Abstract
Many bacteria can exist as surface-attached aggregations known as biofilms. Presented in this unit are several approaches for the study of these communities. The focus here is on static biofilm systems, which are particularly useful for examination of the early stages of biofilm formation, including initial adherence to the surface and microcolony formation. Furthermore, most of the techniques presented are easily adapted to the study of biofilms under a variety of conditions and are suitable for either small- or relatively large-scale studies. Unlike assays involving continuous-flow systems, the static biofilm assays described here require very little specialized equipment and are relatively simple to execute. In addition, these static biofilm systems allow analysis of biofilm formation with a variety of readouts, including microscopy of live cells, macroscopic visualization of stained bacteria, and viability counts. Used individually or in combination, these assays provide useful means for the study of biofilms.
Keywords: biofilm, bacterial adhesion, microscopy, static biofilm assay, attachment, microcolony, microtiter dish
There are a variety of systems available for examining the formation of bacterial biofilms. In this unit, several approaches are described that are useful for studying, in particular, the earlier stages of formation of these communities. Static biofilm systems may be preferable to chemostat or continuous-flow methods for a number of reasons. First, static assays are particularly useful for examining early events in biofilm formation, in some cases detecting biofilm formation in <60 min. For example, these assays can be used to identify signals that modulate the transition from a planktonic to a biofilm mode of existence. A second major advantage of these systems is the simplicity of the protocols: these assays can be executed primarily using common laboratory equipment. Furthermore, several of the assays outlined below have a relatively high throughput and can easily be adapted to study a variety of biofilm formation conditions, making them excellent tools for performing genetic screens. Such assays can also be adapted to screen for small molecules that modulate biofilm formation (Junker and Clardy, 2007). In addition, simple changes in the configurations of these screens (e.g., media used, medium replacement regimens, incubation time, washing vigor) allow examination of different aspects of these bacterial communities, such as formation or dispersal.
The static nature of these systems does have some drawbacks. Because most of the cultures are neither continuously supplied with fresh medium nor aerated, there may be limitation of nutrients, and an inability to easily generate mature biofilms may be encountered. Continuous-flow and chemostat systems for the production of mature biofilms are described in other units in this chapter, and can address these limitations.
Presented in this unit are four basic protocols for the growth and analysis of biofilms in static systems. The microtiter plate biofilm assay (see Basic Protocol 1) is a useful method for assessing bacterial attachment by measuring the staining of the adherent biomass. Because it utilizes a 96-well plate format, it is suitable as a tool for screening large numbers of bacterial strains or species. The air-liquid interface (ALI) assay (see Basic Protocol 2) is complementary to the microtiter plate biofilm assay in that it provides a mechanism for direct microscopic viewing of the live attached microbes. A colony-based biofilm system is also described (see Basic Protocol 3), with this system being especially useful for monitoring cell death in biofilms treated with antimicrobial agents. Finally, the Kadouri system (see Basic Protocol 4) is a “low-flow” system that can serve as a bridge between the static assays discussed in this unit and continuous-flow systems.
CAUTION: Follow all appropriate biosafety requirements relevant to the microorganism under investigation. Refer to UNIT 1A.1 and other pertinent resources (see APPENDIX 1B) for instructions on safe handling of microorganisms.
BASIC PROTOCOL 1: MICROTITER PLATE BIOFILM ASSAY
This experimental system, whose most common format is often referred to as the 96-well plate assay, is a simple high-throughput method used to monitor microbial attachment to an abiotic surface. While popularized in the mid-to-late 1990s (Mack et al., 1994; O’Toole et al., 1999), the assay in its typically used form is derived from a protocol published by Christensen et al. (1985). Examples of bacteria studied in this manner are listed in Table 1B.1.1, although in theory, the protocol could be applied to any bacterial and fungal species amenable to growth in the prescribed format. In brief, cells are grown in microtiter dishes for a desired period of time, and then the wells are washed to remove planktonic bacteria. Cells remaining adhered to the wells are subsequently stained with a dye that allows visualization of the attachment pattern. This surface-associated dye can also be solubilized for semiquantitative assessment of the biofilm formed.
Table 1B.1.1.
Typical Conditions for Developing and Performing Microtiter Plate Biofilm Assays
| ORGANISM | INCUBATION TEMPERATURE (°C) | SOLVENT FOR SOLUBILIZATION OF STAINED BIOFILMS | REFERENCE |
|---|---|---|---|
| Agrobacterium tumefaciens | 28 | 100% dimethyl sulfoxide (DMSO) | Danhorn et al., 2004 |
| Escherichia coli | 25 | 80% ethanol/20% acetone | O’Toole et al., 1999 |
| Pseudomonas aeruginosa | 25–37 | 95% ethanol | O’Toole et al., 1999 |
| Pseudomonas aeruginosa | 25–37 | 30% acetic acid | Zegans et al., 2009 |
| Pseudomonas fluorescens | 25–30 | 95% ethanol | O’Toole et al., 1999 |
| Staphylococcus aureus | 37 | 33% glacial acetic acid | Stepanovic et al., 2001 |
| Streptococcus mutans | 37 | 95% ethanol or 100% DMSO | O’Toole et al., 1999 |
| Vibrio cholerae | 25–30 | 100% DMSO | O’Toole et al., 1999 |
Materials
Bacterial strains of interest
Appropriate media for bacteria under study (APPENDIX 2C)
70% ethanol
0.1% (w/v) crystal violet in water
Solvent (e.g., 30% v/v acetic acid in water; see Table 1B.1.1 for other options) for solubilizing dye and biofilm biomass
96-well microtiter plates, not tissue culture–treated (Becton Dickinson catalog no. 353911) with lids (Becton Dickinson catalog no. 353913)
96-prong inoculating manifold, sterile (DanKar Scientific)
Small trays (e.g., large pipet tip boxes) sufficient in size to hold 96-well microtiter plates
Optically clear flat-bottom 96-well plates, nonsterile
Plate reader or spectrophotometer
For small sample numbers (<20 strains or species):
-
1a
Inoculate each bacterium of interest in a 3-to-5-ml culture and grow to stationary phase.
-
2aDilute cultures 1:100 in the desired media. Pipet 100 μl of each diluted culture into each of four wells in a fresh microtiter plate which has not been tissue culture treated. Cover plate and incubate at optimal growth temperature for the desired amount of time.
- Lids for the microtiter dishes may be reused. Clean lids with 70% (v/v) ethanol and air-dry prior to each experiment.
- The time course for attachment varies depending on the organism and must be determined empirically, although when using this system, many organisms commonly studied will form a biofilm within 48 hr.
For large screens (>20 strains or species):
-
1b
Inoculate cells directly into sterile microtiter plates filled with 100 μl of the appropriate medium per well, using four wells per strain or species. Incubate overnight.
