Abstract
Chicken growth performance provides direct economic benefits to the poultry industry. However, the underlying genetic mechanisms are unclear. The objective of this study was to identify candidate genes associated with chicken growth and investigate their potential mechanisms. We used RNA-Seq to study the breast muscle transcriptome in high and low tails of Recessive White Rock (WRRh, WRRl) and Xinghua chickens (XHh, XHl). A total of 60, 23, 153 and 359 differentially expressed genes were detected in WRRh vs. WRRl, XHh vs. XHl, WRRh vs. XHh and WRRl vs. XHl, respectively. GO, KEGG pathway and gene network analyses showed that CEBPB, FBXO32, FOXO3 and MYOD1 played key roles in growth. The functions of FBXO32 and FOXO3 were validated. FBXO32 was predominantly expressed in leg muscle, heart and breast muscle. After decreased FBXO32 expression, growth-related genes such as PDK4, IGF2R and IGF2BP3 were significantly down-regulated (P < 0.05). FBXO32 was significantly (P < 0.05) associated with carcass and meat quality traits, but not growth traits. FOXO3 was predominantly expressed in breast and leg muscle. In both of these tissues, the FOXO3 mRNA level in XH was significantly higher than that in WRR chickens with normal body weight (P < 0.05). In DF-1 cells, siRNA knockdown of FOXO3 significantly (P < 0.01) inhibited the MYOD expression and significantly up-regulated (P < 0.01 or P < 0.05) the expression of growth-related genes including CEBPB, FBXO32, GH, GHR, IGF1R, IGF2R, IGF2BP1, IGF2BP3, INSR, PDK1 and PDK4. Moreover, 18 SNPs were identified in FOXO3. G66716193A was significantly (P < 0.05) associated with growth traits. The sites C66716002T, C66716195T and A66716179G were significantly (P < 0.05) associated with growth or carcass traits. These results demonstrated that FOXO3 is a candidate gene influencing chicken growth. Our observations provide new clues to understand the molecular basis of chicken growth.
Introduction
Chicken growth, an important economic trait, is determined by genetic, nutritional and environmental factors. Heritability estimates showed that chicken growth could be enhanced by genetic improvement [1, 2]. This trait is controlled by multiple genes. At present, many studies have been performed to find genetic factors affecting growth. Candidate genes and quantitative trait loci (QTLs) such as GH, IGFBP2 and GHSR have been identified [3, 4]. Recently, genome-wide associate study (GWAS) analysis found that two FOXO1A single-nucleotide polymorphisms (SNPs) were strongly associated with chicken growth [5]. However, the genetic mechanisms of chicken growth are unclear, and a more systematic picture of the genes responsible for this trait is needed. Recently, next generation sequencing provided an important opportunity for the genome-wide characterization of genes and pathways involved in growth [6–8].
In this study, two chicken breeds, Recessive White Rock (WRR) and Xinhua (XH), were used for RNA-Seq. WRR is a typical fast-growing breed that is known for its large body size and thick myofibers. XH is a Chinese native slow-growing breed which is characterized by small body size. The different growth speeds of these two breeds led to distinct growth performance at 7 weeks of age. Both breeds were used for studies on chicken growth and fat deposition traits [1, 9]. With the use of a population derived from reciprocal crosses between these two breeds, a quantitative trait loci (QTL) on chromosome 1 was identified to be related to chicken growth traits by Genome-wide association study [5]. Previous study by Methylated DNA immunoprecipitation-sequencing showed that growth-related genes exhibited altered DNA methylation between WRR and XH [10]. Therefore, in the present study, we intended to use two-tail samples of these two breeds at 7 weeks of age to study the gene expression differences between fast- and slow-growing broilers in a genome-wide level and then to identify candidate genes for chicken growth. In our study, we performed RNA-Seq and differentially expressed genes (DEGs) were randomly selected to conduct qPCR experiments to validate the RNA-Seq results. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene network analyses were performed on the DEGs. Subsequently, the chicken FOXO3 and FBXO32 genes were selected for in vivo and in vitro studies to investigate their potential mechanisms functioned on growth.
Results
Assemble and blast analysis of reads from RNA-Seq
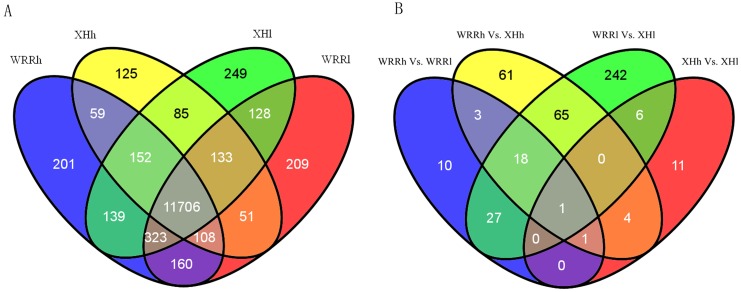
From RNA-Seq, we obtained 44139971, 36937542, 39046772 and 69669990 Illumina reads for WRRh, WRRl, XHh and XHl, respectively, giving rise to total residues of 4286130490, 3586054428, 3758078838 and 6698804887 bp, respectively (Table 1). All sequencing data have been submitted to NCBI Gene Expression Omnibus (GEO) database with the accession number GSE72424 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72424). More than 70.5% of the total reads were mapped to the chicken genome. In total, 13828 genes were detected in the four samples, including 12848 in WRRh, 12818 in WRRl, 12419 in XHh and 12915 in XHl (Fig 1A). Of these genes, 11706 genes were identified in all four samples, while 201, 209, 125 and 249 genes were found exclusively in WRRh, WRRl, XHh and XHl, respectively (Fig 1A). The sequence length distribution showed that 83% of genes identified in the four samples had a length less than 4000 bp, while no more than 0.5% of genes were longer than 10000 bp (S1 Fig).
Table 1. Data generated from RNA-seq.
| Sample 1 | Total number of reads | Total residues (bp) | Total mapped reads | Percentage of mapped reads 2 |
|---|---|---|---|---|
| WRRh | 44139971 | 4286130490 | 32685128 | 74.0% |
| WRRl | 36937542 | 3586054428 | 27449227 | 74.3% |
| XHh | 39046772 | 3758078838 | 27532015 | 70.5% |
| XHl | 69669990 | 6698804887 | 49272817 | 70.7% |
1WRRh, WRRl, XHh and XHl indicated the group of Recessive White Rock with high body weight, Recessive White Rock with low body weight, Xinhua Chickens with high body weight and Xinhua Chickens with low body weight, respectively.
2Percentage of mapped reads in total reads.
Fig 1. Numbers of expressed genes and differentially expressed genes: results of RNA-Seq.
A: Expressed genes among four samples. B: Differentially expressed genes among four contrasts including WRRh Vs. WRRl, XHh Vs. XHl, WRRh Vs. XHh and WRRl Vs. XHl. WRRh, WRRl, XHh and XHl indicated the group of Recessive White Rock with high body weight, Recessive White Rock with low body weight, Xinghua with high body weight and Xinghua with low body weight, respectively. The gene or differentially expressed gene number was shown at the top of each figure section.
DEGs among the four groups
A comparison of gene expression among the four samples showed that there were 60 DEGs (fold changes ≥ 2; q value < 0.05) between WRRh and WRRl (WRRh vs. WRRl), 23 DEGs between XHh and XHl (XHh vs. XHl), 153 DEGs between WRRh and XHh (WRRh vs. XHh), and 359 DEGs between WRRl and XHl (WRRl vs. XHl) (Fig 1B and S1 Table). LAC_CHICK (Ig lambda chain V-1 region) was found in all four comparisons. LAC_CHICK and TTR (transthyretin) were commonly identified in WRRh vs. WRRl and XHh vs. XHl (S2 Table). Moreover, 84 genes, including some crucial to chicken growth such as FBXO32, FOXO3, MYOD1, PDK4, PNPLA2 and SMYD1, were commonly identified in WRRh vs. XHh and WRRl vs. XHl (S3 Table). Among all these DEGs, 6, 10, 11 and 12 genes were uniquely expressed in one of the two samples in each comparison of WRRh vs. WRRl, XHh vs. XHl, WRRh vs. XHh and WRRl vs. XHl, respectively (S4 Table). DEG directionality analysis showed that the number of up-regulated genes was higher than the number of down-regulated genes in both WRRh vs. WRRl and XHh vs. XHl, while there were a greater number of down-regulated genes than of up-regulated genes in both WRRh vs. XHh and WRRl vs. XHl (S2 Fig).
qPCR validation of DEGs obtained from RNA-Seq
To confirm the DEG results obtained from RNA-Seq, four genes (RPL29, PDK4, FOXO3 and LAPTM5) were randomly selected to carry out qPCR using the same RNA samples used for RNA-Seq. The qPCR results showed general agreement with the RNA-Seq results in terms of the direction of expression in each comparison (Table 2).
