Abstract
While the incidence of esophageal and GE junction adenocarcinomas have increased at an alarming rate in the past 30 years, little improvement has been made in treatment strategies. Previous studies have shown that many upper gastrointestinal adenocarcinomas exhibit ERBB2 amplification. In cancers proven to have similar amplification, such as breast, ERBB2-targeted therapies have dramatically improved overall survival and disease-free rates of survival. This study uses siRNA to knockdown ERBB2 in gastrointestinal adenocarcinoma cell lines to evaluate cell viability, apoptosis, and changes in cell cycle. A cell line with a baseline amount of ERBB2 (Seg-1) and two upper gastrointestinal adenocarcinoma cell lines with known amplification of ERBB2 [OE19 (esophageal) and MKN45 (gastric)] were treated with 120 pmol of one of two independent ERBB2 siRNAs or control siRNA for 6 hours. We demonstrate that knockdown of ERBB2 in esophageal and gastric cancer cell lines with known ERBB2 amplification effectively decreases ERBB2 protein levels and decreases cell viability mainly via apoptotic pathways. ERBB2-directed therapy may be of benefit in the subset of patients with gastrointestinal adenocarcinomas exhibiting amplification of ERBB2.
Introduction
The incidence of esophageal adenocarcinoma and esophagogastric junction tumors has increased six-fold since 1970, with an overall alarming increase of 300 to 400 percent in the past 30 years. 1–3 As shown in a SEER (Surveillance, Epidemiology, and End Results) database review by Devesa et al, the annual rate of esophageal adenocarcinoma alone increased by more than 350% from 1974 to 1994.3 This rate of increase exceeds that of all other cancers and the mortality associated with these types of cancers remains high due to local invasion and lymph node metastasis at time of initial diagnosis. In fact, the 5 yr survival is dismal even in the subset of patients who undergo appropriate surgical intervention. Despite the drastic increase in incidence, there has been little improvement or changes in treatment strategies. The mainstay of treatment is surgery if the primary tumor is resectable combined either neoadjuvant or adjuvant chemotherapy, usually consisting of 5-fluorouracil, cisplatin, and epirubicin. Therefore, the development of other treatment strategies is critical.
Several studies have evaluated genes that are commonly deleted or amplified in upper GI adenocarcinomas. In the case of esophageal adenocarcinoma, ERBB2 has been shown to be the most commonly amplified proto-oncogene and is overexpressed in approximately15–40% of the tumors that have been evaluated. 4–10 Similar amplification of ERBB2 has also been noted in GE junction and gastric cardia adenocarcinomas.11–12
The ERBB2 gene (c-erb-B2, HER-2/neu), located on chromosome 17q12, is a member of the epidermal growth factor receptor family and is a transmembrane tyrosine kinase receptor. In normal cells, ERBB2 plays a key role in cellular growth factor signal transduction and is also involved in the regulation of cell growth, survival and differentiation. The ERBB2 oncogene can be activated by point mutations, gene amplification or over-expression. It is hypothesized that high ERBB2 expression promotes spontaneous dimerization of the ERBB2 receptor, thus causing constitutive ERBB2 activation and downstream signaling16. Amplification of the ERBB2 gene and/or over-expression of its corresponding protein have been detected in 20–30% of human breast cancers. HER2 over expression has been found to be an independent prognostic predictor of overall survival and time to relapse, perhaps due to the increased association with estrogen receptor negativity and early metastasis of these tumors. In this subset of patients, ERBB2-targeted therapies have significantly improved overall survival and disease-free rates of survival.13–14 Under current standards of clinical care, patients with ERBB2 amplified tumors receive trastuzumab (Herceptin) in combination with standard chemotherapy.
