Abstract
Advances in flow cytometry and other single-cell technologies have enabled high-dimensional, high-throughput measurements of individual cells and allowed interrogation of cell population heterogeneity. Computational tools to take full advantage of these technologies are lacking. Here, we present COMPASS, a computational framework for unbiased polyfunctionality analysis of antigen-specific T-cell subsets. COMPASS uses a Bayesian hierarchical framework to model all observed functional cell subsets and select those most likely to exhibit antigen-specific responses. Cell-subset responses are quantified by posterior probabilities, while subject-level responses are quantified by two novel summary statistics that can be correlated directly with clinical outcome, and describe the quality of an individual’s (poly)functional response. Using three clinical datasets of cytokine production we demonstrate how COMPASS improves characterization of antigen-specific T cells and reveals novel cellular correlates of protection in the RV144 HIV vaccine efficacy trial that are missed by other methods. COMPASS is available as open-source software.
Introduction
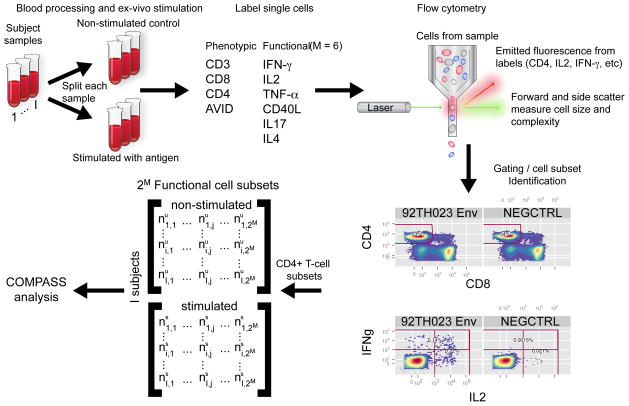
Recent technological advances in both flow and mass cytometry assays have transformed the field of immunology by enabling dozens of parameters to be quantified at the single-cell level in a high-throughput fashion. Increasing numbers of studies and clinical trials now rely on these assays to provide multi-parameter single-cell measurements, and functional evaluation is shifting from the analysis of single markers to these multidimensional measurements. In particular, single-cell analyses by intracellular cytokine staining (ICS) – a type of flow cytometry assay (Figure 1) – have become important tools to characterize subsets of antigen (Ag)-specific T cells capable of simultaneously producing multiple effector cytokines and other functional markers, termed polyfunctional T cells1. Polyfunctional T cells have been shown to play an important role in protective immunity and non-progression of diseases, and to correlate with better clinical outcomes in certain settings2–4. Vaccination in humans can generate broad T-cell cytokine responses5,6, thus, polyfunctional T-cell subsets are attractive potential biomarkers; however, effective statistical tools for analyzing the complexity of these immune responses are lacking.
Figure 1.
Overview of an ICS experiment. Blood samples are drawn from I subjects. A sample is split into aliquots that are subject to stimulation with antigen or are left non-stimulated as negative controls. After stimulation, whole PBMCs are labeled with fluorophore-conjugated antibodies against phenotypic (e.g. CD4, CD3, CD8, AVID/Live/Dead) and functional (e.g. IFNγ, IL2, TNFα, CD40L, IL17, IL4) markers (cells are permeabilized and intra-cellularly labeled for functional markers). The single-cell expression of each marker on each labeled cell is measured via flow cytometry, wherein cells pass in single-file through a flow cell and lasers of different wavelengths excite the fluorophores on the markers. A series of filters and detectors measure the emitted photons from the different fluorophores, providing a measure of intensity proportional to the amount of each protein expressed by each cell. After acquisition, data are processed and distinct cell populations of interest identified via a process termed ‘gating’, which describes identifying thresholds in multivariate space that classify each marker as either ‘positive’ (expressed) or ‘negative’ (not expressed). A COMPASS analysis assumes the ‘gating’ is given, and the COMPASS tool summarizes the number of cells expressing different combinatorial functional markers for specific phenotypic cell subsets (e.g. the number of CD4+ T cells expressing different combinations of cytokines). For M markers, this produces an I by 2M matrix of counts. COMPASS simultaneously models the counts for the paired stimulated and non-stimulated samples for each subject across all combinatorial functional cell subsets.
Although many analytic tools exist for cytometry-based assays7–9, very few tools have been developed specifically for the analysis of high dimensional ICS data. Existing strategies are in their infancy and remain basic and low dimensional, ranging from ad-hoc rules based on fold-changes10, Hotelling’s T2 statistics11, and 2×2 contingency tables12,13, to simple graphical displays of summary statistics7. In most ICS assays, the frequencies (and thus cell counts) of Ag-specific subsets are very small (e.g., less than 0.1% of total T cells), rendering statistical analysis difficult. This problem only worsens as these cells are further partitioned based on co-expression of multiple cytokines. In addition, multiple comparisons across subjects and cell subsets must be taken into consideration and can reduce statistical power. Due to the lack of proper analytical tools, studies have been limited to formal statistical comparisons of only a few functions, and most of the work on polyfunctionality has been phenomenological.
To address some of these limitations, Finak et al.14 have proposed a framework based on mixtures of beta-binomials named MIMOSA (MIxture MOdels for Single-cell Assays). MIMOSA rigorously analyzes count data derived from ICS assays. However, MIMOSA was mainly developed for univariate analysis of cell subsets, such as cells expressing a single function or a specified combination of functions. Although MIMOSA includes a multivariate version, Finak et al. we were solely interested in making positivity calls irrespective of the qualitative aspect of the response, and as such the output is still univariate (probability of response). More importantly, in order to apply the MIMOSA framework to multivariate data, the authors had to make the assumption that there is measurable antigen-specific response across all functional subsets, but this assumption is simply incorrect in practice. Different antigens usually induce very different functional profiles and many of the possible functional cell subsets are not expected to be associated with antigen-specificity. MIMOSA cannot jointly model all subsets to identify distinct Ag-specific responses, yet this is particularly important as the number of definable subsets grows exponentially with the number of cytokines analyzed. As an example, an ICS experiment measuring 7 functions can define 128 Boolean cell subsets, but only a fraction of those are expected to be biologically relevant (e.g., not all combinations respond to a specific antigen). A possible solution would be to model and test each subset separately but this is computationally intensive, ignores the dependence between subsets, and leads to extreme multiple testing problems. In the interest of decreasing the number of variables while taking into account the degree of functionality, Larsen et al. have introduced a Polyfunctionality Index (PI)15 that aims to facilitate statistical analysis and correlation with clinical outcome by summarizing the polyfunctional profile into a single number. However, the PI uses empirical proportions, which are known to be extremely noisy when cell counts are small, and combines information from all cell subsets, including non-specific ones, thus masking real signal. Indeed, many of the functional subsets that are not antigen-specific will often exhibit a significant background level, and the inclusion of these will make the defined index more variable. This still remains true even after background correction (using a negative-control) due to sampling and assay variability. In addition, by weighting the subsets by their magnitude, low-magnitude (but potentially important) subsets will be down weighted significantly. As such, the PI falls short of describing the true breadth of polyfunctionality, ultimately limiting its clinical utility (as we will show later). An ideal framework for the analysis of T cells should identify and quantify changes in (possibly rare) Ag-specific cell subsets and permit the definition of different qualities of a (poly)functional response such as summary statistics that can be correlated with outcomes of interest.
