Time-lapse atomic force microscopy imaging is used to visualize the initial stages of fibronectin fibrillogenesis directly in living cells with high resolution. This approach provides new structural and mechanistic details, such as a stepwise extension mechanism and an accelerating effect of extracellular Mn2+ on early FN fibrillogenesis.
Abstract
Fibronectin (FN) is an extracellular matrix protein that can be assembled by cells into large fibrillar networks, but the dynamics of FN remodeling and the transition through intermediate fibrillar stages are incompletely understood. Here we used a combination of fluorescence microscopy and time-lapse atomic force microscopy (AFM) to visualize initial stages of FN fibrillogenesis in living fibroblasts at high resolution. Initial FN nanofibrils form within <5 min of cell–matrix contact and subsequently extend at a rate of 0.25 μm/min at sites of cell membrane retraction. FN nanofibrils display a complex linear array of globular features spaced at varying distances, indicating the coexistence of different conformational states within the fibril. In some cases, initial fibrils extended in discrete increments of ∼800 nm during a series of cyclical membrane retractions, indicating a stepwise fibrillar extension mechanism. In presence of Mn2+, a known activator of integrin adhesion to FN, fibrillogenesis was accelerated almost threefold to 0.68 μm/min and fibrillar dimensions were increased, underlining the importance of integrin activation for early FN fibrillogenesis. FN fibrillogenesis visualized by time-lapse AFM thus provides new structural and mechanistic insight into initial steps of cell-driven FN fibrillogenesis.
INTRODUCTION
Fibronectin (FN) is a large dimeric glycoprotein and an abundant component of the extracellular matrix (ECM) in different tissues, where it mediates integrin-dependent cell attachment and matrix cross-linking (Schwarzbauer and DeSimone, 2011). FN also plays an indispensable role during development, wound healing, and matrix repair (Grinnell, 1984a). A hallmark of FN is the cell-mediated reorganization of FN dimers into fibrils, which activates a range of its biological functions (Mao and Schwarzbauer, 2005; Singh et al., 2010). Fibrillar FN networks stably anchor cells to the matrix environment, but FN fibrillogenesis also often coincides with large morphogenetic changes during embryonic development in conjunction with cell movement. For instance, FN fibrils can guide cell migration during development (Winklbauer and Keller, 1996; Marsden and DeSimone, 2001; Davidson et al., 2004) and may immobilize growth factor gradients (Nagel et al., 2004) and contribute to tissue patterning (Sakai et al., 2003; Larsen et al., 2006).
Despite the well-established physiological importance of fibrillar FN, the molecular mechanisms leading to FN assembly and the ultrastructure of the resulting FN fibrils are incompletely understood. Both plasma and cellular FN, two closely related isoforms, are initially secreted in a compact, inactive conformation. The conversion of globular FN molecules into the extended, active conformation is driven by cellular contraction forces transmitted by integrin receptors at focal adhesion sites (Dzamba et al., 1994; Christopher et al., 1997). Extension of the FN molecule exposes different FN–FN binding sites (Singh et al., 2010), allowing FN molecules to interact laterally and form fibrils. Fibrillar extension initially occurs linearly and unidirectionally (Winklbauer and Stoltz, 1995) but probably involves lateral addition of FN molecules along the entire fibril. Accumulated FN fibrils are further stabilized inside fibrillar networks via noncovalent interactions (Chen and Mosher, 1996), producing a deoxycholate-insoluble FN matrix (McKeown-Longo and Mosher, 1983). FN fibrils are highly elastic, stretching up to fourfold during matrix remodeling (Ohashi et al., 1999), and domain unfolding may provide an additional mechanism for cell-mediated fibrillar extension. FN consists of two nearly identical disulfide-bonded monomers containing repeating type I, II, and III domains (Hynes, 1985). Whereas type I and II domains are stabilized by intramolecular disulfide bonds, type III domains lacks these bridges and can unfold under an applied force (Craig et al., 2001; Oberhauser et al., 2002; Gao et al., 2003). Important insight into FNIII domain unfolding has been provided by fluorescence resonance energy transfer (FRET) spectroscopy in cell culture studies and in vitro stretching experiments (Baneyx et al., 2001, 2002; Smith et al., 2007). Free cysteine labeling experiments support domain unfolding of in vitro–stretched fibrils (Bradshaw and Smith, 2011) but indicate that only a limited subset of FNIII domains may unfold in cell-derived fibrils (Lemmon et al., 2011).
Mature FN fibrils may reach a diameter up to the micrometer level (Singer, 1979) and probably contain several hundred FN molecules. Nevertheless, the exact molecular conformation and arrangement of FN dimers in fibrils is incompletely understood. Electron microscopy (EM) images of cell-derived FN matrices revealed fibrillar building blocks with diameters as small as 5–15 nm (Chen et al., 1978; Dzamba and Peters, 1991) and thus close to the size of one or two extended FN molecules. In vitro–assembled FN fibrils contain 14-nm-thick fibrils, which appear to consist of 7-nm-thick substructures (Ulmer et al., 2008), supporting the idea that two FN dimers constitute the molecular building block. Immunogold labeling experiments suggest a stagger between neighboring FN molecules of 84 nm in early fibrils and 42 nm in mature fibrils (Dzamba and Peters, 1991), whereas a recent superresolution microscopy study found an average periodicity of ∼95 nm (Fruh et al., 2015).
The growth of individual FN fibrils has been observed on the blastocoel roof of Xenopus embryos, demonstrating the highly dynamic nature of FN remodeling and providing important early insight into the rate of fibril elongation (Winklbauer and Stoltz, 1995). Dynamic rearrangement of complex FN networks has also been visualized in embryonic Xenopus explants (Davidson et al., 2008) and in cell culture time-lapse experiments using fluorescent FN fusion proteins (Ohashi et al., 1999) or fluorescently labeled FN (Pankov et al., 2000). However, the initial stages of FN fibrillogenesis occur on the submicrometer level and are therefore difficult to resolve using conventional light microscopy. EM has provided high-resolution images of individual FN molecules as well as cell-generated fibrils but requires sample drying and staining/coating, which not only may affect the structure of FN fibrils but also make this technique incompatible for observing fibrillogenesis in living cells. The majority of high-resolution studies investigating cell-induced fibrillogenesis have thus been performed on fixed and stained samples, providing snapshots of different stages of FN fibrillogenesis, rather than visualizing the entire process from initial precursors to fully formed fibrils. Ideally, cell-induced FN fibrillogenesis would be studied using label-free, high-resolution imaging techniques in real time. AFM is a powerful technique for obtaining high-resolution images under physiological conditions and has been used to study the structure of individual FN molecules (Lin et al., 2000; Chen et al., 2007) and visualize different stages of FN fibrillogenesis in fixed cells (De Jong et al., 2006). Of importance, AFM can track the remodeling of individual ECM macromolecules by living cells with near-molecular resolution (Friedrichs et al., 2007), but this unique property has so far not been applied to study cell-induced FN fibrillogenesis.
In this work, we used time-lapse AFM imaging in combination with fluorescence microscopy to visualize the dynamics of early FN fibrillogenesis in living REF52 fibroblasts. By performing live-cell time-lapse AFM imaging, we visualize initial steps of cell-induced FN fibril formation, revealing a step-like pattern of early FN extension. Furthermore, we demonstrate an important influence of Mn2+ on early FN fibrillogenesis dynamics. From AFM volume images, we also estimate the number of FN molecules incorporated into FN fibrils at different stages of early fibrillogenesis. Together these results provide a novel time-resolved picture of initial and intermediate steps of FN fibrillogenesis.
RESULTS
Investigating FN fibrillogenesis by total internal reflection microscopy
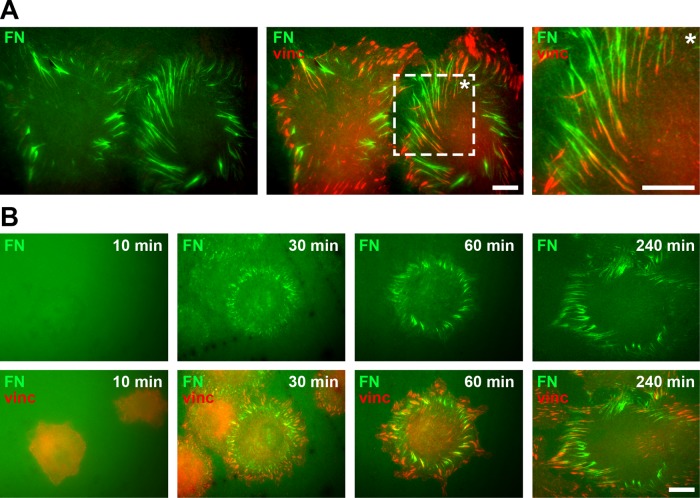
Fibroblasts have a well-characterized role in remodeling FN in different tissues (Grinnell, 1984a; Singer et al., 1984). Fibroblasts such as the REF52 (rat embryonic fibroblast) cell line also provide useful model systems to study cell-induced FN fibrillogenesis in tissue culture experiments. In these experiments, fluorescently labeled FN monomers can be preadsorbed to glass surfaces, which are then remodeled into fibrillar structures after cell seeding at integrin adhesion sites (Avnur and Geiger, 1981). In agreement with these previous reports, REF52 cells remodeled a homogeneous coating of fluorescently labeled FN (FN-AF488) into fibrillar structures near vinculin-positive focal adhesions within 4 h of seeding (Figure 1A). Large FN fibrils often formed radial arrays at the cell periphery. The association of theses fibrils with focal adhesions at the rear suggested that they had formed as a result of traction forces exerted by focal adhesion translocation toward the cell center—for example, during cell membrane retraction or cell migration.
