Abstract
Background
Group A Streptococcus (GAS) M protein is an important virulence factor and potential vaccine antigen, and constitutes the basis for strain typing (emm-typing). Although >200 emm-types are characterized, structural data were obtained from only a limited number of emm-types. were aim to evaluate the sequence diversity of near-full-length M proteins from worldwide sources and analyse their structure, sequence conservation and classification.
Methods
GAS isolates recovered from throughout the world during the last two decades underwent emm-typing and complete emm gene sequencing. Predicted amino acid sequence analyses, secondary structure predictions and vaccine epitope mapping were performed using MUSCLE and Geneious software.
Results
1086 isolates from 31 countries were analysed, representing 175 emm-types. emm-type is predictive of the whole protein structure, independent of geographic origin or clinical association. Findings of an emm-type paired with multiple, highly divergent central regions were not observed. M protein sequence length, the presence or absence of sequence repeats, and predicted secondary structure was assessed in the context of the latest vaccine developments.
Conclusions
Based on these global data, the M6 protein model is updated to a three representative M protein (M5, M80, M77) model, to aid in epidemiological analysis, vaccine development and M protein-related pathogenesis studies.
Keywords: Streptococcus pyogenes, M protein, Virulence, Epidemiology, Typing, Vaccine
Introduction
Amongst bacterial pathogens afflicting humans, group A streptococcus (GAS) is a leading cause of global morbidity and mortality [1]. Colonisation of the respiratory tract or skin by this organism can lead to diseases that manifest in different body sites, and require different modalities of treatment. Of these Rheumatic Heart Disease (RHD) and serious streptococcal invasive diseases are associated with the greatest global mortality. Much of the GAS associated mortality occurs in low income regions and populations [2].
The M protein is a fibrillar coiled-coil dimer that extends from the bacterial cell wall, and is considered an archetypal Gram-positive surface protein [3]. The M protein is a key virulence factor and major target for GAS vaccine development. M protein inhibits phagocytosis of GAS in the absence of opsonising antibodies, promotes adherence to human epithelial cells and helps the bacterium overcome innate immunity [4]. The multifunctional nature of this protein is also evident from its interactions with numerous host proteins [4], occurring along the entire length of the surface exposed portion of M protein.
Most of the M protein sequence consists of heptad repeat motifs in which the first and fourth amino acids are typically hydrophobic, and are core stabilizing residues within the coiled coil [5]. Heterogeneity in the amino acid sequence of the N-terminal part of M protein, resulting in antigenic diversity, forms the basis of GAS serotyping which was used for many decades [6, 7]. Serotyping has recently been superseded by nucleotide sequencing of the corresponding region, in a scheme called emm-typing [6, 8]. emm-type based surveillance studies show that the diversity of strains circulating in low income settings far exceeds that in high income settings [9, 10]. emm-typing relies on sequencing a small variable portion (10-15%) of the complete emm gene. As a consequence, emm-typing is not informative of the sequence, predicted conformational structure, or functional domains of the remainder of the M protein molecule, which remains largely uncharacterised at the global level.
Another typing method, called emm pattern-typing distinguishes distinct chromosomal architectures (patterns A-C, D and E) based on the presence and arrangement of emm and emm-like genes within the GAS genome [11]. Specific emm-types correlate well with specific emm patterns [12]. emm pattern also correlates well with tissue tropism, although several exceptions have been described [13]. Pattern A-C strains are usually associated with throat infections, pattern D strains are mainly recovered from superficial skin infection (impetigo), while pattern E represents a “generalist” group associated with both tissue sites. Although representing only a small proportion of emm-types, pattern A-C strains have been the most extensively studied [4]. Much of our understanding of M protein structure and function is based on early work on the M6 protein, an emm pattern A-C type [7]. The prototypical M6 protein contains several internal repeat sequences called ‘A’, ‘B’, ‘C’, and ‘D’ repeats. Much less is known of the structure of many other M proteins, particularly those belonging to emm patterns D and E [14].
