Abstract
This study was conducted in order to evaluate the transmission of caprine lentivirus to sheep using different experimental groups. The first one (colostrum group) was formed by nine lambs receiving colostrum from goats positive for small ruminant lentiviruses (SRLV). The second group (milk group) was established by nine lambs that received milk of these goats. Third was a control group, consisting of lambs that suckled colostrum and milk of negative mothers. Another experimental group (contact group) was formed by eight adult sheep, confined with two naturally infected goats. The groups were monitored by immunoblotting (IB), enzyme-linked immunosorbent assay (ELISA), agar gel immunodiffusion (AGID) and nested polymerase chain reaction (nPCR). All lambs that suckled colostrum and milk of infected goats and six sheep of the contact group had positive results in the nPCR, although seroconversion was detected only in three of the exposed animals, with no clinical lentiviruses manifestation, in 720 days of observation. There was a close relationship between viral sequences obtained from infected animals and the prototype CAEV-Cork. Thus, it was concluded that SRLV can be transmitted from goats to sheep, however, the degree of adaptation of the virus strain to the host species probably interferes with the infection persistence and seroconversion rate.
Keywords: cross-infection, goats, lambs, small ruminant lentivirus
Introduction
Small ruminant lentiviruses (SRLV) are retroviruses causing caprine arthritis-encephalitis (CAE) in goats and maedi-visna (MV) in sheep (Blacklaws, 2012). Also belonging to the genus Lentivirus are the human (HIV), simian (SIV), feline (FIV), bovine (BIV) immunodeficiency virus and the equine infectious anemia virus (EIAV) (Leroux et al., 2010).
CAE and MV are progressive diseases, with different clinical manifestations. In goats, the arthritic form is more common, occurring lameness and increasing the joints diameter (Lara et al., 2005). The respiratory form is more common in sheep, which have dyspnea and exercise intolerance (Angelopoulou et al., 2005). Besides, animals may show progressive weight loss, indurative mastitis (Gregory et al., 2009a) and encephalomyelitis can occur in lambs and goatlings (Benavides et al., 2007).
For years, isolated lentiviruses in ovine were called maedi-visna virus (MVV) and in caprine, caprine arthritis-encephalitis virus (CAEV), being considered as specific to each species. However, phylogenetic analyzes and findings of cross-infection demonstrated that there are different genotypes and lentiviral subtypes, able to infect both goats and sheep (Shah et al., 2004a, 2004b; Angelopoulou et al., 2005; Reina et al., 2006; Gjerset et al., 2009; Leroux et al., 2010; Pisoni et al., 2010; Kuhar et al., 2013).
Therefore, questions about the spreading and evolution of SRLV in their hosts remain to be answered, aiming the implementation of health programs for goats and sheep, which should also consider the possibility of interspecies transmission (Gjerset et al., 2009). In this sense, the aim of this study was to evaluate the transmission of caprine lentivirus to sheep, under different exposure conditions.
Material and Methods
The study was conducted at the Brazilian Agricultural Research Corporation - Goats and Sheep, in Sobral, Ceará, Brazil, with the approval of the Ethics Committee on Animal Use of Acaraú Valley State University, under the number 001.12.
Aiming to evaluate the interspecies transmission of the virus from colostrum and milk, three experimental groups were composed of male and female lambs, crossbreds of locally adapted breeds, predominantly Santa Inês and Morada Nova, children of mothers and breeders of sheep flock free of SRLV, monitored by agar gel immunodiffusion (AGID), enzyme-linked immunosorbent assay (ELISA), immunoblotting (IB) and nested polymerase chain reaction (nPCR), for two years at seven different times.
The first group (colostrum) was established with nine lambs subjected to artificial feeding of colostrum collected from SRLV positive goats, during the first 24 hours of life. The amount of colostrum fed was approximately 11% of body weight in the first eight postpartum hours, seeking the adequate transfer of passive immunity (Simões et al., 2005). Additional feedings were still offered until 24 hours after birth, after which the animals began to suckle directly on their mothers. During this period of 24 hours after delivery, sheep were milked and teats insulated in order to prevent natural suckling. The average volume of positive colostrum intake to SRLV, by lamb, during the first 24 hours of life, was 537.78 mL, representing 19.36% of the average weight at birth of 2777.78 g.