FIGURE 1:
Cell-induced FN fibrillogenesis. (A) TIRF microscopy images of REF52 cells incubated on a homogeneous coating of Alexa 488–labeled FN (FN-AF488, green) for 4 h and stained for vinculin (red) to visualize focal adhesions. A magnified view (rightmost image) corresponding to a boxed area of the merge image demonstrates the close association of FN fibrils and focal adhesions at the cell periphery. Scale bars, 10 μm. (B) Dynamics of cell-induced FN fibrillogenesis. REF52 cells were incubated on FN-AF488 (green) for 10, 30, 60, or 240 min, fixed, and stained for vinculin (red). Scale bar, 10 μm.
To assess the general dynamics of FN fibrillogenesis in REF52 cells, we visualized different stages of fibrillogenesis by total internal reflection fluorescence (TIRF) microscopy in cells fixed 10, 30, 60, and 240 min after seeding (Figure 1B). After 10 min of incubation, cells began to spread and assemble adhesion sites, but only few and small fibrillar FN structures are visible, indicating that adhesion-site assembly precedes FN fibril formation. After 30 min, fibrils become apparent in radial arrays at the cell periphery, which then continue to grow in size (60 min). After 240 min, cells have extensively remodeled the FN layer below and next to the cell body. The fluorescence pictures visualized the dynamics of FN fibril formation over the first 4 h of cell spreading and verified the active role of focal adhesions in this process.
Investigating FN fibrillogenesis by AFM in combination with fluorescence microscopy
Analyzing fluorescence microscopy images provides a reliable measure of lateral and longitudinal fibrillar dimensions at later stages of extension. However, at the earliest stages of fibrillogenesis (≤10 min), fibril dimensions approach the resolution limits of conventional optical microscopy (∼200 nm), preventing accurate quantification from fluorescence images. Moreover, FN fibrils are three-dimensional structures, but fluorescence images provide no accurate information regarding the fibril height. To obtain additional insight into FN fibril structure, we decided to use high-resolution AFM imaging to visualize FN fibrils at different stages of fibrillogenesis. AFM is a surface-sensitive imaging technique generating high-resolution images of a biological sample under physiological conditions. When operated in contact mode, AFM simultaneously records “deflection” (high-contrast images indicating local height changes) and “height” (representing true sample height) images. However, AFM images provide no direct information regarding the molecular identity of sample features. The identification of FN structures in AFM images therefore requires complementary fluorescence labeling of FN and an integrated AFM and optical microscopy platform.
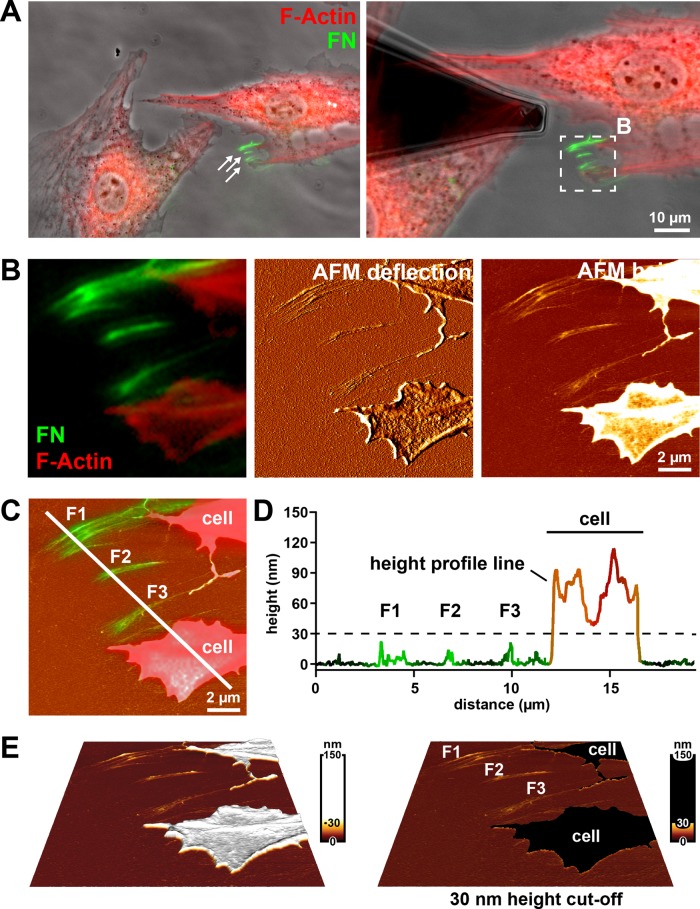
To test the combined AFM and optical microscopy approach, we first analyzed mature FN fibril structure in fixed cells after 4 h of incubation on fluorescently labeled FN, while cellular structures were visualized by F-actin staining. Cells stably spreading on cell culture substrates for several hours primarily remodel FN at the basal cell side (Gudzenko and Franz, 2013), and these fibrils are consequently hidden beneath the cell and inaccessible to the AFM tip. However, FN fibrils forming at the cell periphery during membrane retraction are exposed and amenable to AFM. FN fibrils in the cell vicinity were first identified in overview light microscopy images (Figure 2A). A smaller region containing several arrays of FN fibrils near the cell edge was then imaged at increased resolution by AFM (Figure 2B). Comparison of AFM “height” and “deflection” images with the corresponding region of the fluorescence image indicated excellent overall structural agreement but enhanced structural detail in the AFM images. To correlate the arrangement of fibrillar FN and cellular structures in the light microscopy images with the AFM height information, we overlaid the AFM height image with the merged FN/F-actin fluorescence image (Figure 2C). Superimposing the fluorescence intensity signal on a height profile generated along a line crossing both FN fibrils and cellular protrusions revealed clear height differences between both structures (Figure 2D). Whereas FN fibril height ranged consistently <30 nm, F-actin–positive cellular structures never reached 40 nm in height within cell protrusions and typically extended 70–200 nm at the cell edge. Apparently, a minimum peripheral cell height of ∼70 nm reflects the combined height of the basal and apical plasma membranes and the interjacent cortical cytoskeleton. In addition, focal adhesions, which preferentially form at the cell periphery in REF52 cells (Figure 1, A and B), extend 120–180 nm above the substrate in these cells (Franz and Muller, 2005) and also contribute to the total cell height at the cell edge. Given the clear height difference, applying a 30-nm height cut-off reliably separates fibrillar FN structures from cellular structures (Figure 2D) in AFM height images without the need for complementary fluorescence labeling of FN and cells (Figure 2E). An additional example demonstrating that a 30-nm height cut-off separates cellular structures from FN nanofibrils is given in Supplemental Figure S1. Fibrillar structures resembling the FN fibrils identified by AFM near polarized cells could also be detected on scanning electron microscopy (SEM) images (Supplemental Figure S2). However, unambiguous identification of FN fibrils in the cell vicinity was impossible from SEM images because these images contain no height information.
FIGURE 2:
Imaging FN fibrils using combined AFM and fluorescence microscopy (A) Left, phase contrast image with overlaid fluorescence signals for F-actin (red) and FN (green) of fixed Ref52 cells after 4 h of incubation on FN-AF488. The epifluorescence images were collected using an Achroplan 40×/0.80 W water-immersion lens. White arrows indicate assemblies of FN fibrils in the cell vicinity. Right, the same cell group with an AFM cantilever positioned near the fibrillar FN array. The white box indicates an area imaged by AFM and shown at higher magnification in B. (B) Merged fluorescence image (F-actin in red, FN in green) and corresponding AFM height and deflection mode images. (C) AFM height image overlaid with the FN fluorescence channel and the F-actin signal to indicate cellular protrusions. A profile line crosses several fibrillar FN arrays (F1, F2, F3) and a cellular protrusion. (D) Superimposition of height and fluorescence intensity profiles generated along the white line in C. FN fibrils (F1, F2, F3) can be distinguished from cellular protrusion or retraction structures (“cell”) using a 30-nm height cut-off (dashed line). (E) Three-dimensional reconstruction of the AFM height image shown in B, applying either a nonlinear height scale (left) or a 30-nm height cut-off (left), which eliminates all cellular structures (“cell”) from the image but retains all fibrillar FN structures (F1, F2, F3).
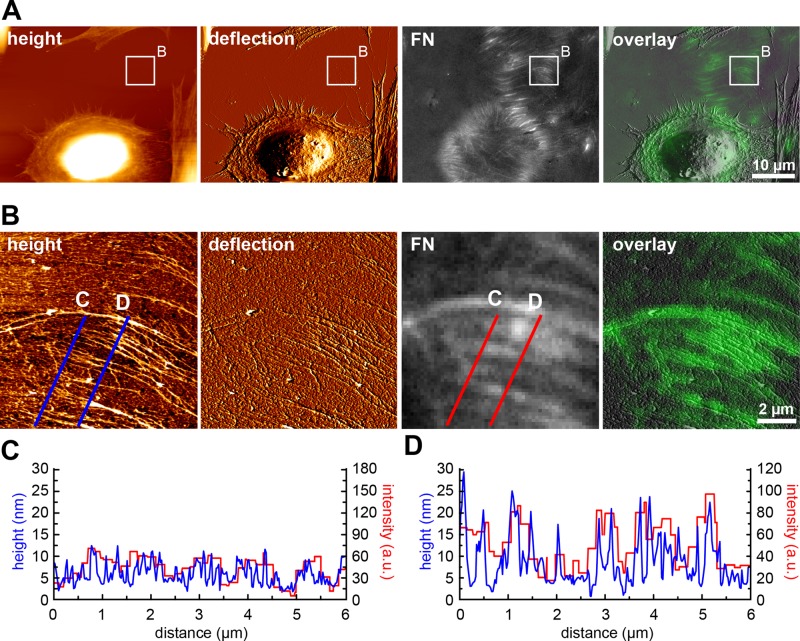
When cell incubation time on FN was extended to 16 h, cells had frequently vacated whole areas of remodeled FN (Figure 3A). These areas displayed no F-actin signal, indicating the absence of potential cellular retraction structures or cellular debris deposited during cell migration (unpublished data). Higher-resolution AFM images of these cell-free areas revealed a complex array of mainly parallel fibrils, which often appeared frayed at one end (Figure 3B). An overlay of AFM and fluorescence images collected from the same area identified the majority of these structures as FN fibrils formed from the initially homogeneous FN layer (Figure 3B). Again, overlay with the corresponding fluorescence image demonstrated good overall structural agreement between the AFM and light microscopy images, but the light microscopy images failed to resolve the FN fibril ultrastructure visible in AFM images. Whereas the fluorescence images suggested a fairly homogeneous distribution of FN building blocks within large fibrils (Figures 1A and 3B), the AFM images revealed a striking suborganization of these structures into several parallel nanofibrils, each typically measuring 30–100 nm in width and spaced apart by 150–300 nm (Figure 3B). The AFM images also demonstrated a gradual transition of frayed nanofibrils ends into thicker and more compact structures in the fibrillar midregion, suggesting progressive fibril bundling. Because the spoke-like array of cell-free fibrils matched the circular arrangement of cell-associated fibrils normal to the cell edge (Figure 1A), we considered the frayed end the cell-distal position, or front end, and the thicker fibril end the cell-proximal position. The majority of nanofibrils were straight, indicating that they had formed under tension.