-
2b
Inoculate biofilm assay plates directly in 100-μl medium per well from the overnight microtiter plate cultures using a sterile 96-prong inoculating manifold. Cover assay plates and incubate at optimal growth temperature for desired amount of time.
-
3Set up four small trays in a series and add 1 to 2 inches of tap water to the last three.
- The first tray is used to collect waste, while the others are used to wash the assay plates.
-
4
Remove planktonic bacteria from each microtiter dish (step 2a or 2b) by briskly shaking the dish out over the waste tray. To wash wells, submerge plate in the first water tray and then vigorously shake out the liquid over the waste tray. Replace water when it becomes cloudy.
-
5
Add 125 μl of 0.1% crystal violet solution to each well. Stain 10 min at room temperature.
-
6Shake each microtiter dish out over the waste tray to remove the crystal violet solution. Wash dishes successively in each of the next two water trays (i.e., the two not used in step 4), and shake out as much liquid as possible after each wash.
- This step will remove any crystal violet that is not specifically staining the adherent bacteria. The wash trays can be reused for a number of plates, but the water should be replaced when its color becomes dark or when the efficiency of the washes is observed to decrease.
-
7Invert each microtiter dish and vigorously tap on paper towels to remove any excess liquid. Allow plates to air-dry.
- At this stage, the staining is stable and the dried plates may be stored at room temperature for at least several weeks.
-
8Add 200 μl of 30% acetic acid (or another appropriate solvent) to each stained well. Allow dye to solubilize by covering plates and incubating 10 to 15 min at room temperature.
- 30% acetic acid appears to more efficiently solubilize the CV stain than the previously recommended 95% ethanol, and does so from a wider range of microbes.
- A number of reagents may be used to solubilize biofilms. Some suggestions are listed in Table 1B.1.1, but solvents should be tested empirically for each organism.
-
9Briefly mix the contents of each well by pipetting, and then transfer 125 μl of the crystal violet/acetic acid solution from each well to a separate well in an optically clear flat-bottom 96-well plate. Measure the optical density (OD) of each of these 125-μl samples at a wavelength of 500 to 600 nm.
- The authors suggest four replicate wells for each strain or species used in step 2. The absorbance values from these replicates are sufficient to determine an average and standard deviation for each strain or species and thus provide a measure of the extent of biofilm formation. The number of replicates can be increased as desired.
- The OD wavelengths are presented as a range to allow for variations in the capabilities of plate readers.
ALTERNATE PROTOCOL 1: DIRECT ENUMERATION OF BACTERIA IN BIOFILMS
The biofilm formed in the microtiter dish can also be quantitated by directly enumerating the bacteria adhering to the surface. This method, while more time consuming, avoids one drawback of the crystal violet assay—crystal violet stains not only cells, but essentially any material adhering to the surface of the plate (for example, matrix components), and therefore, crystal violet staining may overestimate the number of adherent bacteria.
Additional Materials (also see Basic Protocol 1)
PBS (APPENDIX 2A), sterile
Agar plates of appropriate medium
Multichannel pipettor
8-ml plastic tubes with caps
Sonicator (e.g., Sonics and Materials VC-505)
Additional reagents and equipment for counting viable cells (Phelan, 1996)
Inoculate biofilm assay plates as described in Basic Protocol 1, steps 1a and 2a or steps 1b and 2b.
Wash wells six times, each time using a multichannel pipettor to add 100 μl sterile PBS per well and then vigorously shaking out the liquid over a waste container to remove planktonic cells.
Use scissors to cut each individual well from the microtiter plate. Add 100 μl PBS to each well.
- Add each well (i.e., the actual plastic well plus its contents) to a separate 8-ml tube containing 1.9 ml PBS (for a final liquid volume of 2 ml). Cap before and after addition.
- This step is performed so that each sample can be sonicated (step 5) in an 8-ml tube. Microtiter wells are not suitable for use as sonication vessels, as they are too small.
- Insert sonicator microtip and sonicate the contents of each tube for 8 sec at 40% power. Perform viable counts (Phelan, 1996) on the resulting suspensions by plating on agar medium to enumerate bacteria that were attached to the microtiter well surface.
- To verify that the sonication procedure does not reduce cell viability, perform viable counts on a separate culture of planktonic cells before and after sonication. If reduced cell viability is seen, adjust the sonication regimen accordingly.
- If desired, use crystal violet staining followed by microscopy to determine the efficacy of sonication in dislodging attached cells from microtiter well walls. Perform crystal violet assay as in Basic Protocol 1. Lack of a crystal violet ring indicates that the bacterial cells have been removed from the wall of the well.
BASIC PROTOCOL 2: AIR-LIQUID INTERFACE ASSAY
Often, a great deal can be learned about the biofilm formation behavior of a particular bacterium simply by observing the early stages of this process, including attachment and early microcolony formation. This point may be particularly relevant for mutants identified in the 96-well plate assay described above.
The air-liquid interface (ALI) assay provides a simple system for microscopic analysis of biofilm formation over a time range of ~4 to 48 hr (Caiazza and O’Toole, 2004). A 24-well plate is placed at an angle of 30° to 50° relative to horizontal, and stationary-phase cultures are diluted and slowly applied to these wells such that the upper edge of each culture aliquot is positioned at the center of a well’s bottom. The bacteria of interest are allowed to grow in this manner for an appropriate length of time, and then the wells are rinsed gently and viewed by phase-contrast microscopy. See Background Information for a more thorough discussion of this assay.
Materials
Bacterial strains of interest
Appropriate media for bacteria under study (APPENDIX 2C)
Flat-bottom 24-well plate, sterile, with lid
Large paper clips or microcentrifuge tube rack and lab tape
Inverted microscope
- Adjust a sterile, flat-bottom, 24-well plate so that it sits at a 30° to 50° angle from horizontal.
- The method used will depend on the brand of plate and the available materials. One approach is to affix four large paperclips to one end of the plate. The paperclips stably elevate the end of the plate to which they are attached, while the opposite end remains low (see Fig. 1B.1.1). Alternatively, one end of the plate may be rested on a microcentrifuge tube rack (or anything of the desired height) and stabilized with tape to prevent slipping.
- If possible, set up the angled plate in the warm room or incubator to be used for sample culturing, and load the wells there (step 4) to avoid potential disruption of the sample meniscuses upon transport.
Inoculate each bacterium of interest in a 3- to 5-ml culture and grow to stationary phase (or to some other appropriate stage of growth).
- Dilute the stationary-phase (or other appropriate) cultures 1:100 in appropriate media.
- Approximately 300 μl diluted culture will be needed for each well that is to be inoculated in the angled 24-well dish.
- Carefully pipet an aliquot (~300 μl) of each diluted culture into a separate well in the angled 24-well plate such that the upper edge of the aliquot just reaches the center of the bottom of the well. Cover plate with lid and incubate at appropriate temperature for desired period of time.