Table 2. qPCR validation of DEGs obtained from RNA-seq.
| Gene | Contrast | qPCR 1 | RNA-seq 2 |
|---|---|---|---|
| RPL29 | WRRh Vs. WRRl | 1.58 | 1.05 |
| XHh Vs. XHl | 1.09 | 1.04 | |
| WRRh Vs. XHh | 1.42 | 0.90 | |
| WRRl Vs. XHl | 0.98 | 0.89 | |
| PDK4 | WRRh Vs. WRRl | 7.26 | 4.60 |
| XHh Vs. XHl | 3.03 | 1.94 | |
| WRRh Vs. XHh | 4.89 | 13.96 | |
| WRRl Vs. XHl | 2.04 | 5.90 | |
| FOXO3 | WRRh Vs. WRRl | 7.82 | 2.18 |
| XHh Vs. XHl | 0.76 | 0.98 | |
| WRRh Vs. XHh | 11.13 | 6.56 | |
| WRRl Vs. XHl | 1.08 | 2.96 | |
| LAPTM5 | WRRh Vs. WRRl | 11.47 | 2.18 |
| XHh Vs. XHl | 2.18 | 1.39 | |
| WRRh Vs. XHh | 4.63 | 0.32 | |
| WRRl Vs. XHl | 0.88 | 0.20 |
1meant the ratio of expression value (2-ΔΔCt) in the second group to the first group,
2fold change between contrasts presented (the ratio of FPKM in the second group to the first group).
GO and KEGG pathway analysis for DEGs
DEGs were then used for GO analysis to uncover enriched (P < 0.05) biological processes terms in each comparison. A total of 142 biological process terms, including 21 in WRRh vs. WRRl, 6 in XHh vs. XHl, 47 in WRRh vs. XHh and 119 in WRRl vs. XHl, were identified in our study (S5 Table). Of these, 99 were unique terms that appeared only once in all of the four comparisons. Many of the repeated biological process terms were focused on developmental process, regulation of biological process, cell differentiation and cell adhesion. A total of 29 DEGs involved in cell differentiation and proliferation were observed in the four comparisons, including well-known genes affecting chicken growth such as CEBPB, MYH11, MYOD1, NOTCH2 and TGFBR2 (Table 3). Moreover, in comparing XHh vs. XHl and WRRh vs. XHh, the following processes related to muscle development were found: skeletal muscle development, muscle organ development, muscle cell differentiation and muscle tissue development (S5 Table). Six DEGs were included in those processes: ACTC1, FOXP2, LGALS1, MYOD1, XIRP1 and ZFAND5. These genes might be crucial to muscle development. DEGs we identified were significant enriched (P < 0.05 and Benjiamini adjusted P < 0.1) in eight KEGG pathways, with the most influenced pathway being lysosome (Table 4). Furthermore, cell junction-related pathways such as focal adhesion, extracellular matrix (ECM)-receptor interaction and cytokine-cytokine receptor interaction were included. There were 45 DEGs in these four pathways, including TGFBR2 and ITGAV (Table 5).
Table 3. Differentially expressed genes involved in the processes of cell differentiation and proliferation in the four comparisons.
| Gene name | Description | Fold change (P value) |
|---|---|---|
| Actc1 | actin,alpha,cardiac muscle 1; actin,alpha 1,skeletal muscle | 4.6673034 (1.20E-06) |
| ANG | leukocyte ribonuclease A-1; leukocyte ribonuclease A-2 | 3.5635050(7.02E-09) |
| ANXA1 | annexin A1 | 5.0952624(8.32E-09) |
| B-MA1 | B locus M alpha chain 1 | 3.5732686(0.000159) |
| btk | Bruton agammaglobulinemia tyrosine kinase | 3.5233038(0.002987) |
| CD3E | CD3e molecule, epsilon (CD3-TCR complex) | 5.5777273(5.02E-08) |
| CD74 | CD74 molecule,major histocompatibility complex | 2.2219375(0.000123) |
| CEBPB | CCAAT/enhancer binding protein (C/EBP), beta | 3.5283824(1.13E-05) |
| CTGF | connective tissue growth factor | 4.2948028(2.72E-06) |
| CXCR4 | chemokine (C-X-C motif) receptor 4 | 3.9855519(0.000613) |
| Dpysl2 | dihydropyrimidinase-like 2 | 2.2867636(0.000232) |
| EFHD1 | EF-hand domain family, member D1 | 3.2853895(3.68E-05) |
| FABP4 | fatty acid binding protein 4, adipocyte | 5.4548149(3.38E-10) |
| Fkbp1b | FK506 binding protein 1B, 12.6 kDa | 2.9395742(0.000495) |
| FYN | FYN oncogene related to SRC, FGR, YES | 2.9063163(1.53E-05) |
| LGALS1 | lectin, galactoside-binding, soluble, 1 | 8.8038404(1.47E-12) |
| LIPA | lipase A lysosomal acid cholesterol esterase | 9.7054782(4.66E-15) |
| LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | 4.8534220(3.98E-08) |
| MDK | midkine (neurite growth-promoting factor 2) | 8.3954349(1.06E-07) |
| MYH11 | myosin heavy chain 10 non-muscle; myosin heavy chain 11 | 2.3873684(9.77E-07) |
| MYOD1 | myogenic differentiation 1 | 8.3954349(1.06E-07) |
| Notch2 | Notch homolog 2 (Drosophila) | 2.1351327(0.000257) |
| plek | pleckstrin | 5.8407637(1.37E-07) |
| Ptprc | protein tyrosine phosphatase receptor type C | 4.1222595(9.99E-14) |
| sfrp2 | secreted frizzled-related protein 2 | 6.4703796(4.28E-08) |
| Tgfbr2 | transforming growth factor, beta receptor II (70/80kDa) | 2.1053138(0.000330) |
| THY1 | Thy-1 cell surface antigen | 7.3370184(4.45E-12) |
| TXNRD1 | thioredoxin reductase 1 | 3.1127232(0.000165) |
| XIRP1 | xin actin-binding repeat containing 2 | 4.0403702(5.75E-08) |
Table 4. Enriched KEGG pathways for all DEGs identified in the four comparisons.
| No. | Pathways | P value | Benjiamini |
|---|---|---|---|
| 1 | Lysosome | 1.2E-07 | 9.4E-06 |
| 2 | Focal adhesion | 0.0041 | 0.15 |
| 3 | Drug metabolism | 0.0045 | 0.12 |
| 4 | Metabolism of xenobiotics by cytochrome P450 | 0.024 | 0.39 |
| 5 | ECM-receptor interaction | 0.027 | 0.36 |
| 6 | Cytokine-cytokine receptor interaction | 0.03 | 0.33 |
| 7 | Intestinal immune network for IgA production | 0.039 | 0.37 |
| 8 | Glutathione metabolism | 0.048 | 0.39 |
P < 0.05 and Benjiamini adjusted p < 0.1 was regarded as enriched.
Table 5. Differentially expressed genes involved in the five pathways related to growth.