In order to address these needs, we have developed COMPASS (Combinatorial Polyfunctionality analysis of Antigen-Specific T-cell Subsets), which uses a Bayesian hierarchical mixture model to permit the identification of Ag-specific changes across all observable T-cell subsets simultaneously, enabling the definition of subject-specific and cohort-specific Ag-specific T-cell profiles that can be summarized and correlated to outcomes of interest. This method jointly models all cell subsets, regularizes small cell counts, and allows information sharing across all subjects through the calculation of subset-specific posterior probabilities that can be used to automatically detect and quantify Ag-specific subsets. COMPASS uses a novel, computationally efficient Markov Chain Monte Carlo algorithm to explore the space of all possible functional T-cell subsets and to compute the subset-specific posterior probabilities. These probabilities naturally account for multiple testing16 and permit the derivation of a false discovery rate estimate through a direct posterior probability approach17. COMPASS provides two novel scores: the functionality (FS) and the polyfunctionality (PFS) scores that can be correlated with any clinical outcome of interest. These scores summarize, as a single number for each subject, the posterior probabilities of Ag-specific response across cell subsets. Here we apply this new method to three ICS data sets: the RV144 HIV vaccine case-control study18,19, a cross-sectional study of South African adolescents infected with Mycobacterium tuberculosis (MTB)20 and a phase 1b trial from the HIV Vaccine Trials Network21. We use these datasets to demonstrate the ability of COMPASS to identify specific (poly)functional responses that are correlated with clinical outcomes of interest.
Results
COMPASS enables the unbiased quantification of polyfunctional cell subsets
Here we reanalyzed the ICS data generated through the RV144 HIV vaccine case-control study with 92TH023-Env peptide pool ex vivo stimulation as described in the Methods and in a prior publication18. Expression of a set of six functions (IFNγ, TNFα, IL2, IL4, IL17 and CD40L) was measured in CD4+ T cells by ICS (n=226 vaccine and n=36 placebo recipients). We used COMPASS to define 64 functional cell subsets from the Boolean combinations of the individual cytokines expressed in each cell. Only 24 of these had non-negligible (>5 cells in >2 subjects) cell counts and were considered for analysis. Arguably, if there are only two subjects with no more than five cells in any cell subset then it would be difficult to detect antigen-specific response, and we feel that these values should be reasonable for most ICS datasets. In addition, changing these cutoffs to be less stringent had no impact on the significance of the results or conclusions presented here, and is purely for computational convenience to reduce the time needed to fit a COMPASS model to cell subsets that contain no signal. In Figure 2a, a heatmap displays posterior probabilities of Ag-specific responses from COMPASS for these T-cell subsets and demonstrates that while some Ag-specific subsets are universally present in almost all vaccinees (e.g., IL2 & CD40L), others exhibit heterogeneity across subjects (e.g., the four- and five-function subsets). These findings support the need for unbiased polyfunctionality analyses that can provide insights on the possible association between polyfunctional Ag-specific T cells and vaccine efficacy.
Figure 2.
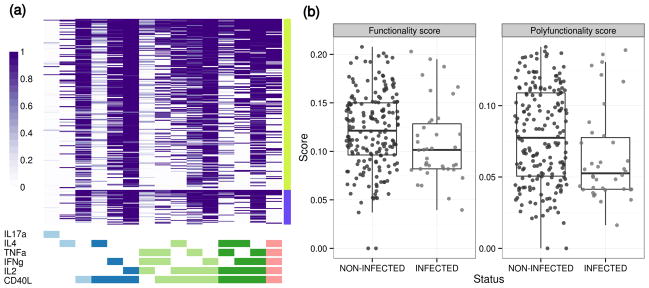
(a) Heatmap of COMPASS posterior probabilities for the RV144 data set. Columns correspond to the different cell subsets modeled by COMPASS (shown are the 15 of 24 subsets with detectable antigen-specific responses that had >5 cells in >2 subjects), color-coded by the cytokines they express (white=“off”, shaded=“on”, grouped by color=“degree of functionality”), and ordered by degree of functionality from one function on the left to five functions on the right. Subsets with maximum posterior probabilities less than 0.005 are removed from the heatmap. Rows correspond to subjects (only shown are 226 vaccine recipients), which are ordered by their status: non-infected and infected, and by functionality score within each group. Each cell shows the probability that the corresponding cell-subset (column) exhibits an Ag-specific response in the corresponding subject (row), where the probability is color-coded from white (zero) to purple (one). (b) Boxplots of functionality and polyfunctionality scores stratified by HIV infection status in RV144 among 226 vaccine recipients. Non-infected individuals have higher scores than infected ones (Wilcoxon test p = 0.03 (FS), p = 0.01 (PFS). Both scores are inversely correlated with infection (Table 1).
ICS assays are often used to identify vaccine responders based on the magnitude of Ag-specific T-cell responses, although current approaches tend to be univariate. COMPASS can identify vaccine responders by computing subject-level response probabilities for each subject (Online Methods). We compared COMPASS with the two standard approaches for designating positivity, Fisher’s exact test12, and MIMOSA14 using the primary endpoint in the original RV144 analysis18 as well as the multivariate MIMOSA and the polyfunctionality index (PI) (Online Methods). A receiver operating characteristic (ROC) analysis shows that COMPASS significantly increases sensitivity and specificity when discriminating between vaccine and placebo recipients compared to all other approaches (Supplementary Figure 1).
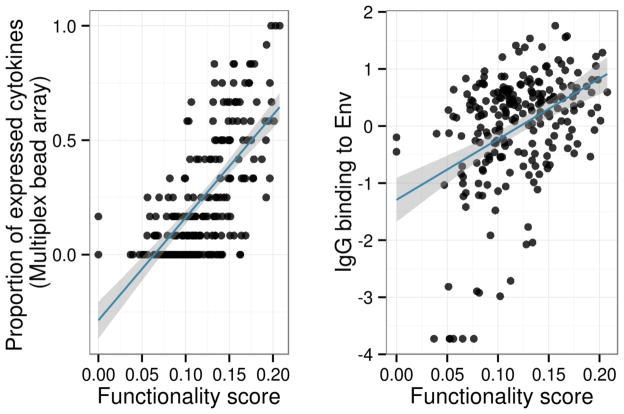
In addition to subject-level response probabilities, COMPASS defines two new scores that summarize a subject’s entire Ag-specific polyfunctional profile into a single numerical value (Figure 2b). The functionality score is defined as the proportion of Ag-specific subsets detected among all possible ones. The polyfunctionality score is similar, but it weighs the different subsets by their degree of functionality, naturally favoring subsets with higher degrees of functionality, motivated by the observations that higher degree function has been correlated with good outcomes in certain vaccine studies5,6 (See Online Methods for details). Figure 3a shows that the functionality score is well correlated with the number of functions (secreted cytokines) as measured by multiplex bead array on the same set of samples from the RV144 case-control study (correlation ρ=0.69, p-value < 2.2×10−16, 95% confidence interval (CI) = (0.62, 0.75)), suggesting that COMPASS detects true Ag-specific responses in T-cell subsets. Out of the 12 cytokines measured by a multiplex bead array, only four were also measured by ICS (IFNγ, TNFα, IL2, IL4). Subjects with high functionality scores also express many other cytokines not measured in the ICS assay. We also tried to restrict the analysis to the four common cytokines but it did not improve the correlation, in fact it decreased slightly to 0.65. Figure 3b shows that the functionality score is also significantly correlated with HIV Env-specific antibody binding in plasma (correlation ρ=0.41, p-value=9.87×10−11, 95% CI = (0.30, 0.52)). In addition, Supplementary Figure 2 shows that the placebo recipients have much smaller FS and PFS scores compared to vaccine recipients, and that the PI is noisier than the FS and PFS scores. All together, these data indicate once again that our FS and PFS scores are good at capturing and summarizing a subject’s response to vaccination. These results were also confirmed with the RV144 pilot data, used to inform the selection of assays to be used for the case-control study, thus providing validation from an independent set of subjects (Supplementary Figures 3–6). Using boxplots of the functionality and polyfunctionality scores for RV144 broken down by infection status (Figure 2b), it can be seen that these two scores vary greatly by subject, and that on average non-infected subjects have higher scores than infected individuals and that these effect are stronger for the polyfunctionality score (Wilcoxon test p = 0.03 (FS), p = 0.01 (PFS)).