To better understand the effect of ERBB2 knockdown in esophageal and gastric adenocarcinomas, we used a transient siRNA model to suppress ERBB2 levels in esophageal and gastric adenocarcinoma cell lines with known ERBB2 amplification. The purpose of this study was to analyze the effect of ERBB2 knockdown on (1) ERBB2 protein expression, (2) cell viability in cells line with different levels of ERBB2 expression, and (3) the mechanism of cell death (apoptosis or cell cycle arrest). We believe that in cell lines with ERBB2 amplification there will be a more significant increase in cell death and apoptosis. We show that ERBB2 knockdown via a siRNA model in upper gastrointestinal (GI) adenocarcinoma cell lines effectively decreases ERBB2 protein levels. Further, ERBB2 suppression significantly decreases cell viability in the cell lines known to have ERBB2 amplification with a corresponding increase in the level of apoptosis. Our data suggest that further research and application of ERBB2-directed therapies may be of benefit in the subset of patient with ERBB2-amplified upper GI adenocarcinoma tumors.
Materials and Methods
Cell Lines
In the present set of experiments, three upper gastrointestinal adenocarcinoma cell lines were chosen due to their known varying amount of ERBB2 and compared. An esophageal adenocarcinoma cell line, OE19, was obtained from the European Collection of Cell Cultures. This cell line has 100-fold amplification of ERBB2.6 A gastric adenocarcinoma cell line, MKN45, was obtained from the Japanese Cancer Research Bank. MKN45 has approximately 10-fold amplification of ERBB2. 6 Finally, a cell line with a baseline level of ERBB2, Seg-1 (a gift from Dr. David Beer, University of Michigan, Ann Arbor), was maintained as previously described. All cell lines were cultured in DMEM, 5% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% amphotericin B. All cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Treatment of cells with siRNA
Two ERBB2 siRNA sequences (5′ CACGTTTGAGTCCATGCCCAA3′ and 5′ AAGTGTGCACCGGCACAGACA 3′) were purchased from Qiagen (Germantown, MD), as well as an All-Star Negative Control siRNA (to determine off-target effects). siRNAs were transfected using the LipofetAMINE 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Prior to starting treatments, optimization experiments were carried out with varying amounts of transfection reagent and siRNA to determine the optimal amount and ratio for each cell line used. Briefly, for a 6 well plate, 6μL LipofectAMINE was diluted in 250μL serum-free antibiotic-free DMEM, mixed gently, and incubated at room temperature for 5 minutes. Simultaneously, a mixture of 120pmol siRNA and 250μL serum-free antibiotic free- DMEM was made and incubated at room temperature for 5 minutes. The LipofectAMINE and siRNA mixtures were then combined and incubated for 20 minutes. Cell viability was performed in 96 well plates using an appropriate ratio of siRNA and LipofectAMINE.
Cells were seeded in 6-well plates at a density of 5×105 cells and then incubated overnight at 37°C for analysis of protein, RNA, or apoptosis. Cells were seeded into 96-well plates at a density of 1×103 cells/well for cell viability measurements. The transfection media described above was then placed on the appropriate wells for 6 hours. After 6 hours, the siRNA was removed and replaced with DMEM containing media. Cells treated with serum-free media served as the control and are referred to as nontreated cells. Cells were then incubated for 3–6 days at 37°C and 5% CO2 until collection for the appropriate study.
Determination of ERBB2 by Western Blot Analysis
Cells grown to 70% confluence were collected by trypsinization, and washed with PBS. Cells were then lysed with lysis buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, complete Protease Inhibitor (Roche, Indianapolis, IN)) for 2 hours at 4°C and cleared by centrifugation for 10 minutes at 10,000g twice. Supernatants were collected and then stored at −80°C. ERBB2 protein expression was analyzed by western blotting by loading 75–100μg samples of total protein from the supernatants onto a 10%SDS-PAGE gel. The separated proteins were transferred onto polyvinyllidene diflouride membranes. Nonspecific binding to the membranes was blocked by 2-hour incubation in a buffer containing TBS-T (10mM Tris-HCl, pH 7.4, 150 NaCl, 0.05% Tween-20) and 5% nonfat dry milk. The ERBB2 protein was subsequently detected by overnight incubation at 4°C with a monoclonal anti-ERBB2 antibody (sc-7301; Santa Cruz; 1:250 dilution) and then 1-hour incubation with a peroxidase-conjugated goat anti-mouse IgG antibody (1:10,000 dilution), both in TBS-T and 5% bovine serum albumin. Bound secondary antibody was visualized by ECL and autoradiography.