| Gene | Description | Gene | Description |
|---|---|---|---|
| actn1 | actinin, alpha 1 | fn1 | fibronectin 1 |
| Atp6v0d2 | ATPase, H+ transporting, lysosomal 38kDa, V0 subunit D2 | FYN | FYN oncogene related to SRC, FGR, YES |
| CCR2 | chemokine (C-C motif) receptor 2 | GNPTAB | N-acetylglucosamine-1-phosphate transferase,alpha and beta subunits |
| CCR5 | chemokine (C-C motif) receptor 5 | GUSB | glucuronidase, beta |
| CD44 | CD44 molecule | il2rg | interleukin 2 receptor, gamma |
| CHAD | chondroadherin | Itgav | integrin, alpha V |
| CLTA | clathrin, light chain (Lca) | LAMP3 | lysosomal-associated membrane protein 3 |
| Col5a1 | collagen, type V, alpha 1 | LAPTM4A | lysosomal-associated protein transmembrane 4 alpha |
| Col6a3 | collagen, type VI, alpha 3 | LAPTM5 | lysosomal associated multispanning membrane protein 5 |
| comP | cartilage oligomeric matrix protein | LGMN | legumain |
| CSF2RA | colony stimulating factor 2 receptor, alpha | lipA | lipase A, lysosomal acid, cholesterol esterase |
| csf3r | colony stimulating factor 3 receptor | LYPLA3 | lysophospholipase 3 |
| CTSA | cathepsin A | PARVB | parvin, beta |
| CTSB | cathepsin B | pdgfra | platelet-derived growth factor receptor, alpha polypeptide |
| CTSC | cathepsin C | Pdgfrb | platelet-derived growth factor receptor, beta polypeptide |
| CTSH | cathepsin H | Pik3cb | phosphoinositide-3-kinase, catalytic, beta polypeptide |
| CTSL2 | cathepsin L2 | Samd13 | deoxyribonuclease II beta;sterile alpha motif domain containing 13 |
| CTSS | cathepsin S | SPP1 | secreted phosphoprotein 1 |
| CXCL12 | chemokine (C-X-C motif) ligand 12 | TCIRG1 | T-cell, immune regulator 1, ATPase, H+ transporting |
| CXCL13L2 | similar to macrophage inflammatory protein-2 | Tgfbr2 | transforming growth factor, beta receptor II (70/80kDa) |
| CXCL14 | chemokine (C-X-C motif) ligand 14 | TLN1 | talin 1 |
| CXCR4 | chemokine (C-X-C motif) receptor 4 | ZYX | zyxin |
| FLNB | filamin B, beta |
Gene network analysis for DEGs
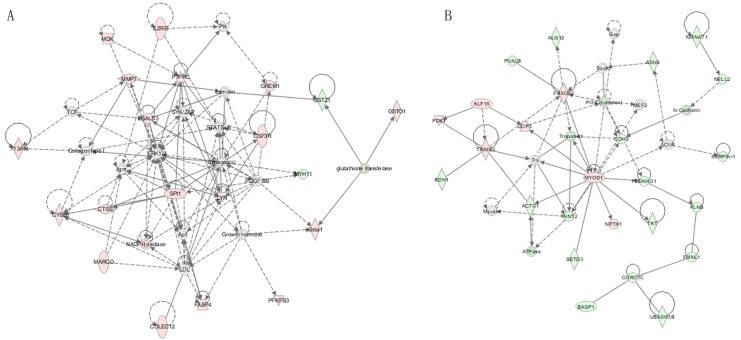
Subsequently, we used Ingenuity Pathway Analysis (IPA, Ingenuity Systems; http://www.ingenuity.com) to investigate the gene networks for DEGs identified in the inner comparisons (WRRh vs. WRRl and XHh vs. XHl). The results showed that the top gene network was cellular movement (Fig 2A). Among the DEGs, FABP4, LGALS3, LYN and SPI1 were central of this network. Network analysis for all DEGs in the cross-breed comparisons (WRRh vs. XHh and WRRl vs. XHl) revealed that the top gene network was skeletal and muscular system development and function (Fig 2B), with MYOD1 and FBXO32 as node genes.
Fig 2. The top gene network in the inner contrasts and across-breed contrasts.
A: The top gene network identified in inner contrasts (WRRh Vs. WRRl and XHh Vs. XHl). B: The top gene network identified in across-breed contrasts (WRRh Vs. XHh and WRRl Vs. XHl). Genes exhibiting up-regulated were shown in red, while gene exhibiting down-regulated were shown in green color. The color intensity indicated the degree of up-/down-regulated. Solid lines and dashed lines indicated direct interaction and indirect interaction, respectively.
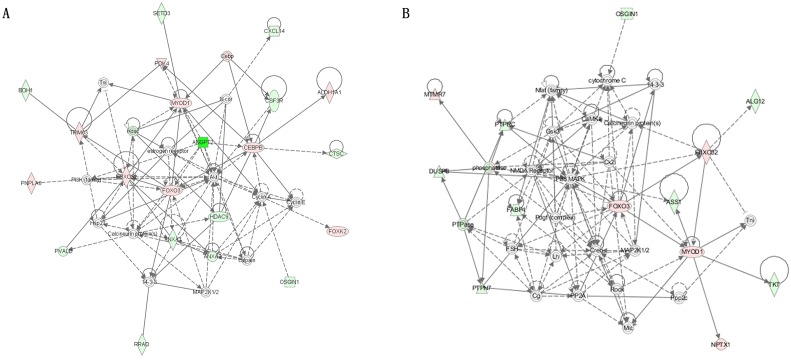
The results of the GO, KEGG pathway and gene network analyses indicated that CEBPB, FBXO32 and MYOD1 might be the key genes related to chicken growth at 7 weeks of age. Further investigation of gene networks involving these three genes showed that FOXO3 might be the crucial gene, interacting with CEBPB, FBXO32, and MYOD1 and to affect growth (Fig 3A and 3B, which show networks compared for WRRh vs. XHh and WRRl vs. XHl, respectively). In WRRh vs. XHh and WRRl vs. XHl, the CEBPB, FBXO32, FOXO3 and MYOD1 mRNA levels were all higher in slow-growing chickens than in fast-growing chickens (Table 6). Furthermore, FBXO32 and FOXO3 showed relatively high fold changes in both of these two comparisons. Therefore, FBXO32 and FOXO3 were selected for functional analysis in this study.
Fig 3. Gene networks involved the FBXO32 and MYOD1 in WRRh Vs. XHh and WRRl Vs. XHl.
A: in WRRh Vs. XHh. B: in WRRl Vs. XHl. Genes exhibiting up-regulated were shown in red, while gene exhibiting down-regulated were shown in green color. The color intensity indicated the degree of up-/down-regulated. Solid lines and dashed lines indicated direct interaction and indirect interaction, respectively.
Table 6. Differential expression of CEBPB, FBXO32, FOXO3 and MYOD1 in contrasts of WRRh Vs. XHh and WRRl Vs. XHl.
| Gene | WRRh FPKM | XHh FPKM | log2Ratio (XHh/ WRRh) | WRRl FPKM | XHl FPKM | log2Ratio (XHl/ WRRl) |
|---|---|---|---|---|---|---|
| CEBPB | 45.09 | 159.10 | 1.82 | 62.82 | 138.74 | 1.14 |
| FBXO32 | 23.94 | 208.70 | 3.12 | 21.52 | 235.93 | 3.45 |
| FOXO3 | 14.13 | 92.67 | 2.71 | 30.76 | 90.89 | 1.56 |
| MYOD1 | 118.89 | 277.56 | 1.22 | 112.65 | 313.63 | 1.48 |
cDNA clone of the chicken FOXO3 gene
Using PCR amplification and 3’ rapid amplification of cDNA ends (3’RACE), a 2,882 bp (or 1,874 bp) cDNA fragment of the chicken FOXO3 gene, including the 1,443 bp CDS and the full-length 3’ UTR (1,439 bp or 431 bp), was obtained (S3 Fig). The chicken FOXO3 showed more than 85.4% identity with its human, mouse, rat, elephant, whale, pig, dog, sheep and cattle, counterparts. However, much lower homology with cFOXO3 was found for fish and xenopus, which were 47.4% and 77.1%, respectively.
FOXO3 and FBXO32 gene mRNA expression in different tissues and between different breeds
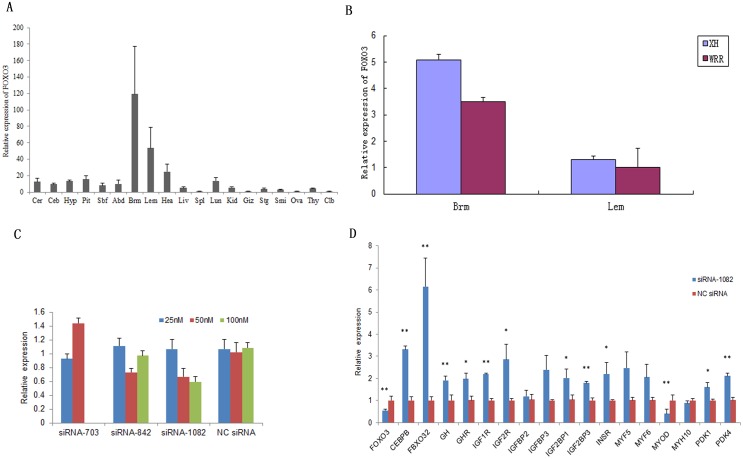
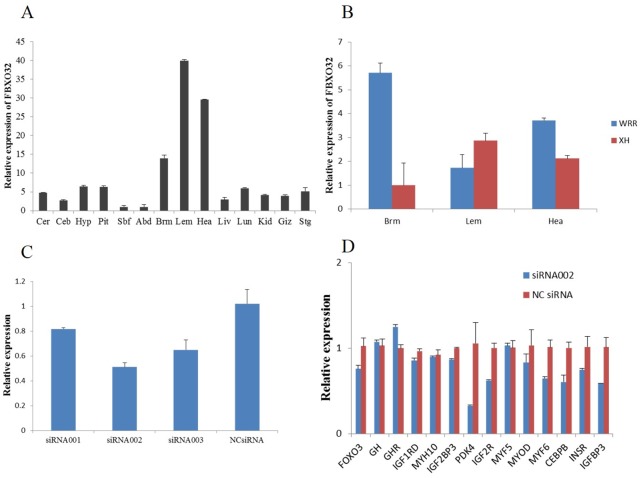
The mRNA expression analysis in various tissues showed that the chicken FOXO3 gene was predominantly expressed in breast muscle and leg muscle tissues, followed by heart, lung and pituitary (Fig 4A). FOXO3 expression was compared between WRR and XH chickens with normal body weight (BW) at seven weeks of age. In breast muscle, its mRNA level was significantly higher in XH chickens (P < 0.01) than in WRR chickens (Fig 4B). In leg muscle, expression in XH was significantly higher (P < 0.05) than in WRR (Fig 4B). The chicken FBXO32 gene was predominantly expressed in leg, heart and breast muscle (Fig 5A). The mRNA level of FBXO32 was significantly higher in breast and heart muscle than in XH (P < 0.05) but lower in leg muscle (P < 0.05) (Fig 5B).