Figure 3.
Functionality score vs. (a) proportion of expressed cytokines as measured by multiplex bead array and (b) IgG antibody binding in RV144. A set of 12 cytokines were measured by multiplex bead array. The proportion of detectable secreted cytokines was calculated in each individual and compared to the functionality score. The functionality score is significantly correlated with overall cytokine production (ρ=0.68, p-value < 2.2×10−16) and antibody binding (ρ=0.50, p-value=3.02×10−10). The fitted regression line from simple linear model is plotted in blue, and the 95% confidence interval is shown in grey.
COMPASS identifies novel cellular correlates of infection risk in RV144 vaccinees
Among the 17 different types of immune assays and their 152 component variables used to assess correlates of infection risk in RV144, 6 assays (including ICS CD4+ T-cell responses) were chosen as primary variables as having optimal statistical power when adjusting for multiple comparisons18 (see Online Methods for primary analysis results). The major findings in a multivariate model including all six primary variables were that gp70-V1V2-specific plasma IgG inversely correlated with infection rate (odds ratio 0.57, p=0.02, q=0.08), and Env-specific plasma IgA was a direct correlate of infection (odds ratio 1.54, p=0.03, q=0.08). No CD4+ T cell cellular correlates were identified in the primary analysis in the original study. In contrast, using the same method as in the original correlate analysis18 (also see Online Methods) but including the functionality and polyfunctionality scores rather than the primary CD4+ T cell endpoint variable, the polyfunctionality score was inversely correlated with infection (p = 0.005, q=0.05, Table 1), as was the functionality score but to a lesser extent (p=0.01, q=0.06, Table 1). This correlation is also supported by the fact that the number of functions (secreted cytokines) as measured by multiplex bead array is also inversely correlated with infection (p=0.013, Supplementary Table 1). However, the multiplex bead array data do not permit polyfunctionality analysis since it is an assay performed on bulk cells, and as such the co-expression of multiple cytokines cannot be measured at the single-cell level. Our COMPASS PFS score suggests that polyfunctional Ag-specific CD4+ T-cells might have played a role in vaccine efficacy. To identify the specific cell subsets that most contributed to this correlation, each subset of interest as shown in Figure 2a was evaluated as a correlate for infection risk. Two Ag-specific T-cell subsets were significantly correlated with infection (q≤0.1, Table 1). It can be seen that the polyfunctional subset expressing CD40L, IL2, IL4, IFNγ and TNFα shows the most significance (OR=0.58, p=0.006, q=0.05) followed by a three-function subset (CD40L, IL2, and IL4, OR=0.62, p=0.01, q=0.06). The 5-degree subset is as significant as the previously reported gp70-V1V2 correlate. Importantly, these subsets were identified in an unbiased way rather than by limiting our analysis to very specific subsets based on expected biological relevance as was done in the original study. Interestingly, both subsets include IL4 and CD40L, a cytokine and functional marker important for the CD4 T cell/B cell interaction. Thus, these particular CD4+ T-cells subsets may contribute the T-cell help necessary for the antibody production detected as a correlate in the primary analysis. All subsets, including non-significant ones, are presented in Supplementary Table 2. In addition, we have also assessed the magnitude of response for all functional subsets (Supplementary Table 3) as well as the PI and MIMOSA scores (Supplementary Table 1) as individual correlates of risk, and none of these variables were significant. The superiority of COMPASS is also supported by an ROC analysis using the predicted infection probabilities from a logistic regression model (Supplementary Figure 7).
Table 1.
Estimated odds ratios for HIV-1 Infection risk for the subset specific responses as determined by logistic regression models that adjust for baseline risk category and gender in the RV144 case-control study. Only subsets with p-values less than 0.05 are shown here. Odds ratio are per one standard deviation for each variable and are adjusted for IgA level, gender and baseline behavioral risk score. Lower and upper limits of the 95% confidence intervals (CI) for the estimated ratios are also shown. Q-values are the FDR adjusted p-values across all 17 considered variables.
| Variable | Odds ratio (95% CI) | p-value | q-value |
|---|---|---|---|
| Functionality score | 0.62 (0.42, 0.91) | 0.014 | 0.06 |
| Polyfunctionality score | 0.57 (0.38, 0.84) | 0.005 | 0.05 |
| IL4+IL2+CD40L+ | 0.62 (0.43, 0.90) | 0.013 | 0.06 |
| TNFα+IFNγ+IL4+IL2+CD40L+ | 0.58 (0.39, 0.86) | 0.006 | 0.05 |
Mycobacterium tuberculosis (MTB) infection induces highly polyfunctional responses
We used COMPASS to investigate polyfunctionality in CD4+ T cells obtained from a South African study of 18 MTB-uninfected and 22 MTB-infected subjects. Subjects were classified as TB-infected as described in the Methods section and in the original study20. T cells were stimulated with MTB-specific22 and non-specific peptide pools, and a set of seven cytokines/functional markers (see Methods) was measured by ICS, with 22 Boolean combinations defined for COMPASS analysis.