- When inoculating each well, it is important to slowly add the culture down the lower wall to avoid wetting the entire bottom of the well. The meniscus of the liquid should pass through the center of the well, which is often the most optically clear part. This will ensure that the air-liquid interface is appropriately positioned for viewing.
- Aspirate culture from the wells and gently wash twice, each time by adding 400 μl sterile medium and then aspirating. After completing these two washes, add ~200 μl medium (i.e., enough to cover the bottom of the well) to each well.
- Washing should remove most of the unattached bacteria from the wells. This is important, as planktonic cells can make viewing more difficult and can continue to colonize surfaces, potentially obscuring the air-liquid interface.
- Lay the plate flat on the stage of an inverted microscope and visualize by phase-contrast or epifluorescent microscopy. Focus just above the center of the bottom of the well to observe bacteria at the air-liquid interface.
- Depending on the bacterial species and conditions, the bacteria may form a well-defined aggregations at the ALI. The ALI will occur wherever the meniscus of the culture was in contact with the plastic, but it will be most visible in the center of the well, where the well tends to be the most optically clear. If desired, the ALI may be circled on the bottom of the plate with a wax pencil prior to microscopic analysis to aid in its identification. Some sample results obtained with Pseudomonas aeruginosa are shown in Figure 1B.1.2.
Figure 1B.1.1.
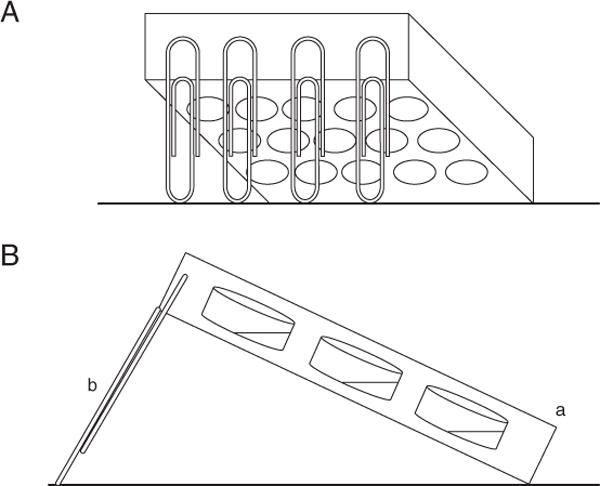
(A) Schematic of angled plate for air-liquid interface (ALI) assay (back view). (B) Side view of ALI assay setup. An angled plate (a) is supported by paper clips (b). The ALI occurs at the center of the bottom of each well, where a meniscus forms.
Figure 1B.1.2.
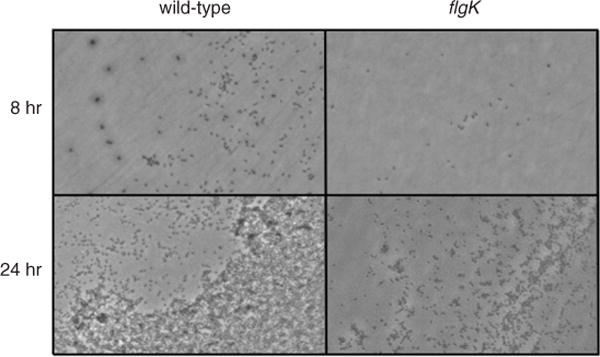
Phase-contrast micrographs taken at the air-liquid interfaces of biofilms formed by the wild-type Pseudomonas aeruginosa PA14 strain and a nonflagellated mutant (flgK), both grown as described in Basic Protocol 2.
ALTERNATE PROTOCOL 2: AIR-LIQUID INTERFACE COVERSLIP ASSAY
If an inverted microscope is not available, a plastic or glass coverslip can be placed in a well of a in a 24-well plate such that the air-liquid interface of the culture is in the approximate center of the coverslip. After an appropriate incubation, the coverslip is rinsed, stained with crystal violet, and visualized by conventional microscopy using an upright microscope.
Additional Materials (also see Basic Protocol 2)
0.1% (w/v) crystal violet in water
Flat-bottom multiwell (e.g., 12-well) plates, sterile, with lids
Glass or plastic coverslips (e.g., Fisherbrand unbreakable 22-mm2 coverslips; Fisher Scientific catalog no. 12-547)
Conventional, upright microscope
Inoculate each bacterium of interest in a 3- to 5-ml culture and grow to stationary phase (or to some other appropriate stage of growth).
Dilute the stationary-phase (or other appropriate) cultures 1:100 in appropriate media.
- Use each diluted culture to fill a well in a flat-bottom multiwell plate to approximately half of its capacity.
- Almost any size multiwell dish is acceptable for use in this assay, as coverslips are available in a variety of sizes. Plastic coverslips, such as those listed above, can easily be cut to the appropriate size with a pair of scissors.
Insert a coverslip into each well so that it is held at a 90° angle relative to the bottom of the well (i.e., perpendicular to the bottom of the well) so that the meniscus of the medium is at the center of the coverslip. Cover plate and incubate at appropriate temperature for desired period of time.
Remove each coverslip from its well and rinse off nonadherent cells by dipping in sterile medium.
Stain bacteria by submerging coverslips in 0.1% crystal violet for 10 min.
Rinse off excess dye by dipping each coverslip in two successive water baths, and then allow coverslips to air-dry.
Visualize the bacteria at the air-liquid interface on each coverslip by microscopy.
BASIC PROTOCOL 3: COLONY BIOFILM ASSAY
A colony biofilm is grown on a semipermeable membrane that sits on an agar plate (Fig. 1B.1.3). The bacteria in this type of biofilm can be given a new supply of nutrients simply by relocating the membrane-grown cells to a fresh agar plate. One therefore can easily change the carbon source or type of drug treatment while avoiding the need to wash the cells. The colony biofilm has been especially useful in the study of the antibiotic-resistant properties of cells. The method provided below is an example of the use of a colony biofilm to study the effects of antibiotic treatment.
Figure 1B.1.3.
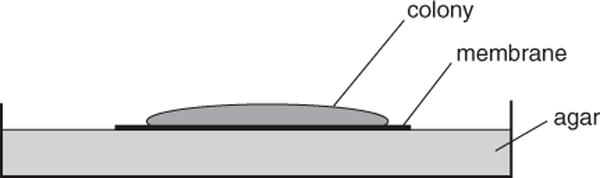
Side view of a colony biofilm assay.