Although there is increasing interest in GAS vaccine development by global health authorities, including the World Health Organisation, a GAS vaccine remains unavailable. Three M protein-based GAS vaccines are poised to enter, or are progressing through, human clinical trials. One vaccine candidate incorporates amino terminal, M-type determinants from multiple M-proteins [15], while the others consist of more highly conserved sequence from the C repeat region (CRR) [16-19]. Given the clinical relevance of M protein in molecular epidemiology and GAS virulence, and its importance to vaccine development, a comprehensive unified view of M protein is needed. In this study we fill this knowledge gap by characterizing the complete surface-exposed portions of a large number of M proteins from strains recovered from geographical regions throughout the world.
Materials and Methods
Study profile
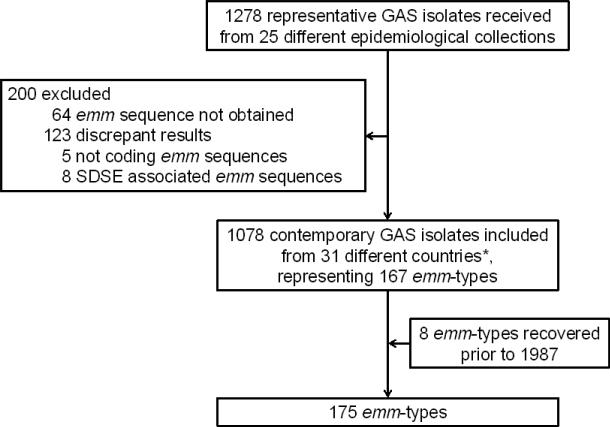
Globally distributed GAS isolates recovered during two recent decades (from 1987 to 2008) by the 25 partners of the M-protein study groups were included in the study. Each partner provided bacterial isolates, or genomic DNA representatives of each emm-type in their collection. Most isolates (n=835; 77%) are unique representatives of a particular emm-type per country of isolation. In some cases, two or three isolates of the same emm-type were included if they were collected in different regions of large countries such as USA, Canada, Brazil, and Australia. With one exception, continents and countries were classified according to the geoscheme created by the United Nations Statistics Division [20]; isolates recovered from Hawaii (USA) were artificially included in Oceania because of geographical proximity to the Pacific Islands with which Hawaii shares similar GAS epidemiology [9, 10]. Clinical data was also provided with most isolates. Eight emm-types could not be recovered during the two last decades in our dataset (Figure 1; Table 1); the sequence of those particular emm-types were obtained and described, but were not included in data analysis.
Figure 1. Study profile.
* List of countries with respective number of isolates in brackets: Argentina (5), Australia (137), Belgium (46), Brazil (105), Canada (69), Chile (5), Czech Republic (17), Germany (50), Egypt (39), Ethiopia (4), Fiji (55), India (51), Israel (67), Japan (12), Kenya (1), Malaysia (1), Mali (58), Mexico (7), New Zealand (1), Norway (19), Papua New Guinea (2), Portugal (21), Romania (22), Russia (15), South Africa (22), Sweden (45), Taiwan (37), The Gambia (1), United Kingdom (22), USA (138, including 83 in mainland and 55 in Hawaii), Venezuela (1). Geographical origin is unknown for 3 isolates. SDSE, Streptococcus dysgalactiae subspecies equisimilus. The eight emm-types recovered prior to 1987 are as follows: emm-types 17, 34, 37, 38, 46, 47, 51 and 72.
Table 1.
emm pattern groupings for 184 emm-types.