Quantitative real-time PCR for ERBB2 mRNA levels
The expression of ERBB2 was examined in upper GI adenocarcinoma cell lines after they were treated with ERBB2 siRNA on post-transfection day 3 using real-time quantitative PCR. RNA was extracted using the Qiagen RNeasy Mini Kit, according to manufacturer’s instructions and treated with DNase-I on the column. Total RNA (1μg) was then reverse transcribed. Using the Qiagen Quantitect SYBR green PCR kit, real-time PCR was performed using an Applied Biosystems 7300 real-time PCR system. The ERBB2 primers used were from Qiagen. All data were normalized to the housekeeping gene 18S (Qiagen).
Determination of cell viability
Cell viability was determined using MTS assay (CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay from Promega, Madison, WI). Cells were seeded into a 96-well plate at a density of 1×103 cells/well and allowed to adhere overnight. After treatment with siRNA, cells were incubated for 3–6 days. Next, 20μL of MTS-PMS solution was added to each well. Plates were incubated at 37°C for 2 hours, after which the absorbance at 490nm was measured.
Measurement of Apoptosis
Phosphatidylserine externalization was analyzed using the Guava Nexin kit. Phosphatidylserine externalization is a marker of apoptosis that can be measured by staining cells for Annexin-V and 7-aminoactinomycin D. Briefly, 5×105 cells were washed with PBS and adjusted in 1x binding buffer to a concentration of 5×105/mL. To 40μl of cell suspension, 5μL of Annexin V-phycoerythrin and 5μL 7-aminoactinomycin D were added and incubated for 20 min at room temperature. Samples were analyzed (2,000 events) on a Guava PCA flow cytometer according to manufacturer’s instructions.
Determination of Cell cycle
Approximately 1 × 106 cells were resuspended in 500 μl of propidium iodide solution (20 μg/ml propidium iodide, 0.2 mg/ml RNase, 0.2 mg/ml EDTA, 0.5% NP-40) and incubated at room temperature for 1 h. Samples were analyzed (2,000 events) on a Guava PCA flow cytometer according to manufacturer’s instructions.
Statistical Analysis
Values are expressed as the mean ±SE and statistical differences between groups were assessed with the unpaired Student’s t test. A value of p < 0.05 is considered to be significant. All experiments were performed at least three independent times.
Results
siRNA transfection efficiency was measured at approximately 75–80% for each cell line.
Effect of ERBB2 siRNA on ERBB2 protein expression and mRNA levels
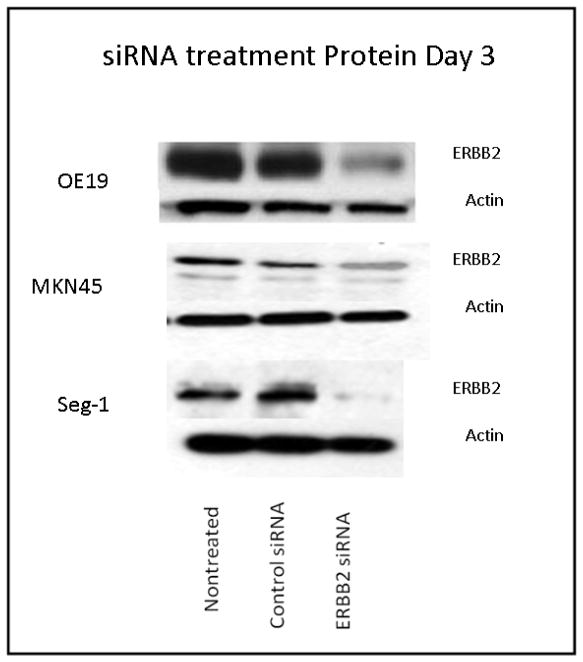
Transfection with ERBB2 siRNA decreased mRNA levels significantly at 72 hrs and ERBB2 remained suppressed at 50% until day 5 post-transfection in all cell lines (data not shown). ERBB2 protein levels decreased by 80% with siRNA treatment in OE19, with similar results seen in the other two cell lines (Fig 1). Results were reproduced with both ERBB2 siRNA sequences three times.
Figure 1. ERBB2 protein levels after treatment with ERBB2 siRNA.
