This study examines whether active transport to the inner nuclear membrane, as shown for yeast membrane proteins Heh1 and Heh2, is conserved in metazoans. In support of this, the nuclear localization signal of metazoan Pom121 shares biochemical, structural, and functional properties with those of Heh1 and Heh2, and a Heh2-derived reporter protein targets to the inner membrane in Hek293T cells.
Abstract
Endoplasmic reticulum–synthesized membrane proteins traffic through the nuclear pore complex (NPC) en route to the inner nuclear membrane (INM). Although many membrane proteins pass the NPC by simple diffusion, two yeast proteins, ScSrc1/ScHeh1 and ScHeh2, are actively imported. In these proteins, a nuclear localization signal (NLS) and an intrinsically disordered linker encode the sorting signal for recruiting the transport factors for FG-Nup and RanGTP-dependent transport through the NPC. Here we address whether a similar import mechanism applies in metazoans. We show that the (putative) NLSs of metazoan HsSun2, MmLem2, HsLBR, and HsLap2β are not sufficient to drive nuclear accumulation of a membrane protein in yeast, but the NLS from RnPom121 is. This NLS of Pom121 adapts a similar fold as the NLS of Heh2 when transport factor bound and rescues the subcellular localization and synthetic sickness of Heh2ΔNLS mutants. Consistent with the conservation of these NLSs, the NLS and linker of Heh2 support INM localization in HEK293T cells. The conserved features of the NLSs of ScHeh1, ScHeh2, and RnPom121 and the effective sorting of Heh2-derived reporters in human cells suggest that active import is conserved but confined to a small subset of INM proteins.
INTRODUCTION
The gateway between the cytoplasm and the nuclear interior is formed by the nuclear envelope (NE)–embedded nuclear pore complex (NPC) through which bidirectional transport between the two compartments occurs. Small solutes and proteins readily diffuse through the NPC, whereas diffusion of larger macromolecular complexes is slow or prevented. The presence of a nuclear localization signal (NLS) or nuclear export signal recognized by transport receptors permits transport using an energy-dependent mechanism (Chook and Suel, 2011) and also allows the passage of larger molecules. Specific interactions of these transport receptors with phenylalanine-glycine repeats of nucleoporins (Nups) filling the central channel of the NPC mediate the transport of soluble cargoes (Fiserova et al., 2010; Peleg and Lim, 2010; Yang, 2013), and receptor–cargo binding and release are dictated by a gradient of RanGTP across the NPC (Kalab et al., 2002; Fried and Kutay, 2003; Madrid and Weis, 2006; Cook et al., 2007).
In postmitotic cells and cells undergoing a closed mitosis, inner nuclear membrane (INM)–localized integral membrane proteins also have to find their way through the NPC. Upon synthesis, polytopic membrane proteins are first incorporated into the endoplasmic reticulum (ER) membrane and travel via the interconnected outer nuclear membrane (ONM) and pore membrane (which lines the NPC) to the INM. The INM contains a unique set of integral membrane proteins (Schirmer et al., 2003). In contrast to the extensively studied energy-dependent import mechanisms for soluble proteins, little is known about active transport of INM membrane proteins.
At first, accumulation of INM proteins in metazoans was explained by diffusion and retention (Powell and Burke, 1990; Ellenberg et al., 1997). Here membrane proteins diffuse between the ONM and the INM, while their extralumenal domain slides through the lateral channel of the NPC, and are retained (and accumulated) at the INM by interaction with chromatin or nuclear proteins, most notably lamins (Soullam and Worman, 1993; Furukawa et al., 1998; Lin et al., 2005). This mode of import is limited to membrane proteins with an extralumenal domain small enough to fit the lateral channel, which has a width of ∼10 nm (Hinshaw et al., 1992; Ohba et al., 2004; Bui et al., 2013). Later it was shown that metabolic energy is required for the translocation of a group of integral membrane proteins to the INM (Ohba et al., 2004). The dynamics of 15 NE transmembrane proteins in the NE-ER network showed that ATP- and Ran-dependent translocation mechanisms are distinct and not used by all inner nuclear membrane proteins (Zuleger et al., 2011).
In Saccharomyces cerevisiae, Heh2 and Src1/Heh1 are actively transported to the INM by karyopherins 60 and 95 (Kap60/Kap95), the yeast homologues of importin α and importin β, respectively (King et al., 2006). The NLSs of Heh1 and Heh2 are separated from the transmembrane domain by a region that is intrinsically disordered (ID; Meinema et al., 2011). This ID linker region and an NLS are required and sufficient to accumulate a membrane protein at the INM in the absence of retention, in that a reporter protein consisting of just these domains is mobile in the INM (Meinema et al., 2011).
Importin α consists of a tandem array of 10 Armadillo (ARM) repeats that together form a superhelical structure with a major NLS binding pocket between ARM repeats 2–4 and a minor NLS binding pocket between ARM repeats 7 and 8 (Conti et al., 1998). In both binding sites, five contact points with NLSs are identified, P1–P5 at the major binding site and P1′–P5′ at the minor binding pocket (Conti and Kuriyan, 2000; Fontes et al., 2003; Giesecke and Stewart, 2010; Marfori et al., 2012; Roman et al., 2013). An N-terminal autoinhibitory domain of importin α, called the importin β–binding domain (IBB domain; reviewed in Lott and Cingolani, 2011), and the NLSs compete for the same binding pocket on importin α. Once importin β binds to the IBB domain, the binding sites on importin α are exposed and able to interact with an NLS (Gorlich et al., 1996; Moroianu et al., 1996; Lott et al., 2010). The NLS of Heh2 (residues 102–131) is distinctive from other NLSs in that it binds to full-length importin α in the absence of importin β (King et al., 2006). Further, the Heh1 and Heh2 NLS binds importin α in the absence of importin β, and both adopt an IBB-like fold while bound to Kap60 (Lokareddy et al., 2015).
NLS sequences are predicted or described in many metazoan INM proteins, but whether a similar metabolic energy– and Ran and Kap-dependent import mechanism as described in yeast applies is not known. Indeed, these (potential) importin α/β interacting sequences could also act in nuclear envelope reformation, in which importin α and importin β act in mitotic spindle formation and NE assembly. Here the Ran guanine exchange factor regulator of chromosome condensation (RCC1) generates GTP-bound Ran GTPase (RanGTP) and is chromatin associated, resulting in high concentrations of RanGTP locally around chromosomes (Kalab et al., 2002; Zhang et al., 2002a; Hutchins et al., 2009). RanGTP induces the dissociation of the importin α/β from NLS-containing proteins that function as spindle assembly factors (e.g., NUMA, TPX2, and the kinesin XCTK2), which are inactive when bound to importin α (Gruss et al., 2001; Schatz et al., 2003; Ems-McClung et al., 2004). How importin β contributes to the reformation of the NE is not completely elucidated, but in vitro studies with cell-free systems made from Xenopus laevis egg extracts show that it might be involved in recruitment of FxFG-containing Nups (Zhang et al., 2002b) and lamin-B receptor (LBR)-containing membrane vesicles to the decondensing chromatin (Ma et al., 2007). Other integral membrane proteins contribute to the reformation of the NE as well, via direct interaction with DNA (Ulbert et al., 2006), or by recruitment mediated by importin α/β.
Here we studied five INM proteins that contain putative NLSs and ID linker domains: Sun2, Lem2, LBR, Lap2β, and Pom121. We tested the ability of these putative NLSs to target a membrane protein to the inner membrane in yeast and found that only the NLS of Pom121 does. Structural, biochemical, and in vivo studies reveal that the first two boxes of positive residues (residues 290–320) in the NLS region of Pom121 (residues 290–484) share with Heh1 and Heh2 an IBB-like fold when bound to importin α. Pom121NLS290-326 is able to restore the INM localization of Heh2Δh2NLS and rescues cellular sickness of Heh2Δh2NLS in a strain lacking Nup84 as well. Consistent with the evolutionary conservation of these NLSs, we show that the NLS of Heh2 supports INM localization of a membrane protein in HEK293T cells. This suggests that active import of INM proteins may be conserved but confined to a small subset of the inner nuclear membrane proteins.
RESULTS
Pom121NLS mediates INM import of a membrane reporter protein in yeast
In S. cerevisiae, it was shown for the membrane protein Heh2 that import to the INM depends on the presence of an NLS (King et al., 2006) that adopts an IBB-like fold (Lokareddy et al., 2015) and on an intrinsically disordered linker region (L) that creates distance between the NLS and the transmembrane (TM) segment (Meinema et al., 2011); this domain composition is collectively called NLS-L-TM. Several metazoan INM proteins with described or putative NLSs also encode regions that are predicted to be intrinsically disordered, based on predictions from FoldIndex (Prilusky et al., 2005). We found a putative NLS-L-TM signature in Homo sapiens Sun2, LBR, and Lap2β and Mus musculus Lem2, and we studied whether the NLSs in these domains are sufficient to support active INM import in baker’s yeast. The choice of baker’s yeast is related to its closed mitosis, meaning that the NE stays intact during anaphase, and in this way the sole route to the INM is via the NPCs. In metazoan systems, it is more difficult to distinguish INM import via the NPC from other (NLS-mediated) targeting mechanisms that may occur when the NE is disintegrated.