| emm pattern | emm-types | Number of emm-types for set of 184 (%) |
|---|---|---|
| A-C | 1, 1-2, 1-4, 3, 5, 6, 12, 14, 17*, 18, 19, 23, 24, 26, 29, 30, 37*, 38/40*, 39, 46*, 47*, 51*, 54, 55, 57, st1RP31, st412, st465, st818, st3765, st4119, st7323, st854, st980584 (stHK), stCK401, stil62, stmd216, stn165, stNS90 | 39 (21) |
| D | 32, 33, 34*, 36, 41, 42, 43, 52, 53, 54, 56, 59, 64, 65/69, 67, 70, 71, 72*, 74, 80, 81, 83, 85, 86, 91, 93, 95, 97, 98, 99, 100, 101, 105, 108, 111, 115, 116, 119, 120, 121, 122, 123, st38, st62, st204, st221, st369, st809, st854, st1967, st2037, st2105, st2461, st2861UK, st2911, st2917, st2926, st2940, st3757, st3850, st5282, st6030, st7395, st7700, stCK249, stD432, stD631, stD633, stNS1033, stxh1 | 70 (38) |
| E | 2, 4, 8, 9, 11, 13, 15, 22, 25, 27, 28, 44/61, 48, 49, 50/62, 58, 60, 63, 66, 68, 73, 75, 76, 77, 78, 79, 82, 84, 87, 88, 89, 90, 92, 94, 96, 102, 103, 104, 106, 107, 109, 110, 112, 113, 114, 117, 118, 124, st106M, st212, st213, st804, st833, st1207, st1389, st1731, st2147, st2460, st2463, st2904, st6735, st7406, st11014, stknb1, stMTH81, stNS292, stNS554, sts104 | 68 (37) |
| REA | st211, st1815 | 2 (1) |
| ND | 31, st22, st1692, st1969, st9505, stil103, stpa57 | 7 (4) |
Several identical emm-types were originally assigned two numbers: emm-type 44 is identical to emm-type 61 (emm-type 44/61), emm-types 50 and 62 (50/62), emm-types 65 and 69 (65/69), emm-types 38 and 40 (38/40). 223 emm-types are listed in the CDC database (September 18, 2012) [21]. REA, rearranged emm pattern (atypical amplification patterns). ND, not determined.
emm-types from strains isolated prior to 1987. emm-types not included in this study, but whose emm pattern grouping was previously established [31], are indicated in italics. Isolates of emm-types 54 and st854 (underlined) are associated with more than one emm pattern group. Note that emm-types 7, 10, 16, 20, 21, 35, and 45 do not exist.
Molecular typing
PCR amplification and sequencing of emm genes was performed as previously described [14]. The alignment of the forward and reverse emm sequences was performed using the CodonCode Aligner® version 3.7 software with default parameters and were all manually checked. emm-type was determined by BLASTn analysis using the CDC emm-type database containing 223 emm-types [21]. After translation, the predicted amino acid sequences of all M proteins were trimmed from the first amino acid (AA) of the predicted mature protein to the first AA of the D repeat near the COOH-terminal end [14]. The size of mature M proteins (from the first NH2-terminal residue to the Thr of the LPXTG sortase motif) was calculated by adding 54 or 73 residues respectively to the sequence we obtained for the M proteins of patterns E or A-C and D, as described previously [4]. The emm pattern of at least one isolate of each of the 168 emm-types was experimentally determined following the PCR mapping methodology previously described [22] or deduced from previous publications [12, 14].
Bioinformatics
Multiple alignments of trimmed amino acid sequences belonging to the same emm-types were performed using the MUSCLE algorithm with default parameters. The presence of repeat sequences was detected by using T-reks with 3 different percentage similarity (Psim) thresholds (1, 0.9, and 0.7) and extensive manual analysis [23]. M protein annotation and structure prediction was performed with Geneious® 5.6 for one representative of each emm-type.
Statistical analysis
Two-tailed student's T-test were performed using Stata 12 software.
Results
Study population
The final dataset included 1086 GAS isolates representing 175 different emm-types recovered from 31 countries on six continents (Figure 1). Thus, this collection includes 78% of the emm-types described to date [21]. Twenty percent of the 175 emm-types belong to emm pattern A-C, while the remaining are distributed evenly among patterns D and E (Table 1). The number of isolates examined per emm-type ranged from 1 to 25 (mean 6.5) (Table S1). Clinical manifestations were reported for 1019 isolates: invasive diseases (n=365; 35.8%), pharyngitis (n=338; 33.2%) and skin infections (n=233; 22.9%; includes impetigo, wound infections and other unspecified skin infections). The remainder were associated with oropharyngeal carriage (n=46; 4.5%), non-suppurative sequelae (n=13; 1.3%) and other types of infections (n=24; 2.4%).