All cell lines showed protein suppression with siRNA treatment. Membranes were stripped and reprobed for actin to show equal protein loading (n = 3).
The effect of ERBB2 siRNA on Gastrointestinal Adenocarcinoma Cell Viability
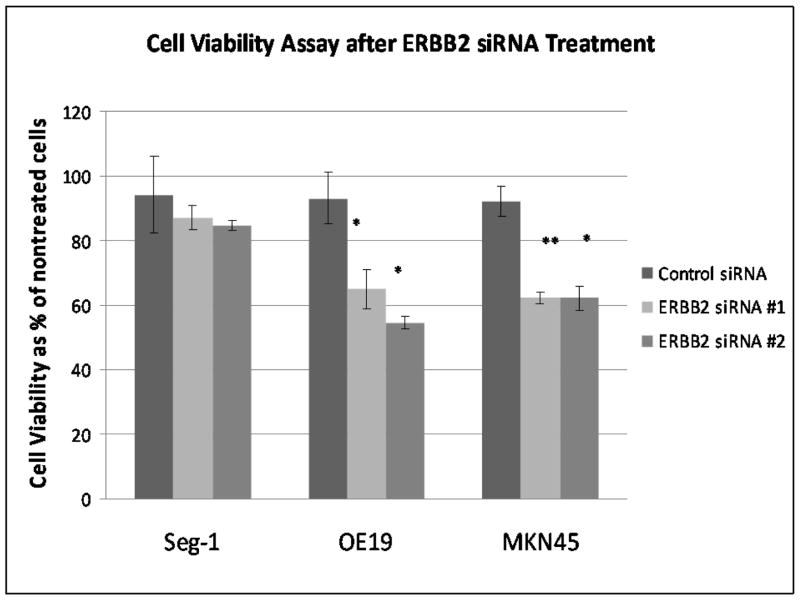
The effect of ERBB2 siRNA on the viability of the upper GI adenocarcinoma cancer cells was examined after 6 hour incubation with siRNA and incubation with serum containing media for 72 hours post-infection. The presence of ERBB2 siRNA significantly reduced cell viability of the two cell lines with known ERBB2 amplification as shown by cell death of 40% in MKN45 cells and 50% in OE19 cells (Table 1; Figure 2). The cell line with a normal ERBB2 levels, Seg-1, exhibited no significant reduction in cell viability when compared to non-treated cells and to cells treated with control siRNA (Table 1; Fig. 2).
Table 1. Cell Viability assay of upper GI adenocarcinoma after ERBB2 siRNA treatment Day 3.
Effect of ERBB2 siRNA treatment on upper GI adenocarcinoma cell viability. After 6 hour transfection with ERBB2 siRNA, cell viability was significantly decreased in OE19 and MKN45, with no significant change in Seg-1. n=4.
| Cell Line | Nontreated Cells | Control siRNA | ERBB2 siRNA #1 | ERBB2 siRNA #2 |
|---|---|---|---|---|
| Seg-1 | 100(±9.50)% | 94.2(±11.90)% | 87.23(±3.69)% | 84.58(±1.63)% |
| OE-19 | 100(±11.24)% | 93.06(±8.09)% | 64.91(±5.90)%** | 54.42(±2.02)%** |
| MKN45 | 100(±10.10)% | 92.23(±4.64)% | 62.23(±1.76)%** | 62.16(±3.56)%** |
p<0.001 when compared to control siRNA
Figure 2. Cell Viability assay of upper GI adenocarcinoma after ERBB2 siRNA Day 3.
