Septins often form filaments and rings at the neck of cellular appendages. Assembly of these structures must be coordinated with membrane remodeling. In budding yeast, the Rho1 GTPase and its effector, Pkc1, play a role in septin ring stabilization during budding at least partly through phosphorylation of the bud neck–associated F-BAR protein Syp1.
Abstract
In many cell types, septins assemble into filaments and rings at the neck of cellular appendages and/or at the cleavage furrow to help compartmentalize the plasma membrane and support cytokinesis. How septin ring assembly is coordinated with membrane remodeling and controlled by mechanical stress at these sites is unclear. Through a genetic screen, we uncovered an unanticipated link between the conserved Rho1 GTPase and its effector protein kinase C (Pkc1) with septin ring stability in yeast. Both Rho1 and Pkc1 stabilize the septin ring, at least partly through phosphorylation of the membrane-associated F-BAR protein Syp1, which colocalizes asymmetrically with the septin ring at the bud neck. Syp1 is displaced from the bud neck upon Pkc1-dependent phosphorylation at two serines, thereby affecting the rigidity of the new-forming septin ring. We propose that Rho1 and Pkc1 coordinate septin ring assembly with membrane and cell wall remodeling partly by controlling Syp1 residence at the bud neck.
INTRODUCTION
Septins are conserved cytoskeletal proteins that bind and hydrolyze GTP. With the exception of higher plants, they are found in most eukaryotes, where multiple septins generally assemble into symmetric linear oligomeric complexes. These then polymerize into nonpolar filaments and supramolecular structures such as rings, gauzes, and arcs (reviewed in Beise and Trimble, 2011; Weirich et al., 2008).
Septins are believed to act as cortical organizers by forming lateral diffusion barriers that compartmentalize the plasma membrane into separate domains and shape cellular architectures (reviewed in Barral, 2010; Caudron and Barral, 2009; Mostowy and Cossart, 2012). Septin depletion or mutations disrupt normal morphogenesis and differentiation of sperms, dendrites, and cilia (Hu et al., 2010; Kissel et al., 2005; Tada et al., 2007; Xie et al., 2007; Hu et al., 2010; Kuo et al., 2012) and are linked to a variety of human diseases, such as male sterility, cancer, and neurodegenerative disorders (reviewed in Hall and Russell, 2012; Peterson and Petty, 2010).
In most cases, septins localize to the base of cellular projections, where they demarcate subcellular boundaries. This is the case in the budding yeast Saccharomyces cerevisiae, in which septins form a ring at the bud neck—the constriction between mother and future daughter cell (Byers and Goetsch, 1976; Haarer and Pringle, 1987; Kim et al., 1991). In the filamentous fungi Aspergillus nidulans and Ashbya gossypii, a similar pattern is seen during hyphal branching (Westfall and Momany, 2002; Helfer and Gladfelter, 2006). Furthermore, septins are found at the base of other cellular appendages, such as cilia, dendritic spines, and the flagellum of spermatozoa (Steels et al., 2007; Tada et al., 2007; Xie et al., 2007; Hu et al., 2010). Thus septin rings are intimately associated with membranes in areas of membrane curvature, which are likely fragile because they are inherently subject to mechanical stress.
Septins associate with membranes through a highly conserved polybasic region at their N-terminus (Zhang et al., 1999; Casamayor and Snyder, 2003). Mammalian septin-4 specifically binds phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2; Zhang et al., 1999). In yeast, PI(4,5)P2 is enriched in membrane areas of polarized growth and the bud neck (Garrenton et al., 2010) and stimulates formation and organization of septin filaments, which are in turn essential for cell viability (Bertin et al., 2010; McMurray et al., 2011). Indeed, in S. cerevisiae, septins are required for cytokinesis by tethering to the division site (i.e., the bud neck) most proteins involved in this process, including components of the contractile actomyosin ring (reviewed in Oh and Bi, 2010; Weirich et al., 2008). Septins are found at the cleavage furrow in fission yeast (Berlin et al., 2003; Tasto et al., 2003), Caenorhabditis elegans (Nguyen et al., 2000), Drosophila (Neufeld and Rubin, 1994; Fares et al., 1995), and mammalian cells (Kinoshita et al., 1997), suggesting that they contribute to cytokinesis in many organisms.
In migrating cell types such as T-lymphocytes, septins form an annular corset at the cell cortex to rigidify the cell periphery, protect it against mechanical stress, and promote the persistence of migration (Tooley et al., 2009). Thus septins function in a number of mechanical processes at the cell cortex and may therefore respond to mechanical cues. However, the mechanisms driving septin ring assembly at the right positions and in coordination with membrane reorganization are not known. Investigating how fungal septin rings respond to alterations in cell wall organization and the resulting changes in turgor pressure is likely to be very instructive about how septins are regulated in response to mechanical stress.
In fungi, septin localization is dynamic during development and the cell cycle (Caviston et al., 2003; Dobbelaere et al., 2003; Gonzalez-Novo et al., 2008; McMurray and Thorner, 2008; Hernandez-Rodriguez et al., 2012). In budding yeast, septins are first recruited to the presumptive bud site as an unorganized cap, which is rapidly transformed into a cortical septin ring in late G1. At the time of bud emergence, the septin ring expands into an hourglass-like structure referred to as septin collar, which spans the whole bud neck. Immediately before cytokinesis, the collar splits into two distinct rings that sandwich the contractile ring (reviewed in Oh and Bi, 2010; Weirich et al., 2008). Fluorescence recovery after photobleaching (FRAP) experiments indicate that septin structures are highly dynamic before bud emergence. On bud emergence, the septin collar at the bud neck is conversely very rigid, whereas at cytokinesis the split septin rings become dynamic again (Caviston et al., 2003; Dobbelaere et al., 2003). In mammalian cells, septin rings remain dynamic throughout cytokinesis, rapidly exchanging septin subunits with the cytoplasm (Schmidt and Nichols, 2004). The dynamics of mammalian septins at other submembrane ring structures is not known.
In budding yeast, the Cdc42 GTPase of the Rho family plays a crucial role in the recruitment of septins to the presumptive bud site (Cid et al., 2001; Iwase et al., 2006), and cycles of Cdc42 GTP binding and hydrolysis are required for septin collar formation (Gladfelter et al., 2002; Caviston et al., 2003). Polarized exocytosis sculpts the septin ring by driving the insertion of membrane into the emerging bud, thereby hollowing the septin cap into a ring (Okada et al., 2013). Several protein kinases, such as Cla4, Gin4, and Elm1, as well as septin-interacting proteins, such as Bni5, locate to the bud neck in a septin-dependent manner and promote septin collar stabilization (Longtine et al., 1998; Barral et al., 1999; Lee et al., 2002). Finally, the protein phosphatase PP2A bound to its B′ regulatory subunit, Rts1, promotes timely septin disassembly after cytokinesis (Dobbelaere and Barral, 2004). How these regulators directly or indirectly promote septin ring assembly and disassembly at the bud neck is not clear.
A key question is how septin dynamics is coupled to the dramatic membrane rearrangements occurring during the formation of cellular protrusions. We set out to address this question using budding yeast as model system. We took advantage of our previous finding that the ubiquitin ligases Dma1 and Dma2 are essential together with the PAK kinase Cla4 for septin ring formation and stabilization. Indeed, dma1 dma2 cla4 triple mutants show pronounced septin localization defects and cytokinesis failure and are unviable (Fraschini et al., 2004; Merlini et al., 2012). We used this observation to set up a genetic screen aiming at the identification of novel regulators of septin dynamics. Here we report for the first time that the conserved Rho1 GTPase, its target protein kinase C, and the F-BAR protein Syp1 that induces membrane curvature promote robust septin recruitment and likely coordinate membrane and septin reorganization during budding.
RESULTS
A genetic screen for novel regulators of septin dynamics identifies the Rho1 GTPase and its target, Pkc1
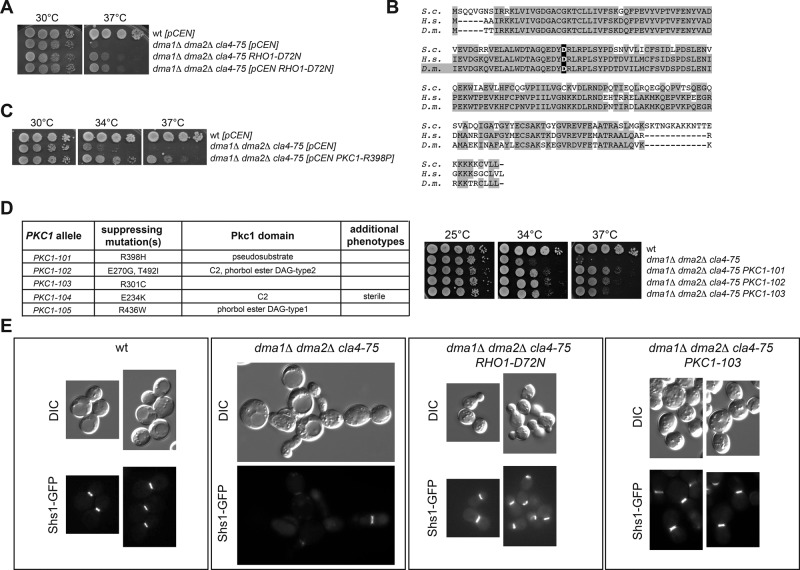
Because dma1 dma2 cla4 triple-mutant cells are not viable and show strong septin organization defects (Fraschini et al., 2004; Merlini et al., 2012), we wondered whether the lack of viability of these cells was mainly due to the collapse of septin structures. We rationalized that if this were the case, extragenic suppressors of the lethality of dma1 dma2 cla4 cells would most likely identify regulators of septin dynamics (see Materials and Methods). To test this possibility and identify new regulators of septin dynamics, we isolated 44 independent suppressors that restored cell viability in the total absence of DMA1, DMA2, and CLA4. Genetic tests classified 11 of the suppressing mutations as dominant, 10 as semidominant, and 19 as recessive. Four suppressors were sterile, hampering the assessment of their dominance/recessivity. FACS analysis of DNA contents showed that several suppressors completed cytokinesis properly (unpublished data), supporting the idea that septin organization was restored. We subsequently focused our attention on one of the best dominant suppressors of the temperature sensitivity of dma1Δ dma2Δ cla4-75 triple-mutant cells (Figure 1A). This suppressor indeed restored septin ring assembly and position at the bud neck, as determined using the septin Shs1 fused to green fluorescent protein (GFP) as reporter and demonstrated by the fact that it restored cytokinesis in the complete absence of DMA1, DMA2, and CLA4 (Figure 1E and Supplemental Figure S1B). Thus, restoring septin organization appeared to rescue the lethality of the dma1 dma2 cla4 triple mutant. We concluded that our suppressors might indeed identify new regulatory pathways of septin function.
FIGURE 1:
Rho1 and Pkc1 hyperactivation suppresses the temperature sensitivity of dma1Δ dma2Δ cla4-75 cells. (A, C, D) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates and incubated at the indicated temperatures for 2 d. The table in D lists the PKC1 alleles isolated in our genetic screen as suppressors of the lethality of dma1Δ dma2Δ cla4Δ cells. (B) Amino acid sequence alignment of Rho1/RhoA from budding yeast (S.c.), human (H.s.), and fruit fly (D.m.). The aspartate in bold is the conserved residue changed to asparagine in the RHO1-D72N allele. (E) Representative images of cells with the indicated genotypes and expressing Shs1-GFP after shift to 37°C for 6 h.
The foregoing strong suppressor mutation mapped in the RHO1 gene, which encodes the yeast counterpart of the conserved GTPase RhoA (Qadota et al., 1994). In metazoans, RhoA is required for cytokinesis through assembly and contraction of the contractile actomyosin ring (reviewed in Glotzer, 2005; Pollard and Wu, 2010; Green et al., 2012). The suppressor mutation was due to a single nucleotide substitution replacing the aspartic acid in position 72, which is located inside the highly conserved switch II of the guanine nucleotide–binding domain of Rho GTPases (Figure 1B), by asparagine (RHO1-D72N). This finding prompted us to sequence RHO1 in all the other dominant suppressors. Strikingly, three additional suppressors carried exactly the same RHO1-D72N mutation (unpublished data).
The RHO1-D72N allele likely causes Rho1 hyperactivation, at least toward some of its effectors. Indeed, similar to RHO1-D72N, an episomal plasmid bearing the GTP-locked RHO1-G19V allele, but not the dominant-negative RHO1-D125A allele, suppressed the temperature sensitivity of dma1Δ dma2Δ cla4-75 triple-mutant cells (Supplemental Figure S1C). Furthermore, deletion of LRG1 and SAC7, which encode the main GTPase-activating proteins (GAPs) for Rho1 down-regulation during cytokinesis (Yoshida et al., 2009), partially suppressed the lethality of dma1Δ dma2Δ cla4-75 cells at high temperatures (Supplemental Figure S1D). In addition, we found that one of the recessive suppressors isolated in our genetic screen carried a premature stop codon in LRG1 (LRG1-L67stop). Conversely, eliminating the Sac7 paralogue Bag7 (Supplemental Figure S1D) or mutating the catalytic residue of the Rho GAP Bem2 (Marquitz et al., 2002; unpublished data) did not rescue the temperature sensitivity of dma1 dma2 cla4-75 triple-mutant cells. Finally, since the yeast polo-like kinase, encoded by the CDC5 gene, recruits and activates Rho1 at the cell division site for cytokinesis (Yoshida et al., 2006), we asked whether the RHO1-D72N allele could partially rescue the temperature-sensitive growth defects of cdc5 mutants. This was indeed the case (Supplemental Figure S1E), suggesting that the Rho1-D72N protein is likely hyperactive.