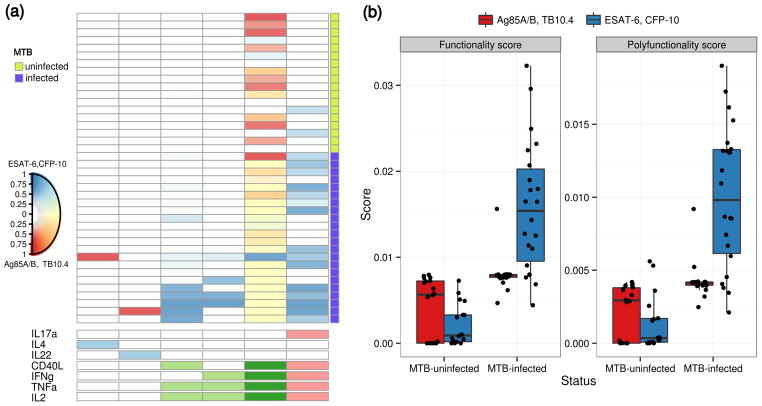
Figure 4a shows a heatmap of posterior probabilities of Ag-specificity with subjects color-coded by the specificity of the peptide stimulation (MTB-specific and MTB non-specific). Figure 4b shows boxplots of the functionality and polyfunctionality scores broken down by TB infection status and stimulation. As expected, CD4+ T-cell responses to MTB-specific proteins were largely absent from MTB-uninfected subjects. Ex vivo stimulation with MTB-specific antigens induces strong Ag-specific responses spread across polyfunctional subsets of degree three and five in MTB-infected subjects. In contrast, MTB-uninfected subjects have weak responses in these subsets. The appearance of the five-degree subset (IL17α, CD40L, IFNγ, TNFα and IL2) in some MTB-uninfected persons may represent a more sensitive marker of infection or may simply be a false positive result. Further study will be required to distinguish among these possibilities. In contrast, the response to non-specific mycobacterial peptides is concentrated over one subset expressing IFNγ, IL2, TNFα and CD40L, and is independent of a subject’s MTB status. This response to non-specific mycobacterial proteins is likely due to BCG vaccination or environmental exposure to non-tuberculous mycobacteria.
Figure 4.
(a) Heatmap of COMPASS posterior probabilities for the TB data set. Columns correspond to the different cell subsets modeled by COMPASS (shown are the six of 22 and 19 subsets with detectable antigen-specific response that had >5 cells in >2 subjects), color-coded by the cytokines they express (white=“off”, shaded=“on”, grouped by color=“degree of functionality”), and ordered by degree of functionality from one function on the left to five functions on the right. Subsets with maximum posterior probabilities less than 0.1 are removed from the heatmap Rows correspond to 40 subjects, which are ordered by level of interferon-gamma release as measured by Quantiferon Test (QFT). Subjects with positive QFT test results are labeled as TB positive. Each cell shows the probability that the corresponding cell-subset (column) exhibits an Ag-specific response in the corresponding subject (row), where the probability is color-coded from white (zero) to blue (one) for MTB-specific stimulation and red (one) for MTB-non-specific. A mix of blue and red (i.e. yellow) indicate a response to both stimulations. (b) Boxplots of functionality and polyfunctionality scores stratified by TB positive and TB negative for both TB-specific and non TB-specific stimulations.
As can be seen in Figure 4a, COMPASS detects MTB-specific responses for some cell subsets that do not include IFNγ, suggesting that such analysis may add value to standard QFT analyses in identifying patients with MTB infection. Given this observation, we assessed whether COMPASS could be used to classify MTB-uninfected and MTB-infected subsets and how this would compare to a classification based on marginal IFNγ response alone, essentially a surrogate for the QFT. Supplementary Figure 8 shows an ROC analysis comparing COMPASS to various summary statistics based on the IFNγ response including MIMOSA, Fisher’s exact test and background corrected empirical proportions of IFNγ+ cells, as well as the multivariate MIMOSA and the PI. These data demonstrate that using COMPASS’ polyfunctional profile to classify subjects as MTB infected vs. uninfected provides increased sensitivity and specificity compared to using IFNγ alone or the PI. The lack of total concordance between the QFT and ICS IFNγ response may be explained by differences between fresh whole blood and frozen PBMC used for the two assays, the lack of TB7.7 in the ICS assay, and measurement of IFNγ expression (ICS) vs secretion (ELISA).
COMPASS identifies functional differences between vaccine regimens missed by traditional analyses
As a third example, we applied COMPASS to ICS data from a clinical trial of candidate HIV vaccines23 to determine if a polyfunctionality analysis could unveil differences missed in the primary analysis (see Methods). The combination of heterologous vectors has proven to be a good strategy to increase immune responses to vaccination24, which is particularly relevant for HIV where there is not yet an effective, licensed vaccine. Bart et al.21 showed that priming with rAd5 followed by NYVAC boost resulted in increased percentage of Ag-specific T cells producing IL2 and/or IFNγ compared to NYVAC followed by rAd5, but that higher doses of rAd5 prime did not lead to any increased response. Figure 5a shows a heatmap of the posterior probabilities for all CD4+ T-cell Boolean subsets considered. While this heatmap supports the findings of Bart et al., it also shows that increasing dosage of rAd5 led to a decrease in overall response, which is nicely summarized by the FS and PFS scores (Figures 5b). Although T3 and T4 have comparable FS and PFS scores, it is notable from Figure 5a that very few subjects in T4 produce Ag-specific T cells co-expressing IL4/IFNγ/TNFα/CD40L compared to T3. This is confirmed when the probability of response for this subset is plotted separately (Supplementary Figure 9). This suggests that a difference in dosage can lead to substantial qualitative differences in immune responses after vaccination (p-value = 0.004 for Fisher’s exact test on proportions of responders between T3 and T4).
Figure 5.
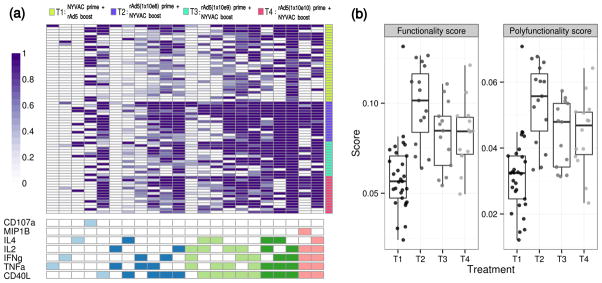
(a) Heatmap of COMPASS posterior probabilities for the HVTN078 data. Columns correspond to the different cell subsets modeled by COMPASS (shown are the 22 of 26 subsets with detectable antigen-specific response and having >5 cells in >2 subjects), color-coded by the cytokines they express (white=“off”, shaded=“on”, color=“degree of functionality”), and ordered by degree of functionality from one function on the left to five functions on the right. Rows correspond to 71 subjects ordered by treatment group: T1: NYVAC+Ad5, T2–T4: increasing doses of Ad5+NYVAC, and by functionality score within each group. Each cell shows the probability that a given cell-subset (column) exhibits an Ag-specific response in the corresponding subject (column), where the probability is color-coded from white (zero) to purple (one). (b) Boxplots of functionality and polyfunctionality scores for CD4+ T cells stratified by treatment groups.
Discussion
These three examples highlight the ability of COMPASS to reveal differences in the quality of an immune response that were not evident using standard approaches. It remains uncertain as to whether specific cellular subsets of unique polyfunctional profiles have clinical significance, for example, in terms of mediating protection from disease. The RV144 data set provided an opportunity to investigate association between subset detection and a clinical endpoint, HIV infection. Indeed, two vaccine-induced polyfunctional CD4+ T-cell subsets not identified in the prior analyses were shown to associate with decreased risk of HIV infection. Although the original cellular endpoint in RV144 was not designed to detect polyfunctional responses, current tools used to assess polyfunctionality are inadequate and miss this association. The TB study also allowed for association of cellular subsets with a clinical outcome, presence or absence of MTB infection. In this case, the diagnostic methods for determining infection are not definitive, and our approach has the potential for augmenting diagnostic methods. The third example applied in the context of a phase I clinical trial does not have a clinical endpoint, but the striking difference in one four-function subset between two identical vaccine regimens that differ only in dose raises questions about the functional significance of this subset, or at a minimum, the potential of this subset as a future correlate of vaccine efficacy.