FIGURE 3:
Analyzing mature FN fibril structure in cell-free regions vacated by migrating cells. (A) AFM height and deflection images (left), corresponding FN fluorescence image, and an overlay of AFM deflection mode and fluorescence images. Cells were fixed after 16 h of incubation on the FN layer. (B) A cell-free region indicated by the white rectangle in A scanned by AFM at higher resolution to visualize FN fibril ultrastructure. (C) Superimposition of height (blue) and fluorescence intensity (red) profiles generated along the corresponding C lines across a thin, distal section of an array of FN fibrils in panel B. (D) Superimposition of height (blue) and fluorescence intensity (red) profiles generated along the corresponding D lines across a proximal, thicker section of an array of FN fibrils in B. The AFM profiles (in blue) display higher spatial resolution than the corresponding fluorescence intensity profiles (in red).
To investigate the correlation between fluorescence and AFM images further, we extracted height (blue lines) and fluorescence intensity (red lines) profiles along lines traversing the fibrillar arrays at different positions (Figure 3B). We then plotted corresponding height and intensity profile lines together in single diagrams (Figure 3, C and D). Overall, height and fluorescence intensity signals correlated both at the fibril front (cell distal orientation; Figure 3C) and in the central region, where fibrils reached their greatest width (Figure 3D). However, the FN fibril fluorescence signal was generally of lower spatial resolution, whereas the height profiles from the AFM image contained more structural detail and easily resolved fibrils with widths <200 nm. The AFM cross sections also yielded additional information about height variations along the FN nanofibrils. Fibrils were typically ∼10 nm high at the frayed front end (Figure 3C, blue trace) and ≤30 nm at the fibril middle and rear (Figure 3D, blue trace), indicating gradually increasing fibril height along the cellular pulling direction.
Imaging early FN fibrillogenesis in living cells by time-lapse AFM
Initial steps of FN fibrillogenesis occur on the nanoscale, and standard optical microscopy techniques provided only limited insight (Figure 1B). To monitor the initial steps and dynamics of FN fibril formation at higher resolution, we performed AFM time-lapse scanning of living cells. We performed scanning in contact mode using a low scan force (<1 nN) and moderate scan speeds to minimize a potentially destructive influence of the AFM tip on cell and FN morphology. To maximize the frame rate while maintaining adequate image resolution, we limited scan regions to ≤10 × 10 μm2 at 512 × 512 pixels. Using these conditions and a maximum line scan frequency of 2–4 Hz, we recorded image series containing approximately one image every 2–5 min. Ultraflat cleaved mica disks were used as sample supports in time-lapse experiments to enhance the vertical resolution of the AFM images.
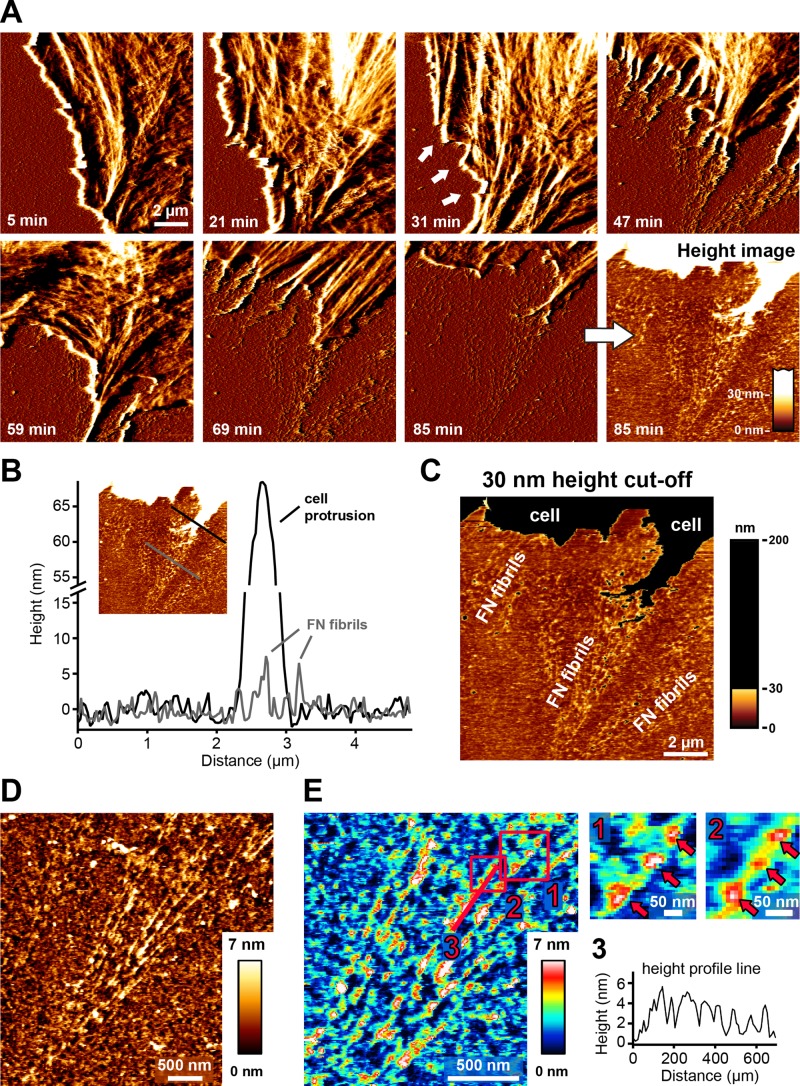
Before commencing live-cell imaging, we incubated cells on FN for 5 min to ensure initial cell attachment and initiation of spreading. Based on images obtained on fixed cells (Figure 1A) and in agreement with previous results (Ohashi et al., 2002), FN fibrillogenesis often initiates near the cell edge. During fibril extension, the distal fibril tip then remains stationary, whereas the opposite end of the fibril shows movement toward the cell center together with translocating integrin receptors (Pankov et al., 2000). Because the immobile fibril end extends beyond the retracting cell edge (Ohashi et al., 2002), FN fibrils are exposed and can be imaged by AFM in regions at the cell periphery. In agreement, AFM time-lapse recordings showed that FN fibrils frequently emerged during membrane retraction (Figure 4A and Supplemental Movie S1). Fibrils were usually aligned in the direction of the retracting membrane, suggesting that fibrils formed as the result of traction forces applied by the retracting cell membrane (Figure 4A, 47 min). Optimal scan results were obtained when the scan direction was roughly parallel to the cell edge and therefore perpendicular to the emerging FN fibrils. During live-cell scanning, the AFM image quality of FN fibrils sometimes appeared degraded (Figure 4A, 47 min) because fragile retracting cellular extensions transiently connected to nascent FN fibrils could not be stably imaged by AFM (Figure 4A, 47 and 69 min). However, once cell membranes had fully moved out of the scan area, tip-sample tracing stabilized, revealing a structurally intact array of FN nanofibrils (Figure 4A, 85 min). These early FN fibrils are typically <10 nm in height (Figure 4B) and could be clearly separated from cellular structures by applying the previously established 30- nm height threshold (Figure 4C). To further demonstrate that the fibrillar structures forming during membrane retraction represent remodeled FN molecules, we performed control time-lapse AFM experiments on FN layers fixed with glutaraldehyde (GA) before cell seeding. Cross-linking by GA efficiently prevents cell-induced FN remodeling, as verified by fluorescence microscopy (unpublished data). Cells spread readily on the fixed FN layer and formed focal adhesions (unpublished data), indicating that chemical cross-linking preserved the adhesion functionality of FN and that cells were exerting traction forces on the substrate. However, cell retraction on fixed FN exposed a smooth FN layer lacking any fibrillar structures and which was indistinguishable from other cell-free areas on the substrate (Supplemental Figure S3A and Supplemental Movie S2). These experiments demonstrated that REF52 cells in an early spreading phase (<5–60 min after seeding) deposit no cellular structures or debris on the FN substrate during membrane retraction. In further control experiments, we also imaged cells on laminin-111, an ECM component not undergoing cell-induced fibrillogenesis. Laminin surfaces were less homogeneous than FN surfaces, but cells also retracted without depositing any fibrillar structures (Supplemental Figure S3B and Supplemental Movie S3), demonstrating that fibril formation was specific to FN. Together the control experiments on laminin and fixed FN demonstrate that the elongated structures forming at the cell edge represent early FN nanofibrils, in agreement with their height profile (<30 nm). For comparison, we also analyzed fibrillogenesis in human foreskin fibroblasts (HFFs). These cells often retracted fully out of the AFM imaging area during a time-lapse experiment, again without leaving behind membranous or other cellular material (Supplemental Figure S4 and Supplemental Movie S4). Like REF52 cells, HFFs efficiently remodeled the FN layer at the cell edge into nanofibrils with a similar height profile (<10 nm). Thus, FN remodeling at the cell edge during cell membrane retraction appears to be a common feature of fibroblast cells.