The second group (milk) was formed with nine lambs subjected to artificial feeding of milk collected from positive goats, during 15 days, from the second week of life. The animals remained with their mothers since birth, naturally suckling colostrum and milk and, during artificial feeding, they were set apart in the morning for individual supply (ad libitum). The average volume of SRLV positive milk ingestion, by lamb, was 1532.78 mL. The third group was the control one, consisting of ten lambs, which suckled colostrum and milk naturally in their negative mothers.
Colostrum and milk fed to animals where delivered from goats naturally infected for SRLV, belonging to Saanen and Anglo-Nubian breeds, tested by IB, ELISA, AGID and nPCR. At 90 days of age, the weaning of lambs happened and after 330 days of experiment, males were removed from groups (three animals from colostrum group, two from milk group and five from control group) due to scarcity of forage caused by a prolonged drought.
In order to analyze the virus horizontal transmission from goats to sheep, another experimental group (contact group) was established by eight adult sheep, originated from the same herd of sheep initially reported, free from SRLV. These animals were confined with two naturally infected goats, positive in the AGID, ELISA, IB and nPCR tests, having arthritis.
Each group remained in isolated stalls, under intensive system and clinical monitoring. Blood samples with and without anticoagulant were performed before the start of the experiment (day 0), and after 1, 7, 15, 30, 50, 70, 90, 120, 150, 180, 210, 240, 270, 300, 330, 390, 480, 520, 570, 630, and 720 days of monitoring to obtain peripheral blood leukocytes (PBL) and serum stored at −20 °C until the time of diagnostic tests. No samples were collected at 24 hours for milk group. Regarding the contact group, the first four samples were collected before the start of the experiment (day 0) and after 30, 60 and 90 days.
The leukocytes were obtained from the processing of blood samples with solution of ammonium chloride at 0.84% (Feitosa et al., 2011). Subsequently, DNA was extracted (Grimberg et al., 1989). The proviral DNA detection was carried out by applying the nPCR (Barlough et al., 1994 modified by Andrioli et al., 2006).
Aiming to confirm infection of sheep by caprine lentivirus strain, PCR amplification products were sequenced on Applied Biosystems® 3500 Genetic Analyzer platform. The sequences obtained were aligned with BR/CNPC-G1, BR/CNPC-G2, BR/CNPC-G3, BR/CNPC-G4 and CAEV-Cork sequence (available in GenBank, numbered in EU300976, EU300977, EU300978, EU300979 and M33677 respectively), using the Clustal W algorithm (Thompson et al., 1994), from the BioEdit Sequence Alignment Editor® (Hall, 1999).
In order to investigate anti-SRLV antibodies, AGID, ELISA and IB tests were applied, considering the methodologies described by Pinheiro et al. (2010), Lima et al.(2013) and Pinheiro et al.(2011), respectively. To this end, antigen was produced from cell culture of caprine synovial membrane, inoculated with standard strain CAEV-Cork (Pinheiro et al., 2006). In the AGID test, the antigen concentrated by ultrafiltration and treated with ethyl ether (Pinheiro et al., 2010) was used. For ELISA, the antigen used was obtained from the pellet of cells treated with sodium dodecyl sulphate, according to the methodology described by Torres et al. (2009). In the IB technique, the antigen produced by ultracentrifugation at sucrose was used (Pinheiro et al., 2006).
Results
All animals in the control group had negative results in nPCR, from birth to 720 days of life. On the other hand, a significant positivity (p < 0.05) was observed in animals exposed to SRLV when compared to unexposed ones (Table 1).
Table 1. Number of positive and negative results obtained in nPCR for small ruminant lentiviruses in different experimental groups.
| Experimental groups | Number of results until 720 days of experiment | |||
|---|---|---|---|---|
|
|
||||
| Positive* | Negative* | Total | ||
| Lambs | Colostrum | 35a | 145b | 180 |
| Milk | 24ab | 153b | 177 | |
| Control | 0c | 190a | 190 | |
| Adults | Contact | 15b | 121b | 136 |
| Total | 74 | 609 | 683 | |
Different letters in the same column indicate significant different values greater or smaller by t-test for proportions (one-tailed test and p < 0.05).
