PubMed] [Google Scholar]
- Becker A, Forster DH, Kneiehl E. Oxacillin resistance screening agar base for detection of methicillin-resistant Staphylococcus aureus . J Clin Microbiol. 2002;40:4400–4401. doi: 10.1128/JCM.40.11.4400-4401.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron M, Dauwalder O, Gouy M, et al. Species identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis. 2011;30:343–354. doi: 10.1007/s10096-010-1091-z. [DOI] [PubMed] [Google Scholar]
- Chah KF, Gómez-Sanz E, Nwanta JA, et al. Methicillin-resistant coagulase-negative staphylococci from healthy dogs in Nsukka, Nigeria. Braz J Microbiol. 2013;45:215–220. doi: 10.1590/S1517-83822014005000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Wang Y, Chen F, et al. A highly pathogenic strain of Staphylococcus sciuri caused fatal exudative epidermitis in piglets. PLoS One. 2007;1:e147. doi: 10.1371/journal.pone.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing: Twenty-first informational supplement. CLSI Document. 2014;34:M100–S24. [Google Scholar]
- Coimbra DG, Almeida AGCS, Junior JBO, et al. Wound infection by multiresistant Staphylococcus sciuri identified by molecular methods. New Microbiol. 2011;34:425–427. [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, et al. Bap, a Staphylococcus aureussurface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis O, Suetens C, Hallin M, et al. Methicillin resistant Staphylococcus aureusST398 in swine farm personnel, Belgium. Emerg Infect Dis. 2009;15:1098–1101. doi: 10.3201/eid1507.080652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzen SM, Santoso S, Santosaningsih D, et al. Evaluation of latex agglutination test and oxacillin resistant screening agar base (ORSAB) medium for the detection oxacillin resistant coagulase negative staphylococci (ORCoNS) (Preliminary study) Med J Indones. 2007;16:228–232. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST) Breakpoint tables for interpretation of MICs and zone diameters. Version 4.0, 2014. 2014. http://www.eucast.org.
- Graveland H, Duim B, van Duijkeren E, et al. Livestock-Associated methicillin resistant Staphylococcus aureus in animals and humans. Int J Med Microbiol. 2011;301:630–634. doi: 10.1016/j.ijmm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Golding GR, Bryden L, Levett PN, et al. Livestock-Associated methicillin resistant Staphylococcus aureus Sequence Type 398 in humans, Canada. Emerg Infect Dis. 2010;16:587–594. doi: 10.3201/eid1604.091435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Sanz E, Torres C, Lozano C, et al. Detection, Molecular Characterization and Clonal Diversity of Methicillin-Resistant Staphylococcus aureus CC398 and CC97 in Spanish slaughter pigs of different age groups. Foodborne Path Dis. 2010;7:1269–1277. doi: 10.1089/fpd.2010.0610. [DOI] [PubMed] [Google Scholar]
- Gómez-Sanz E, Torres C, Lozano C, et al. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp Immun Microbiol Infect Dis. 2013;36:83–94. doi: 10.1016/j.cimid.2012.10.001. [DOI] [PubMed] [Google Scholar]
- Huber H, Ziegler D, Pfluger V, et al. Prevalence and characteristics of methicillin-resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet Res. 2011;7:6. doi: 10.1186/1746-6148-7-6. www.biomedcentral.com/1746-6148/7/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inegol E, Turkyilmaz S. Determination of SCCmec types in methicillin resistant staphylococci isolated from cows and farm workers. Ankara Univ Vet Fak Derg. 2012;59:89–93. [Google Scholar]
- Kadlec K, Schwarz S. Identification of a novel trimethoprim resistance gene, dfrK, in a methicillin-resistant Staphylococcus aureus ST398 strain and its physical linkage to the tetracycline resistance gene tet(L) . Antimicrob Agents Chemother. 2009;53:776–778. doi: 10.1128/AAC.01128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaynarca S, Turkyilmaz S. Methicillin resistance and slime positivity of staphylococci isolated from bovine mastitis. Kafkas Univ Vet Fak Derg. 2010;16:567–572. [Google Scholar]
- Khanna T, Friendship R, Dewey C, et al. Methicillin resistant Staphylococcus aureuscolonization in pigs and pig farmers. Vet Microbiol. 2008;128:298–303. doi: 10.1016/j.vetmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Kloos WE, Bannerman TL. Update on clinical significance of coagulase negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar M, Bardon J, Hanulik V, et al. Resistance to methicillin in coagulase negative staphylococci and its detection. ACTA BRNO. 2010;79:261–267. [Google Scholar]