COMPASS analysis of the tuberculosis dataset revealed two findings that would likely have been missed using standard methods of analyzing multiparameter FACS data. First, we demonstrate the presence of a polyfunctional CD4+ T-cell subset recognizing MTB-specific proteins that does not include IFNγ yet was preferentially detected among MTB-infected subjects. IFNγ release assays are the clinical standard for diagnosis and are still imperfect25, so this observation may have important implications for the design of improved diagnostics. Second, we demonstrated the presence of a T-cell subset that simultaneously produced IL17A, IFNγ, CD40L, IL2, and TNFα in response to MTB-specific proteins among MTB-infected subjects. To our knowledge, this has not been previously reported and it is not clear if such highly-polyfunctional T cells are important for protection from tuberculosis, but the analysis of RV144 data presented here would suggest this is possible. The availability of COMPASS will make it easier to analyze phase 2 studies of TB vaccines currently underway for the association of these subsets with clinical outcomes. Finally, only a small proportion of MTB-infected individuals will eventually develop active tuberculosis, so the stratification of MTB-infected subjects into different T-cell response categories could be utilized to predict which individuals are at highest risk of progression to active TB disease.
The two functionality and polyfunctionality scores introduced in this paper provide unique T-cell response summaries that can be used to quantify a subject’s immune response and be used as a biomarker to be correlated with a given outcome. Although the score definitions are related, and thus the scores expected to be correlated, we believe that the two scores are complementary and provide additional information versus one score alone. For example, even though the correlation between the two scores in the RV144 case-control study is 0.95, we have shown that while the functionality score was better correlated with other cellular and antibody functions as measured by multiplex bead array assay, the polyfunctionality score provided a better correlate of protection. In addition, multivariate response profiles based on the posterior probabilities returned by COMPASS, were helpful in singling out specific subsets that were associated with specific treatment groups or clinical variables. Other published studies have also reported similar findings26–28, reinforcing the need for unbiased, multivariate analyses. It should be noted, however, that since COMPASS analyzes and reports probabilities for all functional subsets, the results could be prioritized and/or summarized in reference to cytokine-based functionalities (e.g. Th1, Th2, Th17, etc).
These findings reinforce the idea that the quality rather than the magnitude of T-cell responses is more important for determining the outcome of infection or response to vaccination29. We think that for the purpose of summarizing the functionality or polyfunctionality over all subsets, it is preferable to define a variable that does not include the magnitude of the response; otherwise the large-magnitude subsets would mask the low-magnitude ones. This is particularly important since the magnitude of high-degree (i.e. polyfunctional) subsets is typically smaller. This being said, our functionality and polyfunctionality scores do take into account the magnitude of response up to a certain point, after which the subsets are treated equally (when the model judges that there is a significant increase upon stimulation and we are certain of the specificity of the response). Also, because our approach is model based, the relationship between the magnitude and the probability of antigen-specificity takes into account the uncertainty in the observed cell counts (Supplementary Figure 10), such that cell subset with greater background will have lower response probabilities, even at comparable magnitude. Although we could use the actual proportion of Ag-specific cells in our calculation, we have found that it did not add any predictive value (Supplementary Figures 11–12, Supplementary Table 4). This could be explained by the fact that the actual magnitude of the proportion is not that important in terms of clinical outcome, i.e. what matters is the quality of the Ag-specific response and not the quantity of Ag-specific T cells, or that the estimates of proportions are too noisy to be meaningful when the overall proportion of Ag-specific cells is small, which is clearly the case for HIV and TB. However, if one really wants to look at the magnitude, we think that it might be better quantified at the subset level, which can easily be done with COMPASS since it models all subsets jointly.
The development of statistically sound methods for the characterization of Ag-specific T cells from single-cell assays is becoming increasingly important as the underlying technologies are improving, leading to a combinatorial explosion of the number of functionally distinct subsets that can be defined from an experiment. As we have shown, standard approaches that ignore the multivariate nature of ICS data can be quite sub-optimal, as they will likely miss rare but important signals. This problem will only get worse as new high-dimensional single-cell technologies such as CyTOF, multiplexed-qPCR and RNA-seq become more widely used. Although our approach was developed primarily for flow cytometry based ICS assays, it is directly applicable to CyTOF-based ICS experiments, and should be generalizable to other assay technologies including multiplexed-qPCR and single-cell RNA-seq.
COMPASS is available as an R package (http://www.github.com/RGLab/COMPASS) and provides an interactive, web-based interface for visualizing all data and results. All data and results presented in this paper can be visualized through a web-tool available at: http://rglab.github.io/COMPASS/.
Online Methods
Data for these studies derive from three clinical protocols that were approved by the relevant institutional review boards. All study participants provided written informed consent for immune response exploratory analyses.
RV144 case-control dataset
HIV negative healthy volunteers enrolled in the RV144 trial (ClinicalTrials.gov registration number NCT00223080) were vaccinated at weeks 0, 4, 12, and 24, and immune responses at week 26 were evaluated as immune correlates of infection risk through a case-control analysis18,19. A total of 246 vaccinated subjects were used for this analysis: 41 subjects who acquired HIV-1 infection (cases) after week 26 and 205 frequency matched subjects who did not become infected over the follow-up period (controls). A total of 17 types of immune assays were run on the case-control samples. In addition, these assays were also performed on random samples from 40 placebo recipients (20 cases and 20 controls). The analysis presented here, focused on assessing poly-functional HIV-1 envelope proteins (Env)-specific T-cell response using ICS, one of the 17 available assays. As a mean to validate our analysis and explore relationships between cellular and antibody functions, we also correlated our ICS responses with two other assays: a multiplex cytokine bead array measuring antigen-specific cytokine secretion in PBMCs, and a binding antibody multiplex assay measuring IgG binding to HIV-1 envelope proteins. After removing missing data due to assay failure, we ended up with 226 vaccinated subjects (38 infected, 188 non-infected) with complete data across the three assay types.
ICS
In this dataset, a set of 6 functions: TNFα, IFNγ, IL4, IL2, CD40L and IL17A were measured at the single-cell level in CD4+ T cells in the presence and absence of stimulation with a peptide pool matching one of the HIV-1 envelope proteins contained in the vaccine (92TH023). Cell level data were extracted from raw data files using the analysis described in Haynes et al. (2012)18, leading to the definition of count data for the 26=64 theoretical Boolean subsets for each subject and stimulation condition. However, the actual number of observed (i.e. non-empty) cell subsets is 59, and we further filtered out subsets that had fewer than six cells in fewer than three subjects, reducing that number to 24.
Multiplex cytokine bead array
A set of 12 cytokines (IFNγ, IL4, IL2, IL5, TNFα, IL10, TNFβ, IL13, MIP1β, GM-CSF, IL3, IL9) were measured using multiplex bead array technology. For each subject and cytokine, the response is defined as the difference in log concentration between the stimulated (92TH023-Env) and un-stimulated samples. For each subject, individual cytokines were called positive/negative using the thresholds defined in the original study18.
IgG total binding
Binding IgG antibodies to the envelope (Env) protein of the HIV virus were measured using a binding antibody multiplex assay. Here, we used the IgG total binding response to Env defined as the mean binding IgG to multiple Env proteins18.