FIGURE 4:
Investigating FN fibrillogenesis by time-lapse AFM in living cells. Suspended REF52 cells were added to FN-coated mica substrates. After 5–10 min of cell spreading, ∼10 × 10 μm2 regions near the cell periphery were scanned continuously by AFM in contact mode. (A) Time-lapse images (deflection channel) of a region at the cell edge. The cell membrane initially protrudes (21 min), but after 31 min, it starts retracting, and FN fibrils appear (white arrows). After several cycles of extension and retraction, the cell membrane finally retracts fully from the imaging area (85 min), revealing an array of FN fibrils (deflection and corresponding height images). The complete time-lapse series is presented in Supplemental Movie S1. (B) Height profile lines generated across a newly formed array of cell-induced FN fibrils (gray line) or across cellular retraction structures (black line). FN fibrils and cellular structures can be distinguished based on their different height profiles. (C) AFM height image to which a 30-nm height cut-off has been applied to isolate FN nanofibrils. (D) High-resolution AFM scan revealing the linear ultrastructure of cell-induced FN nanofibrils. (E) Part of the AFM height image shown in D in a rainbow color scale (left). Pink squares indicate two regions shown at higher magnification on the right. The pink line indicates the position of a height profile shown on the lower right. Pink arrows indicate periodic features on cell-induced FN fibrils.
Ultrastructure of FN nanofibrils
The time-lapse series provided the first direct high-resolution view of the initial stages of cell-induced FN fibrillogenesis. Frequently, cells went through several cycles of membrane retraction and extension, apparently recontacting newly formed fibrils (Figure 4A, 31–69 min). FN nanofibrils emerging as a result of cyclical membrane movement often appeared structurally inhomogeneous along their length (Figure 4C). Higher-resolution scans (5 × 5 μm2 scan area at 512 × 512 pixels) of early cell-induced FN fibrils revealed predominantly linear fibrillar arrangement along the direction of cellular retraction (Figure 4D) but a complex height profile along the fibrils. Bead-like domains appeared to be interspersed by more-elongated, smooth stretches (Figure 4E). The height of the interbead structures was in the range of 1–2 nm, whereas the bead-like structures had a height of 3–6 nm and a width of 12–25 nm. In some regions, three globular domains appeared to be evenly spaced at distances of 60–90 nm (Figure 4E). This was most apparent in thinner fibrils, whereas bigger fibrils usually displayed no obvious periodicity. The presence of regular globular features in thin, cell-induced FN fibrils suggests a regular stagger of FN building blocks in these areas.
Mn2+ promotes early FN fibrillogenesis
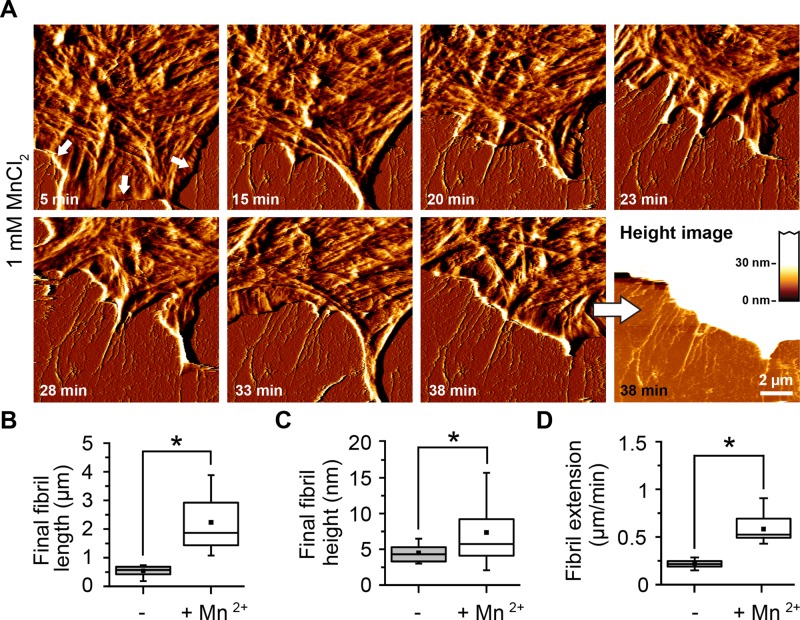
Extracellular Mn2+ increases the affinity of different integrins, including α5β1, to FN (Gailit and Ruoslahti, 1988; Danen et al., 1995; Mould et al., 1995), enhances cell attachment, spreading, and migration on FN (Grinnell, 1984b), and accelerates the FN fibrillogenesis process (Sechler et al., 1997).Traction force experiments showed that integrin activation with Mn2+ increases cytoskeletal tension transmitted onto FN (Lin et al., 2013), stimulating the assembly of a mature, deoxycholate-insoluble FN matrix after several hours of incubation (Sechler et al., 1997; Brenner et al., 2000). However, less is known about whether Mn2+ also affects the formation dynamics and structure of early FN fibrils. To assess the effect of Mn2+ on the morphology of early FN fibrils, we performed live-cell scanning by AFM in the presence of 1 mM Mn2+. In this case, cells began forming fibrils almost immediately (6 min) after cell seeding (Figure 5A and Supplemental Movie S5). After 30 min, FN fibrils had reached an average length of ∼2 μm and a height of ≤20 nm, in contrast to ∼1 μm and ∼6 nm, respectively, in the absence of Mn2+ (Figure 5, B and C). Cells also formed FN fibrils at roughly threefold-higher speeds in the presence of Mn2+ (0.68 ± 0.14 μm/min) than in its absence (0.25 ± 0.02 μm/min; Figure 5D).
FIGURE 5:
Enhanced early fibrillogenesis in presence of Mn2+. Cells were incubated for 5 min on FN in the presence of Mn2+ (1 mM) and then continuously imaged by AFM. (A) FN fibrils are already visible after 5 min (white arrows) and show enhanced growth rates. Scale bar, 1.5 μm; full range of the AFM height scale, 20 nm. (B) Box-and-whisker plot of final fibril length in absence or presence of 1 mM Mn2+. (C) Box-and-whisker plot of final fibril height in absence or presence of 1 mM Mn2+. (D) Velocity of lamellipodium retraction in absence or presence of 1 mM Mn2+. Data (mean ± SD) from nine independent experiments. Statistically significant differences (p < 0.01) are denoted by an asterisk. The complete time-lapse series is presented in Supplemental Movie S2.
Fast FN rearrangement at retracting cell membranes
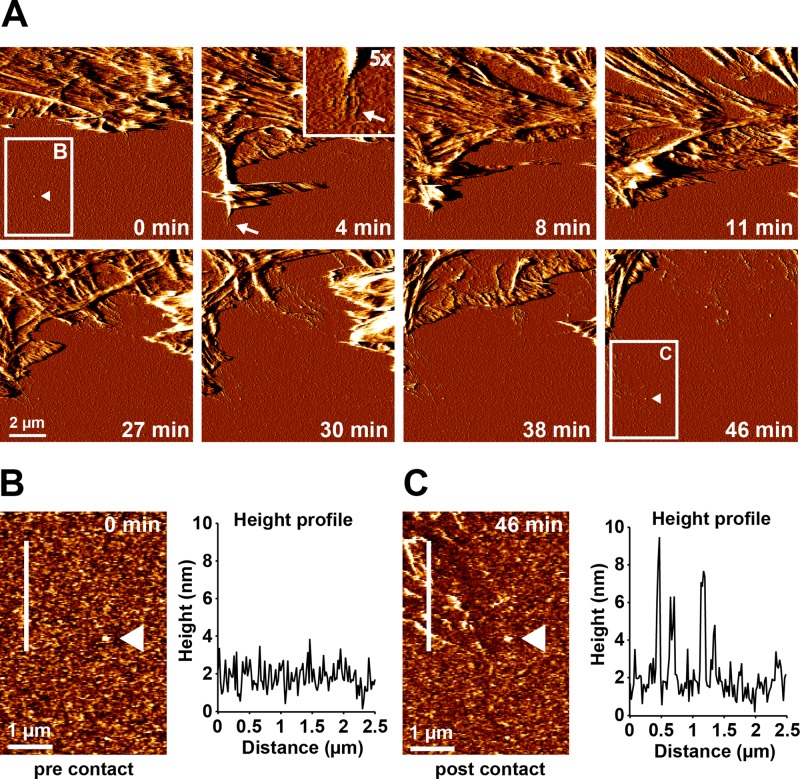
Initial fibrillar FN nanofibrils became visible at sites of active membrane retraction and were usually oriented in the direction of retraction, suggesting that high traction forces typically building up during membrane retraction provide a mechanical mechanism for FN monomer extension and fibrillogenesis. However, we also considered the alternative possibility that FN preforms at the basal cell side while the extended membrane covers the substrate. These preformed FN fibrils would then progressively become uncovered as the cell membrane sheet retracts during AFM scanning. To further clarify this point, we imaged the same FN area before (Figure 6A, 0 min) and after (Figure 6A, 46 min) cells had extended and retracted a membrane sheet at a frame rate of ∼4 min (Supplemental Movie S6). Determination of the height profile of the unmodified FN layer before cell contact (Figure 6B) and of the reorganized FN matrix after cell contact at the same position (Figure 6C) confirmed that its maximal height had increased from 0.5–3 to 4–10 nm after cell contact, consistent with fibrillar remodeling. The AFM time-lapse series furthermore showed that FN fibrils started to appear as soon as membrane retraction commenced within 4 min of cell–substrate contact (Figure 6A, 4 min; see inset at 5× magnification). During the single 4-min interval, the membrane had to extend first before it could retract, leaving substantially less time for the initiation of FN fibrillogenesis. However, given the limited frame rate of AFM scanning, this fast process could not be time-resolved. From these experiments we concluded that the initiation of fibrillogenesis is a fast process occurring on the seconds-to-minute scale and that initial FN fibrils at the cell periphery form as a result of high traction forces created at retracting membranes.