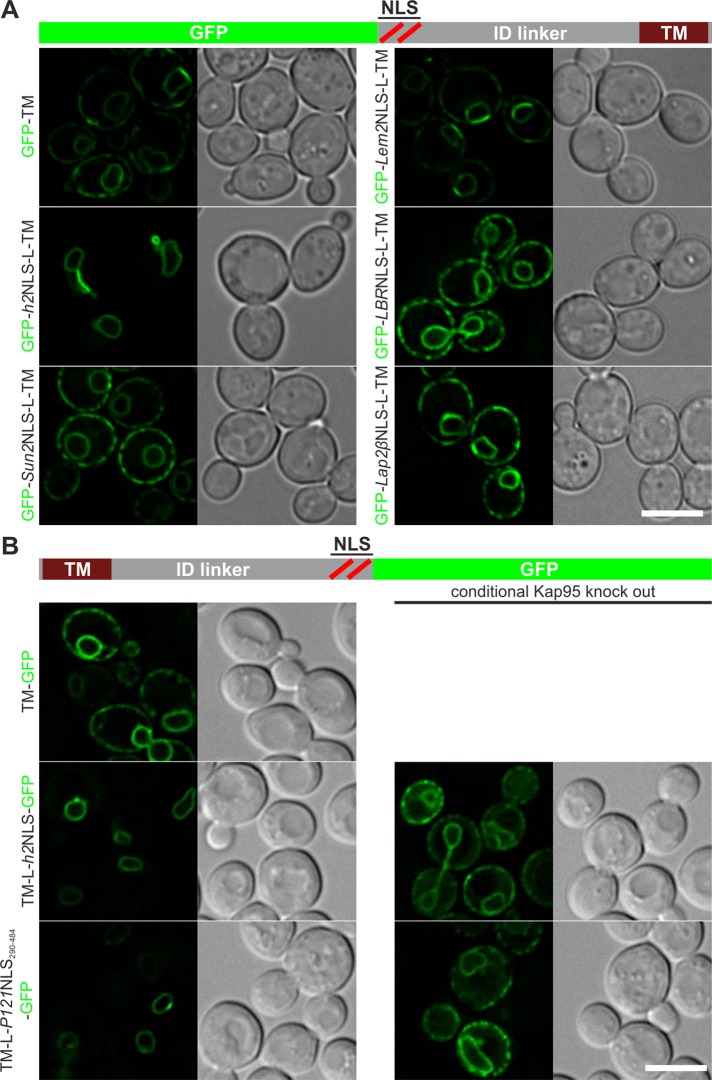
We constructed reporter proteins in which the mammalian NLSs were fused to green fluorescent protein (GFP) and to the ID linker region and first TM segment of Heh2 (Figure 1A, GFP-NLS-L-TM). The localization of the membrane reporter proteins was visualized with fluorescence microscopy, and all four reporters with NLSs derived from Sun2, Lem2, LBR, and Lap2β localized to the NE-ER network as the TM segment alone does (Figure 1A), whereas the same reporter with the Heh2 NLS (h2NLS) localized to the nuclear envelope. We also fused the complete predicted NLS-ID linker region of Sun2, Lem2, LBR, and Lap2β to the TM segment of Heh2, but none of these regions was able to mediate INM import in yeast (unpublished data). This does not negate a contribution of the NLSs to targeting in vivo. Instead, it shows that these mammalian NLSs are not a functional equivalent of the h2NLS, as they are insufficient to support active INM import, leading to a steady-state accumulation of the membrane protein at the INM.
FIGURE 1:
Localization of membrane-embedded reporter proteins encoding mammalian NLS sequences. (A) N-terminal GFP fusions of (putative) NLS regions of Sun2, Lem2, LBR, or Lap2β, the intrinsically disordered linker region (L), and first transmembrane segment (TM) of Heh2 localize to the NE-ER network, like the TM of Heh2 (GFP-TM), and do not mediate nuclear accumulation like the NLS of Heh2 (GFP-h2NLS-L-TM). (B) Whereas a C-terminally GFP fusion of a TM localizes to the NE-ER network (TM-GFP), fusion of this TM to the ID linker region of Heh2 and the NLS regions of Heh2 or Pom121 (TM-L-h2NLS-GFP and TM-L-P121NLS290-484-GFP, respectively) results in strong NE accumulation of the protein. This accumulation is lost in the conditional Kap95 knockout. Scale bars, 5 μm.
Another candidate with an established NLS region that we tested was Pom121 (Doucet et al., 2010; Yavuz et al., 2010; Funakoshi et al., 2011). Pom121 is a single-pass membrane protein with a small lumenal domain at the N-terminus and a large C-terminal extralumenal domain. The NLS region of Pom121, containing four or five boxes of basic residues, is highly conserved among species and encodes at least two bipartite NLSs (Yavuz et al., 2010; Funakoshi et al., 2011). The region between the TM domain and the NLS region is predicted to be at least in part disordered but also includes a more hydrophobic domain involved in targeting (Funakoshi et al., 2011). We did not study this region but instead focused on the NLS. In Pom121, the topology is predicted as TM-L-NLS (Soderqvist and Hallberg, 1994), rather than NLS-L-TM as in Heh1 and Heh2. It was unknown whether reporters with this topology could support import at all, and this was tested first. We show that whereas a control reporter (TM-GFP) localized to the NE-ER network, a reporter with the ID linker and NLS of Heh2 (TM-L-h2NLS-GFP) accumulated at the INM (Figure 1B). To prove that this localization was Kap95 dependent, we studied its localization in a conditional Kap95-knockout strain. In this strain, Kap95 was tagged with FKBP12-rapamycin binding domain (FRB), which forms a stable heterodimer with Pma1-fused FK506-binding protein (FKBP) in the presence of rapamycin, resulting in trapping of Kap95 at the plasma membrane (Haruki et al., 2008) and abolishing Kap60/Kap95-mediated transport over the NPC (Meinema et al., 2011). Indeed, INM localization of TM-L-h2NLS-GFP depends on the presence of Kap95, as addition of rapamycin resulted in the appearance of ER- localized TM-L-h2NLS-GFP (Figure 1B). Consistent with previous findings that the ID linker length between the NLS and the transmembrane segment scales with the level of accumulation at the INM (Meinema et al., 2011), we observed that accumulation is abolished when the length of the ID linker region in TM-L-h2NLS-GFP is reduced to 53 residues (Supplemental Figure S1A; TM-L53-h2NLS-GFP). We conclude that reporters with N- or C-terminal h2NLS and ID linker sorting signals are imported to the INM of yeast in a Kap60/Kap95-dependent manner.
Next, to investigate whether the NLS of Rattus norvegicus (Rn) Pom121 supported INM import, we replaced the NLS of Heh2 for the NLS region of RnPom121 (residues 290–484), including multiple potential NLSs (Figure 1B; TM-L-Pom121NLS290-484-GFP), and determined its subcellular localization. Clearly, the presence of this NLS region also resulted in Kap95-mediated INM localization of the TM reporter protein. Of the mammalian NLSs tested so far, only the NLS of Pom121 was able to mediate Kap60/Kap95-dependent INM targeting of a membrane protein in yeast.