effect of ERBB2 siRNA treatment on upper GI adenocarcinoma cell viability. After 6 hour transfection with ERBB2 siRNA, cell viability was significantly reduced in OE19 and MKN45, with no significant change in Seg-1. n=4. **p<0.001 when compared to control siRNA
The effect of ERBB2 siRNA on Apoptosis in Upper GI Adenocarcinomas
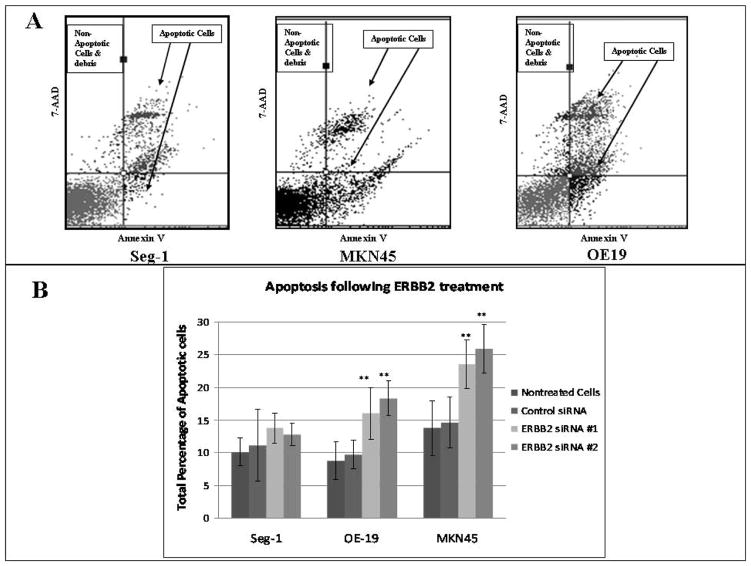
To access the mechanism by which ERBB2 knockdown induces cell death, apoptosis and cell cycle were analyzed. First, ERBB2 siRNA significantly increased apoptosis in the two overexpressing cell lines, with an overall increase in OE19 of 230% when compared to non-treated cells as well as to OE19 cells treated with control siRNA (Table 2, Fig. 3). In MKN45, there was an increase in apoptosis of 155% On the other hand, Seg-1 did not demonstrate an increase in apoptosis.
Table 2. Apoptosis of upper GI adenocarcinoma after ERBB2 siRNA treatment Day 3.
Effect of ERBB2 siRNA treatment on upper GI adenocarcinoma on apoptosis. After 6 hour transfection with ERBB2 siRNA, apoptosis significantly increased in OE19 and MKN45, with no significant change in Seg-1. n=4.
| Cell Line | Nontreated Cells | Control siRNA | ERBB2 siRNA #1 | ERBB2 siRNA #2 |
|---|---|---|---|---|
| Seg-1 | 10.18(±2.06)% | 11.11(±5.42)% | 13.78(±2.34)% | 12.76(±1.74)% |
| OE-19 | 8.82(±2.90)% | 9.75(±2.13)% | 16.05(±3.96)%** | 18.32(±2.68)%** |
| MKN45 | 13.78(±4.12)% | 14.64(±3.91)% | 23.54(±3.66)%** | 25.86(±3.69)%** |
p<0.001 when compared to control siRNA
Figure 3. Apoptosis of GI adenocarcinoma following ERBB2 siRNA treatment post-transfection day 3.
Effect of ERBB2 siRNA treatment on apoptosis. A) Representative flow diagrams of Annexin staining of nontreated cells in each cell line. B) After 6 hours of transfection with ERBB2 siRNA, Annexin-V was significantly increased in OE19 and MKN45, with no change in Seg-1. n=4 **p<0.001 when compared to control siRNA.
The Effect of ERRB2 siRNA on Cell Cycle
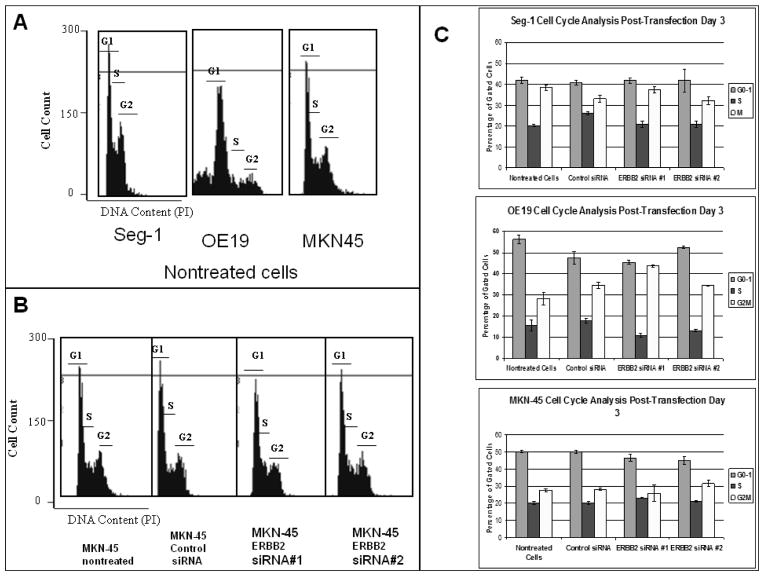
After collection of cells on post-transfection day 3, cells were fixed and stained for cell cycle analysis. There were no significant shifts in cell cycle in all upper GI cancer cell lines evaluated. (Figure 4).