Fig 4. FOXO3 expression in different tissues, different breeds, DF-1 cells and the up/down-regulation of growth-related genes.
A: The mRNA level of FOXO3 in different tissues. Cer: cerebrum, Ceb: cerebellum, Hyp: hypothalamus, Pit: pituitary, Sbf: subcutaneous fat, Abd: abdominal fat, Brm: breast muscle, Lem: leg muscle, Hea: heart, Liv: liver, Spl: spleen, Lun: lung, Kid: kidney, Giz: gizzard, Stg: stomachus glandularis, Smi: small intestine, Ova: vary, Thy: thymus, Clb: cloacal bursa. The horizontal axis indicated different tissues, while the vertical axis indicated 2-ΔΔCt value. B: The expression difference of FOXO3 between XH and WRR. C: Relative expression of FOXO3 in DF-1 cells transfected with different siRNAs. The x-axis described siRNAs and the y-axis showed the relative expression of FOXO3. D: The relative expression changes of growth related genes in DF-1 transfected with siRNA-1082 at 100 nM. **and*indicated P < 0.01 and P < 0.05, respectively.
Fig 5. FBXO32 expression in different tissues, different breeds, DF-1 cells and the up/down-regulation of growth-related genes.
A: The mRNA level of FBXO32 in different tissues in XH chickens. Cer: cerebrum, Ceb: cerebellum, Hyp: hypothalamus, Pit: pituitary, Sbf: subcutaneous fat, Abd: abdominal fat, Brm: breast muscle, Lem: leg muscle, Hea: heart, Liv: liver, Lun: lung, Kid: kidney, Giz: gizzard, Stg: stomachus glandularis. The horizontal axis indicated different tissues, while the vertical axis indicated 2-ΔΔCt value. B: The expression difference of FBXO32 between XH and WRR. C: Relative expression of FBXO32 in DF-1 cells transfected with different siRNAs. The x-axis described siRNAs and the y-axis showed the relative expression of FBXO32. D: The relative expression changes of growth related genes in DF-1 transfected with siRNA002 at 50 nM. **and*indicated P < 0.01 and P < 0.05, respectively.
The expression change of growth-related genes after RNA interference of candidate genes
To find suitable conditions for subsequent RNA interference experiments, we first evaluated the cell activity after transfection. From 5 to 48 h post-transfection, cell activity was not reduced and apoptosis did not occur in either DF-1 cells (S4A Fig) and skeletal muscle cell (S4B Fig). Moreover, the cell density became stronger with the advancement of time. Therefore, the transfected cells were collected at 48 h post transfection for further analysis.
To investigate the interfering efficiency of different small interfering RNAs (siRNAs), three specific siRNAs targeting FOXO3, siRNA-703, siRNA-842 and siRNA-1082 were transfected into DF-1 cells at three different concentrations (25 nM, 50 nM and 100 nM). Cells treated with non-specific siRNA (NC siRNA) were used as a control. The transfection of DF-1 cells with 25 nM siRNA-703 led to a 10% decrease in FOXO3 gene expression, whereas transfection with 25 nM siRNA-842 or siRNA-1082 had no effect on gene expression (Fig 4C). After transfection with 50 nM siRNA-703, gene expression was not decreased. After transfection of siRNA-842 or siRNA-1082 at 50 nM, gene expression decreased 30% (Fig 4C). The best knockdown was mediated by siRNA-1082 transfection at 100 nM, resulting in 41% reduced gene expression (Fig 4C). For FBXO32, siRNA-001, siRNA-002 and siRNA-003 were synthesized and transfected into DF-1 cells at 50 nM which is the recommended concentration for Lipofectamine 2000. NC siRNA was used as a control at 50 nM. The three siRNAs decreased FOXO3 gene expression by 18%, 49% and 35%, respectively (Fig 5C). Therefore, siRNA-002 was used for transfecting DF-1 cells.
Then, 100 nM siRNA-1082 was transfected into DF-1 cells. At 48 h after transfection, qPCR results showed that it effectively (P < 0.01) inhibited the expression of FOXO3 (Fig 4D). FOXO3 expression knockdown resulted in the significant up-regulation of many growth related genes, including CEBPB (P < 0.01), FBXO32 (P < 0.01), GH (P < 0.01), GHR (P < 0.05), IGF1R (P < 0.01), IGF2R (P < 0.05), IGF2BP1 (P < 0.05), IGF2BP3 (P < 0.01), INSR (P < 0.05), PDK1 (P < 0.05) and PDK4 (P < 0.01), while the expression of MYOD was highly significantly down-regulated (P < 0.01). The mRNA levels of IGFBP2, IGFBP3, MYF5, MYF6 and MYH10 were not significantly changed (P > 0.05) (Fig 4D). The IGF1 and IGF2 expression levels were too low to be detected in DF-1 cells. After FBXO32 gene expression was significantly decreased, many growth-related genes, including PDK4, IGF2R and IGFBP3, were significantly down-regulated (P < 0.05). The GH and GHR expression levels tended to increase. (Fig 5D)
Association of SNPs in both of FOXO3 and FBXO32 with chicken growth traits
A total of 18 SNPs were identified in the chicken FOXO3 gene (Table 7). Four SNPs were located in the intron region and 11 were located in the coding region. Among these SNPs in the coding region, only one (G66715314C) was a non-synonymous variation (Ala → Pro). Then, 13 SNPs, C66716195T (rs314403820), G66716193A (rs317912153), A66716179G (rs15379322), A66716172G (rs15379320), T66716119C (rs15379317), A66716041G (rs15379315), C66716002T, G66715897A, C66715807T, C66715738T (rs317670452), G66715672A (rs317919122), A66715543G (rs15379310) and T66715378C (rs15379307), were selected for marker-trait association analyses (Table 8). The minor allele frequency for each SNP was greater than 0.05. Association analysis showed that G66716193A (rs317912153) was significantly (P < 0.05) (P < 0.01) associated with growth traits, including BW at different weeks, shank length at 70 days (SL70), shank length at 84 days (SL84), shank diameter at 70 days (SD70) and the average daily gain (ADG) from 4 to 8 weeks. Birds with the AG genotype had higher BW at various time points than those with the GG genotype. The site C66716002T was significantly (P < 0.05) associated with growth traits including BW at 14 days, 21 days, 77 days and SD70. Conversely, the polymorphic site C66716195T (rs314403820) was significantly (P < 0.05) associated with carcass traits, such as chest width (CW), dressed weight (DW), semi-eviscerated weight (SEW), eviscerated weight (EW) and breast muscle weight (BMW). The site A66716179G (rs15379322) had significant (P <0.05) association with SD70 and abdominal fat pad weight (AFW).
Table 7. Summary of SNPs identified in the chicken FOXO3 gene.
| NO. | SNP 1 | RefSNP | Region | Nucleotide change | AA change 2 |
|---|---|---|---|---|---|
| 1 | G66786154A | rs315777420 | unknown | / | / |
| 2 | 66785514GTTC Indel | / | unknown | / | / |
| 3 | G66785391A | / | unknown | / | / |
| 4 | C66716195T | rs314403820 | Intron | / | / |
| 5 | G66716193A | rs317912153 | Intron | / | / |
| 6 | A66716179G | rs15379322 | Intron | / | / |
| 7 | A66716172G | rs15379320 | Intron | / | / |
| 8 | T66716119C | rs15379317 | Exon | CAT → CAC | H → H |
| 9 | A66716041G | rs15379315 | Exon | CCA → CCG | P → P |
| 10 | C66716002T | / | Exon | GCC → GCT | A → A |
| 11 | G66715897A | / | Exon | TCG → TCA | S → S |
| 12 | C66715807T | / | Exon | AAC → AAT | N → N |
| 13 | C66715738T | rs317670452 | Exon | GAC → GAT | D → D |
| 14 | G66715672A | rs317919122 | Exon | CCG → CCA | P → P |
| 15 | A66715543G | rs15379310 | Exon | ACA → ACG | T → T |
| 16 | T66715378C | rs15379307 | Exon | TTT → TTC | F → F |
| 17 | G66715314C | / | Exon | GCC → CCC | A → P |
| 18 | C66715165T | rs317127440 | Exon | AAC → AAT | N → N |
1The number indicated the position on the chromosome 3.
2AA indicated amino acid.