Pom121NLS resembles the membrane protein NLS of Heh2
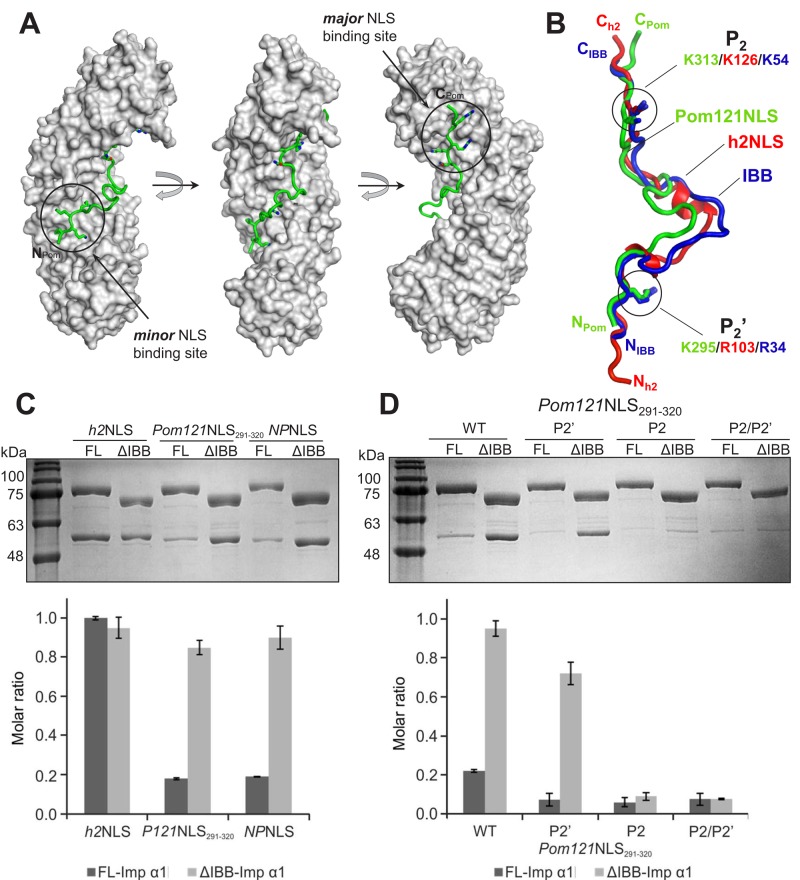
To visualize the interaction of RnPom121NLS with importin α1, we focused on the NLS regions encoded by residues 291–320. We coexpressed in bacteria plasmids encoding ΔIBB-importin α1 and GST-tagged Pom121NLS (residues 291–320), and captured a stoichiometric complex of the two proteins on glutathione beads. Coexpression prevented proteolytic degradation of Pom121NLS291-320 and was instrumental in purifying milligram quantity of homogeneous complex that we used to grow high-quality crystals. The structure of ΔIBB-importin α1 bound to Pom121NLS291-320 was solved by molecular replacement and refined to an Rwork/Rfree of 18.2/20.9% at 1.8-Å resolution (Supplemental Table S1). Pom121NLS291-320 adopts an S-shaped conformation that binds the concave surface of importin α1 Arm core, burying 2960 Å2 of solvent-accessible surface area (Figure 2A). Twenty-two Pom121NLS291-320 residues make close contact with as many as 59 residues in the Arm core, which are mainly clustered at the major (Arm 2–4) and minor (Arm 7 and 8) NLS-binding pockets and with sporadic interactions between Arm 5 and 6. Of note, Pom121NLS’s first basic box makes four strong contacts at the minor NLS pocket, where the basic stretch 294-KKKR-297 occupies positions P1′–P4′, as seen for membrane protein NLS of Heh1 (174-KKRK-177) and Heh2 (102-KRKR-105; Lokareddy et al., 2015). Similarly, at the major NLS-binding pocket, Pom121NLS291-320 inserts four basic side chains at sites P2–P5 (313-KRRR-316) and makes additional contacts at P1 and P6 with N312 and H317, respectively. The average B factor of Pom121NLS291-320 boxes interacting at minor (39.8 Å2) and major (19.7 Å2) NLS-binding pockets is comparable to that of importin α Arm core (28.6 Å2), consistent with the high avidity for this bipartite NLS for importin α. In contrast, the 14-residue spacer between NLS boxes (298-TVAEEDQLHLDGQE-311) has significantly weaker electron density (visible continuously only by blurring the B factor) and adopts a random coiled conformation. Overall Pom121NLS291-320 binding to importin α Arm core is stabilized by 45 hydrogen bonds, seven salt bridges, and a handful of hydrophobic and cation II (Koerner et al., 2003) contacts involving importin α–conserved tryptophans.
FIGURE 2:
Structural and biochemical analysis of Pom121NLS bound to importin α1. (A) Crystal structure of ∆IBB-importin α1 (gray surface) in complex with Pom121NLS291-320 (green) shown in three views rotated clockwise by 90°. (B) Superimposition of ΔIBB-importin α1 bound to Pom121NLS291-320 with ΔIBB-Kap60 bound to the h2NLS (Lokareddy et al., 2015) or as FL-Kap60 (Protein Data Bank ID 1WA5). Pom121NLS291-320 and h2NLS are green and red, respectively, and the IBB domain is blue. For clarity, importin α1 and Kap60 are omitted, and only the NLSs are shown. Residues at P2′ and P2 are shown as sticks. (C) Pull-down analysis and quantification of the interaction of GST-tagged importin α1 lacking the IBB (ΔIBB) or FL immobilized on glutathione beads and incubated with MBP-tagged h2NLS, Pom121NLS291-320, and NPNLS. (D) Pull-down analysis and quantification of the interaction of GST-ΔIBB-importin α1 or FL-importin α1 with WT Pom121NLS291-320 and mutants at P2′, P2, and P2/P2′. Pull downs in C and D are shown as mean ± SD for three experiments.
Structural alignment of Pom121NLS291-320 with NLSs visualized crystallographically in complex with importin α/Kap60 (Supplemental Table S2) revealed the position of critical residues at P2 and P2′. Pom121NLS291-320 inserts a lysine at position P2 (K313) in the major NLS-binding pocket, as observed for the vast majority of classical and nonclassical NLSs (Kalderon et al., 1984). In contrast, unlike most NLSs, which make strong contacts at the minor NLS box (Kosugi et al., 2009; Giesecke and Stewart, 2010; Lott et al., 2011; Chang et al., 2013; Pang and Zhou, 2014; Pumroy et al., 2015), Pom121NLS291-320 inserts a lysine at P2′ (K295) as opposed to an arginine. The structural alignment also showed that Pom121NLS291-320 shares striking structural resemblance to the IBB domain of importin α in its inhibitory conformation and to the Heh2 NLS (h2NLS). Pom121NLS, IBB, and h2NLS have superimposable traces (Figure 2B) and diverge only in the variable region between NLS boxes (residues 299–311 in Pom121NLS291-320), which makes minimal contacts with the Arm core. As observed for h2NLS, Pom121NLS291-320 does not associate directly with importin β (unpublished data), suggesting that this NLS cannot adopt the helical fold essential for IBB association with importin α (Lott et al., 2010).
The striking similarity of Pom121NLS291-320 to the IBB domain prompted us to determine whether Pom121NLS can bypass IBB autoinhibition, as found for h2NLS and, to a lesser extent, h1NLS (King et al., 2006; Lokareddy et al., 2015). To test this hypothesis, we used an on-bead binding assay in which glutathione S-transferase (GST)–tagged full-length (FL)-importin α1 and GST-ΔIBB-importin α1 were immobilized on glutathione beads and incubated with a twofold molar excess of maltose binding protein (MBP)–tagged Pom121NLS291-320, NPNLS (a negative control for IBB displacement), or h2NLS (a positive control for IBB displacement). Of note, MBP-Pom121NLS291-320 was minimally able to overcome autoinhibition, and <20% of the starting material was recovered bound to FL-importin α1 beads after 15 min of incubation (Figure 2C), comparable to the negative control NPNLS, which is autoinhibited by the IBB domain (Lokareddy et al., 2015; Pumroy et al., 2015). In contrast, as previously shown, h2NLS completely bypassed IBB autoinhibition, binding importin α1 Arm core equally in the presence or absence of IBB (Figure 2C). Mutation at P2′ and P2 completely disrupted the interaction of Pom121NLS291-320 with FL-importin α1, indistinguishable from a double mutant at P2′/P2 (Figure 2D). A single mutation at P2 was also sufficient to disrupt Pom121NLS291-320 binding to ΔIBB-importin α1, suggesting the interaction of this NLS with the Arm-core is cemented by the major NLS-binding box, as observed in classical NLS (Figure 2D). Thus Pom121NLS291-320 adopts an IBB-like structure that combines binding determinants seen in the recognition of classical NLSs and a deeper interaction at the minor NLS-binding site as observed for h2NLS.
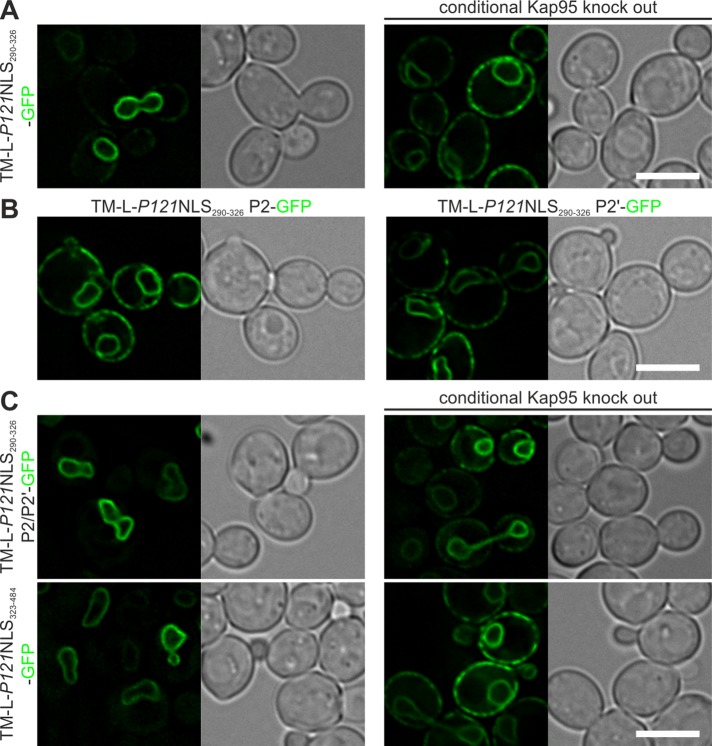
Pom121NLS290-326 is sufficient to mediate INM import of a membrane-embedded protein, and interactions at P2 and P2′ positions are critical
To further study the characteristics of the IBB-like region of Pom121NLS, we created a reporter protein that was fused to only the IBB-like region of Pom121NLS (TM-L-P121NLS290-326-GFP; Figure 3A). TM-L-Pom121NLS290-326-GFP accumulated at the INM, although a fraction of the protein was observed in the ER. As expected, the INM accumulation was abolished upon depletion of Kap95. The introduction of an alanine at P2 or P2′ (Figure 3B; TM-L-P121NLS290-326 P2-GFP and P2′-GFP, respectively) disrupted the INM targeting of the reporter protein. This shows that interaction of the P2 position, as well as of the P2′ position, of Pom121NLS290-326 with importin α is required to mediate efficient INM import of a membrane-embedded protein, consistent with the structural data. Here the IBB-like NLS in the region of Pom121NLS (residues 291–320) differs from what was observed for h2NLS, for which binding of the P2′ position contributed more significantly to INM import of the reporter membrane protein (Lokareddy et al., 2015).