Updated structural model of M proteins
The size of the predicted mature form of M protein was highly heterogeneous, ranging from 229 to 509 residues. Importantly, M protein length was highly correlated with emm pattern. M proteins of pattern A–C were the longest (average 443 residues; 95% CI 427-463) followed by pattern D (average 360 residues; 95% CI 353-368) while those of pattern E were the shortest (average 316 residues; 95% CI 312-320) (Student's T-test; for 2-way comparisons among all pattern groups, t < 0.001).
emm sequence data, including detailed annotation of sequence repeats, for one representative of each of 175 emm-types are available in GenBank (accession numbers JX028599-JX028772, JX472406). The ‘A’ repeats are defined as amino acid sequence repeats beginning within the first 50 amino-terminal residues of the mature protein (i.e., emm typing region). Similarly, ‘B’ repeats are defined as sequence repeats starting between residue 51 and the beginning of the CRR. The ‘C’ repeats are defined by their homology with a highly conserved 35-residue block (supplementary data S2). Data show that a majority (65%) of M proteins do not possess ‘A’ repeat sequences. However, ‘A’ repeats are more frequent amongst the pattern A-C group, whereby ~50% of M proteins have ‘A’ repeats, than amongst the D and E (33 and 30% respectively). The presence of ‘B’ repeats also correlates with the emm pattern groupings: 57, 51 and 15% of M proteins of patterns A-C, D and E, respectively, possess ‘B’ repeats. When present, 85% of the ‘B’ repeats consist of only two repeat units in tandem (size range, 7 to 62 residues); higher numbers of ‘B’ repeat units were almost exclusively associated with M proteins of the pattern A-C group. Both ‘A’ and ‘B’ repeat sequences originating from different emm-types were rarely found to share extensive sequence homology. On the contrary, all M proteins possess a CRR. The number of ‘C’ repeat units ranges from 1 to 5, with the vast majority of M-types (90%) harboring 3 repeat units.
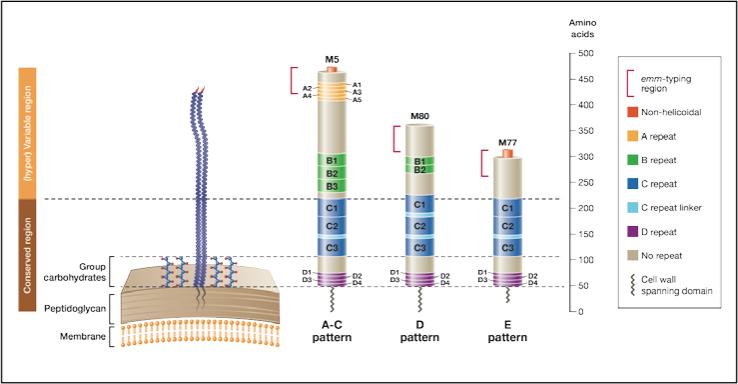
Based on the data obtained in this study, and on information from published literature [4, 9, 10, 13], we propose a new structural model with 3 representative M proteins (Figure 2). M5, M80 and M77 proteins were selected as prototypes for the structural characteristics within each emm pattern group. This model provides the advantage of being far more representative of M proteins from organisms recovered worldwide.
Figure 2. Three representative M proteins model.
Three representative M proteins (M5, M80 and M77) were selected as prototypes for the structural characteristics within each emm pattern group. M protein length and the size of the repeat and non-repeat regions are drawn to scale. Pattern A-C emm-types represent the longest M proteins, with a (hyper)variable portion of about 230 residues. In comparison, pattern D and E proteins possess a (hyper)variable portion of ~ 150 and 100 residues, respectively. The ‘A’ repeats are absent from the vast majority of M proteins belonging to the pattern D and E groups. The ‘B’ repeats are present in most of the pattern A-C and D emm-types, but absent from most of the pattern E emm-types. Thirty-five conserved residues constitute the ‘C’ repeat unit. Consecutive ‘C’ repeat units are sometimes separated by a seven residue unit called ‘C’ repeat linker (See supplementary data S2). Twenty percent of the M proteins (such as M80) do not possess non-helicoidal amino terminus. This proportion is 10%, 19% and 25% amongst the pattern A-C, D and E emm-types respectively. The portion of the protein considered by the emm-typing method is represented.