Table 8. Association of SNPs in the chicken FOXO3 gene with the growth and carcass traits.
| SNP | Traits | P value | Least-square Means ± standard errors (SE) | ||
|---|---|---|---|---|---|
| G66716193A | BW7 (g) | <1×10−4 | 56.86±1.02A(AA/52) | 61.33±0.68B(AG/116) | 58.18±0.60A(GG/220) |
| BW14 (g) | 0.0245 | 120.90±2.17a(AA/55) | 126.00±1.40b(AG/124) | 121.67±1.25a(GG/244) | |
| BW21 (g) | 0.0326 | 206.49±4.23ab(AA/51) | 214.55±2.64a(AG/125) | 205.90±2.38b(GG/239) | |
| BW35 (g) | 0.0455 | 433.45±9.91ab(AA/54) | 446.95±6.38a(AG/122) | 425.48±5.73b(GG/236) | |
| BW42 (g) | 0.0316 | 561.83±13.13ab(AA/55) | 585.67±8.54a(AG/122) | 556.68±7.66b(GG/240) | |
| BW49 (g) | 0.0408 | 691.84±15.63ab(AA/55) | 721.30±10.11a(AG/124) | 689.33±9.02b(GG/243) | |
| BW56 (g) | 0.0104 | 843.66±18.33b(AA/55) | 883.89±11.85Aa(AG/123) | 838.64±10.54Bb(GG/243) | |
| BW70 (g) | 0.0005 | 1124.73±27.26AB(AA/47) | 1169.77±17.74A(AG/94) | 1075.11±15.47B(GG/196) | |
| BW77 (g) | 0.0127 | 1311.23±31.19AB(AA/48) | 1360.08±20.28A(AG/95) | 1279.17±17.67B(GG/197) | |
| BW84 (g) | 0.0373 | 1461.47±40.97ab(AA/36) | 1514.82±25.69a(AG/76) | 1425.07±22.82b(GG/161) | |
| SL70 (mm) | 0.0065 | 81.62±0.77AB(AA/47) | 83.16±0.50A(AG/94) | 81.09±0.43B(GG/196) | |
| SD70 (mm) | 0.0415 | 9.50±0.13ab(AA/47) | 9.55±0.08a(AG/94) | 9.23±0.07b(GG/196) | |
| SL84 (mm) | 0.0403 | 87.76±0.94ab(AA/36) | 89.52±0.59a(AG/76) | 87.66±0.52b(GG/161) | |
| ADG2 (g/day) | 0.0144 | 19.15±0.53b(AA/55) | 20.26±0.35Aa(AG/120) | 18.99±0.30Bb(GG/240) | |
| C66716002T | BW14 (g) | 0.0326 | 122.52±0.93a(CC/332) | 124.75±1.85a(TC/83) | 136.29±5046b(TT/7) |
| BW21 (g) | 0.0361 | 208.33±1.76a(CC/325) | 210.71±3.47a(TC/82) | 234.99±10.25b(TT/7) | |
| BW77 (g) | 0.0454 | 1302.25±13.26a(CC/267) | 1341.32±26.25ab(TC/66) | 1473.21±76.22b(TT/6) | |
| SD70 (mm) | 0.0462 | 9.36±0.054 a(CC/265) | 9.58±0.11ab(TC/65) | 9.92±0.30b(TT/6) | |
| C66716195T | CW (mm) | 0.0131 | 68.20±4.49a(CC/76) | 62.43±5.199b(TC/163) | 62.26±6.55ab(TT/183) |
| DW (g) | 0.0131 | 1347.10±157.76a(CC/76) | 1136.51±182.62b(TC/163) | 1119.53±230.26ab(TT/183) | |
| SEW (g) | 0.0212 | 1227.26±142.5a9(CC/76) | 1046.46±165.07b(TC/163) | 1030.24±208.12ab(TT/182) | |
| EW (g) | 0.0199 | 1097.35±125.03a(CC/76) | 935.36±144.74b(TC/163) | 918.04±182.49ab(TT/182) | |
| BMW (g) | 0.0132 | 91.07±11.39a(CC/76) | 79.69±13.12b(TC/163) | 83.25±16.63ab(TT/183) | |
| A66716179G | SD70(mm) | 0.0286 | 9.49±0.07A(AA/204) | 9.37±0.07a(AG/111) | 9.00±0.17Bb(GG/22) |
| AFW (g) | 0.0478 | 24.94±9.62a(AA/251) | 7.97±12.41b(AG/143) | 1.87±19.63ab (GG/227) | |
BW7, BW14, BW21, BW35, BW42, BW49, BW56, BW70, BW77 and BW84 indicated body weight at 7, 14, 21, 35, 42, 49, 56, 70 and 77 days, respectively. SL70 or SL84 = shank length at 70 or 84 days. SD70 = shank diameter at 70 days. ADG2 meant average daily gain from 4 to 8 weeks. CW = chest width. DW = dressed weight. SEW = semi-eviscerated weight. EW = eviscerated weight. BMW = breast muscle weight. AFW = abdominal fat pad weight. Number in brackets meant the number of birds tested for each genotype. The a,b or A,B values with no common superscripts within a column indicated the comparison was differed significantly (P < 0.05 or P < 0.01).
SNPs of chicken FBXO32 were obtained from the NCBI SNP database. The 5’flank region is considered the promoter region of many genes. A total of 14 SNPs in the 5’flank region of FBXO32 were chosen and confirmed by sequencing (Table 9). All of these SNPs, including A137669051T (rs314992955), C137669076T (rs313186471), C137669094T (rs741279417), C137669311G (rs733740424), C137669446T (rs16155707) A137669484C (rs737463671), A137669551G (rs316423136), A137669618G (rs731278593), C137669630T (rs315617235), A137669638T (rs313162537), C137669668G (rs317677782), A137669706G (rs316476530), A137669723C (rs313990885), A137669732G (rs735987765), were selected for marker-trait association analyses (Table 10). Association analysis showed that C137669094T (rs741279417) was significantly (P < 0.05) (P < 0.01) associated with carcass traits and meat quality traits, including small intestine length (SIL) and breast muscle fleshcolor (BMF). The site C137669311G (rs733740424) was significantly (P < 0.05) associated with BMF. The site C137669446T (rs16155707) was significantly associated with chest angle (CA) (P < 0.01) and head width (HW) (P < 0.05). The site A137669484C (rs737463671) was significantly (P < 0.05) associated with CA. The site A137669551G (rs316423136) was significantly (P < 0.05) associated with CW. The site A137669618G (rs731278593) was significantly (P < 0.05) (P < 0.01) associated with SL at various weeks. The site C137669668G (rs317677782) was significantly (P < 0.05) (P < 0.01) associated with leg muscle fleshcolor (LMF) and BMF. Moreover the site A137669723C (rs313990885) and A137669732G (rs735987765) was significantly (P < 0.05) associated with BMF.
Table 9. Summary of SNPs identified in the 5' region of chicken FBXO32 gene.
| NO. | SNP 1 | RefSNP | Region | Nucleotide change |
|---|---|---|---|---|
| 1 | A137669051T | rs314992955 | 5'near gene | / |
| 2 | C137669076T | rs313186471 | 5'near gene | / |
| 3 | C137669094T | rs741279417 | 5'near gene | / |
| 4 | C137669311G | rs733740424 | 5'near gene | / |
| 5 | C137669446T | rs16155707 | 5'near gene | / |
| 6 | A137669484C | rs737463671 | 5'near gene | / |
| 7 | A137669551G | rs316423136 | 5'near gene | / |
| 8 | A137669618G | rs731278593 | 5'near gene | / |
| 9 | C137669630T | rs315617235 | 5'near gene | / |
| 10 | A137669638T | rs313162537 | 5'near gene | / |
| 11 | C137669668G | rs317677782 | 5'near gene | / |
| 12 | A137669706G | rs316476530 | 5'near gene | / |
| 13 | A137669723C | rs313990885 | 5'near gene | / |
| 14 | A137669732G | rs735987765 | 5'near gene | / |
1The number indicated the position on the chromosome 2.