FIGURE 3:
The IBB-like region is sufficient for INM targeting, and the Pom121NLS region consists of redundant NLSs. (A) The IBB-like region of Pom121NLS alone in TM-L-P121NLS290-326-GFP is sufficient to induce Kap95-dependent INM accumulation of the reporter protein, and (B) substitution of the lysine residues at the P2 or P2′ positions by alanine residues is sufficient to abolish NE accumulation. (C) Lysine-to-alanine mutation at the P2 and P2′ positions of the IBB-like region of the NLS of Pom121 in the context of the full Pom121NLS cluster (TM-L-P121NLS290-484 P2/P2′-GFP) or removal of the IBB-like region from the full Pom121NLS (TM-L-P121NLS323-484-GFP) did not alter Kap95-dependent INM accumulation of the indicated reporter proteins. Scale bars, 5 μm.
The Pom121NLS region encodes multiple NLSs (Doucet et al., 2010; Yavuz et al., 2010; Funakoshi et al., 2011). Indeed, the substitutions at P2 or P2′ (unpublished data), as well as the combination of these (Figure 3C; TM-L-P121NLS290-484 P2/P2′-GFP), left the Kap95-dependent INM localization of the protein unaltered. In addition, removal of the IBB-like NLS of the NLS region of Pom121 (Figure 3C; TM-L-P121NLS323-484-GFP) did not influence the Kap95-dependent import of the reporter protein. Clearly, besides Pom121NLS291-320, the Pom121NLS region contains at least one alternative importin-binding site that is recognized in yeast and mediates INM import.
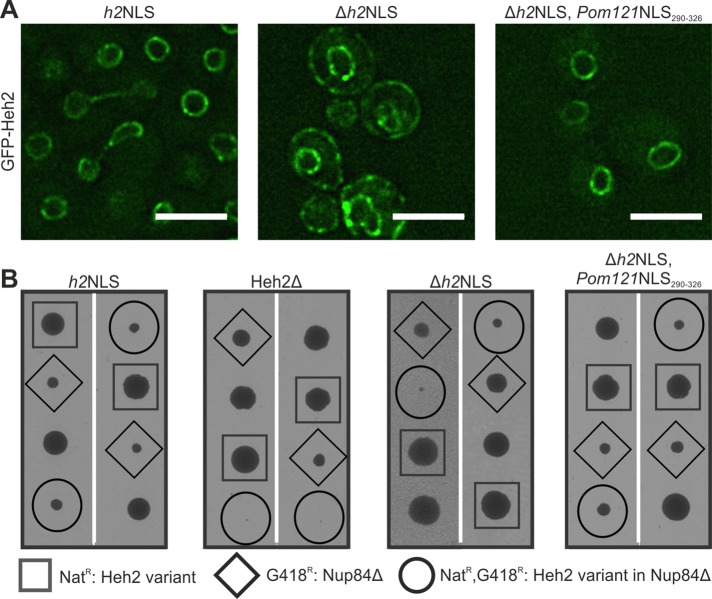
Next we looked for evidence of similarity between the h2NLS and Pom121NLS290-326 in the context of an actual inner membrane protein. First, we addressed whether replacement of the h2NLS in the endogenous Heh2 gene for Pom121NLS290-326 affects the subcellular localization. GFP-Heh2Δh2NLS expressed from its endogenous promoter is mislocalized to the peripheral ER (Figure 4A; GFP-Heh2Δh2NLS). Introducing Pom121NLS290-326 rescues the localization at the NE: the localization of GFP-Heh2(Δh2NLS, Pom121NLS290-326) is indistinguishable from that of GFP-Heh2 (Figure 4A). We also assessed synthetic lethality/sickness of yeast strains that combine mutations of NPC components with mutations in the NLS region of Heh2, based on the previous finding that a double mutant lacking Heh2 and Nup84 is not viable (Yewdell et al., 2011). Strains lacking Nup84 and expressing GFP-Heh2, GFP-Heh2(Δh2NLS, Pom121NLS290-326), or GFP-Heh2(Δh2NLS) were tested, and whereas the double mutant expressing GFP-Heh2(Δh2NLS) was synthetic sick, the double mutants expressing GFP-Heh2 or GFP-Heh2(Δh2NLS, Pom121NLS290-326) grew well (Figure 4B). Overall these studies show the importance and similarity of the Pom121NLS290-326 and the NLS of Heh2 in vivo and in the context of full-length Heh2.
FIGURE 4:
Pom121NLS rescues localization of Heh2ΔNLS and synthetic sickness of Heh2ΔNLS,nup84Δ double mutant. (A) Deconvolved wide-field images of yeast expressing native levels of GFP-Heh2 with wild-type NLS (h2NLS), without the NLS (Δh2NLS), or with Pom121NLS290-326 instead of the h2NLS (Δh2NLS, Pom121NLS290-326). Scale bar, 5 μm. (B) Synthetic sick/lethal interaction using tetrad dissection of nup84Δ expressing wild-type (h2NLS) and mutant variants (Δh2NLS and Δh2NLS, Pom121NLS290-326) or no Heh2 (Heh2Δ). Each tetrad is oriented vertically and represents the meiotic progeny of a heterozygous diploid between GFP-HEH2-NAT/NUP84 and HEH2/nup84::KANMX. Two representative tetrads for each double mutant are shown. The genetic background of each spore is identified by the presence of the NAT or KAN marker, respectively. The double-mutant spore colonies are enclosed in circles, single mutants are enclosed in squares or diamonds, and wild-type strains are not enclosed.
h2NLS, Pom121NLS290-484, and Pom121NLS290-326 mediate nuclear import of a soluble reporter in HEK293T cells
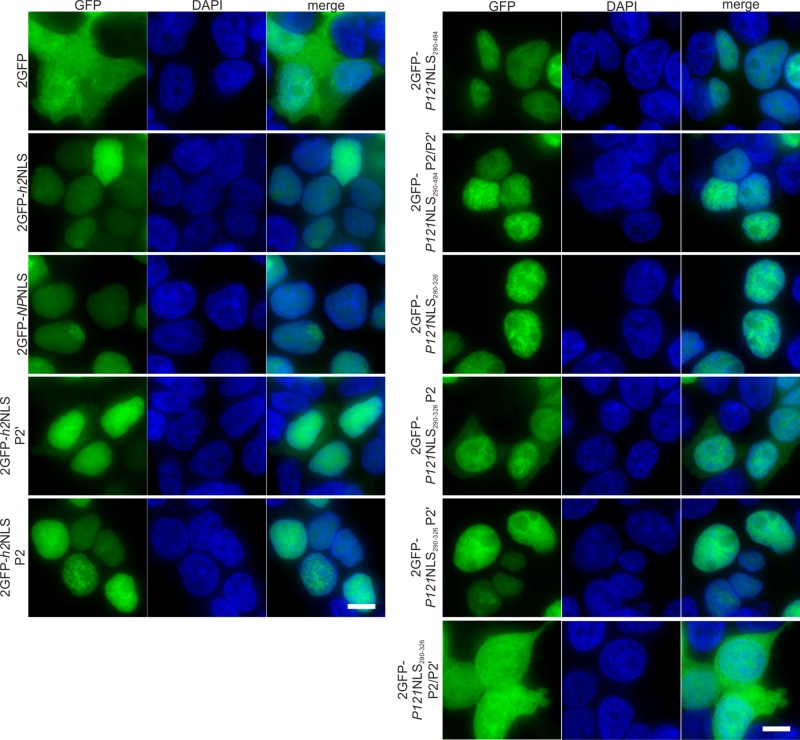
Thus far, our in vivo experiments in yeast reveal that the NLS of Pom121 and the NLS of Heh2 are largely interchangeable. To investigate whether this is also true in mammalian cells, we first performed a localization study in HEK293T cells with a soluble tandem GFP fused to the Pom121NLSs and h2NLS and mutant versions thereof (Figure 5). A tandem GFP (2GFP) protein localized dispersed over the cytoplasm and the nucleoplasm, and fusion to h2NLS, Pom121NLS290-484, or Pom121NLS290-326 caused strong accumulation in the nucleoplasm. The tandem GFP fused to the nucleophosmin NLS (2GFP-NPNLS) was used as a control. Mutant analysis showed that the accumulation of 2GFP with the h2NLS depends more strongly on the lysine at position P2′ than on the residues at P2: ∼30–40% of the cells expressing the P2′ mutant showed cytoplasmic fluorescence, whereas for the wild type and the P2 mutant those percentages were >95%. This dependence on the residues at P2′ rather than P2 was also found in yeast (Lokareddy et al., 2015). A similar mutant analysis of the Pom121NLS290-484 showed that introduction of alanine residues at the P2 and P2′ positions did not affect the localization in Pom121NLS290-484. On the substitution of lysine by alanine at position P2 of 2GFP-Pom121NLS290-326, the accumulation significantly decreased, resulting in 2GFP-NLS residing in the cytoplasm in all cells, whereas replacement of the P2′ did not alter the localization of 2GFP-Pom121NLS290-326. However, the P2′ substitution was additive to the effect of P2 substitution in Pom121NLS290-326 (2GFP-Pom121NLS290-326 P2/P2′) and resulted in complete lack of accumulation of 2GFP in the nucleoplasm in all cells. The fact that P2/P2′ substitutions in Pom121NLS290-484 did not influence this accumulation, whereas it does so in Pom121NLS290-326, is consistent with previous studies (Doucet et al., 2010; Yavuz et al., 2010; Funakoshi et al., 2011) and our conclusions from the localization studies in yeast that the Pom121NLS region has redundant importin α binding sites.