Pilot data
As part of the RV144 case-control study, the 17 assay types used in the case-control study were selected from 32 pilot assay types on the basis of reproducibility, ability to detect post vaccine responses, and uniqueness of responses detected, from which 6 primary assay variables were selected for the correlate analysis. ICS and multiplex bead array were two of the assays types that were evaluated during the pilot phase. All samples used in the pilot phase were from non-infected subjects. For ICS, 36 placebo and 119 vaccine recipient (60 pre and 59 post vaccination) samples were used. For multiplex bead array data, 30 placebo and 98 vaccine recipient (57 pre and 41 post vaccination) samples were used.
TB data set
We obtained cryopreserved PBMC from an epidemiologic study of South African adolescents who were screened for the presence of latent tuberculosis infection (LTBI) using tuberculin skin testing and QuantiFERON-TB GOLD in tube testing of whole blood20. This dataset includes 40 subjects, 22 MTB-infected and 18 MTB-uninfected. Subjects were classified as MTB-infected using both TB skin testing and the Quantiferon test in-tube gold (QTF-gold) that measures IFNγ release in whole-blood after stimulation with ESAT-6, CFP-10 and TB7.7 peptides25. PBMCs were plated at a density of 2E5 per well and stimulated for six hours with either DMSO or peptide pools consisting of 15mers overlapping by 12 peptides for the following mycobacterial proteins: ESAT-6, CFP-10, TB10.4, Ag85A, and Ag85B at a final concentration of 1mcg/ml. Cells were stained using a published panel in which we replaced MIP-1b and CD107a with IL-17a Alexa 700 and IL-22 PE Cy712,30. Analysis of CD3+CD4+ events was performed in FlowJo (TreeStar Inc.) after first gating on single cell events, CD14− events, live cells, and lymphocytes. For the purposes of analysis, we pooled counts obtained after stimulation with ESAT-6 and CFP-10 and defined this as ‘MTB-specific’ because these proteins are known to be absent in Bacillus Calmette-Guérin (BCG) and many environmental mycobacteria. By extension, counts obtained after stimulation with Ag85A, Ag85B, and TB10.4 were pooled and defined as ‘MTB-nonspecific’ because these proteins are present in M. tuberculosis as well as BCG and many environmental mycobacteria. Seven functions were measured at the single-cell level in CD4+ T cells: TNFα, IFNγ, IL4, IL2, CD40L, IL17A and IL22 leading to 128 theoretical subsets. However, the actual number of observed cell subsets was 79, and using the filtering criteria described above we reduced that number to 22 (MTB-specific) and 19 (MTB-non-specific).
HVTN078
HVTN 07821 is a randomized, double-blind phase 1b clinical trial (ClinicalTrials.gov registration number NCT00961883) to evaluate the safety and immunogenicity of heterologous prime/boost vaccine regimens (NYVAC-B/rAd5 vs. rAd5/NYVAC-B) in healthy, HIV-1 uninfected, Ad5 seronegative adult participants. Eighty participants were enrolled into one of four groups receiving different combinations of NYVAC-B (New York Vaccinia [NYVAC] vector containing HIV-1BX08 gp120 and HIV-1IIIB gag-pol-nef at a dose of 1×107 PFU) and rAd5 (HIV-1 recombinant adenoviral serotype 5 [rAd5] vector vaccine VRC-HIVADV014-00-VP [(HIV-1HXB2/NL4-3 Gag-Pol fusion; HIV-192RW020, HIV-1HXB2/Bal and HIV-197ZA012 Env], at three increasing doses [1×108, 1×109, 1×1010 PFU]). We refer to the four different groups as T1, T2, T3 and T4. In the T1 group, NYVAC was the prime with rAd5 as the boost, while subjects in T2–T4 received rAd5 as the prime and NYVAC as the boost with the three increasing doses of the prime. Here we used a subset of the ICS data generated through the trial measuring 7 functions: IFNγ, IL2, IL4, TNFα, MIP1β, CD107a and CD40L in CD4+ T cells in the presence and absence of stimulation with HIV-1 peptides, leading to 128 theoretical subsets in 71 subjects (T1:29, T2:15, T3:13, T4:14). However, the actual number of observed cell subsets is 107, and using the filtering criteria described above we reduced that number to 26 subsets.
Statistical framework for modeling count data
Without loss of generality, we assume cell counts are obtained from I subjects under two conditions: (antigen)-stimulated and un-stimulated as depicted in Figure 1. Let M denote the number of markers measured, then in theory, there are KM=2M possible Boolean combinations defining functional cell subsets, depending on whether the marker is expressed or not. We let K (K ≤ KM) denote the actual number of cell subsets considered for statistical analysis (i.e. after filtering empty and very sparse cell subsets). The observed counts for the K cell subsets in the stimulated and un-stimulated samples are represented by nsik and nuik, k = 1,…, K, i = 1,…, I, respectively, or represented as vectors we have nsi = (nsi1,…,nsiK)′ and nui = (nui1,…,nuiK)′. Then Nsi = Σk nsik and Nui = Σk nuik are the total number of cells for subject i in the stimulated and un-stimulated samples, respectively (Figure 1). Without loss of generality, we order the cell subsets such that the Kth category represents the subset where no cytokines are expressed, i.e. the degree of functionality is zero.
For a given subject i, we jointly model the cell counts under the two conditions using multinomial distributions: (nsi | psi) ~ MN(Nsi, psi ) and (nui | pui) ~ MN(Nui, pui), where psi and pui are the unknown proportions for the stimulated and un-stimulated paired samples, respectively. In order to detect a responding subject, as well as Ag-specific subsets within a subject, we consider two competing hypothesis: H0: pui = psi, and Ha: ∃k ∈ {1, …, K − 1} such that psik > puik. Under the null hypothesis H0, there is no difference in the proportion of cytokine producing cells between the stimulated and unstimulated samples, and thus the proportion vector parameter is shared across the two multinomial models. Under the alternative hypothesis Ha, some subsets show an increase in their proportions. We define the cell subsets that express at least one function and are different under Ha as Ag-specific subsets as the change in proportion is induced by the antigen stimulation. The Kth null category is not considered here, as a change there would only reflect a change in some other category since the proportion vector has to sum to one. This framework allows each subject to be responding in none, some or all of the subsets, and jointly models all the subjects and subsets to allow information sharing to improve the power in detecting rare signals. The ultimate objective is to automatically identify Ag-specific cell subsets for each subject.
Statistical model for detecting antigen specific polyfunctional T-cell subsets
In order to allow the automatic identification of Ag-specific cell subsets for each subject, we introduce a binary indicator, γik associated with each subject i and each category k, such that if the category is Ag-specific, γik = 1, otherwise γik = 0. In other words, when γik = 1, the (unknown) cell proportions (psik, puik) for the stimulated and un-stimulated samples are different, otherwise they are equal. The distribution of cell proportions can be specified easily conditional on the latent indicators, which is shown in Supplementary section: Priors. Our implementation of the COMPASS model uses an optimized Markov chain Monte Carlo (MCMC) algorithm allowing full exploration of the joint posterior distribution (Supplementary section: Posterior sampling). Then statistical inference about responding subsets across subjects, subsets or polyfunctionality degree would be based on the posterior summaries of the latent indicators, such as using the posterior mean, and/or FDR-thresholded posterior probabilities. Our approach uses a Bayesian variable-selection prior, with a subset specific – but common across subjects – prior weight shared and estimated across all subjects. As a result, the inference automatically takes into consideration multiple comparisons16 across subjects.