FIGURE 6:
Fast FN rearrangement at retracting cell membranes. Cells were adhered to a homogeneous FN substrate in the presence of 1 mM Mn2+ for 10 min. Subsequently, a 10 × 10 μm2 area at the cell edge was continuously imaged by AFM in contact mode. (A) Time series of AFM deflection images showing part of a cell lamellipodium next to an uncontacted area on the FN surface. After 4 min, a transient cellular extension first forms and then retracts, inducing the formation of FN nanofibrils in the process (arrow). Inset, magnified view (5×) of the tip of the cellular extension and the associated FN nanofibrils. After several rounds of extension and retraction (8–30 min), the cell gradually retracts out of the imaging area, leaving behind a remodeled FN layer. Higher-resolution AFM height images of the region indicated by the white rectangle in A before cellular contact at time point zero (B) and after complete cell retraction 46 min later (C). A height profile along the white line demonstrates only small (<3 nm) variations in FN height before cellular contact (B), but large (≤10 nm) variations in FN height after retraction, consistent with the formation of FN nanofibrils. White triangles points to a small irregularity in the FN layer used as a positional marker in both images. The entire time-lapse series is presented in Supplemental Movie S3.
Despite the fairly short interaction time between the extended membrane sheet and the FN layer, fibroblasts not only may remodel the precoated plasma FN layer but also deposit endogenous FN or other macromolecules on top. However, the total volume of the FN layer did not change after fibrillar rearrangement (Supplemental Figure S5), demonstrating that fibrils formed through remodeling of the predeposited plasma FN but not by depositing significant quantities of additional molecules.
FN fibrils form during cycles of membrane extension and retraction
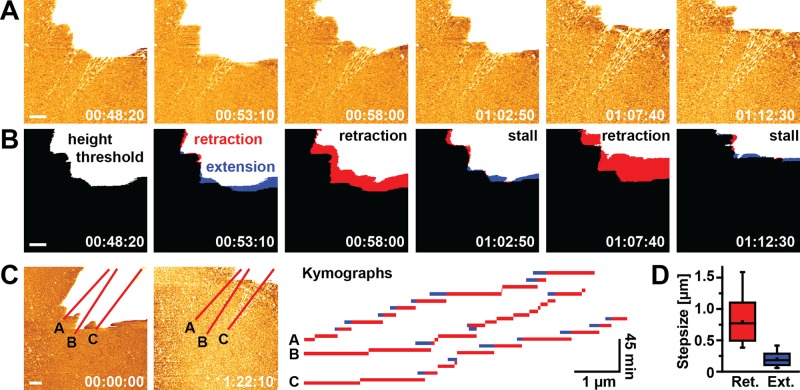
In some cases, membrane retraction was slower (<0.1 μm/min), and we observed a typical pattern involving repeated cycles of membrane retraction, followed by a stall phase or a short membrane extension phase (Figure 7, A and B, and Supplemental Movies S7 and S8). On average, retraction events covered larger distances (817 ± 384 nm, mean ± SD) and occurred more frequently than extension events (mean extension distance 203 ± 124 nm), leading to persistent net displacement of the cell edge. Kymographs taken along emerging FN fibrils (Figure 7C) indicated that retraction/extension cycles occurred at a frequency of 5–10 min. Given the link between FN fibril formation and membrane retraction, cyclical membrane movement may be important for fibrillogenesis and suggests a stepwise extension mechanism of FN fibrils.
FIGURE 7:
Stepwise formation of FN nanofibrils. (A) Several consecutive image frames from an AFM time-lapse series visualizing the formation of FN nanofibrils during membrane retraction of a single cell (top right corner). AFM images were obtained every 290 s. The height scale was adjusted to best visualize newly formed FN nanofibrils, which uniformly allocated the cell body the maximum height scale value (white). (B) Applying a height threshold (>20 nm) to the AFM topographs generated binary images, from which the membrane spread area could be determined. Differential superimposition of two consecutive thresholded image frames identifies areas of membrane retraction (red) or extension (blue). (C) Kymographs generated along three lines across the entire time-lapse movie (total duration 82 min) to visualize the step-like extension patterns of FN nanofibrils. Bars indicate membrane retraction events, and blue segments indicate transient membrane extension relative to the preceding frame. Gaps in the time traces indicate absence of net membrane movement. (D) Quantification of retraction (Ret.) and extension (Ext.) step sizes from the kymographs shown in C. Scale bars 1 μm.
DISCUSSION
In this study, we investigated the initial steps of cell-induced FN fibrillogenesis using AFM and fluorescence microscopy. High-resolution AFM images revealed thin fibrillar FN networks that could not be resolved with conventional fluorescence microscopy. For instance, cell-distal ends of FN nanofibrils often appeared frayed, similar to the loose fibrillar bundling seen on high-resolution SEM images of in vitro–assembled FN fibrils (Ulmer et al., 2008). Initial growth occurred only on the cell-proximal end, but later fibrils displayed increased diameters, suggesting lateral addition of building blocks along the entire fibril length, as suggested previously by fluorescence imaging (Pankov et al., 2000; Ohashi et al., 2002). Several previous AFM studies investigated cellular FN reorganization using paraformaldehyde or glutaraldehyde treatment to stabilize FN structures for scanning (De Jong et al., 2006). However, it was unclear whether chemical cross-linking affects the fibril structure, for instance, by bundling thin neighboring fibrils into larger fibrillar structures. Our AFM images of chemically fixed FN fibrils revealed a comparable nanoarchitecture to FN fibrils imaged in unfixed, living cells, indicating that chemical fixation is not harmful to FN fibril structure. Of importance, however, chemical fixation is not required for high-resolution imaging of FN fibrillogenesis with AFM, and under optimized AFM scanning conditions, cell-driven fibrillogenesis processes could be continuously visualized over several hours without affecting cell or FN morphology.
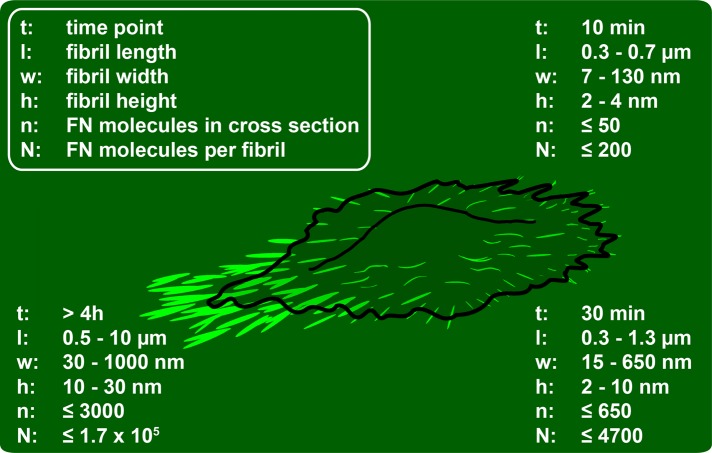
A detailed analysis of AFM and fluorescence images provided additional quantitative information on fibrillar dimensions and enabled us to establish a timeline of structural changes during FN fibrillogenesis, which was divided into three general phases (Figure 8). FN nanofibrils form at the cell periphery after ∼5–10 min of spreading. These initial fibrils are <700 nm long and have a height of ∼2–5 nm (Figure 3C), in agreement with previous EM observations of fibrillar diameters as small as ∼5–10 nm (Chen et al., 1978). During subsequent cell–substrate interaction (<1 h), cells reorganize these initial fibrillar precursors into larger fibrils predominantly at the cell periphery but also more centrally beneath the cell body. At this time, fibrils are oriented mainly perpendicular to the cell edge and reach a length of up to 2 μm, a maximal width of 800 nm, and a height of up to 30 nm. Subsequently, cells may recontact early fibrils several times through cycles of membrane extension and retraction and then either vacate these structures or continue to remodel FN into superfibrils, which can reach ≤6 μm in length.
FIGURE 8:
Time line of early cell-induced FN fibrillogenesis. Fibrillar volumes were approximated from fibrillar height, width, and length values extracted from AFM height images. Width values were corrected for tip convolution (see Supplemental Figure S6). Number of FN molecules per fibril cross section and total molecules per fibril were estimated assuming maximal hexagonal packing of cylindrical FN dimers with a length of 160 nm, a diameter of 3 nm, and a 90-nm stagger between dimer building blocks (see Supplemental Figure S6).
Fibrillogenesis appeared to be tightly associated with plasma membrane retraction. Previous fluorescence microscopy studies demonstrated initiation of fibril formation at focal adhesion sites some 5–10 μm behind the leading edge (Ohashi et al., 2002). The mobile fibrillar end then translocates toward the central region at the basal cell side together with associated α5β1 integrin complexes (Pankov et al., 2000). We also observed some fibril initiation in central parts of the basal cell side (Figure 1A), but these areas cannot be imaged by AFM. It is not clear whether centripetally growing fibrils at the basal cell side and fibrils emerging at retracting membranes share a common formation and extension mechanism, although extension speeds are comparable in both cases (see later discussion). Fibrils emerged at the basal side of retracting membranes without apparent delay, suggesting association with integrin contacts at the extreme cell perimeter. In REF52 cells, membrane retraction typically coincides with a simultaneous translocation of peripheral focal adhesions in the direction of retraction (our previous observation). These translocating focal adhesions typically locate near the very cell edge, as they are the last cellular structures providing significant resistance to membrane retraction. AFM imaging can therefore visualize FN fibrils as they are forming immediately next to the cell edge during membrane retraction and focal adhesion translocation. However, it is possible that that the cell-proximal fibril ends extend some distance underneath the cell if they terminate at more centrally located focal adhesion sites. By fluorescence microscopy, we also observed FN fibrillogenesis at FA located somewhat farther away from the cell edge (Figure 1, A and B). In this case, AFM imaging can reveal the entire fibrillar structure only after cells have completely vacated the respective substrate area.