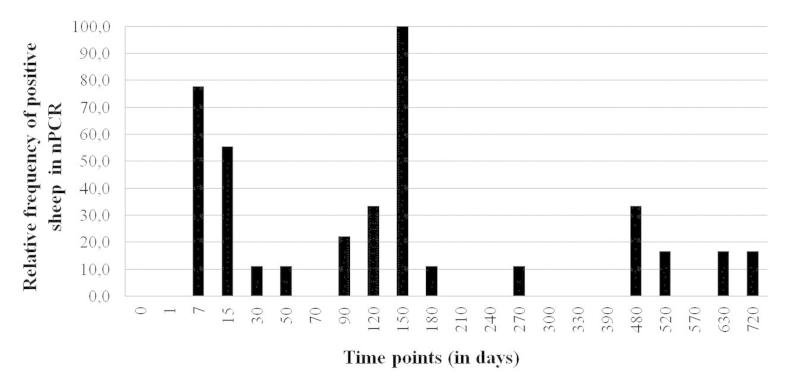
The animals of colostrum group had at least two positive results by nPCR. Seven of the nine (77.78%) animals were positive on the seventh day of life and the other two remaining animals (22.22%) had the first positive result on day 15. There was intermittence between positive and negative tests for the same animal, in different time points, throughout the experiment (Figure 1). From the nine animals in colostrum group, two were twice nPCR positive; three were three times nPCR positive; three were four times and one was ten times. It was not possible to demonstrate significant correlation (p > 0.05) between the amount of colostrum ingested and the number of times the animals obtained positive results in PCR, using Spearman’s correlation coefficient (Table 2).
Figure 1. Results of nPCR for small ruminant lentiviruses, in peripheral blood leukocytes from nine lambs that received colostrum from infected goats within the first 24 hours of life.
Table 2. Correlation between the amounts of colostrum ingested by lamb and the number of positive results in nPCR for small ruminant lentiviruses during 720 days of experiment.
| Animals | Colostrum ingested volume (mL) | Number of positive results |
|---|---|---|
| 1 | 600 | 2 |
| 2 | 330 | 3 |
| 4 | 530 | 3 |
| 6 | 420 | 3 |
| 7 | 530 | 4 |
| 8 | 345 | 2 |
| 9 | 795 | 4 |
| 10 | 740 | 4 |
| 11 | 550 | 10 |
| Average | 537.78 | 3.89 |
Spearman’s correlation coefficient = 0.470 (p > 0.05).
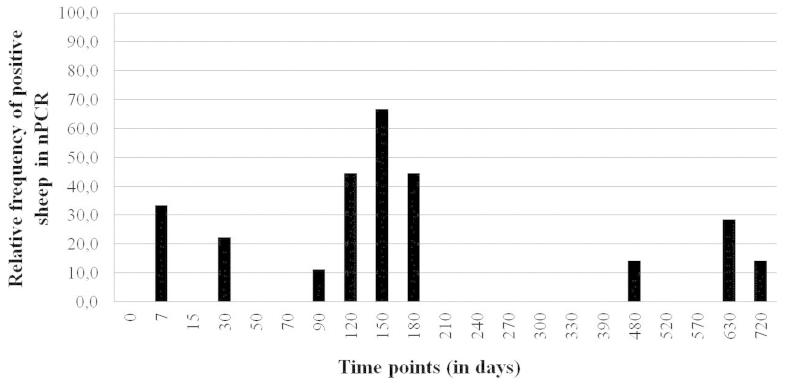
As for the milk group, the first positive results of each animal were observed between 7 and 150 days after the first intake of positive goats’ collected milk. All nine animals of the milk group had at least one positive result in nPCR, also with intermittency between positivity and negativity (Figure 2). From the nine animals, one showed a positive result once; four, twice; two, three times; one, four times and one, five times. Also it was not possible to demonstrate the existence of significant relationship (p > 0.05) between the amount of provided milk and the number of times the animals were nPCR positive, using Spearman’s correlation coefficient (Table 3).
Figure 2. Results of nPCR for small ruminant lentiviruses, in the peripheral blood leukocytes of nine lambs that received milk from infected goats during 15 days.