FIGURE 5:
Localization of different GFP-NLS fusions in HEK293T cells. A tandem GFP fusion (2GFP; (top left) and 2GFP-Pom121NLS290-326 P2/P2′ (bottom right) equilibrate between the cytoplasm and the nucleoplasm. All remaining 2GFP-NLS fusion proteins accumulated in the nucleoplasm in >95% of the cells, with the exception of 2GFP-h2NLS P2′– and 2GFP-Pom121NLS290-326 P2–expressing cells, in which cytoplasm-localized protein was observed in 30–40 and 100% of cells, respectively (100 < n < 200 cells). NPNLS: nucleoplasmin NLS. Scale bar, 5 μm.
We conclude that h2NLS, Pom121NLS290-484, and Pom121NLS290-326 are all able to mediate nuclear accumulation of soluble tandem GFP in HEK293T cells, but there are different dependences of the interactions with the major and minor binding pockets of importin α. Whereas for h2NLS the interaction of the residue at P2′ with importin α contributes mostly to efficient INM import, for Pom121NLS290-326, the interaction of the P2 position with importin α is dominant for effective translocation of a soluble protein to the nucleus.
h2NLS targets a membrane protein to the INM in HEK293T cells
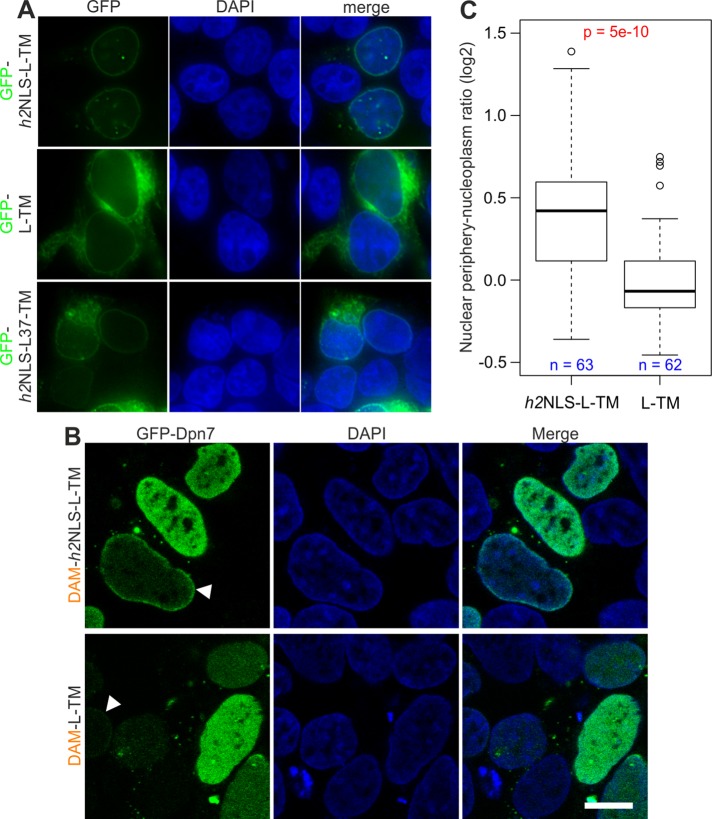
We established that the h2NLS and Pom121NLS290-320 have a similar IBB-like interaction with importin α—that is, both target a soluble protein to the nucleus in HEK293T cells—and that the Pom121NLS mediates nuclear import of reporter and full-length membrane proteins in yeast. We next asked whether h2NLS is able to mediate INM import of a membrane-embedded reporter protein to the nucleus in mammalian cells. To answer this, we expressed GFP-h2NLS-L-TM, encoding Heh93-378, in HEK293T cells. The reporter protein GFP-h2NLS-L-TM was clearly enriched at the nuclear envelope in HEK293T cells (Figure 6A). This enrichment was absent in cells expressing the reporter lacking the h2NLS (GFP-L-TM), which was dispersed over the NE/ER network, demonstrating the importance of the NLS for NE enrichment. The length of the ID linker region also influenced the localization of the reporter protein. A reporter with a shorter linker length of 37 instead of 180 residues, GFP-h2NLS-L(37)-TM, was enriched to the NE to some extent in a few cells, but in the majority of cells, this enrichment was minor or even absent, and the protein localized to the NE/ER network.
FIGURE 6:
h2NLS mediates INM localization of a membrane-embedded protein in HEK293T. (A) The NE accumulation observed for GFP-h2NLS-L-TM is absent for GFP-L-TM, whereas GFP-h2NLS-L(37)-TM accumulates at the NE in a fraction of cells, and in the majority of cells, the reporter is NE/ER localized. Scale bar, 5 μm. (B) Localization of GFP-Dpn7 in HEK293T expressing DAM-h2NLS-L-TM or DAM-L-TM. Arrowheads point at cells where GFP-Dpn7 accumulates at the nuclear periphery. Scale bar, 5 μm. (C) Analysis of indicated number of cells with low GFP-Dpn7 levels shows that the accumulation of GFP-Dpn7 at the nuclear periphery, represented by the ratio of mean fluorescence signal at the nuclear periphery over the mean fluorescence signal in the nucleoplasm, is significantly lower for cells expressing DAM-L-TM than for cells expressing DAM-h2NLS-L-TM. The p values are calculated using Student’s t test.
We wanted to know whether the NE enrichment of the reporter protein GFP-h2NLS-L-TM reflected INM localization of this membrane protein, and therefore we developed an assay based on the visualization of DNA methylation by an Escherichia coli DNA adenine methyltransferase (DAM) fused to the reporter protein: DAM-h2NLS-L-TM. DAM methylates adenine residues when it comes in contact with DNA. A GFP-fused truncation of DpnI (GFP-Dpn7) specifically interacts with adenine-6-methylation (m6A) in the sequence GATC (Gm6ATC; Kind et al., 2013). If an INM-localized DAM fusion methylates adenine residues at the nuclear periphery, like DAM fusion of lamin B1 does (Kind et al., 2013), this would be reflected in the specific localization of GFP-Dpn7 at the nuclear periphery. DAM-L-TM, lacking the h2NLS, served as the control. With a lumenal domain of 54 kDa, this protein may be small enough to pass the NPC passively (Soullam and Worman, 1995; Wu et al., 2002; Ohba et al., 2004) but is not expected to accumulate at the INM because of lack of retention.
Indeed, when we coexpressed DAM-h2NLS-L-TM and GFP-Dpn7 in HEK293T cells, we observed increased fluorescence signal at the nuclear periphery of some cells (Figure 5B). Consistent with Kind et al. (2013), GFP-Dpn7 is homogeneously distributed over the nucleoplasm upon expression of DAM alone (unpublished data). However, localization of GFP-Dpn7 at the periphery was also observed in some cells expressing DAM-L-TM (Figure 6B), although the fraction of cells showing this GFP-Dpn7 localization is lower than for DAM-h2NLS-L-TM–expressing cells.
To determine whether there is a difference between cells expressing DAM-h2NLS-L-TM and DAM-L-TM, we determined the ratio of the average fluorescence signal at the nuclear periphery over the average fluorescence signal in the nucleoplasm (nuclear periphery vs. nucleoplasm, or P/N ratio). We plotted this ratio against the mean nuclear fluorescence signal of GFP-Dpn7 (Supplemental Figure S2A) and observed a correlation between the expression level of GFP-Dpn7 and the P/N ratio. Whereas a clear accumulation of GFP-Dpn7 at the nuclear periphery could be measured in cells with a low GFP-Dpn7 expression level, this accumulation was practically absent in cells with a high expression level. GFP-Dpn7 has a low affinity for unmethylated DNA, and therefore we reasoned that upon high expression of GFP-Dpn7, the DNA reaches a point of GFP-Dpn7 binding saturation, losing the resolution to determine the effect of the reporter-induced DNA methylation at the periphery. Therefore for our analysis we included only cells that have an average nucleoplasmic GFP-Dpn7 signal <90 (arbitrary units), using the same microscope settings between experiments. For cells transfected with DAM-L-TM, no significant difference was observed between P/N ratios of low and high GFP-Dpn7– expressing levels, whereas this difference was significant for cells transfected with DAM-h2NLS-L-TM (Supplemental Figure S2B). For cells expressing low levels of GFP-Dpn7, the mean P/N ratio was significantly higher than in cells expressing DAM-h2NLS-L-TM compared with cells expressing DAM-L-TM (Figure 6B). Because the P/N ratio is a measure for accumulation of GFP-Dpn7 at the nuclear periphery, we conclude that the NE enrichment that we observed for GFP-h2NLS-L-TM indeed reflects NLS- dependent INM localization of this reporter membrane protein.