Frequently, fibrils were seen to extend during repeating cycles of short extension (∼200 nm) and larger membrane retraction (∼800 nm) occurring every 5–10 min. The striking recontacting pattern of nascent fibrils through repeated retraction/extension cycles could have an important role in fibrillar reinforcement. Repeated ligand engagement has been shown to increase integrin α5β1/FN bond lifetime (Kong et al., 2013), and cyclical integrin binding could provide a molecular basis for fibrillar reinforcement. We determined a retraction step size of ∼817 ± 384 nm, but it is unclear whether this correlates with a fibrillar extension mechanism at discrete increments. The slow, steady, stepwise retraction pattern appeared to be linked directly to the process of fibril formation and was different from the recontacting of more mature fibrils, which occurred at high membrane velocities and could be associated with fibrillar reinforcement instead. Cyclic membrane retractions are typical for fibroblast cells. For instance, embryonic mouse fibroblasts display cyclical phases of membrane extension and retraction with a periodicity of ∼25 s and velocities between 20 and 100 nm/s (Giannone et al., 2004). These fibroblasts feature a shorter cycle time and a higher retraction speed than the stepwise fibrillar extension pattern we observed in REF cells. However, because we achieved a maximum scan speed of ∼2–5 min/image, several cycles of membrane extension and retraction may occur during the acquisition of each frame, in which case, time-lapse AFM would underestimate the true rate of membrane cycling.
From AFM time-lapse images, we measured mean fibrillar extension rates of 0.25 (without Mn2+) and 0.68 μm/min (with Mn2+), translating into extension speeds of 4–11 nm/s. Of interest, these speeds are on a similar scale as the speed of integrin translocation (6.5 μm/h, ∼2 nm/s) associated with fibril extension (Pankov et al., 2000). Ohashi et al. (2002) observed similar extension rates of 2–3 nm/s in cells by fluorescence microscopy Pulling rates might vary in different cell systems, which could account for the slightly higher speeds in our system. In addition, we quantitated initial extension rates, whereas fluorescence-based studies primarily investigate remodeling of more mature fibrils once they become optically detectable after several hours or days in tissue culture. Fibrillar extension speeds might gradually decrease as fibrils are increasingly stretched. Important insight into the dynamics of FN fibrillogenesis in other cell systems has also been gained from antibody labeling experiments of the blastocoel roof of Xenopus embryos (Winklbauer and Stoltz, 1995). Here the fastest-growing FN fibrils extended at a speed of 4.7 μm/min (∼80 nm/s), considerably faster than in the fibroblast systems. However, these speeds were determined from the longest FN fibrils, whereas the presence of many smaller FN fibrils indicated that most fibrils either grew more slowly, or, more likely, at the same speed but only for short time periods, similar to what we assume for fibroblast-mediated fibrillogenesis (see later discussion). Nevertheless, during development, FN rearrangement might occur at considerably higher speeds as part of highly dynamic tissue rearrangement processes compared with comparatively slowly moving fibroblasts.
Activating integrins with Mn2+ had a profound effect on early FN fibril formation, leading to accelerated initiation of fibril formation, increased growth rates, and larger final fibrillar dimensions. In contrast to Mn2+-free conditions, in presence of Mn2+, fibrillar cross sections of early fibrils reached their maximum strength after ∼10 min. Integrin activation by Mn2+ thus appears to cause a strong increase in the total number of FN molecules incorporated into fibrils and an acceleration of fibrillar reinforcement. In a previous study using fluorescence microscopy analysis, Sechler et al. (1997) did not detect significant differences in FN matrix assembly in Mn2+-treated or untreated cells after 30 min of incubation, but a significant effect of Mn2+ on FN fibril morphology started after 4 h of incubation. Owing to the limited resolution of conventional fluorescence microscopy, subtle nanoscale structural differences may not be detectable at the earliest stages of fibrillogenesis when fibrils are still small. Here the resolution advantage of AFM can reveal new structural insight into the effect of integrin activation state on early FN fibrillogenesis.
Fibrils forming in presence of Mn2+ were longer, and individual nascent fibrils could be tracked together with the retracting membranes over several AFM frames. In contrast, fibrils forming in absence of Mn2+ were short (∼500 nm) and usually did not show appreciable further growth across scan frames. This suggests that FN fibril extension is completed at a time frame below the acquisition frequency of AFM and that the true extension speed may again have been underestimated due to the limited temporal resolution of AFM. In the presence of Mn2+, fibrils typically measured ∼1–4 μm in length. At the determined pulling rate of ∼0.68 μm/min, fibril growth may progress uninterruptedly for ∼90–350 s. The increased mean fibril length could results from increased integrin bond lifetimes to RGD sites in FN in the presence of Mn2+, as demonstrated by single-molecule studies (Kong et al., 2009).
Early FN nanofibrils occasionally displayed a series of globular structures evenly spaced apart by 60–90 nm. Similar series of nodules interconnected by short, smooth linkers have been observed in cell-derived FN fibrils by cryo-EM (Chen et al., 1997; Peters et al., 1998). These nodules have been proposed to represent discrete units of three or four adjacent FNIII domains (Peters et al., 1998). Of interest, the maximal interbead distance of ∼90 nm corresponds well to the recently determined ∼95-nm periodicity in native fibrils (Fruh et al., 2015).
FN fibrils with beaded periodicity have also been observed using high-resolution AFM scanning of FN adsorbed onto negatively charged mica in a cell-free system (Nelea and Kaartinen, 2010). In this study, extended FN dimers were suggested to interact in an antiparallel manner, so that FNI1-5 domains from one dimer would interact with FIII12-14 domains from an adjacent dimer, leading to the antiparallel juxtaposition of bulky FNIII1-7 domain regions every 60 nm. Compared to the cell-free system used by Nelea and Kaartinen (2010), in cell-induced FN fibrils, the interbead distance varied more widely and the array of globular domains was less regular. A predominantly random distribution of nodules along the fibril length was observed in cell-derived FN fibrils (Peters et al., 1998), suggesting the coexistence of different conformations and different degrees of domain unfolding within fibrils. Similarly, FRET experiments indicate a heterogeneous distribution of stretch-dependent conformations in native FN matrices (Baneyx et al., 2001, 2002; Smith et al., 2007). Of interest, nodules are absent in mature FN fibrils suspended between cells (Chen et al., 1997; Peters et al., 1998), suggesting that increased tension applied to the fibril induces widespread domain unfolding. The abundance of globular domains in our AFM images may therefore indicate that early substrate-attached FN fibrils retain folded units, although their overall straight appearance showed that they had formed under tension. More extensive nodule unfolding may require further adhesion reinforcement, for example, occurring during mature matrix remodeling.
Knowing the number of FN dimer building blocks constituting individual fibrils would be useful for better understanding FN fibril function under physiological conditions. Such information could, for example, generate new insight into how cellular traction forces are shared by individual FN molecules in fibrils. Determining the number of FN molecules populating a fibril requires knowing the fibril volume, which can be estimated from AFM topographs corrected for tip convolution effects and knowledge regarding FN dimer size, molecular arrangement, and packing density. The fully extended FN molecule has a length of ∼120–160 nm (Erickson et al., 1981) and a diameter of ∼2–3 nm (Engel et al., 1981; Erickson et al., 1981; Leahy et al., 1996), but the precise length of FN dimers and their lateral and longitudinal arrangement of FN dimers in native fibrils has long been elusive. A recent study combining site-specific protein labeling with single-molecule localization revealed end-to-end distances of FN molecules between 90 and 160 nm (mean ∼133 nm) in native fibrils (Fruh et al., 2015). The density of FN molecules in native fibrils has not been determined, but transmission electron microscopy images suggest dense packing of FN molecules in cell-stretched fibrils (Singer, 1979; Dzamba and Peters, 1991). We observed suborganization of larger cell-induced fibrils into thinner nanofibrils typically spaced apart by 150–300 nm (Figure 3B). Native fibrils may therefore consist of several loosely spaced, yet densely packed nanofibrils. Assuming maximum-density (hexagonal) packing of fully extended FN dimers in individual nanofibrils, we estimated the maximum number of FN dimers that could populate a transverse fibrillar cross section of early (10 min) and later fibrils (30 min; Supplemental Figure S6 and Supplemental Table S1). Based on a maximum-density packing model, the cross section of the earliest detectable fibrils forming within the first 5–10 min of matrix contact could accommodate ∼11 hexagonally stacked FN molecules on average, whereas the smallest detectable fibrillar structures would contain just one or two FN molecules. In agreement, EM studies showed that FN nanofibrils might contain as few as two or three molecules (Peters et al., 1990, 1998). According to the same model, cross sections of later fibrils (30 min) contain ∼40 FN molecules on average, whereas early fibrils formed in the presence of Mn2+ contain ∼150 FN molecules on average per cross section and the thickest fibrils up to ∼1000 FN molecules. Although these numbers may provide an approximate scale of molecular building blocks that can be accommodated in cell-stretched FN fibrils at maximum packing density, the true number of FN building blocks may be considerably lower due to looser molecular packing or incomplete dimer extension. On the other hand, native FN matrix may contain a significant fraction of FN molecules in which some FIII domains are unfolded (Smith et al., 2007), which may reduce the effective cross-sectional area of the FN molecule and permit even higher stacking densities.
In conclusion, in this work, we visualized the early stages of FN fibrillogenesis using time-lapse AFM imaging of living fibroblasts for the first time. The enhanced resolution of AFM provides information about the conformation of newly created FN nanofibrils before subsequent fibril maturation during tissue remodeling. Future AFM studies may benefit from faster and gentler AFM imaging techniques (Pfreundschuh et al., 2014; Uchihashi et al., 2014), enhancing the temporal resolution when monitoring FN fibrillogenesis. Accurate information about total fibrillar volumes obtained from AFM topographs may also complement future studies investigating the exact conformation and arrangement of FN dimers in fibrils using other microscopy techniques or theoretical models (Gao et al., 2002; Bradshaw et al., 2012) and in this way further enhance our understanding of FN fibril structure and function.