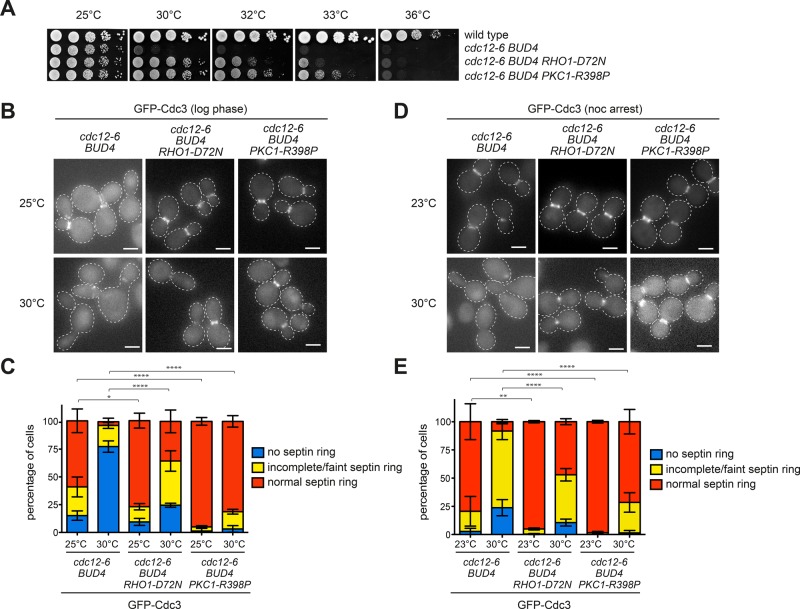
The RHO1-D72N allele also partially suppressed the temperature sensitivity of septin mutants like cdc12-1, which fails to assemble the septin ring, cdc12-6, which disassembles the septin ring upon shift to the restrictive temperature (Barral et al., 2000; Dobbelaere et al., 2003), and shs1Δ, which lacks the only nonessential septin (Figure 2, A–C, and Supplemental Figure S2, A and B), suggesting that septin stabilization may underlie the mechanism of suppression. This conclusion was confirmed by direct inspection of the septin ring marked by GFP-Cdc3 in cdc12-6 cells shifted to the nonpermissive temperature of 30°C. Whereas cdc12-6 cells, either cycling (Figure 2, B and C) or arrested in mitosis by nocodazole treatment (Figure 2, D and E), rapidly disassembled the septin ring after 1-h shift to 30°C, a significant fraction of them could maintain a septin ring at the bud neck upon expression of RHO1-D72N.
FIGURE 2:
Rho1 and Pkc1 hyperactivation rescues the septin ring instability of cdc12-6 mutant cells. (A) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates and incubated at the indicated temperatures for 2 d. (B, C) cdc12-6 BUD4+ cells expressing wild-type RHO1 and PKC1, RHO1-D72N, or PKC1-R398P at their respective genomic loci were grown at 25°C and shifted to the restrictive temperature of 30°C for 1 h. The septin ring is marked by GFP-Cdc3. Average values and error bars (SD) are derived from three independent experiments (n ≥ 100). (D, E) The same strains as in B and C were grown at 23°C, arrested in mitosis by 3 h of treatment with nocodazole, and shifted to 30°C for 1 h to image GFP-Cdc3. Average values and error bars (SD) are derived from three independent experiments (n ≥ 100). Scale bars, 5 μm.
Although constitutively active GTP-locked variants of Rho1 as sole source of Rho1 in the cell cannot support actomyosin ring assembly and viability of yeast cells (Yoshida et al., 2009), RHO1-D72N cells were perfectly viable, assembled a normal actomyosin ring and contracted it with kinetics similar to that of wild-type cells (Supplemental Figure S3). Thus Rho1-D72N might be hyperactive toward some but not all Rho1 effectors.
Because the Rho GTPase Cdc42 is known to promote septin deposition (Gladfelter et al., 2002; Caviston et al., 2003), we tested whether the hyperactive CDC42-D65N allele (Mosch et al., 2001), which carries a synonymic mutation to RHO1-D72N, could suppress the temperature sensitivity of dma1 dma2 cla4-75 triple-mutant or cdc12 single-mutant cells. Expression of CDC42-D65N had no effect on cell division of dma1 dma2 cla4-75 triple- mutant and cdc12-6 single-mutant cells, whereas it was detrimental for cdc12-1 cells growing at the permissive temperature (Supplemental Figure S2, D–F). Consistent with these data, GTP-locked Cdc42 negatively regulates septin polymerization in budding yeast (Gladfelter et al., 2002) and mammalian cells (Joberty et al., 2001).
Five Rho1 effectors are known: protein kinase C (Pkc1), the formins Bni1 and Bnr1, the exocyst component Sec3, and the Fks1/2 glucan synthase (Levin, 2011). Pkc1 is a major regulator of membrane dynamics (Kono et al., 2012) and localizes, like Rho1, to sites of polarized growth and the bud neck (Yamochi et al., 1994; Denis and Cyert, 2005). Furthermore, it is the only bud neck protein that colocalizes with septins also in meiosis (Lam et al., 2014), which makes it an excellent Rho1-target candidate in the regulation of septin stability. Consistently, the PKC1-R398P hyperactive allele, which is mutated in the pseudosubstrate autoinhibitory domain (Nonaka et al., 1995), suppressed the temperature sensitivity of dma1Δ dma2Δ cla4-75 triple-mutant (Figure 1C) and cdc12-1 and cdc12-6 single-mutant cells (Figure 2A and Supplemental Figure S2C). This was not the case for a dominant hyperactive allele of BNI1 (BNI1-V360D; Kono et al., 2012; unpublished data). Furthermore, PKC1-R398P efficiently suppressed septin ring disassembly of cdc12-6 cells at restrictive temperature (Figure 2, B–E). Finally, direct sequencing showed that five dominant suppressors found in our genetic screen carried PKC1 mutations in different protein domains (PKC1-R398H, PKC1-E270G,T492I, PKC1-R301C, PKC1-E234K, and PKC1-R436W; Figure 1, D and E), further corroborating the idea that hyperactivation of the Rho1/Pkc1 pathway stabilizes the septin ring.
Rho1 and Pkc1 stabilize the septin ring
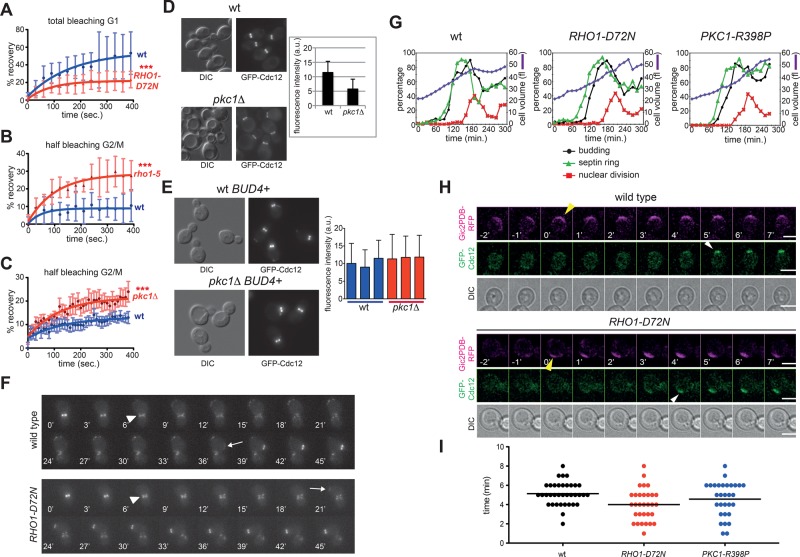
Although the involvement of Rho1/RhoA in the assembly and contraction of the actomyosin ring is well established, no data linked its function to septins. We therefore tested directly the effect of Rho1 and Pkc1 on septin dynamics. The septin ring was visualized by expression of GFP-Cdc12 and its stability analyzed by FRAP. As expected (Caviston et al., 2003; Dobbelaere et al., 2003), after its complete bleaching in late G1, there was efficient fluorescence recovery in wild-type cells (Figure 3A), indicating high turnover of septins at the presumptive bud site (mobile fraction 54 ± 21%, n = 10). In contrast, turnover was lower in RHO1-D72N cells (mobile fraction 25 ± 11%, n = 10), indicating that in these cells the septin ring exchanged less rapidly with the cytoplasmic pool and was more rigid already in G1. We then asked whether RHO1 inactivation destabilizes the septin ring. To this end, we used the temperature-sensitive rho1-5 mutant, which is most likely specifically defective in Pkc1 activation since its temperature sensitivity is efficiently suppressed by PKC1 overexpression (Kamada et al., 1996) and the PKC1-R398P allele (unpublished data). Half of the GFP-Cdc12 signal was bleached in large-budded cells. Consistent with the frozen state of the ring at this cell cycle stage (Caviston et al., 2003; Dobbelaere et al., 2003), turnover at 37°C was low in wild-type cells (mobile fraction 10 ± 6%, n = 12). In contrast, recovery was significantly higher in rho1-5 cells at 37°C (Figure 3B; mobile fraction 31 ± 9%, n = 7), indicating that Rho1 inactivation enhances septin ring turnover.
FIGURE 3:
RHO1 and PKC1 mutations affect septin dynamics. (A) FRAP analysis of the septin ring in G1 cells after complete bleaching of the GFP-Cdc12 fluorescence signal at the presumptive bud site (time 0: postbleach). (B, C) FRAP analysis of the septin ring in budded cells after bleaching of half of the GFP-Cdc12 fluorescence signal at the bud neck (time 0: postbleach). Curves were fitted to monoexponential decay. (D) Wild-type and pkc1Δ cells in the W303 background (bud4) were grown in sorbitol-containing YEPD medium at 30°C and imaged. Fluorescence intensities of GFP-Cdc12 signals at the bud neck were quantified on one single in-focus plane (n ≥ 100). (E) Three independent wild-type and pkc1Δ strains in W303 carrying a wild-type copy of BUD4 (BUD4+) were grown and imaged as in D to quantify fluorescence intensities of GFP-Cdc12 signals at the bud neck (n ≥ 100). (F) Representative movies of wild-type and RHO1-D72N cells expressing GFP-Cdc12. Arrowheads indicate splitting of the septin ring, whereas arrows indicate appearance of a new septin ring. Time 0 is arbitrarily set 6 min before septin ring splitting. (G) Small, unbudded cells of wild-type, RHO1-D72N, and PKC1-R398P cells were isolated by centrifugal elutriation and released in fresh YEPD medium at 25°C (time 0). At the indicated time points, cells were collected for FACS analysis of DNA contents (not shown) and kinetics of cell volume, budding, nuclear division, and septin ring formation. This last was analyzed by indirect immunofluorescence with anti-Cdc11 antibodies. (H, I) Wild-type, RHO1-D72N, and PKC1-R398P cells expressing GFP-Cdc12 and the Gic2PDB-RFP marker of Cdc42 activation were filmed at 30°C to measure the time interval between Cdc42 activation and septin recruitment. The graph (I) represents the distribution of time intervals in each strain (n ≥ 30) and mean times (black bars).
Because the PKC1 gene is essential for viability and cell integrity, to test its possible involvement in septin dynamics, we had to grow pkc1Δ cells in the presence of an osmostabilizer (i.e., sorbitol). Under these growth conditions, PKC1 deletion accelerated the turnover of GFP-Cdc12 at the septin ring in budded cells (Figure 3C; mobile fraction 11.5 ± 6.5%, n = 8, for wild-type and 22 ± 8%, n = 8, for pkc1Δ cells). Thus inactivation of Rho1 and Pkc1 increases the fluidity of the septin ring, whereas RHO1 hyperactivation makes the septin ring more rigid. In agreement with this conclusion, even when incubated at 25˚C and provided with sorbitol, combining PKC1 deletion with the cdc12-1 or cdc12-6 temperature-sensitive mutations dramatically compromised viability (unpublished data), whereas pkc1Δ shs1Δ double-mutant cells were inviable. These data indicate that Pkc1 becomes essential in the presence of septin defects that are normally tolerated by otherwise wild-type cells.
Mutants affecting Rho1 activity were previously shown to assemble septin rings but fail to undergo the normal conversion from single ring into hourglass (Cid et al., 2001). Similarly, we found that PKC1 deletion in the W303 strain background led to thinner septin rings containing lower amounts of GFP-Cdc12 than otherwise wild-type cells (Figure 3D). Because W303 bears a frameshift mutation in the BUD4 gene (Voth et al., 2005), which encodes an anillin-related protein that stabilizes the septin ring during splitting (Wloka et al., 2011; Kang et al., 2013), we asked whether introduction of a wild-type copy of BUD4 restored normal septin levels at the bud neck of pkc1Δ cells. This was indeed the case (Figure 3E), suggesting that Pkc1 and Bud4 play overlapping role(s) in septin ring stability during a normal cell cycle.