The other biofilm systems presented in this unit involve attachment of bacteria to a surface while bathed in liquid medium. This is the only protocol where the substratum to which the bacteria attach themselves is also the avenue for meeting nutritional needs and removing waste. For that reason, this assay is more analogous to growing bacteria on plates rather than in broth. In this protocol, however, the colonies can grow much larger, because they are periodically relocated to fresh plates and thus have a fresh supply of nutrients. An advantage of this system is that the biofilms formed are derived from clonal growth of the original population of bacteria deposited on the filter, whereas in liquid media, motile bacteria may be able to “invade” an aggregation of cells. Likewise, in the system described below, bacteria have limited ability to detach and migrate or drift away from the biofilm formed. The stable and spatially restricted nature of this system makes the colony biofilm especially useful for observing the effects of antimicrobial agents, as changes in cell number can be more easily attributed to cell death rather than detachment.
Materials
Bacterial strain of interest
Appropriate liquid medium for bacterial strain under study (APPENDIX 2C)
Appropriate agar medium with and without antibiotic (or other) supplementation
PBS (APPENDIX 2A), sterile
Forceps, sterilized (e.g., autoclaved or flame-sterilized in 70% v/v ethanol)
Poretics 25-mm-diameter black polycarbonate membranes with pore size of 0.22 μm (GE Osmonics catalog no. K02BP02500)
100-mm-diameter Petri dishes, sterile
UV light source (e.g., UVP 8-W multiple-ray laboratory lamp)
15-ml tubes, sterile, with tightly fitting lids
Vortex mixer
On the night before the colony biofilm assay is to be started, inoculate the bacterium of interest in 3- to 5-ml cultures and grow overnight (i.e., to stationary phase).
- Using sterilized forceps, place 25-mm black polycarbonate membranes with a pore size of 0.22 μm in a sterile Petri plate. Position plate ~30 cm from a UV light source and treat with UV light in a sterile environment (e.g., a biological control hood) for 10 min per side, using the sterilized forceps to flip the plate.
- Here and in all subsequent steps, the Petri plate should be kept covered whenever possible to maintain the sterility of the membranes.
- The number of membranes needed depends on the desired length of the experiment. Two membranes are required for the initial baseline count of cells (in duplicate), and four more are needed for each time point (for duplicate sampling of treated and untreated cells). Additionally, a few extra membranes may be desirable as backups. For example, starting with sixteen membranes per strain for an experiment with a 72-hr antibiotic treatment period will allow duplicate sampling of treated and untreated cells every 24 hr, leaving two extra backup membranes per strain.
Dilute stationary-phase cultures (step 1) in the appropriate medium to an OD600 of 0.05 (~1.64 × 108 cfu/ml).
Place a membrane (step 2) with its shiny side up on an agar medium plate not containing antibiotic, and inoculate with 5 μl diluted culture. Repeat this procedure for all remaining membranes from step 2, inoculating up to six membranes on the same agar plate. When the liquid on the membranes has dried, incubate plates upright at appropriate temperature for 24 hr.
- Using sterile forceps, gently lift each membrane off of its agar plate and transfer to a fresh agar plate at up to six membranes per plate. Incubate plates 24 hr at appropriate temperature.
- Flame-sterilized forceps must be cooled before touching the membranes.
- The entire surface of the membrane should be in direct contact with the agar in the fresh plate. Air bubbles may be removed simply by gently lifting the membrane and repositioning it on the plate.
- Following this step, the colony biofilms will have been grown for a total of 48 hr. Membranes are transferred to new plates every 24 hr hereafter to provide a fresh nutrient supply or to expose the colony biofilms to fresh antimicrobial agents.
- After the incubation in step 5 is complete, sample biofilm growth in the following way.
- Aseptically transfer a membrane to each of two separate 15-ml tubes containing 10 ml sterile PBS, such that each tube contains a single membrane.
- The membrane can be gently folded over onto itself before it is lifted off the plate. Doing so will allow the membrane to fit easily into a 15-ml tube.
- Cap tubes and vortex samples for two pulses of 60 sec each (or as necessary) to detach all bacteria.
- Prepare dilution series of the vortexed samples, and plate each dilution on a separate agar plate. Incubate plates overnight at the appropriate temperature and determine the average number of colony-forming units (cfu) per membrane.
- The counts obtained here will serve as the baseline for future experimentation; this sampling time point is designated T = 0 with respect to the start of experimental treatments.
Transfer half of the remaining membranes (step 5) to fresh agar plates not containing antibiotic, and transfer the other half to agar plates supplemented with antibiotic at the appropriate concentration. Incubate all plates at appropriate temperature for 24 hr.
When incubation is complete, sample biofilm growth (as in step 6) in two untreated membranes and two treated membranes. Transfer the remaining untreated membranes to fresh agar plates not containing antibiotic, and transfer the remaining treated membranes to fresh agar plates supplemented with antibiotic at the appropriate concentration. Incubate plates at appropriate temperature for another 24 hr.
Repeat step 8 until all colony biofilms have been sampled.
BASIC PROTOCOL 4: KADOURI DRIP-FED BIOFILM ASSAY
This system overcomes one limitation of the other static systems presented in this unit by constantly refreshing the bacterial growth medium. Bacterial growth can therefore be maintained for a much longer period, allowing formation of mature biofilms. A 6-well plate is inoculated with culture, and growth medium is pumped into each well through a needle that has been inserted through the plate lid; waste and planktonic cells exit through a second needle and are pumped away from the well. Biofilm formation can be monitored as desired by visualizing the bottom of each well using an inverted microscope. See Background Information for a more thorough discussion of this assay.
Materials
Silicone sealant
Sodium hypochlorite
70% (v/v) ethanol
Bacterial strain of interest
Appropriate liquid medium for bacterial strain under study (APPENDIX 2C)
20-G needles, sterile (Becton Dickinson catalog no. 305175)
Flat-bottom 6-well plates, sterile, with lids
1/16 × 1/16–in. (~0.16 × 0.16–cm) straight and 90°-elbow, barbed, polypropylene fittings (such as those included in Cole-Parmer catalog no. 6365-90)
0.8- to 1.6-mm-i.d. silicone tubing (e.g., Watson-Marlow catalog no. 913.A008.016 or 913.A016.016)
2-liter flasks
Two-hole rubber stopper (no. 8) equipped with a long glass tube (i.e., one that can reach almost to the bottom of a 2-liter flask) and a short glass tube (for gas exchange)
Peristaltic pump (e.g., Watson-Marlow PumpPro) equipped with Marprene manifold tubing (inner diameter, 0.8 mm; e.g., Watson-Marlowe catalog no. 978.0165.000)
Inverted microscope (UNIT 2A.1)
Modify 6-well plate lid
- Heat a 20-G needle in the flame of a Bunsen burner. Push the heated needle through the lid of a 6-well plate such that it enters directly above one side of a well. Remove the needle and repeat this process to make a hole in the lid directly above the opposite side of the same well.
- The holes should be large enough for the needle to pass through freely.
- These holes will serve as ports for connecting a peristaltic pump that will supply growth medium to and carry waste away from the well. If desired, multiple wells may be prepared on the same plate and/or multiple plates may be run from the same pump simultaneously, depending on the number of channels the pump has for tubing.