MATERIALS AND METHODS
Cell culture
All cell culture reagents were obtained from Life Technologies, Darmstadt, Germany, unless stated otherwise. Rat embryonic fibroblasts (REF52) were cultured in DMEM containing 1% penicillin/streptomycin and 10% fetal bovine serum (FBS). Cells were grown in T25 tissue culture flasks in a humidified atmosphere containing 5% CO2 and passaged either every 3–4 d or before reaching confluency. For passaging, cells were rinsed with phosphate-buffered saline (PBS) and incubated with 1 ml trypsin/EDTA at 37°C for 5 min until detachment. Trypsin was inactivated by adding 9 ml of growth medium, and cells were diluted according to their growth rate.
Fibronectin substrates
Lyophilized human plasma FN (Roche, Grenzach-Wyhlen, Germany) was resuspended in sterile water and stored at −20°C. For light microscopy experiments, FN was fluorescently labeled using the Alexa Fluor 488 Protein Labeling Kit at a labeling ratio of 2–5 Alexa molecules/FN molecule as determined by spectrophotometry. Labeled FN (FN-AF488) was placed on a glass-bottom cell culture dish (FD35-100; WPI, Berlin, Germany) at a concentration of 50 μg/ml at room temperature in the dark for 1 h, followed by washing with PBS. For AFM time-lapse imaging, a freshly cleaved mica disk (Indiamart, Noida, India) was FN coated at a concentration of 50 μg/ml at room temperature for 1 h, and the substrate was subsequently washed once with DMEM containing 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES).
Fluorescence microscopy
Cells were seeded on FN-AF488 substrates, incubated in DMEM containing 1% FBS at 37°C in 5% CO2 for 10 min to 24 h, and fixed with 4% paraformaldehyde (PFA) (Carl Roth, Karlsruhe, Germany) for 30 min. TIRF images were acquired using an iMIC microscope (FEI Life Sciences, Munich, Germany) and an APON 60× OTIRF oil immersion lens (Olympus, Hamburg, Germany). Epifluorescence images of FN in combined light microscopy/AFM experiments were obtained using an Achroplan 40×/0.80 W water-immersion lens (Zeiss, Oberkochen, Germany). For visualizing focal adhesions, cells were stained using a mouse monoclonal anti-vinculin antibody (V9131; Sigma-Aldrich, Taufkirchen, Germany).
AFM imaging
AFM imaging was performed using a JPK Nanowizard II AFM (JPK Instruments, Berlin, Germany) mounted on top of an Axio Observer inverted optical microscope (www.zeiss.com). Glutaraldehyde-fixed (1%/PBS) cells were scanned in PBS in contact mode using V-shaped, gold-coated silicon nitride cantilevers (MLCT-C) with a nominal spring constant of 0.06 N/m and tip radius of 20 nm (Bruker, Camarillo, CA). For time-lapse imaging, cells growing in T25 flasks were trypsinized briefly. Immediately after cell detachment, the trypsin solution was inactivated by adding an equal volume of trypsin inhibitor solution (Sigma-Aldrich). Cells were centrifuged at 170 × g for 4 min and resuspended in DMEM containing 25 mM HEPES. Five minutes before beginning of AFM scanning, cells were seeded on FN substrates and imaged in DMEM containing 25 mM HEPES with or without 1 mM MnCl2 in contact mode at a scanning frequency ranging from 2 to 4 Hz. In control experiments, cells were imaged on laminin-111 substrates (50 μg/ml). Cells were further imaged on FN cross-linked using 1% glutaraldehyde/PBS for 10 min. Before cell seeding, unreacted aldehyde groups were quenched by two washes with freshly prepared 1 mg/ml sodium borohydride solution. During imaging, a constant temperature of 37°C was maintained using a temperature-controlled sample holder (PetriDish Heater; JPK Instruments). AFM images were processed using the JPK image processing software (version 4.0.23).
SEM imaging
REF52 cells were cultured on FN-coated mica for 10, 60, or 240 min or overnight and fixed with 1% glutaraldehyde/4% PFA in 0.1 M cacodylate buffer (pH 7.2) at room temperature for 1 h. Afterward, samples were progressively dehydrated in a 30, 50, 70, 90, and 100% ethanol series for 5 min each and critical point-dried (EM cpd300; Leica, Wetzlar, Germany). The samples were sputter-coated with 5-nm platinum and imaged with a LeoSupra55VP scanning electron microscope (Zeiss), using an aperture size of 20 μm and an accelerating voltage of 7.0 kV.
Estimating the number of FN molecules per fibril
Fibrillar dimensions were approximated from height and from length and width (full-width at half-maximum height) values extracted from AFM height images. Lateral fibrillar dimension were corrected for tip convolution effects using the nominal tip radius supplied by the manufacturer (20 nm; also see Supplemental Figure S6). We consistently obtained larger (tip-corrected) width than height values for surface-attached FN nanofibrils and therefore assumed an elliptical fibrillar cross section. We furthermore modeled extended FN dimers as perfect cylinders with a length of 160 nm and a diameter of 3 nm, yielding a cross-sectional area of ∼7 nm2, and assumed maximal-density, hexagonal packing of FN dimers inside fibrils (packing density, 0.9069). For lateral arrangement of FN dimer, we assumed an antiparallel arrangement of fully extended FN dimers with overlap zones of 70 nm and gap zones of 20 nm. Addition of an FN dimer thus leads to a net extension of the fibril by ∼90 nm. Accordingly, the extended molecular building block occupies an effective fibril volume of 1260 nm3, given by multiplying its cross-sectional area (∼7 nm2) by the sum of the extended dimer (∼160 nm) and gap zone (∼20 nm) length.
Acknowledgments
We thank Ramona Ring for tissue culture support, Carina Gonnermann for critically reading the manuscript, and Anna Taubenberger and Joachim Fischer for fruitful discussion and preliminary experiments. We acknowledge financial support from the Deutsche Forschungsgemeinschaft and the State of Baden-Württemberg through the DFG–Center for Functional Nanostructures within subprojects E2.4 and through DFG grant FOR 1756.
Abbreviations used:
- AFM
atomic force microscopy
- AF488
Alexa Fluor 488
- FN
fibronectin
- TIRF
total internal reflection microscopy
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.3190) on July 29, 2015.
REFERENCES
- Avnur Z, Geiger B. The removal of extracellular fibronectin from areas of cell-substrate contact. Cell. 1981;25:121–132. doi: 10.1016/0092-8674(81)90236-1. [DOI] [PubMed] [Google Scholar]
- Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2001;98:14464–14468. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci USA. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw MJ, Cheung MC, Ehrlich DJ, Smith ML. Using molecular mechanics to predict bulk material properties of fibronectin fibers. PLoS Comput Biol. 2012;8:e1002845. doi: 10.1371/journal.pcbi.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw MJ, Smith ML. Contribution of unfolding and intermolecular architecture to fibronectin fiber extensibility. Biophys J. 2011;101:1740–1748. doi: 10.1016/j.bpj.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner KA, Corbett SA, Schwarzbauer JE. Regulation of fibronectin matrix assembly by activated Ras in transformed cells. Oncogene. 2000;19:3156–3163. doi: 10.1038/sj.onc.1203626. [DOI] [PubMed] [Google Scholar]
- Chen H, Mosher DF. Formation of sodium dodecyl sulfate-stable fibronectin multimers. Failure to detect products of thiol-disulfide exchange in cyanogen bromide or limited acid digests of stabilized matrix fibronectin. J Biol Chem. 1996;271:9084–9089. doi: 10.1074/jbc.271.15.9084. [DOI] [PubMed] [Google Scholar]
- Chen LB, Murray A, Segal RA, Bushnell A, Walsh ML. Studies on intercellular LETS glycoprotein matrices. Cell. 1978;14:377–391. doi: 10.1016/0092-8674(78)90123-x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wu Y, Cai J. Atomic force microscopic investigation on the potential early intermediate stages of fibrillogenesis of fibronectin within fibrils. Biochem Biophys Res Commun. 2007;361:391–397. doi: 10.1016/j.bbrc.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zardi L, Peters DM. High-resolution cryo-scanning electron microscopy study of the macromolecular structure of fibronectin fibrils. Scanning. 1997;19:349–355. doi: 10.1002/sca.4950190505. [DOI] [PubMed] [Google Scholar]
- Christopher RA, Kowalczyk AP, McKeown-Longo PJ. Localization of fibronectin matrix assembly sites on fibroblasts and endothelial cells. J Cell Sci. 1997;110:569–581. doi: 10.1242/jcs.110.5.569. [DOI] [PubMed] [Google Scholar]
- Craig D, Krammer A, Schulten K, Vogel V. Comparison of the early stages of forced unfolding for fibronectin type III modules. Proc Natl Acad Sci USA. 2001;98:5590–5595. doi: 10.1073/pnas.101582198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danen EH, Aota S, van Kraats AA, Yamada KM, Ruiter DJ, van Muijen GN. Requirement for the synergy site for cell adhesion to fibronectin depends on the activation state of integrin alpha 5 beta 1. J Biol Chem. 1995;270:21612–21618. doi: 10.1074/jbc.270.37.21612. [DOI] [PubMed] [Google Scholar]
- Davidson LA, Dzamba BD, Keller R, Desimone DW. Live imaging of cell protrusive activity, and extracellular matrix assembly and remodeling during morphogenesis in the frog, Xenopus laevis. Dev Dyn. 2008;237:2684–2692. doi: 10.1002/dvdy.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson LA, Keller R, DeSimone DW. Assembly and remodeling of the fibrillar fibronectin extracellular matrix during gastrulation and neurulation in Xenopus laevis. Dev Dyn. 2004;231:888–895. doi: 10.1002/dvdy.20217. [DOI] [PubMed] [Google Scholar]