Sequence conservation within an emm-type
In order to examine sequence heterogeneity from isolates of the same emm-type originating from different geographic regions, we identified all emm-types recovered from at least five locations. 80 emm-types encompassing 900 isolates fulfilled this criterion (Table S3). Sixty-five (81%) emm-types showed intra-emm-type differences in the size of M-proteins (Table S3). Within each emm-type, an average mean of 69% of isolates belonged to the most common size variant. The most prevalent size variant was used as the basis for comparison to other size variants within each emm-type. Comparisons of the 900 protein sequences revealed 408 insertions or deletions. Indels (i.e. insertions or deletions) were found in similar frequencies across all emm pattern groups (data not shown). As classically observed with coiled-coil proteins, 304 (75%) indels involved a sequence stretch that is a multiple of seven residues, and this heptad periodicity increases from the amino- to carboxy-terminal ends of the protein (Figure 3). These observations suggest that strong selective pressures preserve the coiled-coil structure at the carboxy-terminal end of M protein, whereas the amino-terminal extremity may better tolerate variation in its higher order structure.
Figure 3. Insertion-deletion (indel) characteristics of M proteins belonging to the same emm-type.
Intra-emm-type alignments that include 900 M protein sequences of 80 emm-types reveal 408 indels. More than half of the indels (n=224; 55%) are located in the CRR (C Repeat Region). The remaining indels are equally distributed between regions corresponding to the 50 amino-terminus proximal residues (n=85; 21%; emm-type determinant) and from residue 51 to the beginning of the CRR (n=99; 24%; sub-N-terminal, central region). The number of indels having a heptad periodicity increases from the amino-terminal (36% of indels) to the carboxy-terminal (CRR; 99% of indels) regions of M proteins, whereas half (50%) of the indels from the central region of M protein involve multiples of seven residues.
M proteins assigned to the same emm-type are highly conserved across their surface exposed portions, despite differences in both geographical origins and clinical manifestations (Table S1 and S3). After excluding gaps, M protein sequences of the same emm-type are nearly identical, with an average pair wise identity ranging from 88% to 100% (Supplementary data S3). The median pairwise identity is 99%. Only two M-types, emm14 (pattern A-C) and emm100 (pattern D) exhibit an average pairwise identity <90%. One or two isolates belonging to each of those two emm-types presented an atypical M protein sequence which was vastly different from the others (protein identity between atypical and typical variants ranging from 63 to 77%). Although many emm-types share highly homologous central regions spanning residue 51 to the CRR, it was extremely rare to find a given emm-type paired with multiple, highly divergent sub-N-terminal domains. Thus the emm-type of an M protein is largely predictive of the structure of the full-length protein, indicating that the emm-typing method is far more informative than previously appreciated.
M protein conserved vaccine epitopes
The highly conserved nature of the CRR signifies that CRR-based vaccines can potentially target a wide range of M proteins [16, 24]. One such vaccine candidate, J14, consists of 14 amino acid residues derived from CRR [17]. In this study, 42 J14 variants were identified, including 17 newly recognized variants (Supplementary data S4). Seven J14 variants accounted for 89% of all J14 variants recovered from the 1078 isolates (Supplementary data S5). To prevent bias due to over-representation of particular emm-types, we also examined the distribution of J14 variants in single representatives of each emm-type with similar results. Specific J14 variants clearly segregate with emm pattern. For example, M proteins belonging to pattern A-C almost exclusively contain variant J14.0 in their third C-repeat unit, pattern D proteins contain a mix of both variant J14.0 and J14.1 whereas J14.0 is absent from pattern E proteins (data not shown).
Discussion
This study is the most comprehensive analysis of globally distributed GAS M proteins undertaken. The data provide a significant increase in our understanding of the M protein structure as a whole, a new understanding of the biological relevance provided by older typing tools such as emm typing and emm pattern determination and insights for the development of future GAS vaccine formulations.
Approximately 75% of emm-types belong to the pattern D and E groups. emm-types of pattern D and E are also frequently recovered in epidemiologic settings where there is a high GAS-associated mortality burden and a very high diversity of circulating emm-types [9, 10, 12, 14, 25]. Despite their epidemiologic relevance, these emm-types have not been as extensively characterised as those of the pattern A-C group. The structure of M6 protein served well in the past as representative of M proteins. However with increased knowledge of the structure of additional M proteins, it has become evident that extrapolations based on M6 protein are limited. First, M6 is a pattern A-C emm-type, which collectively account for only ~20% of emm-types. Secondly, M6 protein has five ‘A’ and five ‘B’ repeats and is non-representative of even the pattern A-C group because half of pattern A-C emm types lack ‘A’ repeats altogether and most possess fewer ‘B’ repeat units. Third, the size of M6 protein is smaller than most of the A-C pattern emm-type. Our data also demonstrate that ‘A’ repeats are rarely found in the pattern D and E emm–types while ‘B’ repeats are sometimes present, but usually as a single tandem repeat.