- Larsen J, Imanishi M, Hinjoy S, et al. Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS One. 2012;7:e31245. doi: 10.1371/journal.pone.0031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. Methicillin- (oxacillin) resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl Environm Microbiol. 2003;69:6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima LF, Lira AC, Coutinho HDM, et al. Antimicrobial resistance in staphylococci isolated from canine pyoderma. Comunicata Scientiae. 2012;3:181–185. [Google Scholar]
- Lin Y, Barker E, Kislow J, et al. Evidence of Multiple Virulence Subtypes in Nosocomial and Community-Associated MRSA Genotypes in Companion Animals from the Upper Midwestern and Northeastern United States. Clin Med Res l. 2011;9:7–16. doi: 10.3121/cmr.2010.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellmann A, Becker K, von Eiff C, et al. Sequencing and staphylococci identification. Emerg Infect Dis. 2006;12:333–336. doi: 10.3201/eid1202.050962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppliger A, Moreillon P, Charriere N, et al. Antimicrobial resistance of Staphylococcus aureus acquired by pig farmers from pigs. Appl Environm Microbiol. 2012;78:8010–8014. doi: 10.1128/AEM.01902-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simor A, Goodfellow J, Loiue L, et al. Evaluation of a new medium, oxacillin resistance screening agar base, for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. J Clin Microbiol. 2001;39:3422. doi: 10.1128/JCM.39.9.3422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan S, Peterson LR, Fisher LM. Ciprofloxacin resistance in coagulase-positive and -negative staphylococci: role of mutations at serine 84 in the DNA gyrase A protein of Staphylococcus aureus and Staphylococcus epidermidis . Antimicrob Agents Chemother. 1991;35:2151–2154. doi: 10.1128/aac.35.10.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanovic S, Jezek P, Vukovic D, et al. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J Clin Microbiol. 2003;41:5262–5264. doi: 10.1128/JCM.41.11.5262-5264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata M, Yonezawa M, Matsubara N, et al. Antibacterial activity of quinolones against coagulase-negative staphylococci and the quinolone resistant-determining region of the gyrA genes from six species. J Antimicrob Chemother. 1997;40:383–386. doi: 10.1093/jac/40.3.383. [DOI] [PubMed] [Google Scholar]
- Vanderhaeghen W, Vandendriessche S, Crombe F, et al. Characterization of methicillin-resistant non-Staphylococcus aureus staphylococci carriage isolates from different bovine populations. J Antimicrob Chemother. 2013;68:300–307. doi: 10.1093/jac/dks403. [DOI] [PubMed] [Google Scholar]
- Voss A, Loeffen F, Bakker J, et al. Methicillin resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar JA, Yue H, Pritchard J, et al. Unexpected sequence types in livestock associated Methicillin resistant S. aureus (MRSA): MRSA ST9 and a single locus variant of ST9 in pig farming in China. Vet Microbiol. 2009;139:405–409. doi: 10.1016/j.vetmic.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Wallet F, Stuit L, Boulanger E, et al. Peritonitis due to Staphylococcus sciuri in a patient on continuous ambulatory peritoneal dialysis. Scand J Infect Dis. 2000;32:697–698. doi: 10.1080/003655400459667. [DOI] [PubMed] [Google Scholar]
- Weese JS. Methicillin resistant Staphylococcus aureus in animals. Inst Lab Anim Res J. 2010;51:233–244. doi: 10.1093/ilar.51.3.233. [DOI] [PubMed] [Google Scholar]
- Yonezawa M, Takahta M, Banzawa-Futakuchi N, et al. DNA gyrA mutations in quinolone-resistant clinical isolates of Staphylococcus haemolyticus . Antimicrob Agents Chemother. 1996;40:1065–1066. doi: 10.1128/aac.40.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell C, Resch M, Rosenstein R, et al. Characterization of toxin production of coagulase-negative staphylococci isolated from food and starter cultures. Int J Food Microbiol. 2008;127:246–251. doi: 10.1016/j.ijfoodmicro.2008.07.016. [DOI] [PubMed] [Google Scholar]
- Ziebuhr W, Krimmer V, Rachid S, et al. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]