In agreement with the idea that Rho1 and Pkc1 stabilize the septin ring, we found that their hyperactivation shortened the interval between septin ring splitting and formation of a new septin ring—that is, the cell cycle window characterized by the highest septin dynamics. Time-lapse video microscopy of wild-type and RHO1-D72N cells expressing GFP-Cdc12 showed that the time between septin ring splitting and formation of a new septin ring was on average 21 ± 6.9 min in RHO1-D72N cells (n = 100) and 34 ± 11.5 min in wild-type cells (n = 72; Figure 3F). To confirm these results, we elutriated small, unbudded cells from wild-type, RHO1-D72N and PKC1-R398P cell cultures and released them in fresh medium at 25°C. At various times, cell samples were collected, and cell volume (used as internal reference), budding, septin ring formation, and nuclear division were quantified. In all three strains, bud emergence occurred at the same mean cell volume (34 fl), suggesting that this process is not grossly affected by RHO1 and PKC1 hyperactivation. Under these conditions, wild-type cells formed septin assemblies concomitant to budding and S-phase entry (starting at 90 min after release). In contrast, in the RHO1-D72N and PKC1-R398P mutant cells, septin ring recruitment took place at a smaller cell volume and was advanced by ∼15 min relative to budding and S-phase entry (Figure 3G). Of importance, in these mutants septin deposition still occurred after activation of Cdc42, as shown by time-lapse video microscopy of cells expressing the fluorescent Cdc42 biosensor Gic2PBD–red fluorescent protein (Okada et al., 2013; Figure 3H). However, the time interval between Cdc42 activation and septin recruitment was slightly shorter in the mutants relative to wild-type cells (Figure 3I).
Collectively these data indicate that Rho1 and Pkc1 contribute to septin ring stabilization.
Osmotic stabilization of the cell wall restores septin organization in a septin mutant
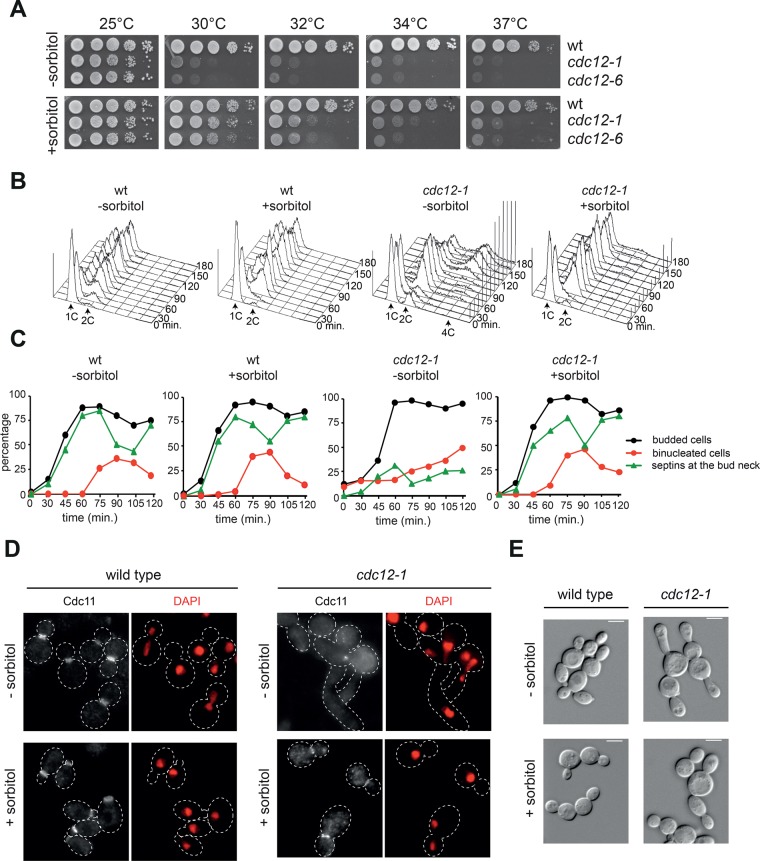
Rho1 and Pkc1 are involved in the cell wall integrity (CWI) pathway, which responds to a variety of cell wall stresses (e.g., osmotic stress) and activates a mitogen-activated protein kinase (MAPK) cascade response. This in turn induces a specific transcriptional program regulating the expression of cell wall enzymes and reorganization of the actin cytoskeleton (Levin, 2011; Supplemental Figure S4A). We therefore asked whether osmotic support, through addition of sorbitol to the growth medium, might also promote septin ring stabilization by counteracting the intracellular turgor pressure. The temperature sensitivity of cdc12-1 and cdc12-6 cells was indeed partially suppressed by sorbitol (Figure 4A). To investigate whether sorbitol could also stabilize the septin ring of cdc12-1 cells, we grew wild- type and cdc12-1 cells at 23°C, arrested them in G1, and released them at 34°C in rich medium either lacking or containing sorbitol. FACS analysis of DNA contents at different time points after release showed that in the absence of sorbitol, cdc12-1 cells displayed pronounced cytokinetic defects leading to polyploidy, which were efficiently rescued by sorbitol (Figure 4B). Strikingly, whereas in the absence of sorbitol septin rings failed to assemble in most cdc12-1 cells, in the presence of sorbitol they assembled at the bud neck in a high fraction of them with kinetics similar to wild type (Figure 4, C and D). Furthermore, sorbitol efficiently rescued also the elongated-bud phenotype of cdc12-1 cells at 34°C (Figure 4E). Thus, similar to hyperactivation of the Rho1/Pkc1 pathway, counteracting osmotic stress may stabilize the septin ring.
FIGURE 4:
Hyperosmotic medium rescues the lethality and cytokinetic defects of septin mutants. (A) Serial dilutions of strains with the indicated genotypes were spotted on YEPD plates and incubated at the indicated temperatures for 2 d. (B–E) Wild-type and cdc12-1 cells were grown in YEPD at 25°C in the absence or presence of sorbitol, arrested in G1 by α-factor, and released in fresh medium at 34°C (time 0). At the indicated time points, cell samples were collected for FACS analysis of DNA contents (B) and kinetics of budding, nuclear division, and septin deposition at the bud neck by indirect immunofluorescence of the septin Cdc11 (C). Distribution of septins (D) and cell morphology (E) in representative cells at t = 120 min after release.
Rho1 and Pkc1 likely have targets other than the cell wall integrity pathway in septin stabilization
Because Rho1 and Pkc1 are involved in the CWI pathway (Supplemental Figure S4A), we wondered whether hyperactivation of the MAPK cascade downstream of Pkc1 was involved in the suppression of the lethality of dma1 dma2 cla4 triple-mutant cells by RHO1 and PKC1 dominant alleles. Whereas hyperactivation of the most upstream MAPKKK in this pathway, using the BCK1-20 allele, weakly suppressed the temperature sensitivity of dma1Δ dma2Δ cla4-75 triple-mutant cells, hyperactivation of the downstream MAPKK and MAPK by the dominant MKK1-S386P or overexpression of Slt2 (Irie et al., 1993) did not (Supplemental Figure S4B). Thus Rho1- and Pkc1-mediated suppression of the septin defects caused by simultaneous inactivation of Dma1, Dma2, and Cla4 is not caused solely by hyperactivation of the MAPK pathway, if at all. Consistently, the RHO1-D72N allele that we isolated in our genetic screen did not hyperactivate the MAPK cascade after thermal stress, as judged by phosphorylation of the MAPK Slt2 (Supplemental Figure S4C). Although our data do not rule out a possible contribution of the CWI pathway on septin ring stability, they do suggest that Rho1 and Pkc1 stabilize the septin ring at least partly independently of the MAP kinase pathway.
The F-BAR protein Syp1 is a novel Pkc1 phosphorylation target
High-throughput analysis of the budding yeast kinome revealed a physical interaction between Pkc1 and Syp1 (Breitkreutz et al., 2010), a membrane protein of the F-BAR family that has been implicated in endocytosis and septin dynamics at the bud neck (Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). SYP1 deletion partially suppressed the temperature sensitivity of cdc12-1 and dma1Δ dma2Δ cla4-75 triple-mutant cells (Supplemental Figure S5A), suggesting that it might be a relevant target of Pkc1 in septin regulation.
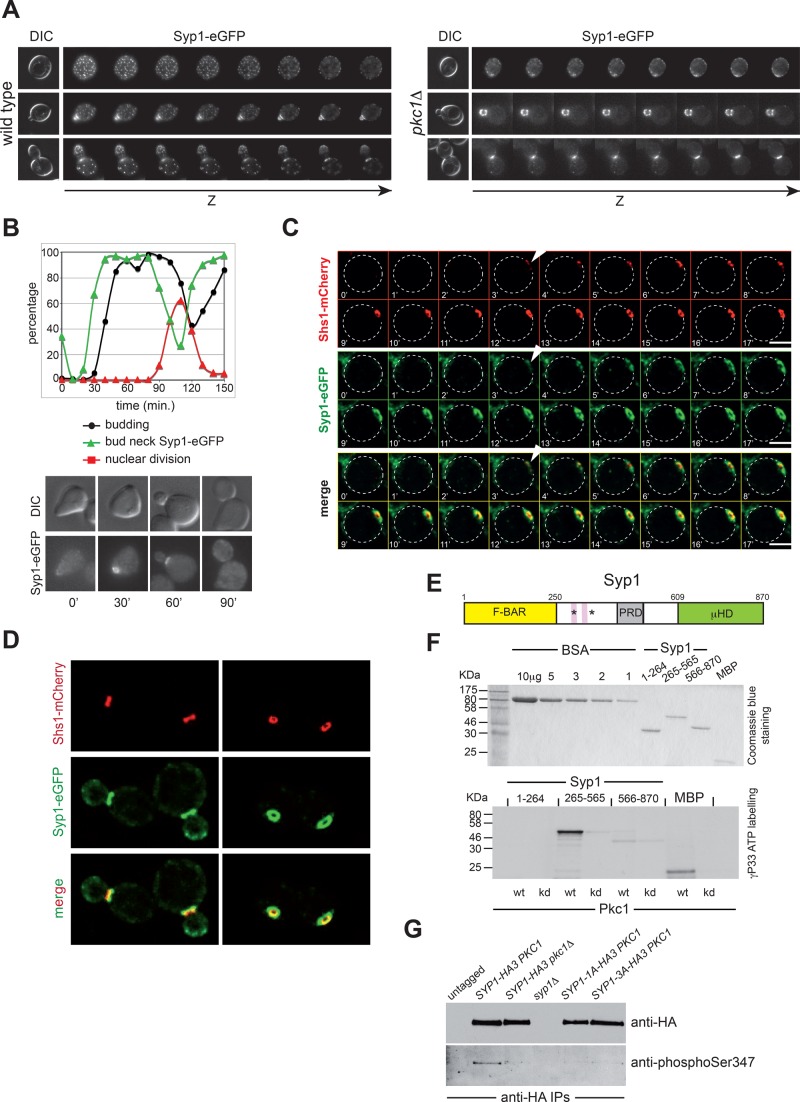
PKC1 inactivation severely compromised the organization of Syp1 in patches while enhancing its bud neck localization (Figure 5A), suggesting that Pkc1 controls the turnover of Syp1 at membranes and its displacement from the bud neck. To gain insights into the relationship between the bud neck localization of Syp1 and that of septins, we first analyzed the localization of Syp1–enhanced GFP (eGFP) during the cell cycle, starting from cells synchronized in G1 by α-factor and released in fresh medium at 25°C. Consistent with published data (Qiu et al., 2008; Reider et al., 2009), Syp1-eGFP was recruited to the bud neck in late G1, before bud emergence. Its levels then decreased at the bud neck around the time of anaphase entry, marked by the increase in binucleate cells (Figure 5B). Time-lapse video microscopy of cells expressing at the same time Syp1-eGFP and Shs1-mCherry revealed that Syp1 was recruited to the bud neck concomitant to septins (Figure 5C). Syp1 rings were larger than septin rings and asymmetrically localized to the mother side of the bud neck in small/medium-budded cells (Figure 5D), similar to the phosphatase complex Bni4/Glc7 (DeMarini et al., 1997; Kozubowski et al., 2003) and the SUMO E3 ligases Siz1 and Siz2 (Johnson and Blobel, 1999; Johnson and Gupta, 2001). Deletion of PKC1 or DMA1 and DMA2 did not appear to affect the pattern of Syp1 localization at the bud neck during the cell cycle (Supplemental Figure S5, B and C).
FIGURE 5:
Pkc1 phosphorylates Syp1 and controls its bud neck localization. (A) Z-stack serial images of wild-type and pkc1Δ cells expressing Syp1-eGFP at different cell cycle stages (top: G1, unbudded; middle: S phase, small budded; bottom: mitosis, large budded). Spacing between sequential planes is 0.3 μm. (B) Wild-type cells expressing Syp1-eGFP were arrested in G1 by α-factor and released in fresh YEPD medium at 25°C. At the indicated time points, cells were collected for FACS analysis of DNA contents (not shown) and kinetics of budding, nuclear division, and Syp1-eGFP localization. (C) Wild-type cells expressing Syp1-eGFP and Shs1-mCherry were filmed at room temperature with 1-min time lapse. Z-stacks (31 planes at 0.2-μm spacing) were maximum projected and deconvolved with Huygens. Arrowheads indicate the appearance of septin and Syp1 rings. (D) Images of wild- type cells expressing Syp1-eGFP and Shs1-mCherry show that Syp1 rings are larger in diameter than Shs1 rings and located on mother side of the bud neck. (E) Schematic representation of the Syp1 protein showing the N-terminal F-BAR domain (yellow), the middle, unstructured region (white) containing the proline-rich domain (gray), and the muniscin-homology domain (green) at the C-terminus. Pink boxes indicate the two phospholipid-binding motifs, and asterisks mark the position of the serines phosphorylated by Pkc1. (F) The three domains of Syp1 were purified from bacteria (purified proteins are shown on top after Coomassie blue staining of an acrylamide gel) and subjected to in vitro phosphorylation by wild-type or kinase-dead (kd) Pkc1 immunoprecipitated from yeast cells ([γ-33P]ATP labeling, bottom). (G) Syp1-HA3 was immunoprecipitated from protein extracts obtained from cycling cultures of strains with the indicated genotypes and probed by Western blot with a phospho-specific antibody raised against phosphorylated Ser-347, as well as with anti-HA antibodies.