The emm-typing region, despite its short length, is largely predictive of the whole M protein sequence independent of clinical association or geographical origin. This finding suggests that emm-typing can be used to infer not only the N–terminal portion of the protein, but the entire surface exposed portion as well. M proteins are multifunctional, having roles in preventing phagocytosis, mediating adherence to host cells, and intracellular invasion, often through binding to human host products such as fibrinogen, factor H, albumin, IgA and IgG, plasminogen and others [26]. Many host proteins are bound to distinct regions or domains within M proteins. Therefore, the emm-typing system may be predictive of a unique array of biological functions for each emm-type.
In fungi, size variation generated by intragenic tandem repeats within surface protein genes allows for rapid adaptation to the environment and/or evasion of the host immune system [27]. In GAS, as previously described for M6 organisms [28], size differences in M protein from different isolates is also a common feature, largely a result of differences in the number of sequence repeat units. The M6 protein size mutants display heterogeneity in their antigenic and opsonogenic epitopes [28]. Our data show that intra-emm-type size variation occurs for most emm-types, and it is evenly distributed across the three emm pattern groups (data not shown). The significant differences in M protein length observed between the three emm pattern groups, and the close uniformity of M protein length within each emm pattern group, have not been previously described. Combined with knowledge that the emm pattern groups are associated with different clinical manifestations [13], it is tempting to speculate that M protein length underlies distinct functional attributes, perhaps related to a capacity to bind different subsets of host proteins. Future studies can assess whether M protein size is an attribute that impacts the virulence potential and clinical manifestations of GAS.
Our study is relevant to M protein-based vaccine design. A new multivalent antigen containing amino-terminal fragments from 30 M proteins showed unexpected in vitro cross protection against isolates expressing M proteins not included in the immunizing antigen [15]. Extensive cross-protection was not demonstrated with a 26-valent antigen produced previously by the same laboratory. The primary difference in the composition of the two vaccines is a significant increase in the number (from 11 to 18) of sequences representing M proteins belonging to pattern E, in the 30-valent vaccine. Antibodies elicited by the 30-valent vaccine were tested against isolates belonging to 40 emm-types. Rates of cross protection, measured by an opsonophagocytosis assay, differed by pattern group (pattern A-C, 60%; pattern D, 45%; and pattern E, 84%) [15]. The underlying mechanism for cross protection against non-vaccine types is not yet understood. However, these data suggest that the emm-types belonging to the different pattern groups might differ in their ability to induce cross-protective antibodies.
Another vaccine approach utilizes protective epitopes within the highly conserved CRR, and aims to confer broad protection against all GAS strains [19, 24]. This and other studies have shown that there are many variants of J14 sequences [17, 29]. Our current study confirms that virtually all C3 repeat units harbor J14.0 or J14.1 variants and that a small number of J14 variants are predominant within a global collection of isolates.
M protein size and structure are characteristic of the emm pattern group to which they belong. The pattern classification, based on the content of emm gene forms and their chromosomal arrangement, was also recognized as a strong marker of preferred tissue sites of infection by GAS [13, 29, 30]. From this exhaustive study, we propose that three M proteins - M5, M80, M77 - belonging to the three emm pattern groups A-C, D and E, respectively, best represent the structures and possible host-pathogen interactions mediated by M proteins. This new model is likely to be a valuable tool for epidemiological, molecular and vaccine studies.