Table 10. Association of SNPs in the chicken FBXO32 gene with the carcass and meat quality traits.
| SNP | Traits | P value | Least-square Means ± standard errors (SE) | ||
|---|---|---|---|---|---|
| C137669094T | SIL(cm) | 0.0463 | 143.61±1.68a(TT/123) | 141.46±4.27ab(CC/11) | 135.85±2.41b(TC/46) |
| BMF | 0.0073 | 56.79±0.96a(TT/124) | 49.91±3.29b(CC/11) | 54.70±1.48ab(TC/46) | |
| C137669311G | BMF | 0.0110 | 56.44±1.32(CC/81) | 55.36±1.26(CG/56) | 55.90±1.56(GG/39) |
| C137669446T | CA(°) | 0.0046 | 60.83±0.58A(CC/170) | 66.55±3.20B(CT/3) | 69.45±2.46B(TT/8) |
| HW(mm) | 0.0476 | 30.79±0.19a(CC/163) | 27.70±0.94b(CT/3) | 28.74±0.72b(TT/8) | |
| A137669484C | CA(°) | 0.0415 | 61.17±0.60b(CC/144) | 61.09±1.25b(AC/27) | 64.73±1.95a(AA/7) |
| A137669551G | CW(mm) | 0.0218 | 66.97±0.83b(GG/130) | 73.330±1.46a(AG/30) | 70.97±1.84a(AA/19) |
| A137669618G | SL63(mm) | 0.0263 | 79.28±1.01b(GG/47) | 87.10±2.09a(GA/4) | 82.08±2.88ab(AA/3) |
| SL70 (mm) | 0.0433 | 83.26±0.60b(GG/118) | 87.05±2.09a(GA/13) | 82.37±2.33b(AA/13) | |
| SL77 (mm) | 0.0070 | 89.24±1.23B(GG/47) | 99.03±3.31A(GA/4) | 92.58±3.36B(AA/3) | |
| SL84 (mm) | 0.0041 | 89.77±1.43B(GG/103) | 95.52±2.04A(GA/12) | 89.57±2.61B(AA/11) | |
| C137669668G | LMF | 0.0140 | 65.98±0.74a(GG/123) | 64.33±0.19a(GC/43) | 58.26±3.01b(CC/12) |
| BMF | 0.0072 | 57.13±0.87A(GG/123) | 54.25±1.46AB(GC/43) | 50.12±2.87B (CC/12) | |
| A137669638T | HW(mm) | 0.0314 | 30.56±0.26ab (TT/64) | 30.94±0.22a (TA/59) | 30.44±0.27b(AA/46) |
| A137669723C | BMF | 0.0128 | 55.99±1.20b(CC/70) | 55.15±1.22b(AC/59) | 59.92±1.49a(AA/47) |
| A137669732G | BMF | 0.0147 | 55.99±1.20ab(GG/70) | 55.21±1.27b (GA/59) | 56.47±1.47a(AA/46) |
SL63, SL70, SL77 or SL84 indicated shank length at 63, 70, 77 or 84 days, respectively. CA = chest angle. SIL = small intestine length. HW = head width. CW = chest width. BMF = breast muscle fleshcolor. LMF = leg muscle fleshcolor. The a,b or A,B values with no common superscripts within a column indicated the comparison was differed significantly (P < 0.05 or P <0.01).
Discussion
During postnatal growth, skeletal muscle increase occurs mainly due to muscle hypertrophy accompanied by satellite cell proliferation to incorporate new myonuclei into existing myofibers [11]. In this study, the comparison with the highest number of DEGs resulted in the most GO biological process terms, and terms of development process, cell differentiation and cell adhesion were present in multiple comparisons. Lysosome was the most significantly enriched pathway. Lysosomes are a membrane-bound cell organelles that contain acid hydrolase enzymes and are interlinked with endocytosis, phagocytosis and autophagy. Previous research showed that the autophagy-lysosome system was a key player in regulating protein degradation in skeletal muscle [12, 13]. Moreover, other enriched pathways were cell junction-related pathways (focal adhesion, ECM-receptor interaction and cytokine-cytokine receptor interaction), suggesting that pathways critical to maintaining the integrity of tissues might be involved in chicken growth at 7 weeks of age. Focal adhesion complexes serve as mechanical linkages to the ECM and are the signaling centers of numerous intracellular pathways related to cell motility, proliferation and differentiation [14]. ECM components play integral roles in the formation of the muscle satellite cell niche, and their specific interactions with satellite cells can regulate cell adhesion, migration, differentiation, proliferation and self-renewal [15]. Cytokines such as IL-6 are key regulators of cell growth, proliferation, differentiation and apoptosis [16]. Some cytokines produced by myofibers and peritendinous tissue are, termed myokines [17]. Myokines activate of intracellular signaling cascades by binding to their specific receptors, thereby regulating metabolism in skeletal muscle [18].
Importantly, we identified, three crucial transcription factors, CEBPB, FBXO32 and MYOD1, that might be related to chicken growth. Previous research revealed that CEBPB regulated multiple genes in response to GH [19]. Moreover, CEBPB was an activator of adipogenesis and acted as an inhibitor of myogenesis [20, 21]. As one well-known myogenic regulatory factor, MYOD1 has been extensively studied, and its effect on growth has been well-demonstrated [22, 23]. It can trans-differentiate many cell types to muscle cells and is a key regulator of myogenesis [23–25]. Several recently published reports suggested that MYOD1 could modulate and facilitate the assembly of muscle enhancers [26, 27]. Furthermore, CEBPB, FBXO32 and MYOD1 can interact with each other [20, 28]. In this study, network analysis indicated that FOXO3 was a central gene interacting with CEBPB, FBXO32 and MYOD1.
FOXO3 is a member of the Forkhead box class O (FOXO) transcription factor family. Like other members of this family, FOXO3 was demonstrated to play a crucial role in many species, from lower animals to mammals. It performs a variety of cellular functions, including cell growth and differentiation, cell-cycle control, energy metabolism, DNA damage repair, response to oxidative stress, and apoptosis [29–33]. In mammals, FOXO3 was relatively ubiquitously expressed, consistent with our findings in chicken [34]. However, the mammalian FOXO3 was predominantly expressed in heart, brain, kidney and ovary, whereas the chicken FOXO3 was highly expressed in breast muscle and leg muscle tissues [34]. A previous study suggested that FOXO3 contributes to cell growth in striated muscle [33]. Our expression comparison between fast-growing and slow-growing breeds also confirmed its important function in skeletal muscle. It is possible that the higher expression of CEBPB, FBXO32 and FOXO3 in slow-growing XH chickens contributed to their lower growth performance, whereas the higher level of MYOD1 would partly counteract these effects. Moreover, a series of genes involved in the somatotropic axis, including GH, GHR, IGF1R, IGF2R, IGF2BP1 and IGF2BP3, was effectively up-regulated by FOXO3 knockdown after siRNA interference, suggesting that FOXO3 might play an important role in growth. In particular, some genes remarkably up-regulated by FOXO3 knockdown, including INSR, PDK1 and PDK4, were present upstream of the IGF1/FOXO signal transduction pathway, which would further promote FOXO3 inhibition [33, 34]. Recent studies suggested that MYOD was a direct target of FOXO3 in myoblasts [35]. In vivo and in vitro, FOXO3 was demonstrated to play an important role in activating MYOD transcription [35]. In this study, FOXO3 knockdown resulted in a significant down-regulation of MYOD, whereas expression of the other MRFs (MYF5 and MYF6) was unaffected, consistent with previously published studies [35]. These results indicated that FOXO3 could interact with MYOD but not the others MRFs. Conversely, previous research identified FOXO3 as a major activator of FBXO32 expression, a factor associated with skeletal muscle protein degradation [36, 37]. In contrary, we found that FOXO3 knockdown strongly increased FBXO32 expression. Nevertheless, our data revealed that FOXO3 down-regulation induced a dramatic down-regulation of CEBPB, which is an inhibitor of myogenesis [20]. The up/down-regulation of FBXO32 and CEBPB was inconsistent with the reported inhibitory roles of FOXO3 on muscle growth. Therefore, further study is needed to investigate protein expression changes. The human FOXO3 contains a sequence of 95 amino acids forming a forkhead DNA-binding domain motif (148–257 amino acids). It also possesses a nuclear localization sequence (NLS, 249–251 and 269–271 amino acids), a nuclear export sequence (NES, 386–396 amino acids) and a conserved C-terminal transactivation domain [38]. The sequences of the chicken FOXO3 gene showed high identity with the previously cloned mammalian FOXO3. The chicken FOXO3 had the same amino acids as the human FOXO3 in the NLS and NES. It shared high homology with human FOXO3 in the forkhead DNA-binding domain, with a six amino acid difference.
FBXO32, also known as Atrogin 1 or MAFbx, is a skeletal and cardiac muscle-specific F-box containing protein that was shown to be associated with the maintenance of muscle mass [22, 25]. FBXO32 is a muscle-specific gene that has a key function in muscle atrophy [39]. Previous research on feed deprivation showed that as the muscle degraded, FBXO32 expression increased [40]. Although we did not find any association between SNPs of FBXO32 and chicken growth traits, our findings showed that it was highly expressed in chicken leg muscle, heart and breast muscle tissues, which is in accordance with its role. FBXO32 was regulated by multiple transcription factors. A previous study suggested that FOXO3 can act on the promoter of FBXO32, mediating FBXO32 transcription and muscle cell atrophy [41]. After FBXO32 expression significantly decreased, growth related genes including PDK4, IGF2R and IGFBP3 were significantly down-regulated. Previous published reports showed that FBXO32 and PDK4 may interact in skeletal muscle metabolism [42, 43]. The expression levels of GH and GHR had tendency to increase, indicating that FBXO32 may have influence on muscle growth at the mRNA level.