Obtain two straight 1/16 × 1/16–in. (~0.16 × 0.16–cm) barbed polypropylene fittings (i.e., the type used to connect two pieces of tubing). Using a sharp razor blade, carefully cut off one of the two male ends on each fitting to give a modified fitting with one male end and a flat base (see Fig. 1B.1.4).
- Set the flat base of each modified fitting on top of the 6-well plate lid (step 1) such that the hole in the fitting lines up with one of the holes in the lid, and then pass a 20-G needle through both the fitting and the hole in the lid to maintain the alignment. Secure each fitting to the lid of the plate using a silicone sealant, and let the sealant dry completely (see Fig. 1B.1.4).
- Once constructed, this modified lid can be sterilized with ethanol or UV irradiation and reused multiple times with fresh plates in subsequent experiments.
Figure 1B.1.4.
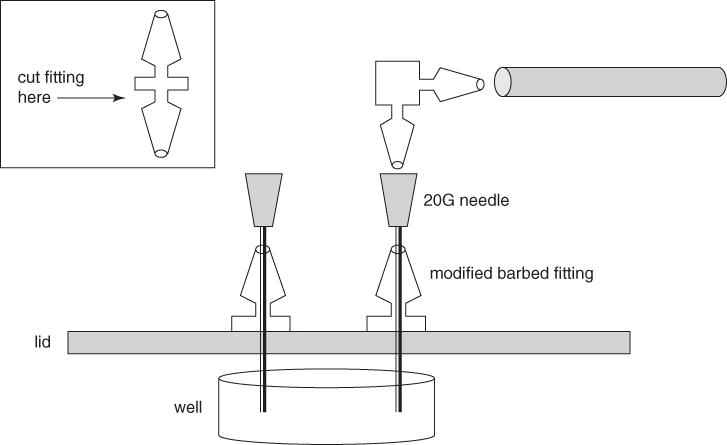
Side view of modified 6-well plate lid for Kadouri system.
Set up pump apparatus
Taking into account the Marprene manifold tubing already installed at the various ports on the peristaltic pump, cut lengths of silicone tubing to sizes appropriate for the following connections (Fig. 1B.1.5): (1) medium reservoir to pump; (2) pump to 6-well plate; (3) 6-well plate back to pump; and (4) pump to waste receptacle. Attach 90°-elbow fittings to the tube ends that are to be connected to the modified plate lid.
Figure 1B.1.5.
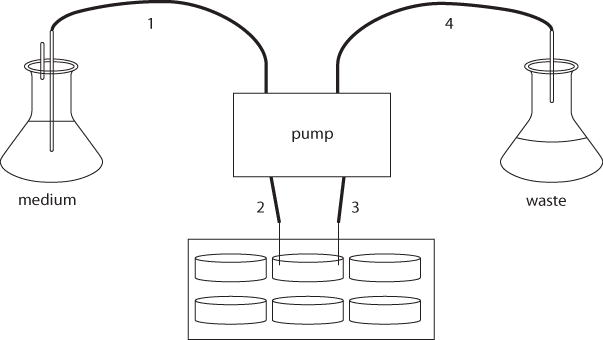
Diagram of Kadouri drip-fed biofilm system. Fresh culture medium is pumped onto a biofilm grown in the bottom of a well in a 6-well plate, while planktonic bacteria and spent medium are removed through a needle placed on the opposite side of the well. Numbers are explained in Basic Protocol 4, step 4.
Prepare to run the system
-
5Fill a 2-liter flask with 500 ml water and fit with a no. 8, two-hole, rubber stopper equipped with one long and one short glass tube. Cover the tube ends with foil. Fill separate 2-liter Erlenmeyer flasks with 1.5 liters water, 500 ml water, and 1.5 liters of medium appropriate for growing the biofilm of interest.
- The 500-ml portion of autoclaved water in the stoppered flask will be used to adjust the liquid flow rate in step 8. The other 500-ml portion of autoclaved water will serve as the solvent for the hypochlorite solution used to sterilize the pumping system in step 10, and the 1.5-liter portion of autoclaved water will be used to rinse the hypochlorite solution out of the system in step 11.
-
6
Submerge the silicone tubing from step 4, not including the Marprene manifold tubing already installed on the pump, in a beaker containing distilled water and cover with foil. Autoclave along with the flasks prepared in step 5.
-
7Connect the plate lid to the medium reservoir line and the waste line as follows (also see Fig. 1B.1.5).
- Using polypropylene fittings, connect one of the intake lines on the pump to the silicone tubing that will eventually lead to the medium reservoir. Connect the corresponding outflow line to the end opposite the 90° elbow on one of the silicone tubes that will eventually connect to the modified plate lid.
- Connect another outflow line on the pump to the silicone tubing that will eventually lead to the waste receptacle. Connect the corresponding intake line to the end opposite the 90° elbow on the other silicone tube that will eventually connect to the modified plate lid (Fig. 1B.1.5).
- Attach a sterile 20-G needle to the elbow fitting on each of the two silicone tubes that are to be connected to the modified plate lid.
-
8Adjust the growth medium influx and efflux rates in the following way.
- Place a sterile, capped microcentrifuge tube in a rack and make two small, diametrically opposed holes in the lid using a 20-G needle heated over a flame.
- Pipet ~1 ml of flow-adjustment water (step 5) into the open tube, close the lid, and then insert the 20-G needles from step 7 through the holes in the lid.
- Return the two-hole rubber stopper to the flask containing the flow-adjustment water. Connect the silicone tubing that will eventually serve as the growth medium intake line to the long glass tube on the stopper, and place the open end of the appropriate silicone tube in the waste receptacle.
- Start the peristaltic pump at a flow rate of 12 ml/hr (2.0 rpm on a Watson-Marlow PumpPro). Adjust the tension on the manifold tubing such that the water level in the microcentrifuge tube is steady or slightly decreasing.
- The purpose of this step is to adjust the liquid flow such that the well will neither dry out nor overflow during the actual experiment. Increasing the flow rate and observing for a period of time will make subsequent recognition of steady or slightly decreasing water levels easier.
-
9
Stop the pump, remove the 20-G needles from the microcentrifuge tube, and insert them into the modified fittings on the 6-well plate lid.
-
10To 500 ml sterile H2O (step 5), add sodium hypochlorite to a final concentration of 0.05% (v/v). Remove the two-hole rubber stopper (still connected to the eventual growth medium intake line) from the flask containing the flow-adjustment water, squirt 70% ethanol around its sides, and transfer it to the flask containing the sodium hypochlorite solution. Restart the pump and run the sodium hypochlorite solution through the system for at least 3 hr at a flow rate of 12 ml/hr.
- Rinsing with 70% ethanol solution will help ensure sterility each time the rubber stopper is transferred to a new flask. In addition, flushing with sodium hypochlorite sterilizes the pump’s Marprene tubing, which cannot be autoclaved.