Supplementary Material
Acknowledgements
The contributing members of the M protein study group (in addition to the authors of this paper) include Michael Batzloff and Rebecca Towers from Australia; Herman Goossens and Surhbi Malhotra-Kumar from Belgium; Luiza Guilherme and Rosangela Torres from Brazil; Donald Low and Allison Mc Geer from Canada; Paula Krizova from Czech Republic; Sawsan El Tayeb from Egypt; Joe Kado from Fiji; Mark van der Linden from Germany; Guliz Erdem from Hawai; Alon Moses and Ran Nir-Paz from Israel; Tadayoshi Ikebe and Haruo Watanabe from Japan; Samba Sow and Boubou Tamboura from Mali; Bard Kittang from Norway; José Melo-Cristino and Mario Ramirez from Portugal; Monica Straut from Romania; Alexander Suvorov and Artem Totolian from Russia; Mark Engel, Bongani Mayosi and Andrew Whitelaw from South Africa; Jessica Darenberg and Birgitta Henriques Normark from Sweden; Chuan Chiang Ni and Jiunn-Jong Wu from Taiwan; Aruni De Zoysa and Androulla Efstratiou from UK; Stanford Shulman and Robert Tanz from USA.
We also would like to sincerely acknowledge Bernard Beall for proofreading of the MS and for managing the very useful emm-typing database from CDC.
Funding
This work was supported by the European Society for Clinical Microbiology and Infectious Diseases, European Society for Paediatric Infectious Diseases, Fonds National de la Recherche Scientifique (Belgium), Fonds Brachet and Fondation Van Buuren (Belgium), Australian National Health and Medical Research Council, National Institutes of Health (AI-065572). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Contributors
PRS was the primary coordinator of data collection, analysis, and writing. DJM, LVM, and KSS supervised data collection, analysis, and writing. TV and PAD were primarily involved in laboratory experiments and data collection. DB, JG, ACS, and JRC were primarily involved in data collection, analysis and writing. All authors contributed substantially to the preparation of the paper.
This study was presented in part at the 29th annual meetings of the European Society for Pediatric Infectious Diseases in The Hague, The Netherlands in June 7-11, 2011; at the 7th World Congress of the World Society for Pediatric Infectious Diseases in Melbourne, Australia in November 16-19, 2011; at the Australian Society for Microbiology 2012 Annual Scientific Meeting in Brisbane, Australia in July 1–4, 2012 and at 52th ICAAC meeting in San Francisco, USA in September 9-12, 2012.
Conflict of interest
No conflict of interest.
References
- 1.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group a streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 2.Parks T, Smeesters PR, Steer AC. Streptococcal skin infection and rheumatic heart disease. Current opinion in infectious diseases. 2012;25:145–153. doi: 10.1097/QCO.0b013e3283511d27. [DOI] [PubMed] [Google Scholar]
- 3.Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smeesters PR, McMillan DJ, Sriprakash KS. The streptococcal m protein: A highly versatile molecule. Trends Microbiol. 2010;18:275–282. doi: 10.1016/j.tim.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 5.McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P. Coiled-coil irregularities and instabilities in group a streptococcus m1 are required for virulence. Science. 2008;319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham MW. Pathogenesis of group a streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischetti VA. Streptococcal m protein: Molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facklam R, Beall B, Efstratiou A, et al. Emm typing and validation of provisional m types for group a streptococci. Emerg Infect Dis. 1999;5:247–253. doi: 10.3201/eid0502.990209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group a streptococci: Systematic review and implications for vaccine development. Lancet Infect Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- 10.Smeesters PR, McMillan DJ, Sriprakash KS, Georgousakis MM. Differences among group a streptococcus epidemiological landscapes: Consequences for m protein-based vaccines? Expert Rev Vaccines. 2009;8:1705–1720. doi: 10.1586/erv.09.133. [DOI] [PubMed] [Google Scholar]