We then asked whether Syp1 could be phosphorylated by Pkc1. Syp1 carries an N-terminal F-BAR domain, followed by a central, unstructured region and a C-terminal μHD domain that interacts with the endocytic protein Ede1 and the Rho1 activator Mid2. In addition, the region spanning amino acids 265–365 carries a liposome-binding domain and two lysine-rich motifs that bind PI(4,5)P2 and contribute to Syp1 association with the plasma membrane (Reider et al., 2009; Figure 5E). We purified from bacterial cells the N-terminal (amino acids [aa] 1–265), the central (aa 265–565), and the C-terminal (aa 566–870) domains of Syp1 and subjected them to in vitro kinase assays using wild-type or kinase-dead Pkc1 immunoprecipitated from yeast cell extracts, using myelin basic protein (MBP) as control substrate (Antonsson et al., 1994). As shown in Figure 5F, Pkc1, but not its kinase-dead version, efficiently phosphorylated the central region of Syp1, as well as MBP. Mass spectrometric analysis of this truncated protein identified two Pkc1-dependent phosphorylation sites on Ser-347 and Ser-389 or Ser-390 (Supplemental Table S1). Both are located in proximity of the two lysine-rich motifs (Figure 5E) and are hence likely to regulate Syp1 interaction with membranes. These phosphosites were previously mapped in vivo, along with many additional phosphorylation sites (Li et al., 2007; Albuquerque et al., 2008; Holt et al., 2009; Bodenmiller et al., 2010; Breitkreutz et al., 2010). Of interest, phosphorylation of Ser-347 was found to increase in response to osmotic stress (Soufi et al., 2009) and rapidly decrease in sorbitol-containing medium (Kanshin et al., 2015). In spite of that, using antibodies specific for phosphorylated Ser-347, we could detect low levels of Syp1 Ser-347 phosphorylation in wild-type but not in pkc1Δ cells grown in sorbitol-containing medium (Figure 5G), suggesting that Syp1 is a substrate of Pkc1 also in vivo. Thus Pkc1 phosphorylates Syp1 directly on Ser-347 and probably Ser-389/Ser-390 in vitro and in vivo and regulates its subcellular localization. Syp1 is the second bona fide substrate of Pkc1 in budding yeast, the first being Bck1 (Levin et al., 1994).
Syp1 phosphorylation by Pkc1 increases its turnover at the bud neck and contributes to the robustness of septin deposition
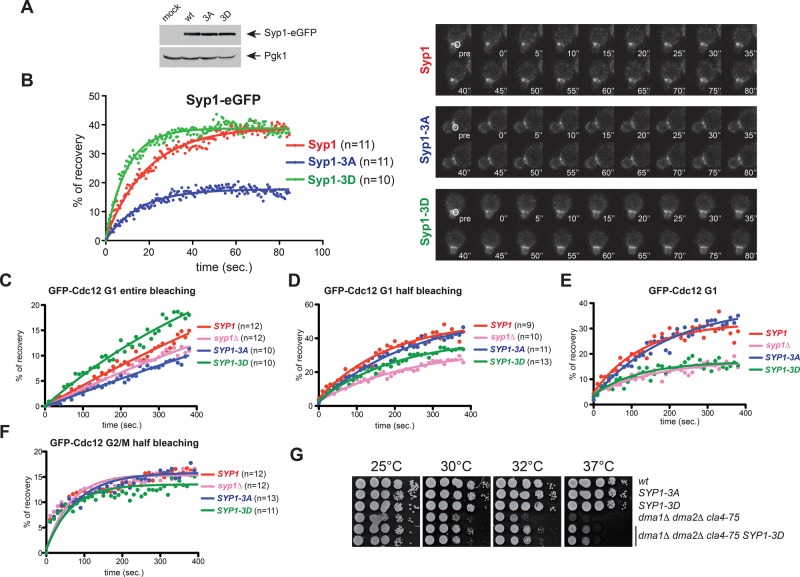
Pkc1-dependent phosphorylation of the central region of Syp1 would introduce negative charges in an otherwise basic domain, which could interfere with Syp1 interaction with membranes. Consistently, in pkc1Δ mutant cells Syp1 localization at the bud neck increased compared with wild-type cells at the cost of patches normally observed at the rest of the cortex (Figure 5A). These data suggest that Pkc1 inactivation stabilizes Syp1 at the bud neck. To address the physiological role of the Pkc1-dependent phosphorylation sites, we mutated Ser-347, Ser-389, and Ser-390 to alanine to abolish phosphorylation and to aspartate to mimic constitutive phosphorylation. The mutant alleles were then integrated in the yeast genome as the only copy of SYP1 in the cells and will be referred to as SYP1-3A and SYP1-3D, respectively. The corresponding mutant proteins tagged with eGFP were expressed at levels similar to those of their wild-type counterpart (Figure 6A) and recruited to the bud neck with the same timing during the cell cycle (unpublished data). We then assessed by FRAP whether they affected Syp1 turnover at the bud neck (Figure 6B). On bleaching of half of the Syp1-eGFP ring in small/medium-budded cells, ∼39% of the molecules recovered (mobile fraction), with a half-recovery time of 13.63 ± 0.59 s. A similar mobile fraction was observed for Syp1-3D-eGFP, but its half-recovery time decreased to 6.77 ± 0.77 s, indicating a faster turnover. In stark contrast, the mobile fraction of Syp1-3A-eGFP was reduced to 18% (half-recovery time of 10.79 ± 0.5 s), indicating that the protein is more stably bound to the cortex. Therefore, Pkc1-dependent phosphorylation stimulates the turnover of Syp1 at the bud neck, consistent with the effect of PKC1 inactivation on Syp1 localization.
FIGURE 6:
Syp1 phosphorylation by Pkc1 promotes Syp1 turnover at the bud neck and rigidity of the septin ring during its formation. (A) Steady-state levels of Syp1-, Syp1-3A-, and Syp1-3D-eGFP in logarithmically growing cells. (B) FRAP analysis of Syp1-eGFP at the bud neck. Half of the Syp1 ring was bleached in small-budded wild-type, SYP1-3A, and SYP1-3D cells, and recovery of fluorescence was measured every 0.5 s. Time 0 indicates the first time point after bleaching. Curves were fitted to monoexponential decay. (C–F) Cells with the indicated genotypes and expressing GFP-Cdc12 were analyzed by FRAP after bleaching of the entire (C) or half septin ring (D, F) in either G1 cells (C, D) or G2/M cells (F). Images were taken every 10 s. Time 0 corresponds to the first frame after bleaching. Curves were fitted with a one-phase association function (C, D) or with a two-phase association function (F). The goodness of the fit was based on 95% confidence intervals and R2 values. Kinetics of recovery after entire bleaching (C) was subtracted from those after half-bleaching (D) to obtain curves of recovery deriving only from septin dynamics inside the ring (i.e., recovery from the unbleached half of the ring; E). (G) Serial dilutions of stationary-phase cultures of strains with the indicated genotypes were spotted on YEPD plates and incubated for 2 d at the indicated temperatures.
We then assessed the effects of SYP1 phosphomutant alleles on septin dynamics by analyzing by FRAP the recovery rates of GFP-Cdc12 at the bud neck. Recovery of GFP-Cdc12 after bleaching of the entire septin ring is indicative of septin recruitment from the cytoplasm, whereas recovery after half-bleaching of the ring is the result of both lateral diffusion from the unbleached half and septin recruitment from the cytoplasm. We measured recovery of GFP-Cdc12 in wild-type SYP1, syp1Δ, SYP1-3A, and SYP1-3D cells in late G1 after bleaching of both entire and half septin rings (Figure 6, C and D). To assess specifically the internal fluidity of the septin ring and have a more precise measurement of the lateral diffusion only, we subtracted the averaged values of recovery upon entire bleaching from those obtained after half-bleaching. Strikingly, the septin ring was significantly more rigid in syp1Δ and SYP1-3D cells than in wild-type and SYP1-3A cells (Figure 6E), suggesting that Pkc1-dependent phosphorylation and removal of Syp1 stabilizes the septin ring in late G1. Hence Syp1 localization to the bud neck in G1 promotes septin ring fluidity during its assembly. Consistent with these ideas, the SYP1-3D allele partially suppressed the temperature sensitivity of the dma1Δ dma2Δ cla4-75 triple-mutant cells (Figure 6G). No significant differences were found between wild-type and SYP1 mutants in the curves of recovery after half-bleaching in budded cells (i.e., in S/G2/M; Figure 6F). Thus phosphorylation of Syp1 may affect septin dynamics most specifically before or at the time of bud emergence, that is, when the plasma membrane remodels at the bud neck and concomitant with septin structures being under mechanical stress.
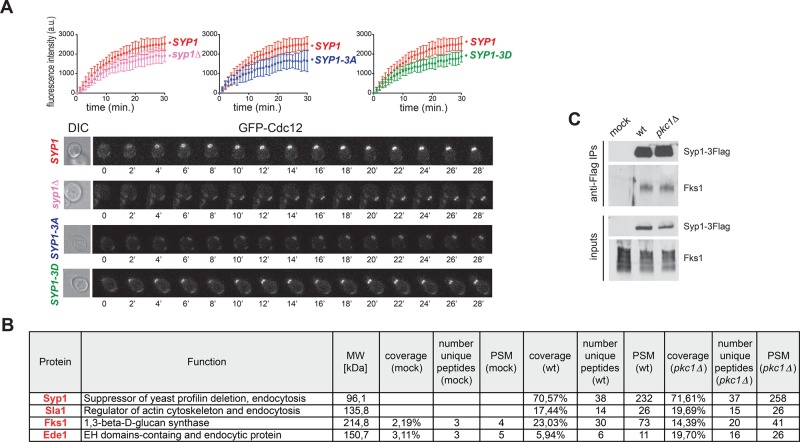
Having found that septin ring fluidity is affected by Syp1 phosphorylation prompted us to test whether SYP1 phosphomutants undergo normal septin ring assembly in late G1. To this end, we quantitatively measured deposition of GFP-Cdc12 at the presumptive bud site by time-lapse video microscopy of cells released from G1 into the cell cycle at 30°C. The syp1Δ, SYP1-3A, and SYP1-3D single-mutant cells all recruited GFP-Cdc12 at the future bud site more slowly than do wild-type cells (Figure 7A), suggesting that that proper turnover of Syp1 at the bud neck upon phosphorylation by Pkc1 is required for timely building of a robust septin ring.
FIGURE 7:
Syp1 contributes to fast septin recruitment at the bud neck and interacts with Fks1. (A) Kinetics of septin recruitment at the presumptive bud site were calculated by measuring the fluorescence intensity of GFP-Cdc12 over time (frames every 1 min) in cells with the indicated genotypes arrested in G1 by α-factor and released into fresh medium at 30°C at time 0. Wild type, n = 9; syp1Δ, n = 18; SYP1-3A, n = 10; SYP1-3D, n = 15. (B) Mass spectrometric analysis of Flag immunoprecipitates from cell extracts of wild-type cells expressing either untagged Syp1 (mock) or Flag-tagged Syp1 (Syp1-3xFlag) and carrying either wild-type PKC1 or PKC1 deletion (pkc1Δ). Cells were grown to exponential phase in sorbitol-containing medium to keep pkc1Δ cells alive. The table contains only a partial list of Syp1-interacting proteins that were uncovered by mass spectrometry. Unique peptides are peptides with different amino acid sequence. PSM (peptide-spectrum match) represents the total number of peptides identified for each protein. (C) Syp1-3Flag was immunoprecipitated from wild-type and pkc1Δ cells grown in YEPD containing sorbitol. Immunoprecipitates were separated by SDS–PAGE electrophoresis and analyzed by Western blot with anti-Fks1 and anti-Flag antibodies.