-
11Turn off the pump and aseptically transfer the two-hole rubber stopper (still connected to the eventual growth medium intake line) to the flask containing 1.5 liters of sterile water for rinsing. Restart the pump and flush the system with this water for 3 hr or overnight, adjusting the flow rate as necessary to equalize the inflow and ouflow of liquid.
- This rinsing step removes the sodium hypochlorite from the system and helps ensure that the input and output flow are balanced.
Grow and monitor biofilm
-
12
Inoculate the bacterium of interest in a 3- to 5-ml culture and incubate overnight at appropriate temperature.
-
13
Dilute the stationary-phase culture 1:100 in appropriate medium, and inoculate ~3 ml of this diluted culture into the appropriate well of a new 6-well plate (i.e., a well whose position on the plate will allow the bacteria to receive growth medium through the modified plate lid). Allow bacteria to adhere to the well for 3 hr or as appropriate for the organism of interest.
-
14
At the end of the adhering period, pipet excess medium from the well and replace with ~3 ml fresh medium.
-
15
Turn off the pump and transfer the two-hole rubber stopper (still connected to the growth medium intake line) to the flask containing 1.5 liters sterilized medium.
-
16
Transfer the modified plate lid (still connected to the peristaltic pump through two 20-G needles) to the 6-well plate from step 14. Pass medium into the well of interest by restarting the pump at a rate of 10 ml/hr, and maintain this flow throughout biofilm growth.
-
17
Establish biofilm by incubating the system at appropriate temperature and replenishing the growth medium reservoir flask as necessary.
-
18Monitor biofilm growth at desired intervals in the following way.
- Pause the pump, transfer the modified lid to another sterile 6-well plate, and place a new sterile lid over the biofilm-containing plate for transport.
- Gently remove planktonic cells from the biofilm-containing well by pipetting off the liquid medium, and then wash the well twice, each time by adding 1.0 ml sterile medium and aspirating.
- Cover the bottom of the well with a small amount of fresh medium to keep the biomass hydrated, and then view through an inverted microscope.
- When viewing is complete, reconnect the plate to the pump system (through the modified plate lid) and resume the flow of growth medium.
Dismantle system and prepare for reuse
-
19
Upon completion of experiment, detach the modified 6-well plate lid and 20-G needles from the elbow fittings on the silicone tubing, and then use a small (~15-cm) piece of silicone tubing to bridge the two elbow fittings.
-
20
Sterilize the system by flushing with 0.05% sodium hypochlorite as in step 10, and then rinse by flushing with sterile water as in step 11.
-
21Dismantle all tubing and coil it gently into the bottom of a 1-liter beaker. Cover with distilled water, cover beaker with foil, and autoclave to sterilize.
- After autoclaving, the tubing may be reused for future experiments; however, before setting up a new system, be sure to examine the entire length of each piece of tubing for clogs or holes that could cause pressure build-ups or leaks.
COMMENTARY
Background Information
Microtiter Plate Assay
Permutations of the microtiter dish biofilm assay (see Basic Protocol 1) have been in use for a number of years, due to their ease of use and the adaptability of the general protocol to a variety of applications. This assay is equally well suited to the analysis of tens or hundreds of strains or species and is particularly useful as the basis of a genetic screen for mutants defective in surface attachment capabilities. Once conditions have been determined in which the wild-type strain forms a biofilm, a library of mutants can be screened to identify those that differ in terms of the extent or kinetics of attachment. Additionally, slight modifications to this system would allow one to look at the influence of other variables that may affect biofilm formation, such as nutritional provision (e.g., the carbon or nitrogen source used or the availability of phosphate or iron) or the presence of antibiotics or detergents. It is important to keep in mind that due to the indirect nature of biofilm assessment in this system, it is desirable to pair this assay with another method to further examine biofilm formation. For example, using direct microscopic observation of the biofilm to confirm the crystal violet staining pattern is suggested.
Air-Liquid Interface Assay
This technique (see Basic Protocol 2) emerged from a simple adjustment to the typical practice of growing biofilms in multiwell plates. Many bacteria are limited to or prefer aerobic growth and therefore are most prone to forming biofilms at the interface between the medium and air. By tilting the 24-well plate such that the air-liquid interface (ALI) falls on an optically clear portion of the plate, the potential for microscopic analysis is improved. This system makes an excellent companion to the microtiter dish assay (see Basic Protocol 1), as both are useful at many of the same time points and stages of biofilm formation. This assay also allows the visualization of live bacteria without the need for crystal violet staining; therefore, real-time interactions of the bacteria with the surface can be monitored (Caiazza and O’Toole, 2004). The ALI assay can be extended to later time points by replacing the growth medium periodically, but for an accurate study of a mature biofilm, a system in which the medium is continually replaced, such as the Kadouri system (see Basic Protocol 4) or a flow cell–based system (UNIT 1B.2), is recommended.
Colony Biofilms
Colony biofilms (see Basic Protocol 3) have typically been used for the purpose of determining antibiotic resistance (Anderl et al., 2000; Walters et al., 2003). Such systems are well suited to studies of the viability of cells within a biofilm in response to various assaults, such as exposure to antimicrobial agents or UV light.
Kadouri System
The Kadouri drip-fed biofilm system (named for the postdoctoral researcher who developed it) is a close relative of the continuous-flow biofilm systems presented in other units in this chapter. Although the medium is constantly being exchanged in this system (see Basic Protocol 4), the shear forces on the bacteria are minimal; therefore, this system may apply less mechanical stress to the biofilms. Planktonic cells in this system are not swiftly swept away but may linger in the well for longer periods. Hence, the system may be more closely representative of an equilibrium state between the attached and planktonic lifestyles. This system is typically considered a “low-flow” scheme and serves as an intermediate regimen between static assays and standard flow cell systems; however, because the system involves continuous feeding with growth medium, it allows observation of the entire process of biofilm formation, from reversible to irreversible attachment and on to microcolony formation, macrocolony formation, and detachment. One advantage this system provides is the potential to generate and easily access a relatively large amount of biomass that can be scraped from the bottom of the well and used for applications such as DNA microarrays and proteomics. This system is also useful for many of the microscopy assays typically done by flow cell–based methods. The 6-well plate format also has the advantage of being easily adaptable to the testing of multiple growth conditions or bacterial species.
Critical Parameters and Troubleshooting
Microtiter Plate Assay
When using the microtiter plate assay, it is important to have a strong positive control (i.e., a wild-type strain) and, if possible, a negative control (e.g., uninoculated medium) on each test plate. Enough variability can emerge in terms of incubation conditions or washing so that intraplate comparisons are preferable, especially when one strives to observe a subtle difference between bacterial species or strains. Additionally, it is important to note that there are factors that can affect the results of this assay other than an organism’s ability to adhere to the surface. For example, a growth defect could easily be mistaken for a lack of attachment capability.