Thus far, many SNPs and QTLs were reported to correlate with growth [3, 4]. In humans, one FOXO3 promoter SNP was associated with human body mass index [44]. In pig, FOXO3 SNPs were closely associated with growth and development traits [45]. To date, this study is the first to scan variations in the chicken FOXO3 gene and then evaluate their effects on chicken growth. In humans, FOXO3 was phosphorylated and acetylated on multiple sites, including K242, K245, K259, K271, K290, K569, S207, S253, S295, S315, S318, S321, S325, S345, S399, S413, S426, S555, S588, S626, S644, T32 and T179 [33, 38, 46]. In this study, we scanned the entire exon 2 of chicken FOXO3, and a total of 11 SNPs were identified in the 1,432 bp exon 2 fragment. These variation sites corresponded to R222, P238, A251, S286, N316, D339, P361, T404, F459, T475 and N530 in human FOXO3. Although these variations were not present in phosphorylation or acetylation sites, C66716002T (corresponding to A251 in human FOXO3), was located in the forkhead DNA-binding domain, which is crucial in regulating the transcription of target genes. This SNP did not cause an amino acid change, but might affect mRNA splicing, stability, or protein folding, and thereby alter protein functions such as DNA-binding [47]. Moreover, it is located in the NLS region. Our association analysis demonstrated that this mutation had a significant effect on growth traits. Among the mutations identified in exon 2 of chicken FOXO3, G66715314C was a non-synonymous variation resulting in a change in the amino acid sequence. It was previously shown that non-synonymous SNPs affect protein functions and protein-protein interactions [48, 49]. Therefore, the G66715314C mutation may have important effects on economic traits, and its association with chicken growth traits needs further study. Conversely, four SNPs found in this study were located near the exon-intron boundaries, which are special regions that usually had important functional roles in protein [50]. Three of these four SNPs were associated with growth traits. Importantly, the G66716193A site, which was 60 bp away from the intron-exon boundary, was significantly associated with most growth traits.
In summary, this study provided a comprehensive transcriptome analysis of breast muscle between fast- and slow-growing chickens. CEBPB, FBXO32, FOXO3 and MYOD1 may play key roles in chicken growth at seven weeks of age. Further expression analysis, siRNA analysis and association analysis demonstrated that chicken FOXO3 is a candidate gene involvement in chicken growth. Our observations provide new clues to understand the molecular basis of chicken growth.
Materials and Methods
Ethics statement
All animal experiments were performed in accordance with and were approved by the Animal Care Committee of South China Agricultural University (Guangzhou, China) (approval ID: SCAU#0011). All efforts were made to minimize animal suffering.
Animals and tissues
Two chicken breeds WRR and XH were used for RNA-Seq in this study. WRR is a breed with a fast growth rate, while XH is a Chinese native breed with a slow growth rate. WRR and XH were provided by Guangdong Wens Foodstuff Company Ltd, Guangdong, China and Zhicheng Avian Breeding Company Ltd, Guangdong, China, respectively. Briefly, birds were fed ad libitum with 16.5% CP and 2800 kcal of ME/kg, with free access to water. BW was measured at seven weeks of age and three female chickens from each of the two-tail samples of WRR and XH were selected based on it. The four groups WRR with high weight (WRRh), WRR with low weight (WRRl), XH with high weight (XHh) and XH with low weight (XHl) were generated. Their average BW7 values were 1064.0 ± 11.1, 695.0 ± 24.4, 305.8 ± 23.3 and 207.6 ± 11.1 g, respectively. At 7 weeks of age, chickens were killed by stunning followed by exsanguination. Breast muscles were collected and stored at -80°C until RNA extraction.
Five XH chickens at 7 weeks of age were used for expression analysis of FOXO3 and FBXO32 in different tissues. A total of 19 tissues, cerebrum, cerebellum, pituitary, hypothalamus, heart, liver, spleen, lung, kidney, breast muscle, leg muscle, muscular stomach, glandular stomach, ovary, duodenum, subcutaneous fat, abdominal fat, thymus and bursa of Fabricius, were collected from each chicken.
Four XH and four WRR chickens with normal BW were used for the expression analysis between different breeds by qPCR. Breast muscle and leg muscle tissues were dissected from those 8 birds. All those birds were obtained from the Chicken Breeding Farm of South China Agricultural University, Guangdong, China.
An F2 full sibling hybrid population, from a WRR and XH cross as described previously, was used for association analysis between SNPs and chicken growth or carcass traits [51]. Genomic DNA was extracted from EDTA-anticoagulated blood and then used for genotyping.
RNA extraction and RNA-Seq
Total RNA from various tissues and DF-1 cells was isolated by TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and then treated with DNase (Promega, Madison, WI, USA) following the manufacturer’s instructions. RNA quality and concentration were evaluated by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Subsequently, three breast muscle RNAs of the same group were mixed in equal amounts. In this way, four pooled samples (WRRh, WRRl, XHh and XHl) were generated and then were sent to Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China) for RNA-Seq. cDNA libraries were constructed according to Illumina's protocols and then each library was sequenced on a single line of Illumina Hiseq 2000 (Illumina, San Diego, CA, USA) to obtain paired-end 101-bp reads.
Bioinformatic analysis of RNA-Seq
For raw data from RNA-Seq, we first removed reads containing adaptors, unknown bases and low quality bases to obtain high quality reads. Then, the clean reads of the four samples were aligned to the chicken reference genome (http://asia.ensembl.org/info/data/ftp/index.html) by TopHat software, with no more than 2 bp mismatches. The mapped reads were used for further transcriptome annotation and expression calculation with using FPKM (Fragments Per Kilobase of transcript per Million mapped reads). All expressed genes were subjected to GO analysis by AmiGO2 (http://amigo.geneontology.org/amigo), with Bonferroni-adjusted P value < 0.05. With the use of Cuffdiff (http://cufflinks.cbcb.umd.edu/), genes with greater than 2-fold changes between two samples and a q value < 0.05 were regarded as DEG. All those DEGs were subjected to GO analysis and KEGG pathway enrichment analysis with the DAVID Functional Annotation Tool (http://david.abcc.ncifcrf.gov/) using a 0.05 cutoff for the P value and Benjiamini adjusted P < 0.1 cutoff for the Benjamini adjusted p-value. Moreover, all DEGs underwent gene network analysis by IPA.
cDNA synthesis and qPCR
Primers P1 to P5 (S6 Table) were used for the validation of RNA-Seq data by qPCR. For the FOXO3 expression analysis in different tissues and breeds, total RNA was reverse transcribed using a PrimeScript RT reagent Kit with gDNA Eraser (Takara Co., Japan) to synthesize the first-strand cDNAs. qPCR was conducted in a total volume of 20 μL: 10 μL Bestar Real time PCR Master Mix (SYBR Green) (DBI Bioscience, Germany), 0.5 μL of each primer (10 μM), 8.0 μL of RNase-free water and 1 μL of cDNA on a BIO-RAD CFX96 system (Bio-Rad, USA). P4 was used for qPCR of FOXO3 and β-actin (P1) was used as the internal control (S6 Table). All reactions were run in triplicates. The relative expression level was calculated by the 2-ΔΔCt method. Where ΔΔCt corresponded to the difference between ΔCt measured for the mRNA level of each sample and ΔCt measured for the mRNA level of the reference sample, ΔCt = Cttarget gene−Ctreference gene.
Cloning of the chicken FOXO3 gene
Total RNA was extracted from the breast muscle of an XH chicken. According to the predicted partial cDNA sequences of the chicken FOXO3 gene (NCBI accession number: XM_001234495.3), a pair of primers (P6) was designed and primer-directed RT-PCR was used to amplify a partial cDNA sequence and part of the 3’UTR region (S6 Table). The products were subcloned into the pMD18-T vector (TaKaRa Biotechnology Co Ltd, Dalian, China) and then sequenced on Applied Biosystem model 3730 sequencer. Based on the obtained sequences, P7 and P8 (S6 Table) were designed for 3’ RACE. With the use of the SMART RACE cDNA amplification kit (Clontech Laboratories, Mountain View, CA), a breast muscle cDNA library was prepared and the 3’ ends of the cDNA were amplified by specific forward primers and random reverse primers. Sequence analysis was conducted using DNASTAR software (http://www.dnastar.com). The homology of FOXO3 among different species, including human (NP_001446.1), mouse (NP_062714.1), rat (NP_001099865.1), elephant (XP_003404353.1), whale (XP_004264796.1), pig (NP_001129431.1), dog (XP_003639448.1), sheep (NP_001254818.1), cattle (NP_001193012.1), chicken, Xenopus (NP_001086418.1) and fish (XP_003449931.1), was analyzed with MEGA 6 software (http://www.megasoftware.net/).