Subject-level response probabilities
ICS assays are often used for determining vaccine responders based on the magnitude of Ag-specific T-cell responses. Current approaches for positivity tend to be univariate; a single cell-subset needs to be defined as an endpoint (e.g. cells expressing IL2 and/or IFNγ). Because COMPASS uses a Bayesian approach to jointly model all cell subsets, any posterior summary of interest is readily available. COMPASS can classify vaccine responders or MTB-infected individuals by computing subject-level response probabilities, defined as the probability that at least two (disjoint) cell subsets exhibit an Ag-specific response in that subject. The rationale is that antigen stimulation in subjects with antigen-specific cells should induce changes in a variety of cell subsets, while non-specific responses are expected to be sporadic and noisy. Note that even though our subject-level response probabilities are computed based on two or more responding cell subsets, we still define the alternative hypothesis Ha (described above) as at least one difference, since COMPASS can return posterior probabilities for each subset, i.e. we really consider all possible alternative models.
Polyfunctionality and Functionality Score Definition
We introduce the functionality (FS) and Polyfunctionality PFS scores to summarize the response across the different cell subsets for each subject. The benefits of a single-number summary of subject-level response have been outlined elsewhere10. Primarily, it greatly facilitates statistical analysis, comparisons across treatment groups and correlation with outcome measures. We define the FS as the posterior mean of the average number of antigen-specific cell subsets among all measured subsets, irrespective of the degree of functionality:
Where γ̂ik is the posterior mean of γik estimated using MCMC (See Supplementary Text). The FS ranges from zero to one and measures the proportion of distinct cell subsets that are expressed for a given subject among all possible subsets.
The PFS is the mean of the posterior probabilities, weighted by the degree of functionality of the corresponding subset, and normalized by the total number of possible cell subsets that could be observed, given the number of markers considered:
Where, d(k) is the degree of functionality for cell subset k. The PFS ranges from zero to one. The normalization by the theoretical number of possible cell subsets facilitates the comparison of FS and PFS across different data sets, provided they utilize the same markers. It should be noted that the FS and PFS – based on the posterior probabilities – do not directly take into account the frequencies (i.e. magnitudes) of Ag-specific cells. The posterior probability summarizes all the evidence that a cell subset is Ag-specific by comparing the proportion of cytokine positive cells in the stimulated sample to the corresponding proportion in the control sample. Once there is enough evidence that the subset is indeed Ag-specific (i.e. the probability is one), the actual proportion of Ag-specific cells, which can be estimated as the difference in proportion between the stimulated and un-stimulated samples, is no longer relevant. In other words, two cell subsets with a probability of one will be treated equally in our approach, except for the degree of functionality that factors into the PFS even though they may have differing magnitude. Although, we could use the actual proportion of Ag-specific cells in our calculation (Supplementary text), we have found that it did not improve our analysis, and that in fact it decreased some of the correlation reported here (Supplementary Figures 11–12, Supplementary Table 4).
The PFS will assign a higher score to subjects with Ag-specific cell subsets of higher degree, whereas the FS will assign a higher score to subjects that exhibit Ag-specificity in more cell subsets irrespective of their degree of functionality. The two scores are complementary in the sense that the PFS emphasize the quality (i.e. the polyfunctionality) of Ag-specific cell subsets, whereas, the FS looks at the quantity of Ag-specific cell subsets. As an example, if we are considering 6 markers, two subjects may have the same PFS if one has a single degree 6 Ag-specific cell subset (PFS=0.286), while the other has Ag-specificity in all subsets with degree < 4 (PFS=0.286). The FS would distinguish between these, assigning score of 0.016 to the first subject, and a score of 0.651 to the latter.
Polyfunctionality index
The polyfunctionality index was calculated as described in Larsen et al15 using both uncorrected and background-corrected cell frequencies with the tuning parameter q set to 1, which is the equivalent of the PFS score where all subsets are weighted by their degree of functionality. Basically, for the uncorrected PI score we simply used the stimulated frequencies, psik, whereas for the corrected PI score, we used the background corrected frequencies defined as max(psik-puik,0). The two scores are referred to as PI and PI corrected in Supplementary Text. We have also tried other tuning parameter values including q=1.2 and q=2, and the results did not improve.
Correlate of risk analysis
All immune variables identified here were assessed as correlates of infection risk (CoR) by using the statistical methods specified in the original correlates study18. Briefly, for each immune biomarker, logistic regression accounting for the sampling design was used to estimate the odds ratio (OR) of infection, controlling for gender and baseline behavioral risk, and IgA levels.
RV144 primary analysis results
Six assays from the 17 different types of immune assays in the original RV144 study were selected as primary endpoints for optimal statistical power when adjusting for multiple comparisons. These selected primary variables included Env-specific plasma IgA, Env- specific plasma IgG binding avidity, gp70-V1V2-specific plasma IgG, neutralizing antibodies, ADCC (antibody dependent cell-mediated cytotoxicity) and Env-specific CD4+ T-cells. The major findings in a multivariate model including all six primary variables were that gp70-V1V2-specific plasma IgG inversely correlated with infection rate (odds ratio 0.57, P=0.03, q=0.08), and Env-specific plasma IgA was a direct correlate of infection (odds ratio 1.54, P=0.03, q=0.08).
Supplementary Material
Acknowledgments
This work was funded by NIH grants [R01 EB008400], and grants [UM1 AI068635] and [UM1 AI068618] to the HIV Vaccine Trials Network (HVTN) and the Statistical Data Management Center (SDMC), the Human Immunology Phenotyping Consortium (HIPC) [U19 AI089986], and the Collaboration for AIDS Vaccine Discovery [OPP1032325]. The work was also supported in part by an Interagency Agreement Y1-AI-2642-12 between the U.S. Army Medical Research and Material Command (USAMRMC) and the National Institutes of Allergy and Infectious Diseases and by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense. We also acknowledge Glenna Peterson for technical assistance with all assays related to the TB dataset.
Footnotes
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Department of Defense.