DISCUSSION
With the discovery of active import of the membrane proteins Heh1 and Heh2 (King et al., 2006; Meinema et al., 2011) to the INM in baker’s yeast, a main question was whether active transport of membrane proteins is yeast specific or conserved in higher eukaryotes. The domain composition of an NLS followed by an ID linker and a transmembrane domain, NLS-L-TM, is required and sufficient to mediate importin α/β–, FG-Nup–, and RanGTP-dependent import of Heh2 and Heh1 in baker’s yeast (Meinema et al., 2011). Indeed, except for Heh1 and Heh2, no other INM proteins have been reported to traffic to the INM by an active import mechanism.
Here we screened metazoan genomes for genes that encode a putative NLS-L-TM domain composition. We identified four candidates—Sun2, Lem2, LBR, and Lap2β—and one for which the reversed topology (TM-L-NLS) was predicted, Pom121. The (putative) NLS-L or NLS regions of Sun2, Lem2, LBR, and Lap2β were not sufficient to mediate INM targeting of membrane reporter proteins in baker’s yeast, but replacement of the h2NLS in TM-L-h2NLS-GFP for the NLS region of Pom121 (Pom121NLS290-484) resulted in efficient accumulation at the INM, which depended on Kap60/95. The inability to mediate INM accumulation in yeast for the “NLS regions” of Sun2, Lem2, LBR, and Lap2β obviously does not contradict their interaction with importin α and/or β, as previously demonstrated for LBR (Ma et al., 2007) and Sun2 (Turgay et al., 2010; Tapley et al., 2011), and these signals could still promote INM localization but require mammalian-specific factors (such as lamins) to be retained once they reach the INM. Their insufficiency to drive accumulation in yeast merely categorizes them as being distinct from the Heh1, Heh2, and Pom121 NLSs, which are sufficient to support active import and accumulation of a membrane protein in yeast.
These experiments identify the Pom121 NLS as an NLS that supports active import of a membrane protein in yeast and also for the first time show that the NLS and linker regions can be N- or C-terminal of the transmembrane segment. We show that, as is the case for a C-terminal sorting signal, import depends on an NLS that interacts with importin α/β separated from a transmembrane domain by an intrinsically disordered region of sufficient length. This argues against the option that active import is related to the biogenesis of tail-anchored proteins and that membrane insertion may occur after nuclear import.
Having identified the NLS of Pom121 as an NLS that supports active import of a membrane protein in yeast, we address its similarities with the Heh1 and Heh2 NLSs. Crystallographic analysis of the binding interface of the Pom121NLS with importin α revealed that of the five boxes of positively charged residues (Funakoshi et al., 2011) in the NLS region (residues 290–484), the first two (P1′–P4′ [294-KKKR-297] and P2–P5 [313-KRRR-316]) adopt a similar fold as the IBB domain of importin β bound to importin α. The NLS encoded by Heh1 and Heh2 adopts an IBB-like fold with intimate interactions in the minor and major binding sites (Lokareddy et al., 2015). The tight interactions of Pom121NLS291-320 with importin α are important for INM targeting, as mutagenesis of the residues interacting with the P2 and P2′ positions of the Kap60 binding site in the Pom121NLS290-326 (K313 for binding P2 or K295 for binding P2′) results in lack of NE accumulation in yeast. These mutant NLSs are strong enough to accumulate a soluble cargo in human cells, hinting that the IBB-like features of the NLS may present a unique requirement for supporting INM import as compared with soluble import. The importance of the h2NLS for Heh2’s function at the INM is clear from the mislocalization and synthetic sickness of the double mutant Nup84Δ, Heh2Δh2NLS. Additional proof for the conserved properties of the h2NLS and the Pom121NLS290-326 come from experiments in which we assess their functionality in the context of native levels of Heh2. The subcellular localization of Heh2 in yeast is unaltered when its NLS is replaced for the Pom121NLS290-326, and in addition, the Pom121NLS suppresses the synthetic sickness of Nup84Δ, Heh2Δh2NLS.
Pom121 is the first metazoan INM protein identified to have a Heh1/Heh2/IBB-like NLS. The shared properties could be related to several factors, one being a shared role in nuclear import, which we investigated. We expressed yeast-derived reporter proteins in HEK293T cells to minimize effects of selective retention on localization, so that the readout of our assays (steady-state subcellular localization) would most likely reflect the dynamic equilibrium between rates of import and efflux. We observed clear NLS-dependent enrichment of the proteins at the NE. The dependence of linker length is less pronounced than in yeast, for which no accumulation is observed with a short linker of 37 residues. To confirm that the increased concentration at the NE indeed reflected an accumulation at the INM, we adapted an assay based on the visualization of methylated DNA at the nuclear periphery and showed that the NE accumulation indeed reflected the presence of DAM fusions of the membrane proteins at the INM.
Taken together, these data confirm NLS-dependent residence of the membrane proteins at the INM in HEK293T cells. However, when human cells are used, it is uncertain whether proteins traffic through the NPC or become INM localized during mitosis when the NE is reassembled onto the decondensing chromatin. Given the strong structural and biochemical similarities between the NLS of Heh1, Heh2, and Pom121, the ability of Pom121NLS to support INM import in yeast, and the NLS-dependent localization of membrane proteins in HEK293T cells, we conclude that the mechanism is likely conserved for Pom121 but not for LBR, Sun2, and Lap2β.
Two very recent studies (Boni et al., 2015; Ungricht et al., 2015) report on the determinants for membrane targeting in mammalian cells using LBR, Sun2, and Lap2β as model substrates. The major determinants are the number and permeability of the NPCs, availability of binding sites at the INM, and kinetics of diffusion through the membranes of ER. Both studies convincingly show that a diffusion-retention model of INM protein transport in mammalian cells explains the measured kinetics of targeting in wild-type and mutant cells and under conditions of energy depletion. We here show that the NLSs in Heh1, Heh2, and Pom121 are distinct from the sorting signals in LBR, Sun2, and Lap2β and therefore suggest that targeting of Heh1, Heh2, and Pom121 does not follow this paradigm but instead is importin dependent, with higher tolerance for large extralumenal domains.
Although our genetic studies confirmed for the first time the in vivo importance of INM sorting of Heh2, the exact function of active import of Heh1, Heh2, and Pom121 remains to be determined. Intrigued by the elite position of Pom121, Heh1, and Heh2 in this respect, we speculate about why specifically these three proteins may require an active transport mechanism. A first consideration is that these proteins have large extralumenal domains, which may preclude fast enough passage through the lateral channels. Passive diffusion of Pom121 through the NPC is also less likely because of the FG repeats in its C-terminus, which promote interactions with components of the NPC. A second consideration is that the property of Pom121, Heh1, and Heh2 to accumulate efficiently at the INM might be essential for their role in the synthesis of new, functional pores. Heh1 and Heh2 are not stable components of the NPC, but they are involved in the biogenesis of intact NPCs in the NE (Yewdell et al., 2011; Webster et al., 2014). The synthetic sickness of the double mutant lacking Nup84 and expressing Heh2ΔNLS may be related to the role of Heh2 in surveillance of NPC assembly (Webster et al., 2014). Pom121 is dispensable for the assembly of NPCs in emerging NEs after mitosis (Funakoshi et al., 2011), when “prepores” are seeded on mitotic chromosomes (Rasala et al., 2008), but during interphase, Pom121 is essential for the insertion of new pores into the intact NE membrane (Doucet et al., 2010; Talamas and Hetzer, 2011). To play an early role in NPC formation, Pom121 should not only localize to NPCs, but also reside at the INM. Indeed, a fraction of Pom121 is targeted to regions in the INM lacking intact NPCs (Funakoshi et al., 2011). It was shown previously that Pom121 contains several bipartite NLSs, which interact with importin α/β and are required for efficient targeting of the protein to the NE/NPC (Doucet et al., 2010; Yavuz et al., 2010; Funakoshi et al., 2011), although postmitotic NPC insertion was independent of the presence of the NLSs (Funakoshi et al., 2011).
The identification of the interchangeability of the Pom121, Heh1, and Heh2 NLSs for nuclear traffic in yeast and humans cells provides the first experimental evidence in support of an active import mechanism to the INM in metazoans and warrants future investigation into its biological function.