Table 3. Correlation between the amount of ingested milk by the lamb and the number of positive results in nPCR for small ruminant lentiviruses during 720 days of experiment.
| Animals | Amount of ingested milk (mL) | Number of positive results |
|---|---|---|
| 21 | 1,450 | 3 |
| 22 | 1,690 | 2 |
| 23 | 1,260 | 3 |
| 24 | 2,990 | 5 |
| 25 | 1,810 | 4 |
| 27 | 1,785 | 2 |
| 28 | 1,190 | 1 |
| 29 | 880 | 2 |
| 30 | 740 | 2 |
| Average | 1,532.78 | 2.67 |
Spearman’s correlation coefficient = 0.656 (p > 0.05).
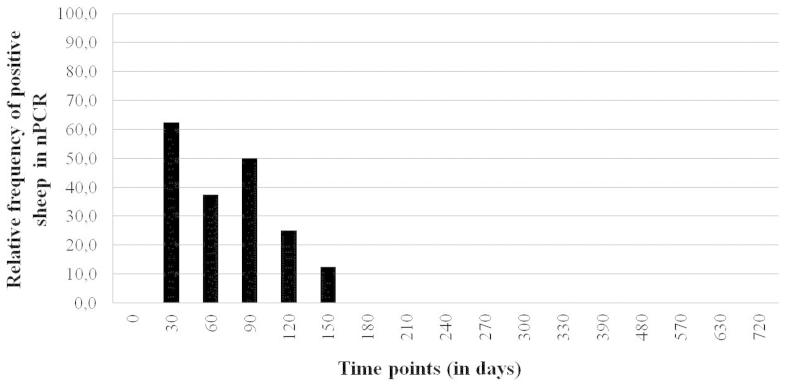
For contact group, from the eight sheep confined with infected goats, six (75%) were positive in nPCR at least twice and the two remaining (25%) had negative results in all evaluated time points. In five animals (62.5%), proviral DNA was detected at 30 days of confinement and the other positive animal was found after 90 days of experiment. From the sixth month of observation, positive results were no longer obtained (Figure 3).
Figure 3. Results of nPCR for small ruminant lentiviruses, in peripheral blood leukocytes from eight sheep raised confined together with infected goats, during 720 days.
Sequences obtained from PCR amplified products showed that animals of colostrum, milk and contact groups acquired viruses highly related with the caprine lentiviral prototype CAEV-Cork and BR/CNPC sequences (BR/CNPC-G1, BR/CNPC-G2, BR/CNPC-G3 and BR/CNPC-G4). These BR/CNPC sequences represent the circulating strain in flocks of goats used as the sources of infection for sheep.
Concerning seroconversion detection, for lambs of the colostrum group, the results obtained between 1 and 120 days of age were disregarded, due to the occurrence of colostral anti-SRLV antibodies, detected by IB, ELISA and AGID, in the period from 1 to 70 days after birth. At 90 and 120 days, there were no more positive results. The IB technique detected the seroconversion of three animals, two from the colostrum group and one from the milk group. From these, two were also positive for ELISA tests and one for AGID (Table 4). There was no clinical manifestation of small ruminant lentiviruses during the evaluated period.
Table 4. Immunoblotting (IB), enzyme-linked immunosorbent assay (ELISA) and agar gel immunodiffusion (AGID), of three lambs infected with small ruminants lentiviruses, from colostrum (animals 2 and 11) and milk (animal 24) from positive goats.
| Animals | Tests | Time points (in days) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| 90 to 210 | 240 | 270 | 300 | 330 | 390 | 480 | 520 | 570 | 630 | 720 | ||
| 2 | IB | − | − | − | − | − | − | + | − | − | − | − |
| ELISA | − | − | − | − | − | − | − | − | − | + | + | |
| AGID | − | − | − | − | − | − | − | − | − | − | − | |
| 11 | IB | − | + | + | + | + | + | + | + | + | + | + |
| ELISA | − | + | + | + | + | + | + | + | + | + | + | |
| AGID | − | − | − | − | − | + | + | + | + | + | − | |
| 24 | IB | − | − | − | − | + | + | − | − | − | − | − |
| ELISA | − | − | − | − | − | − | − | − | − | − | − | |
| AGID | − | − | − | − | − | − | − | − | − | − | − | |
(−) negative; (+) positive.