The microtiter plate suggested for the studies described in this protocol is U-shaped, nonsterile, and relatively inexpensive. Nonsterile plates work well with fast-growing organisms such as E. coli or pseudomonads, which easily outcompete contaminants in the 8 to 10 hr it takes to complete the biofilm assay. For slow-growing or fastidious organisms, however, it is recommended that sterile plates be used. For nonmotile organisms, such as staphylococci, sterile, flat-bottom plates are recommended.
ALI Assay
Although the ALI assay involves a relatively simple system, there are a few important points necessary for successful execution. The ALI should be a well-defined boundary, so care must be taken to avoid wetting the entire bottom of the well during inoculation. Setting up the angled plate in an incubator or warm room and inoculating it in place is a simple way to prevent disturbances to the system. Placement of the air-liquid interface at the center of the bottom of the well will provide the best opportunity for obtaining sharp images by phase-contrast microscopy. In addition, the wells should never be allowed to dry out. If this is a problem, consider adding more medium in the initial setup, decreasing the incubation time, or placing the entire system inside a humidified chamber, which can be improvised by placing some dampened paper towels in the bottom of a plastic container with a lid.
Occasionally, there may be difficulty in locating the air-liquid interface for microscopic examination. There are a number of situations that could lead to this problem. If the initial inoculum has a low cell count and the incubation time is short, there may be an insufficient number of bacteria adhered to the surface at the time of viewing, and this may make identification of the ALI more difficult. If this is the case, consider starting with a less dilute inoculum or increasing the incubation time. Alternatively, circling the ALI with a wax pencil may make it easier to locate under the microscope. Stringent washing of the wells is useful for removing any unattached bacteria; this wash step will help prevent colonization of the formerly dry portion of the well after the whole well has been bathed in medium. Even under the best washing conditions, bacteria can detach from surfaces and colonize other portions of the well. For this reason, it is important to work quickly and analyze plates on a microscope immediately after they have been prepared for observation. If there are many strains or species to analyze, it may be advisable to distribute them over a number of plates so that only a few need be prepared for viewing at one time.
Colony Biofilms
One important aspect of this assay is the predetermination of appropriate experimental conditions (e.g., antibiotic concentration). The resistance of a bacterium to antimicrobial agents in a colony biofilm may differ significantly from the resistance of the same bacterium in a different biofilm system, such as the microtiter plate biofilm assay system described earlier in this unit (see Basic Protocol 1). It is recommended that the organism be tested on various concentrations of antibiotic to find appropriate experimental conditions.
The initial inoculation of the colony biofilm is an important step that can affect the success of the experiment. It is important to ensure that each membrane receives an equal amount of cells and that there is only a single point of inoculation in the center of the membrane. To prevent any of the culture from spraying elsewhere on the membrane, touch the drop of liquid at the end of the pipet tip gently to the membrane before depressing the pipet plunger to its final stop. A single, carefully centered spot will ensure the greatest experimental reproducibility, as the counting is no longer accurate if the bacteria have grown off the edges of the membrane. In addition, complete and uniform removal of the bacteria from the membrane is important for accuracy. Empirically testing different conditions to maximize removal of bacteria from the membrane during the vortexing step while preserving viability is recommended.
Kadouri System
Two common problems that are experienced in this system are overflow and drying out of the wells. Both can be addressed by very carefully calibrating the influx and efflux rates (see Basic Protocol 4, step 8). If the medium is evaporating too quickly from the system, consider increasing the influx rate to compensate. Likewise, ensuring that there is sufficient medium in the reservoir is especially crucial when the system will be left unattended for long periods. Due to the nature of biofilm-forming bacteria, it is also important to carefully monitor the tubing prior to each experiment. The waste tubes are especially prone to being colonized, and the colonizing bacteria can completely block the flow, causing pressure to build up and rupture the junctions between pieces of tubing. If blockage is found, the piece of tubing must be trimmed to remove the clogged portion, or else the entire section must be replaced.
This system is designed for observation of biofilms forming on the bottoms of the wells, but depending on the conditions and the strain or species used, the cells may in addition (or instead) form a biofilm, known as a pellicle, at the air-liquid interface. To prevent this, the environment at the bottom of the plate should be made as hospitable as possible for the bacteria. A common problem is the lack of oxygen at the bottom of the well. This may be overcome by increasing the flow rate in the system to deliver more oxygenated medium to the cells while more rapidly removing unattached bacteria. In some cases, adjusting the composition of the medium (e.g., by providing an alternative electron acceptor) will decrease the requirement for oxygen. As with all of the systems presented in this unit, optimal conditions must be determined empirically for each organism.
After the setup is complete, this system requires little daily attention as the cells grow. It should be monitored periodically (twice a day) for overflow or evaporation of the media in the wells and for potential rupture of tubing, and also to ensure that there is an ample supply of medium in the reservoir. However, biofilms may take over a week to develop to the desired stage (as determined by microscopic observation), and this should be taken into consideration when planning experiments.
Anticipated Results
Microtiter Plate Assay
Using the microtiter plate assay is an effective way to monitor bacterial attachment to an abiotic surface over time. In general, if this assay were performed over several time points, one would expect to see a progressive increase, possibly followed by a decrease, in attached biomass. The eventual decrease in crystal violet staining is presumed to occur because the lack of nutrients may stimulate the bacteria to detach from the surface (Sawyer and Hermanowicz, 1998; Hunt et al., 2004). The time course of biofilm growth must be determined empirically for each organism and set of conditions used. The staining pattern also varies from one organism to another. For example, when P. aeruginosa is grown with only oxygen as an electron acceptor, a ring of staining forms at the air-medium interface. In contrast, when this species is grown on arginine, a carbon source used aerobically and anaerobically by P. aeruginosa, the entire well shows positive crystal violet staining. Examples of typical results with this assay for P. aeruginosa are shown in Figure 1B.1.6.
Figure 1B.1.6.
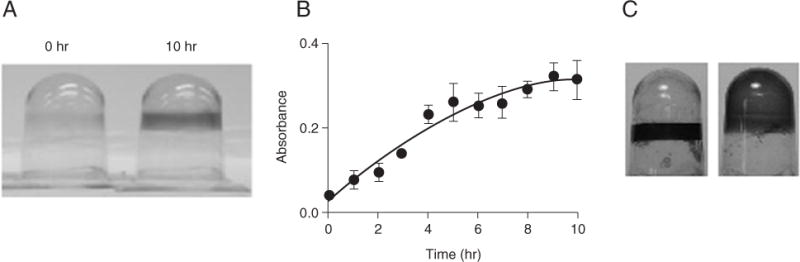
Microtiter plate biofilm assay of Pseudomonas aeruginosa. (A) Crystal violet–stained wells from a microtiter dish at 0 and 10 hr postinoculation. The bacteria were grown on glucose supplemented with casamino acids. Wells were inverted to facilitate photography. (B) Quantification of staining at various time points over 10 hr, based on absorbance readings made at 600 nm. (C) The wells show formation of a biofilm when P. aeruginosa is grown on LB (left; requires aerobic growth) or on arginine (right; can be utilized anaerobically).