MATERIALS AND METHODS
Molecular cloning
All plasmids are constructed according to standard molecular cloning techniques and listed in Supplemental Tables S3 and S5. All constructs generated in this study were fully sequenced to ensure the correctness of the DNA sequence. Details can be provided on request. In all constructs used in this study, the linker sequences originated from Heh2, the sequence encoding the TM in constructs with topology N-TM-Clumen originated from TM1 from Heh2, and the sequence encoding the TM in constructs with topology Nlumen-TM-C was encoded by a synthetic gene (encoding a reversed orientation of the amino acids of Heh2 TM). The exact encoded amino acid sequences and their origin are provided in Supplemental Tables S6 and S7. The gene encoding human importin α1 was cloned as FL and ΔIBB in vectors pET28a (Novagen, Merck Millipore, Darmstadt, Germany) and pGEX-6P (GE Healthcare) as preciously described (Pumroy et al., 2015). Murine ΔIBB-importin α1 used for crystallization was expressed and purified as described (Lott et al., 2010). The NLS sequence of RnPom121 (residues 291–320) was cloned between BamHI and XhoI restriction sites in vectors pGEX-6P and in an engineered pET28 that also contains an N-terminal MBP (pET28-MBP). NPNLS and h2NLS were also cloned in pET28-MBP as previously described (Lokareddy et al., 2015). Alanine mutations at position P2, P2′, and P2/P2′ of pET28-MBP- Pom121NLS291-320 were generated by site-directed mutagenesis.
Yeast cultivation
All reporter proteins were expressed in S. cerevisiae KAP95AA strain (Meinema et al., 2011). Strains were grown at 30ºC in synthetic dropout medium lacking histidine and supplemented with 0.01% adenine and 2% glucose. For induction of the GAL1 promoter, glucose was replaced by raffinose and 0.1% galactose was added for 2 h; full-length expression was confirmed by Western blot (Supplemental Figure S3).
Depletion of cytosolic Kap95-FRB was induced by the addition of 2 μg/ml rapamycin to the cell culture for 15 min.
Synthetic lethality screening
DNA constructs encoding GFP-tagged, full-length Heh2, GFP-tagged Heh2(Δh2NLS), and GFP-tagged Heh2(Δh2NLS,Pom121NLS291-320) were fused to the NAT marker and the flanking regions as encoded immediately upstream and downstream the HEH2 open reading frame. The linear DNA constructs were subsequently integrated in BY4742 heh2::KAN by homologous recombination, thereby replacing the KAN marker for GFP-HEH2-NAT (Supplemental Table S4). Transformants were selected and analyzed for expression of the GFP-tagged proteins by fluorescence microscopy. For assessing the synthetic sick and lethal phenotypes of double mutants, BY4741 nup84::KAN (Mata) was mated with the GFP-Heh2 variant expressing strains AS1 (GFP-HEH2-NAT, Matα), AS2 (GFP-HEH2(Δh2NLS)-NAT, Matα), AS5 (GFP-HEH2(Δh2NLS,Pom121NLS291-320)-NAT, Matα), and AS4 (GFP-NAT, Matα). Diploids were selected on yeast extract/peptone/dextrose (YPD) containing G418 and Nat. After sporulation, tetrads were dissected, and the genotype of each spore was determined by replica plating on YPD containing Nat and YPD containing G418.
Transfection HEK293T cells for microscopy
HEK293T cells were grown in DMEM (Invitrogen) supplemented with antibiotics and 10% fetal calf serum at 37ºC and 5% CO2. Cells were grown on coverslips and transiently transfected using FuGENE HD Transfection reagent (Promega). Subsequently the cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized with 0.1% Triton X-100 in PBS, stained with 1μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and mounted with Vectashield (Vector Labs, Burlingame, CA).
Fluorescence microscopy
Imaging of localization of GFP-fused reporter proteins in yeast and HEK293T cells was done on a DeltaVision Deconvolution Microscope (Applied Precision), using InsightSSI Solid State Illumination of 488 nm (GFP) and 358 nm (DAPI). Detection was done with a CoolSNAP HQ2 camera. For yeast, an Olympus UPLS Apo 100× oil objective with 1.4 numerical aperture (NA) was used. Pixel size was 64 × 64 × 200 nm. For HEK293T an Olympus UPLS Apo 40× oil objective with 1.3 NA was used. Pixel size was 215 × 215 × 500 nm.
DAM methylation assay
HEK293T cells were cultured as described. Cells were cotransfected with 0.8 μg of plasmid encoding DAM-fused reporter proteins (pIND-DAM-V5 (Vogel et al., 2006), pAK49, and pAK50, respectively), 0.8 μg of enhanced GFP–DPN7 (Kind et al., 2013), and 0.8 μg of pVg-RxR (Invitrogen) using 7.5 μl of TransIT-2020 transfection reagent (Mirus) for 24 h. Expression of DAM-fusion proteins was induced by the addition of 5 μM ponasterone (Sigma-Aldrich, St. Louis, MO) for 24 h. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100 and 0.5% bovine serum albumin, stained with 1 μg/ml DAPI, and mounted with Vectashield.
Imaging of eGFP-Dpn7 and DAPI was done on a Leica TCS SP5 laser scanning confocal microscope using a 488-nm argon gas laser for GFP, a 405-nm diode laser for DAPI, and a 63×/1.4 NA oil immersion objective. Detection was performed with a photomultiplier tube. Bidirectional line scanning was used with a line frequency of 700 Hz, and four frames were averaged. Settings were kept constant. Data were analyzed using the ZEN2010B software package (Carl Zeiss, Jena, Germany).
Biochemical techniques
FL- and ΔIBB-importin α1 was expressed and purified as preciously described (Lott et al., 2011; Pumroy et al., 2015). The ΔIBB-importin α1:Pom121NLS291-320 complex was formed by coexpressing plasmids pET28a-ΔIBB-importin α1 and pGEX-Pom121NLS291-320 in E. coli strain BL21-CodonPlus (DE3)-RIL (Stratagene) for 6 h at 30°C. GST-Pom121NLS291-320 bound to ΔIBB-importin α1 was purified on glutathione beads (GenScript), and after cleaving off the GST with PreScission Protease, the complex was purified over a Superdex 200 column (GE Healthcare) equilibrated in G.F. buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol, and 0.2 mM phenylmethylsulfonyl fluoride). All GST-tagged constructs used in this study were purified as described earlier. All MBP-tagged NLSs (Pom121NLS291-320, NPNLS, h2NLS) were expressed and purified as by Pumroy et al. (2015) and Lokareddy et al. (2015). The IBB-displacement assay on glutathione beads in Figure 2 was carried out and quantified as previously described (Lokareddy et al., 2015; Pumroy et al., 2015). The error bars in the quantification represent the SD of three independent experiments carried out under identical conditions.
Crystallographic studies
Crystals of ΔIBB-importin α1 bound to Pom121NLS291-320 were obtained by mixing equal volume of gel filtration–purified complex at 15 mg/ml with 0.1 M sodium citrate tribasic dihydrate, pH 5.6, 0.7 M sodium citrate tribasic dihydrate, and 25 mM dithiothreitol and equilibrating the droplet against 600 μl of the same precipitant. Crystals were harvested in nylon cryoloops, cryoprotected with 27% ethylene glycol, and flash-frozen in liquid nitrogen. Crystals were diffracted at beamlines X6A and X29 at the National Synchrotron Light Source (Brookhaven National Laboratory, Upton, NY) on a Quantum Q270 and a Quantum-315r charge-coupled device detector, respectively. Data indexing, integration, and scaling were carried out with the HKL2000 (Otwinowski and Minor, 1997). Initial phases were obtained by molecular replacement using Phaser (McCoy et al., 2007) and Protein Data Bank entry 3Q5U as a search model. Atomic models were built using Coot (Emsley and Cowtan, 2004) and refined using phenix.refine (Adams et al., 2002). A continuous trace of Pom121NLS291-320 main chain between residues 299 and 307 was obtained by blurring the B factor by applying a positive B factor correction of 20 Å2 in Coot. The refined B factor of this region is obviously very high (120 Å2). Data collection and refinement statistics are summarized in Supplemental Table S1. The structure was analyzed using the PISA server (Xu et al., 2008) and PyMOL (PyMOL Molecular Graphics System, version 1.5.0.4; Schrödinger LLC).
Accession code
The atomic coordinates and structure factors for ΔIBB-importin α1 bound to Pom121NLS were deposited in the Protein Data Bank with accession code 4YI0.
Supplementary Material
Acknowledgments
This work was supported by a Vidi Grant from the Netherlands Organization for Scientific Research (to L.M.V.) and National Institutes of Health Grant GM074846-01A1 (to G.C.). Research reported here includes work carried out at the Sidney Kimmel Cancer Center X-Ray Crystallography and Molecular Interaction Facility, Thomas Jefferson University (Philadelphia, PA), which is supported in part by National Cancer Institute Cancer Center Support Grant P30 CA56036. We thank Jop Kind and Bas van Steensel (Netherlands Cancer Institute, Amsterdam, Netherlands) for kindly sharing the Dpn7-GFP plasmids and communicating unpublished results. We thank Michael Chang, Bert Poolman, and members of the Veenhoff and Chang laboratories for valuable discussions.