Discussion
The positivity in nPCR for SRLV characterizes the cell-associated viremia, from the observation of proviral DNA (Alvarez et al., 2006). Therefore, the results obtained in this study showed interspecific transmission of goats’ lentivirus to sheep. Previous studies have already demonstrated the occurrence of virus spreading through secretions, even identifying it in the water and in the air, allowing the infection of susceptible animals after prolonged contact with infected animals (Villoria et al., 2013). It is known, also, that colostrum and milk are important means of SRLV elimination, allowing infection of neonates and young animals (Lara et al., 2003; Alvarez et al., 2005; Ravazzolo et al., 2006; Gregory et al., 2009b; Sardi et al., 2012). However, this work emphasizes the importance of these transmission forms to the occurrence of cross-infection.
Comparing data from colostrum, milk and contact groups, there is a cumulative proportion of 100% positive nPCR lambs within 15 days after suckling colostrum from infected goats, while for the group receiving milk, this proportion only reached 100% at 150 days and in the case of horizontal transmission, the maximum 75% of positivity was obtained within 90 days of exposure. This reflects the greater potential for transmission of the infectious agent from the colostrum, since in newborn lamb, due to increased intestinal permeability during the first 48 hours of life, a virus derived of the colostrum reaches the intestine and is able to cross the epithelium infecting macrophages and increasing the lymphatic route (Preziuso et al., 2004).
It is noteworthy that the transfer of colostral passive immunity does not prevent infection. As can be observed in this study, lambs acquiring colostral anti-SRLV immunoglobulins became infected by lentivirus precisely by ingestion of positive goats’ colostrum. This is due to the fact that the cell-associated virus can also be transmitted (Preziuso et al., 2004; Hermann-Hoesing et al., 2007) and, also, to other viral scape mechanisms from the immune system, related to the antigenic variability as a result of transcription errors of viral RNA and gene recombination (Pisoni et al., 2007, 2010; L’Homme et al., 2011).
After the first proviral DNA detection, animals from the colostrum, milk and contact groups did not have continuous positive results in PCR. Alvarez et al. (2006) have reported intermittency when verifying that most lambs which received colostrum from positive sheep did not obtained positive results in blood samples continuously. Similarly, Paula et al.(2009) observed intermittency of positivity in blood and semen samples of goats inoculated with CAEV-Cork strain.
The intermittency can be justified by the lower number of infected SRLV blood cells (Blacklaws, 2012). Larger quantities are observed, for example, in lymph nodes, reservoirs for the virus and in the mammary gland, the main site of replication (Ravazzolo et al., 2006). Thus, negative results may happen due to the absence of the provirus or to the insufficient number of infected cells in the blood sample at any given moment.
It must also be considered that in normal range counts of sheep leukocytes, the absolute values of monocytes, targets of SRLV, may vary from 0 to 750 cells/μL, which is equivalent in relative values, to the range between 0 and 6% (Pugh, 2002). Therefore, there is a variation in the number of circulating cells from animals and even for the same animal at different times.
Animal 11, from colostrum group, was the one with more positive results in nPCR, the first one who seroconverted and the only one with positive results also in AGID test, corroborating with Alvarez et al. (2006) who observed a positive correlation between the number of times the animal was nPCR positive with seroconversion. On the other hand, in this study, while 24 sheep have shown positive results in nPCR, only three seroconverted and clinical signs were not observed. It is known, however, that the time required for seroconversion after SRLV infection is variable (Lara et al., 2003; Paula et al., 2009; Rachid et al., 2013) and that some animals simply do not seroconvert (Alvarez et al., 2005).
One of the factors related with the lack of seroconversion or late seroconversion concerns the infectious agent pathogenicity. This involves the restriction of replication and the viral latency in monocytes and progenitor cells in the bone marrow, in the form of proviral DNA integrated into the cellular genome. Thus, the virus remains hidden (Blacklaws, 2012) and the infection persists without the immune system activation (Brellou et al., 2007; Ravazzolo et al., 2006).
Other authors also suggest the possibility of transient virus acquisition occurring due to unknown viral and immunological determinants that can lead to the non-persistence of the infection. In this case, over time, the animals fail to show positive results in nPCR and in serological tests (Herman-Hoesing et al., 2007). Considering these information, it is possible to state that data obtained in this study demonstrated the occurrence of occult and/or transient infection in ovine infected by caprine lentivirus.