References
- 1.De Rosa SC, Lu FX, Yu J, et al. Vaccination in humans generates broad T cell cytokine responses. [Accessed February 28, 2014];J Immunol. 2004 173(9):5372–80. doi: 10.4049/jimmunol.173.9.5372. Available at: http://www.ncbi.nlm.nih.gov/pubmed/15494483. [DOI] [PubMed] [Google Scholar]
- 2.Attig S, Hennenlotter J, Pawelec G, et al. Simultaneous infiltration of polyfunctional effector and suppressor T cells into renal cell carcinomas. Cancer Res. 2009;69:8412–8419. doi: 10.1158/0008-5472.CAN-09-0852. [DOI] [PubMed] [Google Scholar]
- 3.Rodrigue-Gervais IG, Rigsby H, Jouan L, et al. Dendritic cell inhibition is connected to exhaustion of CD8+ T cell polyfunctionality during chronic hepatitis C virus infection. [Accessed February 21, 2014];J Immunol. 2010 184(6):3134–44. doi: 10.4049/jimmunol.0902522. Available at: http://www.ncbi.nlm.nih.gov/pubmed/20173023. [DOI] [PubMed] [Google Scholar]
- 4.Ciuffreda D, Comte D, Cavassini M, et al. Polyfunctional HCV-specific T-cell responses are associated with effective control of HCV replication. [Accessed February 21, 2014];Eur J Immunol. 2008 38(10):2665–77. doi: 10.1002/eji.200838336. Available at: http://www.ncbi.nlm.nih.gov/pubmed/18958874. [DOI] [PubMed] [Google Scholar]
- 5.Precopio ML, Betts MR, Parrino J, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204(6):1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darrah PA, Patel DT, De Luca PM, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13(7):843–850. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]
- 7.Roederer M, Nozzi JL, Nason MX. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytom Part A. 2011 doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aghaeepour N, Finak G, Hoos H, et al. Critical assessment of automated flow cytometry data analysis techniques. Nat Methods. 2013;10(5):445–445. doi: 10.1038/nmeth0513-445c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lo K, Brinkman RR, Gottardo R. Automated gating of flow cytometry data via robust model-based clustering. Cytom Part A. 2008;73(4):321–332. doi: 10.1002/cyto.a.20531. [DOI] [PubMed] [Google Scholar]
- 10.Trigona WL, Clair JH, Persaud N, et al. Intracellular staining for HIV-specific IFN-gamma production: statistical analyses establish reproducibility and criteria for distinguishing positive responses. J Interf {&} cytokine Res Off J Int Soc Interf Cytokine Res. 2003;23(7):369–377. doi: 10.1089/107999003322226023. [DOI] [PubMed] [Google Scholar]
- 11.Nason M. Patterns of Immune Response to a Vaccine or Virus as Measured by Intracellular Cytokine Staining in Flow Cytometry: Hypothesis Generation and Comparison of Groups. J Biopharm Stat. 2006;16(4):483–498. doi: 10.1080/10543400600719426. [DOI] [PubMed] [Google Scholar]
- 12.Horton H, Thomas EPE, Stucky JA, et al. Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods. 2007;323(1):39–54. doi: 10.1016/j.jim.2007.03.002.Optimization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Proschan MA, Nason M. Conditioning in 2 × 2 tables. Biometrics. 2009;65(1):316–322. doi: 10.1111/j.1541-0420.2008.01053.x. [DOI] [PubMed] [Google Scholar]
- 14.Finak G, McDavid A, Chattopadhyay P, et al. Mixture models for single-cell assays with applications to vaccine studies. Biostatistics. 2013:1–15. doi: 10.1093/biostatistics/kxt024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen M, Gorochov G, Fastenackels S, Arnaud L, Appay V, Sauce D. Evaluating Cellular Polyfunctionality with a Novel Polyfunctionality Index. PLoS One. 2012;7:e42403. doi: 10.1371/journal.pone.0042403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott JG, Berger JO. Bayes and empirical-Bayes multiplicity adjustment in the variable-selection problem. [Accessed October 18, 2013];Ann Stat. 2010 38(5):2587–2619. Available at: http://projecteuclid.org/euclid.aos/1278861454. [Google Scholar]
- 17.Newton MA, Noueiry A, Sarkar D, Ahlquist P. Detecting differential gene expression with a semiparametric hierarchical mixture method. Biostatistics. 2004;5(2):155–76. doi: 10.1093/biostatistics/5.2.155. [DOI] [PubMed] [Google Scholar]
- 18.Haynes Barton F, Gilbert Peter B, Juliana McElrath M, Zolla-Pazner Susan, Tomaras Georgia D, Munir Alam S, Evans David T, Montefiori David C, Karnasuta Chitraporn, Sutthent Ruengpueng, MD, PhD, Liao Hua-Xin, MD, PhD, DeVico Anthony L., PhD, M Immune-Correlates Analysis of an HIV-1 Vaccine Efficacy Trial — NEJM. [Accessed December 10, 2012];N Engl J Med. 2012 :1275–1286. doi: 10.1056/NEJMoa1113425. Available at: http://www.nejm.org/doi/full/10.1056/nejmoa1113425. [DOI] [PMC free article] [PubMed]
- 19.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 20.Mahomed H, Hawkridge T, Verver S, et al. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. [Accessed November 14, 2013];Int J Tuberc Lung Dis. 2011 15(3):331–6. Available at: http://www.ncbi.nlm.nih.gov/pubmed/21333099. [PubMed] [Google Scholar]
- 21.Bart P, Huang Y, Frahm N, et al. rAd5 prime/NYVAC-B boost regimen is superior to NYVAC-B prime/rAd5 boost regimen for both response rates and magnitude of CD4 and CD8 T-cell responses. [Accessed February 5, 2014];Retrovirology. 2012 9(Suppl 2):O72. Available at: http://www.retrovirology.com/content/9/S2/O72. [Google Scholar]
- 22.Reddy TBK, Riley R, Wymore F, et al. TB database: an integrated platform for tuberculosis research. Nucleic Acids Res. 2009;37(Database issue):D499–508. doi: 10.1093/nar/gkn652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bart P-A, Huang Y, Karuna ST, et al. Higher Ad5 vector priming doses enhance HIV-specific humoral but not cellular responses in a randomized, double-blind phase Ib clinical trial of preventive HIV-1 vaccines. J Clin Invest. 2014 doi: 10.1172/JCI75894. (Under revision) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. [Accessed January 24, 2014];Immunity. 2010 33(4):542–54. doi: 10.1016/j.immuni.2010.09.011. Available at: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3031162&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pai M, Denkinger CM, Kik SV, et al. Gamma Interferon Release Assays for Detection of Mycobacterium tuberculosis Infection. Clin Microbiol Rev. 2014;27(1):3–20. doi: 10.1128/CMR.00034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewer KJ, O’Hara GA, Duncan CJA, et al. Protective CD8+ T-cell immunity to human malaria induced by chimpanzee adenovirus-MVA immunisation. [Accessed February 5, 2014];Nat Commun. 2013 4:2836. doi: 10.1038/ncomms3836. Available at: http://www.nature.com/ncomms/2013/131128/ncomms3836/full/ncomms3836.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overstreet MG, Cockburn IA, Chen Y-C, Zavala F. Protective CD8 T cells against Plasmodium liver stages: immunobiology of an “unnatural” immune response. Immunol Rev. 2008;225:272–283. doi: 10.1111/j.1600-065X.2008.00671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derrick SC, Yabe IM, Yang A, Morris SL. Vaccine-induced anti-tuberculosis protective immunity in mice correlates with the magnitude and quality of multifunctional CD4 T cells. Vaccine. 2011;29(16):2902–9. doi: 10.1016/j.vaccine.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Hawkridge T, Scriba TJ, Gelderbloem S, et al. Safety and immunogenicity of a new tuberculosis vaccine, MVA85A, in healthy adults in South Africa. 2008:544–552. doi: 10.1086/590185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Rosa SC, Carter DK, McElrath MJ. OMIP-014: validated multifunctional characterization of antigen-specific human T cells by intracellular cytokine staining. Cytometry A. 2012;81(12):1019–21. doi: 10.1002/cyto.a.22218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.