Syp1 interacts with and might regulate the activity of the cell wall–remodeling enzyme 1,3-β-d-glucan synthase
To gain insights into the mechanism by which Syp1 modulates septin dynamics, we searched for Syp1-interacting proteins by mass spectrometry. To this end, we tagged Syp1 with a triple-Flag tag (Syp1-3Flag) and immunoprecipitated from wild-type and pkc1Δ cells growing in 1% yeast extract, 2% bactopeptone, 50 mg/l adenine medium supplemented with 2% glucose (YEPD) containing sorbitol. Immunoprecipitated material was then separated by SDS–PAGE and analyzed by tandem mass spectrometry (MS/MS). Although Syp1 was found to interact with septins by protein-fragment complementation assays (Tarassov et al., 2008) and in vitro pull downs (Qiu et al., 2008), we could never find any septins in our Syp1-3Flag purifications (unpublished data). Instead, we found several endocytic factors associated to Syp1, among which were Sla1 and Ede1, which are known Syp1 interactors (Gavin et al., 2002; Reider et al., 2009; Figure 7B and Supplemental Table S2). Surprisingly, among the best hits, we also found Fks1, the major yeast 1,3-β-d-glucan synthase, which is involved in cell wall synthesis and is an effector of Rho1 (Douglas et al., 1994; Qadota et al., 1996). Furthermore, Fks1 has been implicated in endocytosis, along with Rho1 (deHart et al., 2003). We confirmed the association between Syp1 and Fks1 by immunoprecipitation of Syp1-3Flag followed by Western blot analysis with anti-Fks1 antibodies (Figure 7C). Furthermore, similar levels of Fks1 were coimmunoprecipitated with Syp1-3Flag from wild-type and pkc1Δ cells (Figure 7C), indicating that Pkc1 does not affect Fks1 association with Syp1.
Because 1,3-β-d-glucan synthase contributes to proper septin distribution in the pathogenic budding yeast Candida albicans (Blankenship et al., 2010; Badrane et al., 2012), the association between Syp1 and Fks1 suggests that these proteins might cooperate in controlling septin ring formation.
DISCUSSION
Rho1 functions in the control of septin organization
In metazoans and yeasts, the small Rho-family GTPase RhoA/Rho1 is a central regulator of cytokinesis. In all of these organisms, it promotes the assembly and contraction of the actomyosin ring, partly through activation of formins (reviewed in Piekny et al., 2005). Here we show that budding yeast Rho1 has an unanticipated role in cytokinesis by controlling septin dynamics. Indeed, we show, by a number of different approaches, that a novel dominant allele of RHO1, RHO1-D72N, stabilizes the septin ring at the bud neck. The RHO1-D72N mutation replaces an Asp residue located inside the switch II and conserved in all of the GTPases of the Rho family. The switch I and the switch II are GTPase domains that undergo the most dramatic structural rearrangements upon GTP binding and hydrolysis and cooperate to promote GTPase activation. In addition, these regions modulate the interactions of GTPases with their GAPs and effectors (Vetter and Wittinghofer, 2001). Thus the D72N amino acid substitution could modify Rho1 structure, leading to a Rho1 hyperactive variant that could stabilize the interaction with GTP, promote the interaction with a specific guanine nucleotide exchange factor (GEF), inhibit the binding to a GAP, and/or increase the affinity to specific effector(s). Although another Rho GTPase, Cdc42, is required for septin recruitment to the presumptive bud site (Gladfelter et al., 2002; Caviston et al., 2003; Iwase et al., 2006; Okada et al., 2013), a synonymous amino acid change in Cdc42 (Cdc42-D65N; Mosch et al., 2001) does not have the same effect on septin stability, suggesting that Rho1 and Cdc42 play distinct functions in septin organization. The finding that mutations locking Cdc42 in the active, GTP-bound state cause septin misorganization (Gladfelter et al., 2002; Caviston et al., 2003) further supports this conclusion.
Pkc1 as a central effector of Rho1 in the regulation of septin ring stability
We provide several lines of evidence indicating that Pkc1 is a key effector of Rho1 in the regulation of septin recruitment and ring formation at the bud neck. First, Pkc1 hyperactivation through the well-characterized PKC1-R398P allele (Nonaka et al., 1995) phenocopied the effects of RHO1-D72N on septin stability. Second, five different dominant mutations in our initial suppressor screen mapped in the PKC1 gene. Third, PKC1 deletion destabilizes septin collars, as shown by FRAP. Our finding that lack of Pkc1 is lethal for septin mutants even in the presence of an osmostabilizer further corroborates the notion that Pkc1 stabilizes the septin ring. Note that the real contribution of Pkc1 to septin ring stability under normal conditions might be underestimated in our experiments by the addition to the medium of sorbitol, which we show stabilizes septins at the bud neck. In our hands, conditional pkc1 mutant cells (Anastasia et al., 2012; unpublished data) rapidly lose viability and lyse in the absence of osmostabilizers similar to pkc1Δ cells, thus hampering a rigorous assessment of the effect of Pkc1 on septin regulation.
We cannot exclude that Rho1 contributes to septin ring assembly and stabilization also via additional effectors besides Pkc1. For instance, deletion of the formin Bni1 was shown to cause defects in septin organization and larger septin rings (Kadota et al., 2004). However, Bni1 is activated mainly by Rho3 and Rho4 at physiological temperatures (Dong et al., 2003), and its localization requires Cdc42 activity (Jaquenoud and Peter, 2000; Ozaki-Kuroda et al., 2001). In spite of Rho1 and Bni1 displaying two-hybrid interactions, Rho1 appears to activate Bni1 only at high temperatures and through an indirect mechanism that requires Pkc1 (Dong et al., 2003; Kono et al., 2012). Similarly, although Rho1 activates the β-1,3-glucan synthase encoded by the FKS1 and FKS2 genes (Qadota et al., 1996) for the cell wall synthesis (Douglas et al., 1994; Inoue et al., 1996), our data suggest that Fks1 activity might be further regulated by the Pkc1 target, Syp1. Other effectors of Rho1, such as the Sec3 subunit of the exocyst complex (Guo et al., 2001), which mediates polarized targeting and tethering of secretory vesicles (TerBush et al., 1996), have not been implicated in septin regulation. Thus we favor the idea that Pkc1 is a critical effector of Rho1 in the control of septin organization.
Circumstantial evidence suggests that septin regulation by protein kinase C (PKC) might be operational in other systems. Indeed, dendritic spine morphogenesis requires both septins (Tada et al., 2007; Xie et al., 2007) and atypical PKC (aPKC; Zhang and Macara, 2008). Besides promoting biogenesis of primary cilia in epithelial cells (Fan et al., 2004) and motile cilia in sea urchin, aPKC forms a ring at the base of the latter cilium (Pruliere et al., 2011) that is reminiscent of the ring-like septin structure found at the base of cilia (Hu et al., 2010; Kim et al., 2010). Finally, mammalian Sept2 is efficiently phosphorylated in vitro by PKC (Xue et al., 2000). Further studies will help to shed light on the links between PKC and septins in dendrite arborization, ciliogenesis, and sperm morphogenesis.
The F-BAR protein Syp1 and the control of septin dynamics
We identified the membrane protein Syp1 as a novel bona fide target of Pkc1. Syp1 is a member of the Bin-amphiphysin-Rvs (BAR) family of membrane proteins, which sculpt phosphoinositide-rich membranes to promote membrane curvature. Because of their ability to sense and modulate such curvature, BAR proteins are implicated in numerous processes involving membrane reorganization, such as endocytosis, cell migration, and cytokinesis (Suetsugu et al., 2010).
Syp1 exhibits a peculiar localization, forming a ring in late G1 that surrounds the new-forming septin ring and might define a specific membrane domain. Thus, after budding, the Syp1 ring ends up being larger than the septin ring. Of interest, establishment of cell polarity in yeast has been recently linked to the spatial coordination of exocytosis and endocytosis at the presumptive bud site. Specifically, an exocytic cortical pole is encircled by an endocytic ring rich in actin patches that confines exocytosis to a polarity vertex (Jose et al., 2013). How the septin ring is spatially organized in relation to these membrane compartments has not been addressed directly, but the septin ring likely coincides with the endocytic ring, on the basis of their respective diameters and the evidence that exocytosis is required to poke a hole in the septin cap during ring formation (Okada et al., 2013). Being external to the septin ring, the Syp1 ring could in turn influence the rate of local endocytosis and/or septin recruitment by preventing the lateral spreading of these processes.
Syp1 was shown to interact physically with septins and accelerate septin turnover (Qiu et al., 2008). Consistently, we show that lack of Syp1 suppressed the temperature sensitivity of mutants defective in septin ring assembly. Another yeast F-BAR protein, Hof1, controls actomyosin ring dynamics and cytokinesis (Lippincott and Li, 1998; Meitinger et al., 2011). Similar to Syp1, Hof1 localizes to the bud neck early during the cell cycle and affects the integrity of septin assemblies (Lippincott and Li, 1998; Meitinger et al., 2011), suggesting that besides stabilizing the actomyosin ring, it might share a function with Syp1 in septin regulation. Strikingly, we find that simultaneous inactivation of HOF1 and SYP1 is lethal (unpublished data), further strengthening this hypothesis.
BAR proteins, including Syp1, cluster PI(4,5)P2 (Saarikangas et al., 2009) and prevent its lateral diffusion, thus stabilizing lipid microdomains (Zhao et al., 2013). Because PI(4,5)P2 is concentrated at the bud neck (Garrenton et al., 2010) and stimulates septin filament assembly and organization (Bertin et al., 2010), it is tempting to speculate that BAR proteins might play a general role in the control of septin assemblies. The F-BAR protein syndapin/pacsin, which is a known PKC target (Plomann et al., 1998), promotes dendritic spine morphogenesis in neurons (Dharmalingam et al., 2009) and cytokinesis in Drosophila (Takeda et al., 2013). An exciting possibility is that these functions might be linked to a possible role of syndapin/pacsin in coordination of septin dynamics with membrane reorganization.
Here we show that Syp1 is phosphorylated by Pkc1 in vitro and, presumably, in vivo on Ser-347, -389, and/or -390 within a basic domain of the protein that contains two consensus motifs for phospholipid binding outside of the F-BAR domain. This region of the protein was previously shown to contribute to Syp1 localization at the plasma membrane (Reider et al., 2009). Pkc1-dependent phosphorylation of these serines accelerates the turnover of Syp1 at the bud neck. In agreement with the proposed role of Syp1 in promoting septin dynamics (Qiu et al., 2008), our data indicate that increasing Syp1 turnover at the bud neck with the SYP1-3D allele renders septins less mobile within the new-forming septin ring in late G1, that is, during a cell cycle stage in which the septin ring is usually in a fluid state and characterized by free lateral diffusion (Caviston et al., 2003; Dobbelaere et al., 2003).
Unlike PKC1 deletion, the SYP1-3A mutant allele does not obviously affect septin ring stability and does not cause synthetic lethality or sickness to septin mutants (unpublished data), indicating that it impairs septin organization to a lower extent than Pkc1 inactivation. One possible explanation is that additional Syp1 residues besides Ser-347, -389, and -390 might be phosphorylated by Pkc1. Furthermore, Pkc1 might have additional targets besides Syp1 in septin regulation. One intriguing possibility to be investigated in the future is whether Hof1 is also a Pkc1 phosphorylation target.
Possible mechanisms of septin regulation by Syp1 and other BAR proteins
Given their nature as membrane sculptors, an attractive model is that BAR proteins remodel both membrane and septins while membrane curvature changes.
Although the exact mechanism by which Syp1 controls septin dynamics remains to be established, we can envision several possibilities. First, Syp1 might influence the formation of septin filaments or their organization into a ring through direct interaction with septins (Qiu et al., 2008). Second, Syp1 might contribute to organizing septin assemblies at the presumptive bud site by regulating the local concentration of PI(4,5)P2. A third, non–mutually exclusive hypothesis is that Syp1 controls septin recruitment and efficient septin ring assembly through endocytic recycling of septins, which is consistent with the established involvement of Syp1 in endocytosis (Boettner et al., 2009; Reider et al., 2009; Stimpson et al., 2009). For instance, Syp1 could help to focus septin recruitment to the site of highest membrane curvature, that is, the bud neck. Although at a first glance this hypothesis might seem at odds with the different diameters expected for the Syp1 and the endocytic ring involved in polarity establishment (Jose et al., 2013), Syp1 could mediate a qualitatively different endocytosis relative to the one involved in polarity and, unlike the latter, prevent septins from spreading out. In this context, it is interesting to note that Rho1 was implicated in a novel clathrin-independent endocytic pathway that involves actin cables but not patches (Prosser et al., 2011). Moreover, the 1,3-β-d-glucan synthase Fks1, which we show interacts with Syp1, has been implicated in endocytosis independently of its cell wall–remodeling function (Okada et al., 2010). Further work will be required to establish whether the role of Syp1 in septin regulation is linked to its endocytic function.
Syp1 interaction with Fks1, which synthesizes the main cell wall component and is in turn regulated by Rho1 activity, suggests yet another possible mechanism underlying septin regulation. As mentioned earlier, Rho1, Pkc1, and Fks1 are part of the CWI pathway, which responds to a variety of environmental changes, including mechanical stress caused by cell wall defects (Levin, 2011). Signals are initiated at the cell surface by mechanosensors, such as Mid2 and Wsc1, which localize across the membrane and surrounding periplasmic space. Signals are then transduced to the Rho1 GEFs Rom1 and Rom2 at the plasma membrane, triggering activation of Rho1 and its effectors. Our data suggest that the upstream part of the CWI pathway is involved in septin ring stabilization at least partly through Syp1 phosphorylation. Of importance, the CWI pathway is induced not only under stress conditions, but also during the normal cell cycle, concomitant with polarized growth and bud emergence (Zarzov et al., 1996), that is, when cells undergo the most pronounced membrane/cell wall remodeling and are most vulnerable to lysis. Of note, this is also the cell cycle phase when the septin ring undergoes the transition from fluid to frozen state. Thus it would make perfect sense that the same Rho1/Pkc1 pathway couples new cell wall synthesis with septin ring stability. Intriguingly, Syp1 interacts with the mechanosensor Mid2 (Reider et al., 2009), suggesting that it might itself relay mechanical stress signals, acting as a mechanotransducer and/or participating in a Rho1-controlling feedback loop. Glucan synthesis could then stiffen the cell wall and, consequently, stabilize the septin ring. Consistent with this hypothesis, in the pathogenic fungus C. albicans, cell treatment with the glucan synthase inhibitor caspofungin leads to septin mislocalization (Blankenship et al., 2010; Badrane et al., 2012). Our finding that septin defects can be suppressed by addition of the osmostabilizer sorbitol to the medium further supports the idea of intimate cross-talk between cell wall and septin organization. In this scenario, cell wall reorganization by Rho1/Pkc1 would be part of a physiological circuit responding to mechanical stress and stabilizing the cytokinetic machinery either during the normal cell cycle or upon cell wall injury. This mechanism might be conserved, as suggested by recent data showing that mechanical stress stabilizes cytokinetic factors at the cleavage furrow in Dictyostelium cells (Srivastava and Robinson, 2015).