- De Jong KL, MacLeod HC, Norton PR, Petersen NO. Fibronectin organization under and near cells. Eur Biophys J. 2006;35:695–708. doi: 10.1007/s00249-006-0081-7. [DOI] [PubMed] [Google Scholar]
- Dzamba BJ, Bultmann H, Akiyama SK, Peters DM. Substrate-specific binding of the amino terminus of fibronectin to an integrin complex in focal adhesions. J Biol Chem. 1994;269:19646–19652. [PubMed] [Google Scholar]
- Dzamba BJ, Peters DM. Arrangement of cellular fibronectin in noncollagenous fibrils in human fibroblast cultures. J Cell Sci. 1991;100:605–612. doi: 10.1242/jcs.100.3.605. [DOI] [PubMed] [Google Scholar]
- Engel J, Odermatt E, Engel A, Madri JA, Furthmayr H, Rohde H, Timpl R. Shapes, domain organizations and flexibility of laminin and fibronectin, two multifunctional proteins of the extracellular matrix. J Mol Biol. 1981;150:97–120. doi: 10.1016/0022-2836(81)90326-0. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Carrell N, McDonagh J. Fibronectin molecule visualized in electron microscopy: a long, thin, flexible strand. J Cell Biol. 1981;91:673–678. doi: 10.1083/jcb.91.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz CM, Muller DJ. Analyzing focal adhesion structure by atomic force microscopy. J Cell Sci. 2005;118:5315–5323. doi: 10.1242/jcs.02653. [DOI] [PubMed] [Google Scholar]
- Friedrichs J, Taubenberger A, Franz CM, Muller DJ. Cellular remodelling of individual collagen fibrils visualized by time-lapse AFM. J Mol Biol. 2007;372:594–607. doi: 10.1016/j.jmb.2007.06.078. [DOI] [PubMed] [Google Scholar]
- Fruh SM, Schoen I, Ries J, Vogel V. Molecular architecture of native fibronectin fibrils. Nat Commun. 2015;6:7275. doi: 10.1038/ncomms8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gailit J, Ruoslahti E. Regulation of the fibronectin receptor affinity by divalent cations. J Biol Chem. 1988;263:12927–12932. [PubMed] [Google Scholar]
- Gao M, Craig D, Lequin O, Campbell ID, Vogel V, Schulten K. Structure and functional significance of mechanically unfolded fibronectin type III1 intermediates. Proc Natl Acad Sci USA. 2003;100:14784–14789. doi: 10.1073/pnas.2334390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Craig D, Vogel V, Schulten K. Identifying unfolding intermediates of FN-III(10) by steered molecular dynamics. J Mol Biol. 2002;323:939–950. doi: 10.1016/s0022-2836(02)01001-x. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Dobereiner HG, Kieffer N, Bresnick AR, Sheetz MP. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 2004;116:431–443. doi: 10.1016/s0092-8674(04)00058-3. [DOI] [PubMed] [Google Scholar]
- Grinnell F. Fibronectin and wound healing. J Cell Biochem. 1984a;26:107–116. doi: 10.1002/jcb.240260206. [DOI] [PubMed] [Google Scholar]
- Grinnell F. Manganese-dependent cell-substratum adhesion. J Cell Sci. 1984b;65:61–72. doi: 10.1242/jcs.65.1.61. [DOI] [PubMed] [Google Scholar]
- Gudzenko T, Franz CM. Inverting adherent cells for visualizing ECM interactions at the basal cell side. Ultramicroscopy. 2013;128C:1–9. doi: 10.1016/j.ultramic.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Hynes R. Molecular biology of fibronectin. Annu Rev Cell Biol. 1985;1:67–90. doi: 10.1146/annurev.cb.01.110185.000435. [DOI] [PubMed] [Google Scholar]
- Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Demonstration of catch bonds between an integrin and its ligand. J Cell Biol. 2009;185:1275–1284. doi: 10.1083/jcb.200810002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F, Li Z, Parks WM, Dumbauld DW, Garcia AJ, Mould AP, Humphries MJ, Zhu C. Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell. 2013;49:1060–1068. doi: 10.1016/j.molcel.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. J Cell Sci. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- Leahy DJ, Aukhil I, Erickson HP. 2.0 A crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84:155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- Lemmon CA, Ohashi T, Erickson HP. Probing the folded state of fibronectin type III domains in stretched fibrils by measuring buried cysteine accessibility. J Biol Chem. 2011;286:26375–26382. doi: 10.1074/jbc.M111.240028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GL, Cohen DM, Desai RA, Breckenridge MT, Gao L, Humphries MJ, Chen CS. Activation of beta 1 but not beta 3 integrin increases cell traction forces. FEBS Lett. 2013;587:763–769. doi: 10.1016/j.febslet.2013.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Lal R, Clegg DO. Imaging and mapping heparin-binding sites on single fibronectin molecules with atomic force microscopy. Biochemistry. 2000;39:3192–3196. doi: 10.1021/bi991624o. [DOI] [PubMed] [Google Scholar]
- Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–399. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Marsden M, DeSimone DW. Regulation of cell polarity, radial intercalation and epiboly in Xenopus: novel roles for integrin and fibronectin. Development. 2001;128:3635–3647. doi: 10.1242/dev.128.18.3635. [DOI] [PubMed] [Google Scholar]
- McKeown-Longo PJ, Mosher DF. Binding of plasma fibronectin to cell layers of human skin fibroblasts. J Cell Biol. 1983;97:466–472. doi: 10.1083/jcb.97.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould AP, Akiyama SK, Humphries MJ. Regulation of integrin alpha 5 beta 1-fibronectin interactions by divalent cations. Evidence for distinct classes of binding sites for Mn2+, Mg2+, and Ca2+ J Biol Chem. 1995;270:26270–26277. doi: 10.1074/jbc.270.44.26270. [DOI] [PubMed] [Google Scholar]
- Nagel M, Tahinci E, Symes K, Winklbauer R. Guidance of mesoderm cell migration in the Xenopus gastrula requires PDGF signaling. Development. 2004;131:2727–2736. doi: 10.1242/dev.01141. [DOI] [PubMed] [Google Scholar]
- Nelea V, Kaartinen MT. Periodic beaded-filament assembly of fibronectin on negatively charged surface. J Struct Biol. 2010;170:50–59. doi: 10.1016/j.jsb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc Natl Acad Sci USA. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi T, Kiehart DP, Erickson HP. Dual labeling of the fibronectin matrix and actin cytoskeleton with green fluorescent protein variants. J Cell Sci. 2002;115:1221–1229. doi: 10.1242/jcs.115.6.1221. [DOI] [PubMed] [Google Scholar]
- Pankov R, Cukierman E, Katz BZ, Matsumoto K, Lin DC, Lin S, Hahn C, Yamada KM. Integrin dynamics and matrix assembly: tensin-dependent translocation of alpha(5)beta(1) integrins promotes early fibronectin fibrillogenesis. J Cell Biol. 2000;148:1075–1090. doi: 10.1083/jcb.148.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DMP, Chen Y, Zardi L, Brummel S. Conformation of fibronectin fibrils varies: discrete globular domains of type III repeats detected. Microsc Microanal. 1998;4:385–396. doi: 10.1017/s1431927698980369. [DOI] [PubMed] [Google Scholar]
- Peters DM, Portz LM, Fullenwider J, Mosher DF. Co-assembly of plasma and cellular fibronectins into fibrils in human fibroblast cultures. J Cell Biol. 1990;111:249–256. doi: 10.1083/jcb.111.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfreundschuh M, Martinez-Martin D, Mulvihill E, Wegmann S, Muller DJ. Multiparametric high-resolution imaging of native proteins by force-distance curve-based AFM. Nat Protoc. 2014;9:1113–1130. doi: 10.1038/nprot.2014.070. [DOI] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005041. a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechler JL, Corbett SA, Schwarzbauer JE. Modulatory roles for integrin activation and the synergy site of fibronectin during matrix assembly. Mol Biol Cell. 1997;8:2563–2573. doi: 10.1091/mbc.8.12.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer II. The fibronexus: a transmembrane association of fibronectin-containing fibers and bundles of 5 nm microfilaments in hamster and human fibroblasts. Cell. 1979;16:675–685. doi: 10.1016/0092-8674(79)90040-0. [DOI] [PubMed] [Google Scholar]
- Singer II, Kawka DW, Kazazis DM, Clark RA. In vivo co-distribution of fibronectin and actin fibers in granulation tissue: immunofluorescence and electron microscope studies of the fibronexus at the myofibroblast surface. J Cell Biol. 1984;98:2091–2106. doi: 10.1083/jcb.98.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Carraher C, Schwarzbauer JE. Assembly of fibronectin extracellular matrix. Annu Rev Cell Dev Biol. 2010;26:397–419. doi: 10.1146/annurev-cellbio-100109-104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihashi T, Kodera N, Ando T. Development of high-speed AFM and its biological applications. In: Takeyasu K, editor. Atomic Force Microscopy in Nanobiology. Boca Raton, FL: CRC Press; 2014. pp. 143–176. [Google Scholar]
- Ulmer J, Geiger B, Spatz JP. Force-induced fibronectin fibrillogenesis in vitro. Soft Matter. 2008;4:1998–2007. [Google Scholar]
- Winklbauer R, Keller RE. Fibronectin, mesoderm migration, and gastrulation in Xenopus. Dev Biol. 1996;177:413–426. doi: 10.1006/dbio.1996.0174. [DOI] [PubMed] [Google Scholar]
- Winklbauer R, Stoltz C. Fibronectin fibril growth in the extracellular matrix of the Xenopus embryo. J Cell Sci. 1995;108:1575–1586. doi: 10.1242/jcs.108.4.1575. [DOI] [PubMed] [Google Scholar]