- 11.Hollingshead SK, Readdy TL, Yung DL, Bessen DE. Structural heterogeneity of the emm gene cluster in group a streptococci. Mol Microbiol. 1993;8:707–717. doi: 10.1111/j.1365-2958.1993.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 12.McGregor KF, Spratt BG, Kalia A, et al. Multilocus sequence typing of streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J Bacteriol. 2004;186:4285–4294. doi: 10.1128/JB.186.13.4285-4294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bessen DE, Lizano S. Tissue tropisms in group a streptococcal infections. Future Microbiol. 2010;5:623–638. doi: 10.2217/fmb.10.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smeesters PR, Mardulyn P, Vergison A, Leplae R, Van Melderen L. Genetic diversity of group a streptococcus m protein: Implications for typing and vaccine development. Vaccine. 2008;26:5835–5842. doi: 10.1016/j.vaccine.2008.08.037. [DOI] [PubMed] [Google Scholar]
- 15.Dale JB, Penfound TA, Chiang EY, Walton WJ. New 30-valent m protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group a streptococci. Vaccine. 2011;29:8175–8178. doi: 10.1016/j.vaccine.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey M, Batzloff MR, Good MF. Mechanism of protection induced by group a streptococcus vaccine candidate j8-dt: Contribution of b and t-cells towards protection. PLoS ONE. 2009;4:e5147. doi: 10.1371/journal.pone.0005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer M, Georgousakis M, Vu T, et al. Evaluation of novel streptococcus pyogenes vaccine candidates incorporating multiple conserved sequences from the c-repeat region of the m-protein. Vaccine. 2012 doi: 10.1016/j.vaccine.2011.12.115. [DOI] [PubMed] [Google Scholar]
- 18.Guilherme L, Alba MP, Ferreira FM, et al. Anti-group a streptococcal vaccine epitope: Structure, stability, and its ability to interact with hla class ii molecules. The Journal of biological chemistry. 2011;286:6989–6998. doi: 10.1074/jbc.M110.132118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerino MT, Postol E, Demarchi LM, et al. Hla class ii transgenic mice develop a safe and long lasting immune response against streptincor, an anti-group a streptococcus vaccine candidate. Vaccine. 2011;29:8250–8256. doi: 10.1016/j.vaccine.2011.08.113. [DOI] [PubMed] [Google Scholar]
- 20. http://unstats.Un.Org/unsd/methods/m49/m49regin.Htm.
- 21. http://www.cdc.gov/ncidod/biotech/strep/strepblast.htm.
- 22.McDonald MI, Towers RJ, Fagan P, Carapetis JR, Currie BJ. Molecular typing of streptococcus pyogenes from remote aboriginal communities where rheumatic fever is common and pyoderma is the predominant streptococcal infection. Epidemiol Infect. 2007;135:1398–1405. doi: 10.1017/S0950268807008023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorda J, Kajava AV. T-reks: Identification of tandem repeats in sequences with a k-means based algorithm. Bioinformatics. 2009;25:2632–2638. doi: 10.1093/bioinformatics/btp482. [DOI] [PubMed] [Google Scholar]
- 24.Bessen D, Fischetti VA. Synthetic peptide vaccine against mucosal colonization by group a streptococci. I. Protection against a heterologous m serotype with shared c repeat region epitopes. J Immunol. 1990;145:1251–1256. [PubMed] [Google Scholar]
- 25.Smeesters PR, Dramaix M, Van Melderen L. The emm-type diversity does not always reflect the m protein genetic diversity--is there a case for designer vaccine against gas. Vaccine. 2010;28:883–885. doi: 10.1016/j.vaccine.2009.10.137. [DOI] [PubMed] [Google Scholar]
- 26.Bisno AL, Brito MO, Collins CM. Molecular basis of group a streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- 27.Verstrepen KJ, Jansen A, Lewitter F, Fink GR. Intragenic tandem repeats generate functional variability. Nature genetics. 2005;37:986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones KF, Hollingshead SK, Scott JR, Fischetti VA. Spontaneous m6 protein size mutants of group a streptococci display variation in antigenic and opsonogenic epitopes. Proc Natl Acad Sci U S A. 1988;85:8271–8275. doi: 10.1073/pnas.85.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steer AC, Magor G, Jenney AW, et al. Emm and c-repeat region molecular typing of beta-hemolytic streptococci in a tropical country: Implications for vaccine development. J Clin Microbiol. 2009 doi: 10.1128/JCM.00312-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smeesters PR, Vergison A, Campos D, de Aguiar E, Miendje Deyi VY, Van Melderen L. Differences between belgian and brazilian group a streptococcus epidemiologic landscape. PLoS ONE. 2006;1:e10. doi: 10.1371/journal.pone.0000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGregor KF, Bilek N, Bennett A, et al. Group a streptococci from a remote community have novel multilocus genotypes but share emm types and housekeeping alleles with isolates from worldwide sources. J Infect Dis. 2004;189:717–723. doi: 10.1086/381452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.