In conclusion, we have identified a novel pathway controlling septin dynamics in yeast. Given the evolutionary conservation of some of the players, similar pathways might be involved in controlling septin dynamics in other systems, such as dendritic spines and cilia.
MATERIALS AND METHODS
Strains, media, reagents, and genetic manipulations
All yeast strains were derivatives of W303 (ade2-1, trp1-1, leu2-3112, his3-11,15, ura3, ssd1). W303 bears a frameshift mutation in the BUD4 gene (Voth et al., 2005), which encodes an anillin-related protein that stabilizes the septin ring during splitting (Wloka et al., 2011). For some critical mutants, we made BUD4+ versions in W303. A full list of strains and plasmids used in this study is given in Supplemental Table S3. Unless specified, most strains were generated in the original bud4 W303 background. Cells were grown in either synthetic minimal medium supplemented with the appropriate nutrients, YEPD, or YEPD + 1 M sorbitol. α-Factor was used at 2 μg/ml and nocodazole at 15 μg/ml. Standard techniques were used for genetic manipulations (Sherman, 1991; Maniatis et al., 1992). Gene deletions were generated by one-step gene replacement (Wach et al., 1994). One-step tagging techniques (Janke et al., 2004; Sheff and Thorn, 2004) were used to create triple hemagglutinin (3xHA)–, 3xFlag-, and eGFP-tagged Syp1. SYP1 phosphorylation mutants were obtained by site-directed mutagenesis using the QuikChange Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). Integration of SYP1 alleles into the genome was checked by Southern blot.
Mutagenesis with ethyl methanesulfonate and isolation of RHO1-D72N allele
The temperature sensitivity of dma1Δ dma2Δ cla4-75 cells in the W303 background was unaffected by replacing the defective endogenous bud4 allele with wild-type BUD4 (Supplemental Figure S1A). For mutagenesis, a cellular suspension of dma1Δ dma2Δ cla4-75 bud4 cells was treated with 3% ethyl methanesulfonate for 60 min. Cells were plated on YEPD plates at 37°C to isolate thermoresistant suppressors. Temperature-resistant clones were assessed for the ability to lose the cla4-75 allele carried on an episomal centromeric plasmid by replica plating on 5-fluoroorotic acid (5-FOA) plates to select their derivatives lacking the cla4-75 construct. After this secondary screen, 44 suppressors that retained viability in face of the simultaneous lack of Dma1, Dma2, and Cla4 were recovered.
Dominance/recessivity of the suppressing mutations was assessed by crossing suppressors with the parental strain (dma1Δ dma2Δ cla4-75). Derivative diploids were selected and analyzed for their ability to grow at 37ºC and on 5-FOA.
To identify RHO1-D72N as a suppressing allele, we constructed a genomic library from one of the dominant suppressors by partial digestion of its genomic DNA with Sau3A followed by ligation of 8.0- to 12.0-kb DNA fragments into BamHI of Ycplac111. This genomic library was then used to transform the dma1Δ dma2Δ cla4-75 parental strain, and plasmids carrying the RHO1-D72N allele were isolated by virtue of their ability to confer temperature and 5-FOA resistance to dma1Δ dma2Δ cla4-75 cells. Suppression was confirmed after subcloning the RHO1-D72N allele in Ycplac111.
FRAP and fluorescence microscopy
FRAP experiments in Figure 3, A–C, and Supplemental Figure 6, C–F, were performed as previously described (Dobbelaere et al., 2003) on a Zeiss LSM510 (Zeiss, Jena, Germany) confocal microscope. Logarithmically growing cells expressing GFP-Cdc12 were grown overnight in YEPD, resuspended in synthetic complete medium, and spread on 2% agar pads. Half or the entire septin ring was bleached with a sequence of 20–25 iterations at 50% of laser intensity. Fluorescence recovery was recorded over time at 30 or 37°C (experiments in Figure 3B), and fluorescence intensities were analyzed with ImageJ (National Institutes of Health, Bethesda, MD). Background staining in each cell was subtracted. To correct for general bleaching, fluorescence intensities of septin rings were normalized to those of two or three reference cells present in each movie.
FRAP experiments in Figure 6B were performed on a Zeiss LSM780 confocal microscope equipped with a Definite Focus module and controlled by the Zen2010 software (Zeiss). Logarithmically growing cells expressing Syp1-eGFP were grown overnight in synthetic complete medium at 30°C and mounted on Fluorodishes (World Precision Instruments, Sarasota, FL). Half of the Syp1 ring was bleached in small-budded cells with a sequence of five iterations at 100% of laser intensity. Fluorescence recovery was recorded every 0.5 s at 30°C, and fluorescence intensities were analyzed with ImageJ. Background staining in each cell was subtracted. Data were fitted to monoexponential decay with Prism5 (GraphPad Software, San Diego, CA) to calculate the mobile fraction and the half recovery time.
For time-lapse video microscopy in Figure 3F, cells were imaged at 30°C on 2% agar pads of complete synthetic medium. Movies were recorded with a DeltaVision microscope and a 100× oil immersion objective using SoftWoRx software (GE Healthcare, Velizy-Villacoublay, France). Individual Z-stacks contained 10 planes with a step size of 0.6 μm and were binned symmetrically by two. After image acquisition, the movies were projected as maximum intensity projections.
For time-lapse video microscopy in Supplemental Figures S3 and 5C, cells were mounted in synthetic medium on Fluorodishes and filmed at room temperature (∼22°C) with a DeltaVision OMX (GE Healthcare) using a 63× oil immersion objective using SoftWoRx software. Individual Z-stacks containing 31 planes were acquired every 1 min with a step size of 0.2 μm and were binned symmetrically by one. Z-stacks were maximum projected and deconvolved with Huygens (Scientific Volume Imaging, Hilversum, Netherlands).
For time-lapse video microscopy in Figure 3H, cells were mounted in synthetic medium on Fluorodishes and filmed at 30°C with a 100× oil immersion objective mounted on a confocal spinning disk CSU-X1 Andor-Nikon microscope (Nikon, Tokyo, Japan) equipped with an electron-multiplying charge-coupled device (CCD) iXon Ultra camera and controlled by Andor iQ3 software. Individual Z-stacks contained eight planes with a step size of 0.7 μm and were acquired every 1 min. Fluorescence intensities of GFP-Cdc12 were measured with ImageJ on maximum projected images.
For time-lapse video microscopy in Figure 7A, cells were mounted in synthetic medium on Fluorodishes and filmed at 30°C with 63× oil immersion objective mounted on a Zeiss LSM780 confocal microscope controlled by the Zen2010 software. Individual Z-stacks contained eight planes with a step size of 0.7 μm and were acquired every 1 min. Fluorescence intensities of GFP-Cdc12 were measured with ImageJ on maximum projected images.
Nuclear division was scored on cells stained with propidium iodide.
Visualization of septin or Syp1-HA3 rings was performed on formaldehyde-fixed cells using anti-Cdc11 polyclonal antibodies (Santa Cruz Biotechnology, Dallas, TX) or anti-HA monoclonal antibody (12CA5) as previously described (Merlini et al., 2012). Still digital images were taken with an oil 63×, 1.4-0.6 HCX Plan-Apochromat objective (Zeiss) with a CoolSNAP HQ2-1 CCD camera (Photometrics, Tucson, AZ) mounted on a Zeiss AxioimagerZ1/Apotome fluorescence microscope controlled by the MetaMorph imaging system software (Molecular Devices, Silicon Valley, CA).
Protein purification, immunoprecipitations, and kinase assays
Purification of histidine-tagged Syp1 truncated proteins was performed as previously described (Reider et al., 2009). For use in kinase assays, proteins were dialyzed in 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, and 50 mM NaCl and concentrated using centrifugal filter units (10-kDa MWCO; Millipore, Darmstadt, Germany).
For immunoprecipitations of HA-tagged Syp1, pellets from 50-ml yeast cultures (107 cells/ml) were lysed at 4°C with acid-washed glass beads in lysis buffer (50 mM Tris-Cl, pH 7.5, NaCl 150 mM, 10% glycerol, 1 mM EDTA, and 1% NP-40, supplemented with protein inhibitors [Complete; Roche, Basel, Switzerland], 1 mM Na orthovanadate, and 60 mM β-glycerophosphate). Total extracts were cleared by spinning at 12,000 rpm for 10 min and quantified by NanoDrop (Thermo Scientific, Waltham, MA). The same amounts of protein extracts were subjected to immunoprecipitation with an anti-HA affinity resin (Roche). Immunocomplexes were washed three times in lysis buffer and twice in phosphate-buffered saline (PBS) before SDS–PAGE electrophoresis.
Immunoprecipitations of Flag-tagged Syp1 were performed in TBSN buffer (25 mM Tris-Cl, pH 7.4, 100 mM NaCl, 2 mM EDTA, 0,1% NP-40, 1 mM dithiothreitol [DTT]) supplemented with protein inhibitors (Complete) and phosphatase inhibitors (PhosStop; Roche) using protein G Dynabeads (Life Technologies, Carlsbad, CA) cross-linked to the M2 monoclonal anti-Flag antibody (Sigma-Aldrich, St. Louis, MO). Immunocomplexes were washed four times in TBSN and twice in PBS before elution with 0.5 mg/ml 3xFlag peptide in 50 mM Tris-Cl, pH 8.3, 1 mM EDTA, and 0.1% SDS.
Pkc1 immunoprecipitations and kinase assays were performed essentially as described (Watanabe et al., 1994), except that a high-salt buffer was used for cell lysis. Briefly, wild- type or kinase-dead (K853R) HA-tagged Pkc1 (Watanabe et al., 1994) was overexpressed in yeast cells from the inducible GAL1 promoter after galactose induction for 3 h at 30°C. Cells were lysed at 4°C with acid-washed glass beads in lysis buffer (50 mM Tris-Cl, pH 7.5, 1 M NaCl, 1 mM ethylene glycol tetraacetic acid, 1 mM EDTA, and 1%NP-40 supplemented with protease and phosphatase inhibitors [Complete and PhosStop]), and Pkc1 was immunoprecipitated with 12CA5 anti-HA antibodies preadsorbed on protein A–Sepharose. Immunocomplexes were washed twice with lysis buffer and once with 40 mM 3-(N-morpholino)propanesulfonic acid, pH 7.5. Kinase assays were performed as described (Watanabe et al., 1994) using 2 μg of Syp1 or myelin basic protein (MBP) per reaction.
For Western blot analysis, trichloroacetic acid protein extracts were prepared as previously described (Rancati et al., 2005). Proteins transferred to Protran membranes (Schleicher and Schuell, Dassel, Germany) were probed with monoclonal anti-HA 12CA5, anti-GFP (ChromoTek, Planegg-Martinsried, Germany), and anti-Pgk1 (Life Technologies) antibodies or with polyclonal anti-Fks1 antibodies (Santa Cruz Biotechnology). Phosphorylated Slt2 was detected using a monoclonal antibody against phospho-p44/42 MAPK (Cell Signaling, Danvers, MA), and total levels of Slt2 were monitored with anti-Slt2 polyclonal antibodies (Santa Cruz Biotechnology).
Secondary antibodies were purchased from GE Healthcare, and proteins were detected by an enhanced chemiluminescence system according to the manufacturer.
Mass spectrometry analysis
The identification of Syp1 phosphorylation sites was carried out on three independent replicates. In vitro–phosphorylated Syp1 proteins were digested overnight using 1 mg of trypsin in 2 M urea and 50 mM triethylammonium bicarbonate. Resulting peptides were diluted with four volumes of 0.2% trifluoroacetic acid, loaded on C18 Agilent Bond Elut, desalted with two washes of 0.1% formic acid, eluted with 50% acetonitrile/0.1% formic acid, and dried to completeness in a SpeedVac vacuum concentrator (Thermo Scientific).
For interactome analysis, immunoprecipitates were separated on SDS–PAGE gels (12% polyacrylamide, Mini-PROTEAN TGX Precast Gels; Bio-Rad, Hercules, CA) and stained with Page Blue Stain (Euromedex, Souffelweyersheim, France). Gel lanes were cut into six gel pieces and destained with three washes in 50% acetonitrile and 50 mM triethylammonium bicarbonate (TEABC). After protein reduction (with 10 mM DTT in 50 mM TEABC at 60°C for 30 min) and alkylation (55 mM iodoacetamide/TEABC at room temperature for 30 min), proteins were digested in-gel using trypsin (1.2 μg/band, Gold; Promega, Madison, WI) as previously described (Shevchenko et al., 1996).