Abbreviations used:
- DAM
DNA adenine methyltransferase
- ER
endoplasmic reticulum
- FKBP
FK506-binding protein
- FL
full length
- FRB
FKBP12-rapamycin binding domain
- IBB
importin β binding
- INM
inner nuclear membrane
- NE
nuclear envelope
- NLS
nuclear localization signal
- NPC
nuclear pore complex
- Nup
nucleoporin
- ONM
outer nuclear membrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.3301) on July 15, 2015.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Boni A, Politi AZ, Strnad P, Xiang W, Hossain MJ, Ellenberg J. Live imaging and modeling of inner nuclear membrane targeting reveals its molecular requirements in mammalian cells. J Cell Biol. 2015;209:705–720. doi: 10.1083/jcb.201409133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, von Appen A, DiGuilio AL, Ori A, Sparks L, Mackmull MT, Bock T, Hagen W, Andrés-Pons A, Glavy JS, Beck M. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- Chang CW, Counago RM, Williams SJ, Boden M, Kobe B. Distinctive conformation of minor site-specific nuclear localization signals bound to importin-alpha. Traffic. 2013;14:1144–1154. doi: 10.1111/tra.12098. [DOI] [PubMed] [Google Scholar]
- Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol. 1997;138:1193–1206. doi: 10.1083/jcb.138.6.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ems-McClung SC, Zheng Y, Walczak CE. Importin alpha/beta and Ran-GTP regulate XCTK2 microtubule binding through a bipartite nuclear localization signal. Mol Biol Cell. 2004;15:46–57. doi: 10.1091/mbc.E03-07-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J, Richards SA, Wente SR, Goldberg MW. Facilitated transport and diffusion take distinct spatial routes through the nuclear pore complex. J Cell Sci. 2010;123:2773–2780. doi: 10.1242/jcs.070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J Biol Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Clever M, Watanabe A, Imamoto N. Localization of Pom121 to the inner nuclear membrane is required for an early step of interphase nuclear pore complex assembly. Mol Biol Cell. 2011;22:1058–1069. doi: 10.1091/mbc.E10-07-0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K, Fritze CE, Gerace L. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J Biol Chem. 1998;273:4213–4219. doi: 10.1074/jbc.273.7.4213. [DOI] [PubMed] [Google Scholar]
- Giesecke A, Stewart M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J Biol Chem. 2010;285:17628–17635. doi: 10.1074/jbc.M110.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le BN, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–932. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- Hutchins JR, Moore WJ, Clarke PR. Dynamic localisation of Ran GTPase during the cell cycle. BMC Cell Biol. 2009;10:66. doi: 10.1186/1471-2121-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van SB. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
- Koerner C, Guan T, Gerace L, Cingolani G. Synergy of silent and hot spot mutations in importin beta reveals a dynamic mechanism for recognition of a nuclear localization signal. J Biol Chem. 2003;278:16216–16221. doi: 10.1074/jbc.M301137200. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Lin F, Morrison JM, Wu W, Worman HJ. MAN1, an integral protein of the inner nuclear membrane, binds Smad2 and Smad3 and antagonizes transforming growth factor-beta signaling. Hum Mol Genet. 2005;14:437–445. doi: 10.1093/hmg/ddi040. [DOI] [PubMed] [Google Scholar]
- Lokareddy RK, Hapsari RA, van Rheenen M, Pumroy RA, Bhardwaj A. Distinctive properties of the nuclear localization signals of inner nuclear membrane proteins Heh1 and Heh2. Structure. 2015;23:1305–1316. doi: 10.1016/j.str.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J Biol Chem. 2010;285:13769–13780. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Bhardwaj A, Sims PJ, Cingolani G. A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin alpha1. J Biol Chem. 2011;286:28160–28169. doi: 10.1074/jbc.M111.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Cingolani G. The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim Biophys Acta. 2011;1813:1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Cai S, Lv Q, Jiang Q, Zhang Q, Sodmergen, Zhai Z, Zhang C. Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J Cell Sci. 2007;120:520–530. doi: 10.1242/jcs.03355. [DOI] [PubMed] [Google Scholar]
- Madrid AS, Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- Marfori M, Lonhienne TG, Forwood JK, Kobe B. Structural basis of high-affinity nuclear localization signal interactions with importin-alpha. Traffic. 2012;13:532–548. doi: 10.1111/j.1600-0854.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinema AC, Laba JK, Hapsari RA, Otten R, Mulder FA, Kralt A, van den BG, Lusk CP, Poolman B, Veenhoff LM. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A. The binding site of karyopherin alpha for karyopherin beta overlaps with a nuclear localization sequence. Proc Natl Acad Sci USA. 1996;93:6572–6576. doi: 10.1073/pnas.93.13.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Schirmer EC, Nishimoto T, Gerace L. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J Cell Biol. 2004;167:1051–1062. doi: 10.1083/jcb.200409149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pang X, Zhou HX. Design rules for selective binding of nuclear localization signals to minor site of importin alpha. PLoS One. 2014;9:e91025. doi: 10.1371/journal.pone.0091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg O, Lim RY. Converging on the function of intrinsically disordered nucleoporins in the nuclear pore complex. Biol Chem. 2010;391:719–730. doi: 10.1515/BC.2010.092. [DOI] [PubMed] [Google Scholar]
- Powell L, Burke B. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J Cell Biol. 1990;111:2225–2234. doi: 10.1083/jcb.111.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- Pumroy RA, Ke S, Hart DJ, Zachariae U, Cingolani G. Molecular Determinants for Nuclear Import of Influenza A PB2 by Importin alpha Isoforms 3 and 7. Structure. 2015;23:374–384. doi: 10.1016/j.str.2014.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman N, Christie M, Swarbrick CM, Kobe B, Forwood JK. Structural characterisation of the nuclear import receptor importin alpha in complex with the bipartite NLS of Prp20. PLoS One. 2013;8:e82038. doi: 10.1371/journal.pone.0082038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, III, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Soderqvist H, Hallberg E. The large C-terminal region of the integral pore membrane protein, POM121, is facing the nuclear pore complex. Eur J Cell Biol. 1994;64:186–191. [PubMed] [Google Scholar]
- Soullam B, Worman HJ. The amino-terminal domain of the lamin B receptor is a nuclear envelope targeting signal. J Cell Biol. 1993;120:1093–1100. doi: 10.1083/jcb.120.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B, Worman HJ. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamas JA, Hetzer MW. POM121 and Sun1 play a role in early steps of interphase NPC assembly. J Cell Biol. 2011;194:27–37. doi: 10.1083/jcb.201012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley EC, Ly N, Starr DA. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol Biol Cell. 2011;22:1739–1752. doi: 10.1091/mbc.E10-08-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S, Platani M, Boue S, Mattaj IW. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J Cell Biol. 2006;173:469–476. doi: 10.1083/jcb.200512078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungricht R, Klann M, Horvath P, Kutay U. Diffusion and retention are major determinants of protein targeting to the inner nuclear membrane. J Cell Biol. 2015;209:687–704. doi: 10.1083/jcb.201409127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Guelen L, de WE, Peric-Hupkes D, Loden M, Talhout W, Feenstra M, Abbas B, Classen AK, van SB. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res. 2006;16:1493–1504. doi: 10.1101/gr.5391806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster BM, Colombi P, Jager J, Lusk CP. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 2014;159:388–401. doi: 10.1016/j.cell.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Lin F, Worman HJ. Intracellular trafficking of MAN1, an integral protein of the nuclear envelope inner membrane. J Cell Sci. 2002;115:1361–1371. doi: 10.1242/jcs.115.7.1361. [DOI] [PubMed] [Google Scholar]
- Xu Q, Canutescu AA, Wang G, Shapovalov M, Obradovic Z, Dunbrack RL., Jr Statistical analysis of interface similarity in crystals of homologous proteins. J Mol Biol. 2008;381:487–507. doi: 10.1016/j.jmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. Distinct, but not completely separate spatial transport routes in the nuclear pore complex. Nucleus. 2013;4:166–175. doi: 10.4161/nucl.24874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz S, Santarella-Mellwig R, Koch B, Jaedicke A, Mattaj IW, Antonin W. NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 2010;584:3292–3298. doi: 10.1016/j.febslet.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Yewdell WT, Colombi P, Makhnevych T, Lusk CP. Lumenal interactions in nuclear pore complex assembly and stability. Mol Biol Cell. 2011;22:1375–1388. doi: 10.1091/mbc.E10-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Goldberg MW, Moore WJ, Allen TD, Clarke PR. Concentration of Ran on chromatin induces decondensation, nuclear envelope formation and nuclear pore complex assembly. Eur J Cell Biol. 2002a;81:623–633. doi: 10.1078/0171-9335-00288. [DOI] [PubMed] [Google Scholar]
- Zhang C, Hutchins JR, Muhlhausser P, Kutay U, Clarke PR. Role of importin-beta in the control of nuclear envelope assembly by Ran. Curr Biol. 2002b;12:498–502. doi: 10.1016/s0960-9822(02)00714-5. [DOI] [PubMed] [Google Scholar]
- Zuleger N, Kelly DA, Richardson AC, Kerr AR, Goldberg MW. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J Cell Biol. 2011;193:109–123. doi: 10.1083/jcb.201009068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.