Direct Enumeration of Bacteria from Microtiter Plate Assay
When bacteria are grown in the microtiter plate assay, one generally expects to see increased adherent biomass over a period of time followed by a decrease as the bacteria detach or die. The number of cfu eluted from each well is expected to correlate with the amount of crystal violet staining obtained at the corresponding time point.
ALI Assay
For bacterial species that form a ring around the well in the microtiter plate assay, the air-liquid interface should be heavily colonized with attached bacteria in this system. For nonmotile or facultative anaerobic bacteria, the boundary may not be as well defined. Some typical results obtained with the bacterium P. aeruginosa are displayed in Figure 1B.1.2.
ALI Coverslip Assay
The results for this variation of the ALI assay should be very similar to those noted above, with the exception that the crystal violet staining kills the bacteria. Because of this, the stained coverslips are relatively stable and may be stored upwards of a week prior to microscopy.
Colony Biofilms
The colony biofilm protocol allows assessment of the effects of antibiotics (or other experimental treatments) on a colony of bacteria grown on a solid substratum. Typically, it is expected that bacteria within a biofilm will display a greater resistance to bacteriocidal agents than will planktonic bacteria.
Kadouri System
Various stages of biofilm formation can be observed with this system. An example of the formation of a P. aeruginosa biofilm over 48 hr is shown in Figure 1B.1.7.
Figure 1B.1.7.
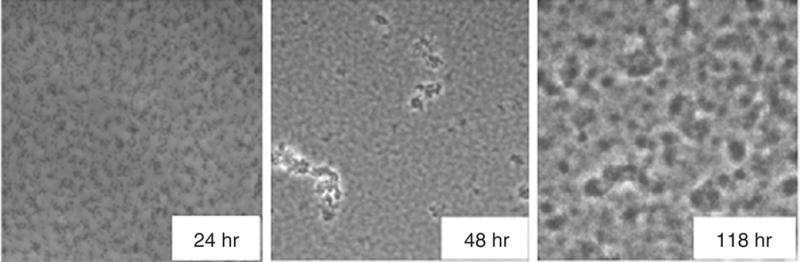
Using the Kadouri system, a biofilm of P. aeruginosa was grown in minimal medium supplemented with arginine at room temperature and photographed by phase-contrast microscopy at 24, 48, and 118 hr. Microcolonies can be observed as early as 24 hr, with large macrocolonies formed by 118 hr.
Time Considerations
Microtiter Plate Assay
After the biofilm plates have been inoculated from stationary-phase cultures, results may be obtained in 2 to 30 hr. This, of course, depends on the desired time course and the size of the experiment. Only a small percentage of this time involves hands-on work, however, including the initial inoculation, staining of plates, and dye solubilization. If only a few bacterial strains or species are being tested, these steps may take as little as an hour in total. Alternate Protocol 1, in which viable counts are performed to enumerate the attached bacteria, will take longer, as more handling is involved and the plated bacteria will need to be incubated for another 12 to 24 hr for the colonies to grow large enough to be counted.
ALI Assay
The ALI assay follows a time course similar to that of the microtiter plate assay. The length of the assay will again depend on the desired time frame to be studied. Washing of plates followed by microscopic analysis takes ~15 min per well. Alternate Protocol 2 is similar to the ALI assay in terms of its time requirements.
Colony Biofilms
During the growth of colony biofilms, the bacteria require little attention, except that the membranes must be transferred to fresh plates every 24 hr. Over the course of an experiment, the separation of bacteria from the membrane and subsequent enumeration are the most labor-intensive steps. An important consideration regarding the experimental timing in this protocol is the requirement for pregrowth of the colony biofilm (i.e., growth prior to the experimental treatment). This requires membranes to be inoculated 48 hr in advance of antibiotic testing.
Kadouri System
The amount of time required to grow a biofilm in this system depends on the desired maturity, the growth conditions (e.g., medium, temperature), and the organism used. In general, it can be expected that a biofilm will be able to reach maturity within a week of inoculation. Setup of the apparatus should be done 24 hr before inoculation to allow sufficient time for bleaching and rinsing of the system prior to use.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grant MCB-9984521 and NIH grant R01AI083256 to G.A.O.
Footnotes
Internet Resources
Microtiter dish biofilm formation assay video by author, George O’Toole. Subscription required.
Contributed by Judith H. Merritt3, Daniel E. Kadouri2, and George A. O’Toole1
Literature Cited
- Anderl JN, Franklin MJ, Stewart PS. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob Agents Chemother. 2000;44:1818–1824. doi: 10.1128/aac.44.7.1818-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, O’Toole GO. SadB is required for the transition from reversible to irreversible attachment during biofilm formation by Pseudomonas aeruginosa PA14. J Bacteriol. 2004;186:4476–4485. doi: 10.1128/JB.186.14.4476-4485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen GD, Simpson WA, Younger JJ, Baddour LM, Barrett FF, Melton DM, Beachey EH. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danhorn R, Hentzer M, Givskov M, Parsek MR, Fuqua C. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J Bacteriol. 2004;186:4492–4501. doi: 10.1128/JB.186.14.4492-4501.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol. 2004;70:7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker LM, Clardy J. High-throughput screens for small-molecule inhibitors of Pseudomonas aeruginosa biofilm development. Antimicrob Agents Chemother. 2007 Oct;51(10):3582–90. doi: 10.1128/AAC.00506-07. Epub 2007 Jul 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- Mack D, Nedelmann M, Krokotsch A, Schwarzkopf A, Heesemann J, Laufs R. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: Genetic identification of a hexosamine-containing polysaccharide intracellular adhesin. Infect Immun. 1994;62:3244–3253. doi: 10.1128/iai.62.8.3244-3253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan MC. Techniques for mammalian cell tissue culture. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smithcand JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons; Hoboken, N.J: 1996. pp. A.3F.1–A.3F.14. [Google Scholar]
- Sawyer LK, Hermanowicz SW. Detachment of biofilm bacteria due to variations in nutrient supply. Water Sci Technol. 1998;37:211–214. [Google Scholar]
- Stepanovic S, Vukovic D, Jezek P, Pavlovic M, Svabic-Vlahovic M. Influence of dynamic conditions on biofilm formation by Staphylococci. Eur J Clin Microbiol Infect Dis. 2001;20:502–504. doi: 10.1007/s100960100534. [DOI] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O’Toole GA. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191:210–219. doi: 10.1128/JB.00797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