Peptides were analyzed by nano-flow HPLC-nanoelectrospray ionization using an LTQ Orbitrap XL mass spectrometer coupled to an Ultimate 3000 HPLC (Thermo Scientific). Preconcentration of samples was performed on-line on a Pepmap precolumn (0.3 mm × 10 mm; Thermo Scientific). A gradient consisting of 2–40% buffer B (100% acetonitrile, 0.1% trfluoroacetic acid; 30 min for phosphorylation analysis, 60 min for interacting protein analysis), 40–80% B (1 min), and 80–0% B (1 min) and equilibrated for 20 min in 0% B was used to elute peptides at 300 nl/min from an Acclaim Pepmap100 C18 capillary (0.075 mm × 150 mm) reverse-phase column (Thermo Scientific). Mass spectra were acquired using a top-5 collision-induced- dissociation, data-dependent-acquisition method. The LTQ-Orbitrap was programmed to perform a Fourier transform (FT) full scan (60,000 resolution) on mass range 400–1400 Th, with the top five ions from each scan selected for LTQ-MS/MS and, in the case of phosphorylation analysis, multistage activation on the neutral loss of 24.49, 32.66, and 48.99 Th. FT spectra were internally calibrated using a single lock mass (445.120024 Th). Target ion numbers were 500,000 for FT full scan on the Orbitrap and 10,000 MSn on the LTQ.
For phosphorylation analysis, Raw files were converted to Mascot generic format (MGF) using raw2MSM, version 1.07, enabling to filter the top eight ions per 100-Da mass windows. MGF files were searched with the Mascot engine, version 2.4, against the SwissProt database, version 2012_07 (536,789 sequences), upon filtering for S. cerevisiae entries (7794 sequences). Modification options were fixed carbamidomethyl on cysteines and variable phosphorylation on serines/threonines and tyrosines. Mass tolerances were 7 ppm for precursor ions and 0.5 Da for fragment ions. MS2 spectra corresponding to phosphopeptides were manually inspected for phosphosite assignment. Ion signals corresponding to phosphorylated peptides were quantified from their ion chromatograms manually extracted using Qual browser, version 2.1 (Thermo Fisher Scientific), with a tolerance of 5 ppm for mass deviation and normalized to signals of their nonphosphorylated counterparts.
For interactome analysis, all MS/MS spectra were searched against the S. cerevisiae database (64,506 sequences, version 2013_03, uniprot.org/) by using Proteome Discoverer software, version 1.4 (Thermo Scientific) with Mascot, version 2.4, and phosphoRS, version 3.0 nodes (Taus et al., 2011). Modification options were fixed carbamidomethyl on cysteines, variable oxidation on methionines, and variable phosphorylation on serines/threonines and tyrosines. Management and validation of mass spectrometry data were performed using Proteome Discoverer software (Thermo Scientific; Mascot significance threshold p < 0.01, with a minimum of one peptide per protein). Site-specific localization of phosphorylation events was determined using the phosphoRS function of Proteome Discoverer. All spectra with pRS score ≥50 and pRS Site Probability ≥0.75 were considered. Furthermore, each phosphopeptide spectrum was manually assessed for quality.
Other techniques
Flow cytometric DNA quantification was performed according to Fraschini et al. (1999) on a Becton-Dickinson FACSCalibur.
Significance of the differences was statistically tested by means of a two-tailed t test, assuming unequal variances. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Supplementary Material
Acknowledgments
We are grateful to E. Bi, G. Braus, G. De Bettignies, M. Hall, T. Hoefken, B. Lapeyre, K. Lee, D. Levin, Y. Ohya, D. Pellman, M. Stark, D. Stillman, R. Tisi, and B. Wendland for sharing reagents; R. Fraschini and M. Venturetti for help with the initial phase of the genetic screen; V. Georget and J. Mateos Langerak for invaluable help with FRAP and video microscopy; the Montpellier RIO Imaging platform for image acquisition and FACS analysis; and D. Liakopoulos, S. Martin, and E. Schwob for critical reading of the manuscript. This work was supported by Grant ANR-09-BLAN-0125-01 from the Agence Nationale pour la Recherche and Grant PJA 20141201926 from the Fondation ARC to S.P. M.A.J. was supported by the Fondation pour la Recherche Médicale.
Abbreviations used:
- CCD, charge-coupled device; CWI, cell wall integrity; EDTA, ethylenediaminetetraacetic acid; e-GFP, enhanced green fluorescent protein; FACS, fluorescence-activated cell sorter; F-BAR, F-Bin–Amphiphysin–Rvs; 5-FOA, 5-fluoroorotic acid; FRAP, fluorescence recovery after photobleaching; GAP, GTPase-activating protein; GEF, guanine nucleotide exchange factor; GFP, green fluorescent protein; GTP, guanosine triphosphate; MAPK, mitogen-activated protein kinase; MAPKK, mitogen-activated protein kinase kinase; MAPKKK, mitogen-activated protein kinase kinase kinase; MBP, myelin basic protein; μHD, muniscin-homology domain; MS/MS, tandem mass spectroscopy; NP-40, Nonidet-P40; PAK kinase, p21-activated kinase; PBS, phosphate-buffered saline; PI(4,5)P2, phosphatidylinositol (4,5)-bisphosphate.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.3245) on July 15, 2015.
REFERENCES
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics. 2008;7:1389–1396. doi: 10.1074/mcp.M700468-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasia SD, Nguyen DL, Thai V, Meloy M, Macdonough T, Kellogg DR. A link between mitotic entry and membrane growth suggests a novel model for cell size control. J Cell Biol. 2012;197:89–104. doi: 10.1083/jcb.201108108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson B, Montessuit S, Friedli L, Payton MA, Paravicini G. Protein kinase C in yeast. Characteristics of the Saccharomyces cerevisiae PKC1 gene product. J Biol Chem. 1994;269:16821–16828. [PubMed] [Google Scholar]
- Badrane H, Nguyen MH, Blankenship JR, Cheng S, Hao B, Mitchell AP, Clancy CJ. Rapid redistribution of phosphatidylinositol-(4,5)-bisphosphate and septins during the Candida albicans response to caspofungin. Antimicrob Agents Chemother. 2012;56:4614–4624. doi: 10.1128/AAC.00112-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral Y. Cell biology. Septins at the nexus. Science. 2010;329:1289–1290. doi: 10.1126/science.1195445. [DOI] [PubMed] [Google Scholar]
- Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- Barral Y, Parra M, Bidlingmaier S, Snyder M. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 1999;13:176–187. doi: 10.1101/gad.13.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beise N, Trimble W. Septins at a glance. J Cell Sci. 2011;124:4141–4146. doi: 10.1242/jcs.087007. [DOI] [PubMed] [Google Scholar]
- Berlin A, Paoletti A, Chang F. Mid2p stabilizes septin rings during cytokinesis in fission yeast. J Cell Biol. 2003;160:1083–1092. doi: 10.1083/jcb.200212016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin A, McMurray MA, Thai L, Garcia G, 3rd, Votin V, Grob P, Allyn T, Thorner J, Nogales E. Phosphatidylinositol-4,5-bisphosphate promotes budding yeast septin filament assembly and organization. J Mol Biol. 2010;404:711–731. doi: 10.1016/j.jmb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner DR, D’Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol. 2009;19:1979–1987. doi: 10.1016/j.cub.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. A highly ordered ring of membrane-associated filaments in budding yeast. J Cell Biol. 1976;69:717–721. doi: 10.1083/jcb.69.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamayor A, Snyder M. Molecular dissection of a yeast septin: distinct domains are required for septin interaction, localization, and function. Mol Cell Biol. 2003;23:2762–2777. doi: 10.1128/MCB.23.8.2762-2777.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Caviston JP, Longtine M, Pringle JR, Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell. 2003;14:4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cid VJ, Adamikova L, Sanchez M, Molina M, Nombela C. Cell cycle control of septin ring dynamics in the budding yeast. Microbiology. 2001;147:1437–1450. doi: 10.1099/00221287-147-6-1437. [DOI] [PubMed] [Google Scholar]
- deHart AK, Schnell JD, Allen DA, Tsai JY, Hicke L. Receptor internalization in yeast requires the Tor2-Rho1 signaling pathway. Mol Biol Cell. 2003;14:4676–4684. doi: 10.1091/mbc.E03-05-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarini DJ, Adams AE, Fares H, De Virgilio C, Valle G, Chuang JS, Pringle JR. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J Cell Biol. 1997;139:75–93. doi: 10.1083/jcb.139.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis V, Cyert MS. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot Cell. 2005;4:36–45. doi: 10.1128/EC.4.1.36-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmalingam E, Haeckel A, Pinyol R, Schwintzer L, Koch D, Kessels MM, Qualmann B. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci. 2009;29:13315–13327. doi: 10.1523/JNEUROSCI.3973-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Gentry MS, Hallberg RL, Barral Y. Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell. 2003;4:345–357. doi: 10.1016/s1534-5807(03)00061-3. [DOI] [PubMed] [Google Scholar]
- Dong Y, Pruyne D, Bretscher A. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J Cell Biol. 2003;161:1081–1092. doi: 10.1083/jcb.200212040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li W, el-Sherbeini M, et al. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-beta-D-glucan synthase. Proc Natl Acad Sci USA. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S, Hurd TW, Liu CJ, Straight SW, Weimbs T, Hurd EA, Domino SE, Margolis B. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr Biol. 2004;14:1451–1461. doi: 10.1016/j.cub.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Fares H, Peifer M, Pringle JR. Localization and possible functions of Drosophila septins. Mol Biol Cell. 1995;6:1843–1859. doi: 10.1091/mbc.6.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Bilotta D, Lucchini G, Piatti S. Functional characterization of Dma1 and Dma2, the budding yeast homologues of Schizosaccharomyces pombe Dma1 and human Chfr. Mol Biol Cell. 2004;15:3796–3810. doi: 10.1091/mbc.E04-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Formenti E, Lucchini G, Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrenton LS, Stefan CJ, McMurray MA, Emr SD, Thorner J. Pheromone-induced anisotropy in yeast plasma membrane phosphatidylinositol-4,5-bisphosphate distribution is required for MAPK signaling. Proc Natl Acad Sci USA. 2010;107:11805–11810. doi: 10.1073/pnas.1005817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Bose I, Zyla TR, Bardes ES, Lew DJ. Septin ring assembly involves cycles of GTP loading and hydrolysis by Cdc42p. J Cell Biol. 2002;156:315–326. doi: 10.1083/jcb.200109062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Novo A, Correa-Bordes J, Labrador L, Sanchez M, Vazquez de Aldana CR, Jimenez J. Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol Biol Cell. 2008;19:1509–1518. doi: 10.1091/mbc.E07-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Guo W, Tamanoi F, Novick P. Spatial regulation of the exocyst complex by Rho1 GTPase. Nat Cell Biol. 2001;3:353–360. doi: 10.1038/35070029. [DOI] [PubMed] [Google Scholar]
- Haarer BK, Pringle JR. Immunofluorescence localization of the Saccharomyces cerevisiae CDC12 gene product to the vicinity of the 10-nm filaments in the mother-bud neck. Mol Cell Biol. 1987;7:3678–3687. doi: 10.1128/mcb.7.10.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall PA, Russell SE. Mammalian septins: dynamic heteromers with roles in cellular morphogenesis and compartmentalization. J Pathol. 2012;226:287–299. doi: 10.1002/path.3024. [DOI] [PubMed] [Google Scholar]
- Helfer H, Gladfelter AS. AgSwe1p regulates mitosis in response to morphogenesis and nutrients in multinucleated Ashbya gossypii cells. Mol Biol Cell. 2006;17:4494–4512. doi: 10.1091/mbc.E06-03-0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Rodriguez Y, Hastings S, Momany M. The septin AspB in Aspergillus nidulans forms bars and filaments and plays roles in growth emergence and conidiation. Eukaryot Cell. 2012;11:311–323. doi: 10.1128/EC.05164-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Milenkovic L, Jin H, Scott MP, Nachury MV, Spiliotis ET, Nelson WJ. A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science. 2010;329:436–439. doi: 10.1126/science.1191054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue SB, Qadota H, Arisawa M, Anraku Y, Watanabe T, Ohya Y. Signaling toward yeast 1,3-beta-glucan synthesis. Cell Struct Funct. 1996;21:395–402. doi: 10.1247/csf.21.395. [DOI] [PubMed] [Google Scholar]
- Irie K, Takase M, Lee KS, Levin DE, Araki H, Matsumoto K, Oshima Y. MKK1 and MKK2, which encode Saccharomyces cerevisiae mitogen-activated protein kinase-kinase homologs, function in the pathway mediated by protein kinase C. Mol Cell Biol. 1993;13:3076–3083. doi: 10.1128/mcb.13.5.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M, Luo J, Nagaraj S, Longtine M, Kim HB, Haarer BK, Caruso C, Tong Z, Pringle JR, Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol Biol Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jaquenoud M, Peter M. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p) Mol Cell Biol. 2000;20:6244–6258. doi: 10.1128/mcb.20.17.6244-6258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G, Perlungher RR, Sheffield PJ, Kinoshita M, Noda M, Haystead T, Macara IG. Borg proteins control septin organization and are negatively regulated by Cdc42. Nat Cell Biol. 2001;3:861–866. doi: 10.1038/ncb1001-861. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- Jose M, Tollis S, Nair D, Sibarita JB, McCusker D. Robust polarity establishment occurs via an endocytosis-based cortical corralling mechanism. J Cell Biol. 2013;200:407–418. doi: 10.1083/jcb.201206081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota J, Yamamoto T, Yoshiuchi S, Bi E, Tanaka K. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:5329–5345. doi: 10.1091/mbc.E04-03-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y, Qadota H, Python CP, Anraku Y, Ohya Y, Levin DE. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- Kang PJ, Hood-DeGrenier JK, Park HO. Coupling of septins to the axial landmark by Bud4 in budding yeast. J Cell Sci. 2013;126:1218–1226. doi: 10.1242/jcs.118521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanshin E, Bergeron-Sandoval LP, Isik SS, Thibault P, Michnick SW. A cell-signaling network temporally resolves specific versus promiscuous phosphorylation. Cell Rep. 2015;10:1202–1214. doi: 10.1016/j.celrep.2015.01.052. [DOI] [PubMed] [Google Scholar]