Figure 4. Cell Cycle Analysis of upper GI adenocarcinoma.
After 6 hour transfection with ERBB2 siRNA, there were no significant shifts in cell cycle phase in all cell lines. A) Representation of cell cycle analysis by flow cytometry of all nontreated cells. The x-axis shows DNA content as determined by propidium iodide (PI) fluorescence. The regions representing G1, S, and G2 are indicated. Each cell line had slightly different cell cycles. B) Cell cycle analysis of all MKN-45 groups. There were no significant shifts in cell cycle phase in any treatment group. Similar results were seen in OE19 and Seg (not shown). C). Analysis and quantitation of the percentage of cells found in each phase of the cell cycle from replicate experiments. The mean and standard deviation are shown. n=3. ** p<0.001 when compared to control siRNA
Discussion
In the present study, ERBB2 siRNA treatment significantly decreased protein levels of ERBB2 in all cell lines, indicating that siRNA is an effective model to evaluate ERBB2 suppression. We found no suppression of ERBB1 or other genes with this model when mRNA levels were evaluated, indicating that the effect is specific to ERBB2 knockdown (data not shown). Further, ERBB2 siRNA treatment significantly decreased cell viability in our two upper GI adenocarcinoma cell lines with known ERBB2 overexpression (OE19 and MKN45), though it did not affect that of a cell line with a normal level of ERBB2 (seg-1). Given that a decrease in cell viability can be the result of an increase in apoptosis, an increase in cell cycle arrest, or a combination of both mechanisms, each pathway was examined. Apoptosis is significantly increased in both OE19 and MKN45, the ERBB2 amplified cell lines, with no change in Seg-1. When we studied cell cycle, there were no significant shifts in cell cycle in any cell line examined, suggesting that cell cycle arrest does not play a role in cell death in the upper GI adenocarcinoma cell lines used. Therefore, we can conclude that the increase in cell death seen in the ERBB2 amplified cell lines is a mainly a result of induction of apoptosis.
Though siRNA technology can be used at proof of principle, there are several limitations that would preclude its use in in vivo or clinical studies. siRNA is a transient model in which the toxicity of the transfection reagent used and the siRNA itself prevents higher transfection efficiencies. Furthermore, siRNA is readily degraded by RNAses. Therefore, a stable transfection model for ERBB2 suppression of upper GI adenocarcinomas. Our lab is currently using lentiviral ERBB2 shRNA vectors to repeat all experiments. Thus far, this lentiviral shRNA model has reproduced similar effects as shown with the siRNA model (data not shown).
The ERBB2 gene is amplified in several tumors including cancers of the breast11,13, prostate17, pancreas18, lung19, and ovary13. Trastuzumab (Herceptin), a humanized monoclonal antibody which binds to the extracellular domain of the HER2 protein, has been used clinically to treat patients with ERBB2-amplified metastatic breast cancers.10 In previous studies by our lab, this drug also inhibited growth of OE19, an esophageal adenocarcinoma with 100-fold amplification of ERBB2. However, in cell lines with normal ERBB2 levels, cell growth was not inhibited. 6 No clinical trials have yet tested trastuzumab in the subset of patients with known ERBB2-amplified upper GI adenocarcinomas. However, there are many obstacles to overcome with the use of herceptin, including the significant side effects (in particular the cardiac toxicity) as well as the cost of treatment (ranging from $20,000–80,000/year). Although herceptin therapy may be an effective strategy in this high-risk population, development of treatment with local administration could potentially bypass these obstacles. Therefore, the future direction of our lab is to generate a stable transfection model with the potential of local administration.