Alvarez et al. (2005) verified, using ELISA, seropositivity of 55% (29/53) for lambs at 300 days of age which received colostrum of positive sheep. Also, these authors pointed out the positive and significant association between positivity percentage and the volume of ingested colostrum that was 200 mL/kg. However, the seroconversion rate observed in this study was lower, though the average amount of colostrum intake was near (193.45 mL/kg). This difference between results can be related to different methodologies of diagnosis, viral load of inocula and adaptation to the host species, since Alvarez et al. (2005) used colostrum from infected sheep while in this study colostrum and milk from infected goats was offered.
This is another question to be discussed regarding the virus and host interaction. It was already found the interspecies transmission and the SRLV heterogeneity in several studies (Shah et al., 2004a, 2004b; Angelopoulou et al., 2005; Pisoni et al., 2005; Gjerset et al., 2009; Leroux et al., 2010; Kuhar et al., 2013), but these studies do not deny that some strains are better adapted to goats and other to sheep (Reina et al., 2006).
The high genetic diversity of SRLV is associated with pathogenicity variation and even when infected with the same virus strain, goats and sheep respond differently to infection, affecting the results of serological and molecular diagnosis (Rachid et al., 2013), and also on clinical findings (Angelopoulou et al., 2005).
In Spain, Pérez et al. (2014) reported an outbreak of arthritis in sheep infected with subtype B2 related to the prototype CAEV. These authors pointed out the transmission of the virus from goats to sheep in a natural way, probably through respiratory route or milk, allowing adaptation to the new host and consequently the development of the pathology. According to Glaría et al. (2009), this adaptation relates with the acquisition of genetic properties that confer changes in viral phenotype.
In this sense, it is possible to justify the low rate of seroconversion observed also by the fact that sheep have been infected with a viral strain poorly adapted to this species, derived from dairy goat herd. Feitosa et al.(2010) characterized this strain as as belonging to subtype B1. Furthermore, the infected sheep are crossbreds of locally adapted breeds, predominantly Santa Inês and Morada Nova, which may also have influenced the results, especially considering the heterosis and individual susceptibility of these animals to infection.
Conclusions
SRLV can be transmitted to sheep from the contact with infected goats and also by ingestion of colostrum and milk from infected goats. However, the tropism to the host species of the viral strain probably interferes in the persistency of the infection and in the seroconversion rate.
Acknowledgments
To the Bahia State Research Support Foundation (FAPESB) for promoting the project and awarding the PhD scholarship, to the National Council for Scientific and Technological Development (CNPq) for research funding and to Brazilian Agricultural Research Corporation - Goats and Sheep, for supporting the implementation of the experiment.
References
- Álvarez V, Arranz J, Daltabuit-Test M, et al. Relative contribution of colostrum from Maedi-Visna virus (MVV) infected ewes to MVV-seroprevalence in lambs. Res Vet Sci. 2005;78:237–243. doi: 10.1016/j.rvsc.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Álvarez V, Daltabuit-Test M, Arranz J, et al. PCR detection of colostrum-associated Maedi-Visna virus (MVV) infection and relationship with ELISA-antibody status in lambs. Res Vet Sci. 2006;80:226–234. doi: 10.1016/j.rvsc.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Andrioli A, Gouveia AMG, Martins AS, et al. Fatores de risco na transmissão do lentivírus caprino pelo sêmen. Pesq Agropec Bras. 2006;41:1313–1319. [Google Scholar]