- Kim HB, Haarer BK, Pringle JR. Cellular morphogenesis in the Saccharomyces cerevisiae cell cycle: localization of the CDC3 gene product and the timing of events at the budding site. J Cell Biol. 1991;112:535–544. doi: 10.1083/jcb.112.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Shindo A, Park TJ, Oh EC, Ghosh S, Gray RS, Lewis RA, Johnson CA, Attie-Bittach T, Katsanis N, et al. Planar cell polarity acts through septins to control collective cell movement and ciliogenesis. Science. 2010;329:1337–1340. doi: 10.1126/science.1191184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Kumar S, Mizoguchi A, Ide C, Kinoshita A, Haraguchi T, Hiraoka Y, Noda M. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997;11:1535–1547. doi: 10.1101/gad.11.12.1535. [DOI] [PubMed] [Google Scholar]
- Kissel H, Georgescu MM, Larisch S, Manova K, Hunnicutt GR, Steller H. The Sept4 septin locus is required for sperm terminal differentiation in mice. Dev Cell. 2005;8:353–364. doi: 10.1016/j.devcel.2005.01.021. [DOI] [PubMed] [Google Scholar]
- Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 2012;150:151–164. doi: 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Kozubowski L, Panek H, Rosenthal A, Bloecher A, DeMarini DJ, Tatchell K. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol Biol Cell. 2003;14:26–39. doi: 10.1091/mbc.E02-06-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo YC, Lin YH, Chen HI, Wang YY, Chiou YW, Lin HH, Pan HA, Wu CM, Su SM, Hsu CC, et al. SEPT12 mutations cause male infertility with defective sperm annulus. Hum Mutat. 2012;33:710–719. doi: 10.1002/humu.22028. [DOI] [PubMed] [Google Scholar]
- Lam C, Santore E, Lavoie E, Needleman L, Fiacco N, Kim C, Neiman AM. A visual screen of protein localization during sporulation identifies new components of prospore membrane-associated complexes in budding yeast. Eukaryot Cell. 2014;13:383–391. doi: 10.1128/EC.00333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PR, Song S, Ro HS, Park CJ, Lippincott J, Li R, Pringle JR, De Virgilio C, Longtine MS, Lee KS. Bni5p, a septin-interacting protein, is required for normal septin function and cytokinesis in Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6906–6920. doi: 10.1128/MCB.22.19.6906-6920.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics. 2011;189:1145–1175. doi: 10.1534/genetics.111.128264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE, Bowers B, Chen CY, Kamada Y, Watanabe M. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell Mol Biol Res. 1994;40:229–239. [PubMed] [Google Scholar]
- Li X, Gerber SA, Rudner AD, Beausoleil SA, Haas W, Villen J, Elias JE, Gygi SP. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- Lippincott J, Li R. Dual function of Cyk2, a cdc15/PSTPIP family protein, in regulating actomyosin ring dynamics and septin distribution. J Cell Biol. 1998;143:1947–1960. doi: 10.1083/jcb.143.7.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, Fares H, Pringle JR. Role of the yeast Gin4p protein kinase in septin assembly and the relationship between septin assembly and septin function. J Cell Biol. 1998;143:719–736. doi: 10.1083/jcb.143.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- Marquitz AR, Harrison JC, Bose I, Zyla TR, McMillan JN, Lew DJ. The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J. 2002;21:4012–4025. doi: 10.1093/emboj/cdf416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Bertin A, Garcia G, 3rd, Lam L, Nogales E, Thorner J. Septin filament formation is essential in budding yeast. Dev Cell. 2011;20:540–549. doi: 10.1016/j.devcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray MA, Thorner J. Septin stability and recycling during dynamic structural transitions in cell division and development. Curr Biol. 2008;18:1203–1208. doi: 10.1016/j.cub.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev. 2011;25:875–888. doi: 10.1101/gad.622411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Fraschini R, Boettcher B, Barral Y, Lucchini G, Piatti S. Budding yeast dma proteins control septin dynamics and the spindle position checkpoint by promoting the recruitment of the Elm1 kinase to the bud neck. PLoS Genet. 2012;8:e1002670. doi: 10.1371/journal.pgen.1002670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Kohler T, Braus GH. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:235–248. doi: 10.1128/MCB.21.1.235-248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13:183–194. doi: 10.1038/nrm3284. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, Rubin GM. The Drosophila peanut gene is required for cytokinesis and encodes a protein similar to yeast putative bud neck filament proteins. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- Nguyen TQ, Sawa H, Okano H, White JG. The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J Cell Sci. 2000;113:3825–3837. doi: 10.1242/jcs.113.21.3825. [DOI] [PubMed] [Google Scholar]
- Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y, Bi E. Septin structure and function in yeast and beyond. Trends Cell Biol. 2010;21:141–148. doi: 10.1016/j.tcb.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Abe M, Asakawa-Minemura M, Hirata A, Qadota H, Morishita K, Ohnuki S, Nogami S, Ohya Y. Multiple functional domains of the yeast l,3-beta-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics. 2010;184:1013–1024. doi: 10.1534/genetics.109.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Leda M, Hanna J, Savage NS, Bi E, Goryachev AB. Daughter cell identity emerges from the interplay of cdc42, septins, and exocytosis. Dev Cell. 2013;26:148–161. doi: 10.1016/j.devcel.2013.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson EA, Petty EM. Conquering the complex world of human septins: implications for health and disease. Clin Genet. 2010;77:511–524. doi: 10.1111/j.1399-0004.2010.01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Plomann M, Lange R, Vopper G, Cremer H, Heinlein UA, Scheff S, Baldwin SA, Leitges M, Cramer M, Paulsson M, et al. PACSIN, a brain protein that is upregulated upon differentiation into neuronal cells. Eur J Biochem. 1998;256:201–211. doi: 10.1046/j.1432-1327.1998.2560201.x. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser DC, Drivas TG, Maldonado-Baez L, Wendland B. Existence of a novel clathrin-independent endocytic pathway in yeast that depends on Rho1 and formin. J Cell Biol. 2011;195:657–671. doi: 10.1083/jcb.201104045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruliere G, Cosson J, Chevalier S, Sardet C, Chenevert J. Atypical protein kinase C controls sea urchin ciliogenesis. Mol Biol Cell. 2011;22:2042–2053. doi: 10.1091/mbc.E10-10-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Anraku Y, Botstein D, Ohya Y. Conditional lethality of a yeast strain expressing human RHOA in place of RHO1. Proc Natl Acad Sci USA. 1994;91:9317–9321. doi: 10.1073/pnas.91.20.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, Watanabe T, Levin DE, Ohya Y. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 1996;272:279–281. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- Qiu W, Neo SP, Yu X, Cai M. A novel septin-associated protein, Syp1p, is required for normal cell cycle-dependent septin cytoskeleton dynamics in yeast. Genetics. 2008;180:1445–1457. doi: 10.1534/genetics.108.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G, Crispo V, Lucchini G, Piatti S. Mad3/BubR1 phosphorylation during spindle checkpoint activation depends on both Polo and Aurora kinases in budding yeast. Cell Cycle. 2005;4:972–980. doi: 10.4161/cc.4.7.1829. [DOI] [PubMed] [Google Scholar]
- Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Baez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarikangas J, Zhao H, Pykalainen A, Laurinmaki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- Schmidt K, Nichols BJ. Functional interdependence between septin and actin cytoskeleton. BMC Cell Biol. 2004;5:43. doi: 10.1186/1471-2121-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5:1337–1346. doi: 10.1039/b902256b. [DOI] [PubMed] [Google Scholar]
- Srivastava V, Robinson DN. Mechanical stress and network structure drive protein dynamics during cytokinesis. Curr Biol. 2015;25:663–670. doi: 10.1016/j.cub.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steels JD, Estey MP, Froese CD, Reynaud D, Pace-Asciak C, Trimble WS. Sept12 is a component of the mammalian sperm tail annulus. Cell Motil Cytoskeleton. 2007;64:794–807. doi: 10.1002/cm.20224. [DOI] [PubMed] [Google Scholar]
- Stimpson HE, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell. 2009;20:4640–4651. doi: 10.1091/mbc.E09-05-0429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol. 2010;21:340–349. doi: 10.1016/j.semcdb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Tada T, Simonetta A, Batterton M, Kinoshita M, Edbauer D, Sheng M. Role of septin cytoskeleton in spine morphogenesis and dendrite development in neurons. Curr Biol. 2007;17:1752–1758. doi: 10.1016/j.cub.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda T, Robinson IM, Savoian MM, Griffiths JR, Whetton AD, McMahon HT, Glover DM. Drosophila F-BAR protein Syndapin contributes to coupling the plasma membrane and contractile ring in cytokinesis. Open Biol. 2013;3:130081. doi: 10.1098/rsob.130081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarassov K, Messier V, Landry CR, Radinovic S, Serna Molina MM, Shames I, Malitskaya Y, Vogel J, Bussey H, Michnick SW. An in vivo map of the yeast protein interactome. Science. 2008;320:1465–1470. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- Tasto JJ, Morrell JL, Gould KL. An anillin homologue, Mid2p, acts during fission yeast cytokinesis to organize the septin ring and promote cell separation. J Cell Biol. 2003;160:1093–1103. doi: 10.1083/jcb.200211126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T, Kocher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Tooley AJ, Gilden J, Jacobelli J, Beemiller P, Trimble WS, Kinoshita M, Krummel MF. Amoeboid T lymphocytes require the septin cytoskeleton for cortical integrity and persistent motility. Nat Cell Biol. 2009;11:17–26. doi: 10.1038/ncb1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Voth WP, Olsen AE, Sbia M, Freedman KH, Stillman DJ. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot Cell. 2005;4:1018–1028. doi: 10.1128/EC.4.6.1018-1028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Chen CY, Levin DE. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J Biol Chem. 1994;269:16829–16836. [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Barral Y. The septin family of GTPases: architecture and dynamics. Nat Rev Mol Cell Biol. 2008;9:478–489. doi: 10.1038/nrm2407. [DOI] [PubMed] [Google Scholar]
- Westfall PJ, Momany M. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol Biol Cell. 2002;13:110–118. doi: 10.1091/mbc.01-06-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wloka C, Nishihama R, Onishi M, Oh Y, Hanna J, Pringle JR, Krauss M, Bi E. Evidence that a septin diffusion barrier is dispensable for cytokinesis in budding yeast. Biol Chem. 2011;392:813–829. doi: 10.1515/BC.2011.083. [DOI] [PubMed] [Google Scholar]
- Xie Y, Vessey JP, Konecna A, Dahm R, Macchi P, Kiebler MA. The GTP-binding protein Septin 7 is critical for dendrite branching and dendritic-spine morphology. Curr Biol. 2007;17:1746–1751. doi: 10.1016/j.cub.2007.08.042. [DOI] [PubMed] [Google Scholar]
- Xue J, Wang X, Malladi CS, Kinoshita M, Milburn PJ, Lengyel I, Rostas JA, Robinson PJ. Phosphorylation of a new brain-specific septin, G-septin, by cGMP-dependent protein kinase. J Biol Chem. 2000;275:10047–10056. doi: 10.1074/jbc.275.14.10047. [DOI] [PubMed] [Google Scholar]
- Yamochi W, Tanaka K, Nonaka H, Maeda A, Musha T, Takai Y. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J Cell Biol. 1994;125:1077–1093. doi: 10.1083/jcb.125.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Bartolini S, Pellman D. Mechanisms for concentrating Rho1 during cytokinesis. Genes Dev. 2009;23:810–823. doi: 10.1101/gad.1785209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Kono K, Lowery DM, Bartolini S, Yaffe MB, Ohya Y, Pellman D. Polo-like kinase Cdc5 controls the local activation of Rho1 to promote cytokinesis. Science. 2006;313:108–111. doi: 10.1126/science.1126747. [DOI] [PubMed] [Google Scholar]
- Zarzov P, Mazzoni C, Mann C. The SLT2(MPK1) MAP kinase is activated during periods of polarized cell growth in yeast. EMBO J. 1996;15:83–91. [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kong C, Xie H, McPherson PS, Grinstein S, Trimble WS. Phosphatidylinositol polyphosphate binding to the mammalian septin H5 is modulated by GTP. Curr Biol. 1999;9:1458–1467. doi: 10.1016/s0960-9822(00)80115-3. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013;4:1213–1223. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.