Acknowledgments
This project was partly supported by NIH/NIDDK R01 DK063615 (Yamamoto).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.http://www.cancer.gov/ncicancerbulletin/NCI_Cancer_Bulletin_051606
- 2.http://www.cancer.gov/cancertopics/pdq/prevention
- 3.Devesa SS, Blot WJ, Fraumeni JF. Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998 Nov 15;83(10):2049–2053. [PubMed] [Google Scholar]
- 4.Metzger R, Schneider PM, Warnecke-Eberz U, et al. Molecular biology of esophageal cancer. Onkologie. 2004;27:200–206. doi: 10.1159/000076913. [DOI] [PubMed] [Google Scholar]
- 5.Lin J, Beer D. Molecular biology of upper gastrointestinal malignancies. Seminars in Oncology. 2004 Aug;31(4):476–486. doi: 10.1053/j.seminoncol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 6.Dahlberg PS, Ferrin LF, Grindle SM, et al. Gene expression profiles in esophageal adenocarcinoma. Ann Thorac Surg. 2004;77:1008–1015. doi: 10.1016/j.athoracsur.2003.09.051. [DOI] [PubMed] [Google Scholar]
- 7.Kuwano H, Kato H, Miyazaki T, et al. Genetic alterations in esophageal cancer. Surg Today. 2005;35:7–18. doi: 10.1007/s00595-004-2885-3. [DOI] [PubMed] [Google Scholar]
- 8.Miller CT, Moy JR, Lin L, et al. Gene amplification in esophageal adenocarcinomas and Barrett’s with high-grade dysplasia. Clin Cancer Res. 2003 Oct 15;9:4819–4825. [PubMed] [Google Scholar]
- 9.Jenkins GJ, Doak SH, Parry JM, et al. Genetic pathways involved in the progression of Barrett’s metaplasia to adenocarcinoma. Br J Surg. 2002;89:824–837. doi: 10.1046/j.1365-2168.2002.02107.x. [DOI] [PubMed] [Google Scholar]
- 10.Chiang P, Beer D, Wei W, et al. Detection of erbB-2 amplification in tumors and sera from esophageal carcinoma patients. Clin Cancer Res. 1999 Jun;5:1381–1386. [PubMed] [Google Scholar]
- 11.Park JB, Rhim JS, Park SC, et al. Amplification, overexpression, and rearrangement of erbB-2 proto-oncogene in primary human stomach carcinomas. Cancer Res. 1989 Dec 1;49:6605–6609. [PubMed] [Google Scholar]
- 12.Tanner M, Hollmen M, Junttila TT, et al. Amplification of Her-2 in gastric carcinoma: association with topoisomerase II-α gene amplification, intestinal type, poor prognosis and sensitivity to trastuzumab. Ann Onc. 2006;16:273–278. doi: 10.1093/annonc/mdi064. [DOI] [PubMed] [Google Scholar]
- 13.Slamon DJ, Godolphin W, Jones LA, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER2-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 14.Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF American Society of Clinical Oncology, and College of American Pathologists. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol. 2007;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 15.Arteaga CL, Baselga J. Tyrosine kinase inhibitors: Why does the current process of clinical development not apply to them? Cancer Cell. 2004 Jun;5:525–531. doi: 10.1016/j.ccr.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 16.Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Experimental Cell Research. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 17.Casimiro M, Rodriguez O, Pootrakul L, et al. ErB-2 induces the cyclin-D1 gene in prostate epithelial cells in vitro and in vivo. Cancer Res. 2007 May 1;67(9):4364–4372. doi: 10.1158/0008-5472.CAN-06-1898. [DOI] [PubMed] [Google Scholar]
- 18.Safran H, Steinhoff M, Mangray S, et al. Overexpression of the Her-2/neu oncogenes in pancreatic adenocarcinoma. Am J Clin Oncol. 2001;24:496–9. doi: 10.1097/00000421-200110000-00016. [DOI] [PubMed] [Google Scholar]
- 19.Hirsch FR, Franklin WA, Veve R, et al. Her2/neu expression in malignant lung tumor. Semin Oncol. 2002;29:51–8. doi: 10.1053/sonc.2002.31523. [DOI] [PubMed] [Google Scholar]