- Angelopoulou K, Karanikolaou K, Papanastasopoulou M. First partial characterization of small ruminant lentiviruses from Greece. Vet Microbiol. 2005;109:1–9. doi: 10.1016/j.vetmic.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Barlough J, East N, Rowe JD, et al. Double-nested polymerase chain reaction for detection of caprine arthritis-encephalitis virus proviral DNA in blood, milk, and tissues of infected goats. J Virol Methods. 1994;50:101–113. doi: 10.1016/0166-0934(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Benavides J, García-Pariente C, Ferreras MC, et al. Diagnosis of clinical cases of the nervous form of Maedi-Visna in 4- and 6- month old lambs. Vet J. 2007;174:655–658. doi: 10.1016/j.tvjl.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Blacklaws BA. Small ruminant lentiviruses: Immunopathogenesis of visna-maedi and caprine arthritis and encephalitis virus. Comp Immunol Microb. 2012;35:259–269. doi: 10.1016/j.cimid.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Brellou GD, Angelopoulou K, Poutahidis T, et al. Detection of Maedi-Visna Virus in the liver and heart of naturally infected sheep. J Comp Pathol. 2007;136:27–35. doi: 10.1016/j.jcpa.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Feitosa ALVL, Teixeira MFS, Pinheiro RR, et al. Phylogenetic analysis of small ruminant lentiviruses from Northern Brazil. Small Ruminant Res. 2010;94:205–209. [Google Scholar]
- Feitosa ALVL, Teixeira MFS, Pinheiro RR, et al. Primeiro isolamento de lentivírus de pequenos ruminantes em caprino naturalmente infectado em rebanho do Rio Grande do Norte, Brasil. Arq Inst Biol. 2011;78:501–505. [Google Scholar]
- Gjerset B, Rimstad E, Teige J, et al. Impact of natural sheep-goat transmission on detection and control of small ruminant lentivirus group C infections. Vet Microbiol. 2009;135:231–238. doi: 10.1016/j.vetmic.2008.09.069. [DOI] [PubMed] [Google Scholar]
- Glaria I, Reina R, Crespo H, et al. Phylogenetic analysis of SRLV sequences from an arthritic sheep outbreak demonstrates the introduction of CAEV-like viruses among Spanish sheep. Vet Microbiol. 2009;138:156–162. doi: 10.1016/j.vetmic.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Gregory L, Birgel EH, Junior, Lara MCCSH, et al. Clinical features of indurative mastitis caused by caprine arthritis encephalitis virus. Braz J Vet Pathol. 2009a;2:64–68. [Google Scholar]
- Gregory L, Lara MCCSH, Villalobos EMC, et al. Detecção do vírus da artrite encefalite caprina em amostras de leite de cabras pela reação em cadeia da polimerase (PCR) e nested-PCR. ARS Vet. 2009b;25:142–146. [Google Scholar]
- Grimberg J, Nowoschik S, Belluscio L, et al. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989;17:83–90. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Herrmann-Hoesing LM, Palmer GH, Knowles DP. Evidence of proviral clearance following postpartum transmission of an ovine lentivírus. Virology. 2007;362:226–234. doi: 10.1016/j.virol.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Kuhar U, Barlic-Maganja D, Grom J. Phylogenetic analysis of small ruminant lentiviruses detected in Slovenia. Vet Microbiol. 2013;162:201–206. doi: 10.1016/j.vetmic.2012.08.024. [DOI] [PubMed] [Google Scholar]
- Lara MCCSH, Birgel EH, Junior, Fernandes MA, et al. Infecção experimental do vírus da artrite-encefalite dos caprinos em cabritos. Arq Inst Biol. 2003;70:51–54. [Google Scholar]
- Lara MCCSH, Birgel EH, Junior, Gregory L, et al. Aspectos clínicos da artrite-encefalite dos caprinos. Arq Bras Med Vet Zootec. 2005;57:736–740. [Google Scholar]
- Leroux C, Cruz JCM, Mornex JF. SRLVs: A genetic continuum of lentiviral species in sheep and goats with cumulative evidence of cross species transmission. Curr HIV Res. 2010;98:94–100. doi: 10.2174/157016210790416415. [DOI] [PubMed] [Google Scholar]
- L’Homme Y, Ouardani M, Lévesque V, et al. Molecular characterization and phylogenetic analysis of small ruminant lentiviruses isolated from Canadian sheep and goat. Virol J. 2011;8:271. doi: 10.1186/1743-422X-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima CCV, Costa JN, Souza TS, et al. Imunodiagnóstico para a artrite-encefalite caprina em rebanhos do semiárido baiano, Brasil. Rev Bras Med Vet. 2013;35:358–364. [Google Scholar]
- Paula NRO, Andrioli A, Cardoso JFS, et al. Profile of the Caprine arthritis-encephalitis virus (CAEV) in blood, semen from bucks naturally and experimentally infected in the semi-arid region of Brazil. Small Ruminant Res. 2009;85:27–33. [Google Scholar]
- Pérez M, Biescas E, Reina R, et al. Small ruminant lentivirus-induced arthritis:clinicopathologic findings in sheep infected by highly replicative SRLV B2 genotype. Vet Pathol Online. 2014 doi: 10.1177/0300985813519654. http://vet.sagepub.com/content/early/2014/01/29/0300985813519654. [DOI] [PubMed] [Google Scholar]
- Pinheiro RR, Olortegui CDC, Gouveia AMG, et al. Desenvolvimento de dot-blot para detecção de anticorpos para o vírus da artrite-encefalite caprina em caprinos. Rev Port Cienc Vet. 2006;101:51–56. [Google Scholar]
- Pinheiro RR, Andrioli A, Gouveia AMG, et al. Avaliação de antígenos para o diagnóstico de lentivírus em rebanho caprino sob programa de controle. Arq Inst Biol. 2010;77:133–137. [Google Scholar]
- Pinheiro RR, Brito RLL, Rodrigues AS, et al. Comunicado Técnico 122. Embrapa Caprinos e Ovinos; Sobral: 2011. Protocolo de immunoblotting para diagnóstico da artrite-encefalite caprina. [Google Scholar]
- Pisoni G, Bertoni G, Puricelli M, et al. Demonstration of coinfection with and recombination by caprine arthritis-encephalitis virus and maedi-visna virus in naturally infected goats. J Virol. 2007;81:4948–4955. doi: 10.1128/JVI.00126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisoni G, Bertoni G, Manarolla G, et al. Genetic analysis of small ruminant lentiviruses following lactogenic transmission. Virology. 2010;407:91–99. doi: 10.1016/j.virol.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Preziuso S, Renzoni G, Allen TE, et al. Colostral transmission of Maedi-visna virus: sites of viral entry in lambs born from experimentally infected ewes. Vet Microbiol. 2004;104:157–164. doi: 10.1016/j.vetmic.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Pugh DG. Sheep and Goat Medicine. Saunders; Philadelphia: 2002. [Google Scholar]
- Rachid A, Croisé B, Russo P, et al. Diverse host-virus interactions following caprine arthritis-encephalitis virus infection in sheep and goats. J Gen Virol. 2013;94:634–642. doi: 10.1099/vir.0.044768-0. [DOI] [PubMed] [Google Scholar]
- Ravazzolo AP, Nenci C, Vogt HR, et al. Viral load, organ distribution, histopathological lesions and cytokine mRNA expression in goats infected with a molecular clone of the caprine arthritis encephalitis virus. Virology. 2006;350:116–127. doi: 10.1016/j.virol.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Reina R, Mora MI, Glaria I, et al. Molecular characterization and phylogenetic study of Maedi Visna and Caprine Arthritis Encephalitis viral sequences in sheep and goats from Spain. Virus Res. 2006;121:189–198. doi: 10.1016/j.virusres.2006.05.011. [DOI] [PubMed] [Google Scholar]
- Sardi SI, Torres JA, Brandão CFL, et al. Early detection of goats infected with lentivirus small ruminant virus by ELISA assay. R Ci Med Biol. 2012;11:35–40. [Google Scholar]
- Shah CA, Böni J, Huder JB, et al. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: evidence for regular sheep-to-goat transmission and world-wide propagation through livestock trade. Virology. 2004a;319:12–26. doi: 10.1016/j.virol.2003.09.047. [DOI] [PubMed] [Google Scholar]
- Shah C, Huder JB, Böni J, et al. Direct evidence for natural transmission of small-ruminant Lentiviruses of subtype A4 from goat to sheep and vice versa. J Virol. 2004b;78:7518–7522. doi: 10.1128/JVI.78.14.7518-7522.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões SVD, Costa RG, Souza PM, et al. Imunidade passiva, morbidade neonatal e desempenho de cabritos em diferentes manejos de colostro. Pesq Vet Bras. 2005;25:219–224. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres JA, Campos GS, Freitas MM, et al. Produção de antígeno viral para o diagnóstico da artrite-encefalite caprina utilizando um teste imunoenzimático (ELISA) R Ci Med Biol. 2009;8:107–114. [Google Scholar]
- Villoria M, Leginagoikoa I, Luján L, et al. Detection of Small Ruminant Lentivirus in environmental samples of air and water. Small Ruminant Res. 2013;110:155–160. [Google Scholar]