Polymorphism identification in FOXO3 and FBXO32 and association analysis with growth traits
With the use of P9, P10 and P11 (S6 Table), ten chickens from the WRR and XH F2 full sibling hybrid population were selected to identify polymorphisms in the coding region and intron region near the exon-intron boundaries of the FOXO3 gene through direct sequencing. Primers P30 (S6 Table) were used for identify polymorphisms in the 5' flank region of the FBXO32 gene. Another ten chickens from the WRR & XH F2 full sib hybrid population were selected to sequence. Sequence analyses were conducted with the software DNASTAR V 3.0 (http://www.biologysoft.com/; Steve ShearDown, 1998–2001 version reserved by DNASTAR Inc., Madison, Wisconsin, USA). Only polymorphisms occurring more than twice were regarded as variations. P10 and P30 was also used for the following association analyses in this population by direct sequencing. Association analyses of polymorphisms with chicken growth and carcass traits were conducted using SAS 8.0 software (SAS Institute Inc, Cary, NC, USA) using the following GLM model:
where Y is an observation of the traits, μ is the overall population mean, G is the fixed effect of genotype, D is the random effect of dam, H is the fixed effect of hatch, S is the fixed effect of sex, and e is the residual error.
siRNA design, cell culture and transfection
siRNA molecules were synthesized by Shanghai GenePharma Co. Ltd (Shanghai, China) with commercial service. Three FOXO3 target sites were selected: siRNA-703 (sense: 5’gccggauggaagaauucaatt3’, antisense: 5’ uugaauucuuccauccggctt3’), siRNA-842 (sense: 5’gcgccguguccauggacaatt3’, antisense: 5’uuguccauggacacggcgctt3’) and siRNA-1082 (sense: 5’gcaccgagcuggaugaugutt3’, antisense: 5’acaucauccagcucggugctt3’). A NC siRNA fragment was produced as the control. Three specific siRNAs targeting FBXO32 were synthesized.: siRNA-001 (sense: 5’ ccuucaacagacuugacuu 3’, antisense: 5’ ggaaguugucugaacugaa 3’), siRNA-002 (sense: 5’ gcaacugaacaacauucaa 3’, antisense: 5’ cguugacuuguuguaaguu3’), siRNA-003 (sense: 5’ ggcuaauccuaucugacaa 3’, antisense: 5’ ccgauuaggauagacuguu 3’).
The chicken fibroblast cell line DF-1 was obtained from the Cell Bank of Committee on Type Culture Collection of Chinese Academy of Sciences and was cultured in Dulbecco’s Modified Eagles Medium (DMEM) (GIBCO BRL, Life Technologies, Invitrogen Corporation, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Hyclone Laboratories, Logan, Utah), 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were cultured and maintained in humidified air at 37°C with 5% CO2. When the DF-1 cells were grown to approximately 50–80% confluence, independent transfections of the three siRNAs and the control NC siRNA were carried out using the transfection reagent Lipofectamine 3000 (Invitrogen Corporation, Carlsbad, CA) following the manufacturer’s protocol. For each siRNA targeting FOXO3, a plasmid concentration gradient (25 nM, 50 nM and 100 nM) was set to investigate which concentration ratio had the highest transfection efficiency. Here, experiments for each gradient were conducted in triplicates. For each siRNA targeting FBXO32, the recommended concentration of Lipofectamine 2000 (50 nM) was transfected into DF-1 cells. At 48 h post-transfection, RNA was extracted from cells to perform qPCR. For each siRNA, the expression of target genes at each concentration was compared to that in cells transfected with NC siRNA. P12 was used for qPCR for FOXO3, P14 was used for qPCR for FBXO32, and β-actin (P1) was used as the control (S6 Table). Eventually, we found that siRNA-1082 at 100 nM had the highest relative efficiency of transfection and it was selected for the following experiments. siRNA-002 for FBXO32 had the highest relative efficiency of transfection and was used for transfecting DF-1 cells. When cells were grown to 50–80% confluence, siRNA-1082 and NC siRNA at 100 nM and siRNA-002 and NC siRNA at 50 nM were transfected into DF-1 cells. RNA was collected at 48 h post-transfection. With the use of qPCR, the expression change was analyzed for growth-related genes including CEBPB, FBXO32, GH, GHR, IGF1R, IGF2R, IGFBP2, IGFBP3, IGF2BP1, IGF2BP3, INSR, MYF5, MYF6, MYOD, MYH10, PDK1 and PDK4 (primers are shown in S6 Table).
Supporting Information
(DOC)
WRRh vs. WRRl, XHh vs. XHl, WRRh vs. XHh and WRRl vs. XHl indicate the comparisons between WRRh and WRRl, between XHh and XHl, between WRRh and XHh and between WRRl and XHl, respectively. In each comparisons, up-regulated indicates that the expression in the second group was higher than that in the first group, while down-regulated indicates that the expression in the first group was higher than that in the second group.
(DOC)
Sequences in the black square frame indicated the sequences absent in the second transcript. Sequences in the CDS are presented in uppercase, while those in the 3’ UTR are presented in lowercase.
(DOC)
A: DF-1 cells. B: Skeletal muscle cells.
(TIF)
(XLS)
(XLS)
(XLS)
(DOC)
(XLS)
(DOC)
Acknowledgments
The authors are grateful to Xinzheng Jia and Ouyang Hongjia in our lab for their technical assistance. This research was supported by the Program for New Century Excellent Talents in University (NCET-13-0803), the National Natural Scientific Foundation of China (31172200; 31472090), and the Foundation for High-level Talents in Higher Education of Guangdong, China.
Data Availability
Relevant data are within the paper and its Supporting Information files. The raw RNA-seq data are available on Gene Expression Omnibus (GEO) at accession number GSE72424 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72424).
Funding Statement
This research was supported by the National Natural Science Foundation of China (31172200; 31472090), the Program for New Century Excellent Talents in University (NCET-13-0803), the Key Technology Research and Development Program of Guangdong Emerging Strategic Industries (2012A020800005), and the Foundation for High-level Talents in Higher Education of Guangdong, China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Claire DH, Paul W, Shen X, Jia X, Zhang R, Sun L, et al. Identification and characterization of genes that control fat deposition in chickens. J Anim Sci Biotechnol. 2013;4:43 10.1186/2049-1891-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flisar T, Malovrh S, Tercic D, Holcman A, Kovac M. Thirty-four generations of divergent selection for 8-week body weight in chickens. Poult Sci. 2014;93:16–23. 10.3382/ps.2013-03464 [DOI] [PubMed] [Google Scholar]
- 3. Darzi NM, Masoudi AA, Vaez TR. Association of single nucleotide polymorphism of GHSR and TGFB2 genes with growth and body composition traits in sire and dam lines of a broiler chicken. ANIM BIOTECHNOL. 2014;25:13–22. 10.1080/10495398.2013.803478 [DOI] [PubMed] [Google Scholar]
- 4. Ahsan M, Li X, Lundberg AE, Kierczak M, Siegel PB, Carlborg O, et al. Identification of candidate genes and mutations in QTL regions for chicken growth using bioinformatic analysis of NGS and SNP-chip data. Front Genet. 2013;4:226 10.3389/fgene.2013.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie L, Luo C, Zhang C, Zhang R, Tang J, Nie Q, et al. Genome-wide association study identified a narrow chromosome 1 region associated with chicken growth traits. PLOS ONE. 2012;7:e30910 10.1371/journal.pone.0030910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schmidt A, Schmid MW, Klostermeier UC, Qi W, Guthorl D, Sailer C, et al. Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLOS GENET. 2014;10:e1004476 10.1371/journal.pgen.1004476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ayers KL, Davidson NM, Demiyah D, Roeszler KN, Grutzner F, Sinclair AH, et al. RNA sequencing reveals sexually dimorphic gene expression before gonadal differentiation in chicken and allows comprehensive annotation of the W-chromosome. GENOME BIOL. 2013;14:R26 10.1186/gb-2013-14-3-r26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mutz KO, Heilkenbrinker A, Lonne M, Walter JG, Stahl F. Transcriptome analysis using next-generation sequencing. Curr Opin Biotechnol. 2013;24:22–30. 10.1016/j.copbio.2012.09.004 [DOI] [PubMed] [Google Scholar]
- 9. Liang SS, Ouyang HJ, Liu J, Chen B, Nie QH, Zhang XQ. Expression of variant transcripts of the potassium channel tetramerization domain-containing 15 (KCTD15) gene and their association with fatness traits in chickens. Domest Anim Endocrinol. 2015;50:65–71. 10.1016/j.domaniend.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 10. Hu Y, Xu H, Li Z, Zheng X, Jia X, Nie Q, et al. Comparison of the genome-wide DNA methylation profiles between fast-growing and slow-growing broilers. PLOS ONE. 2013;8:e56411 10.1371/journal.pone.0056411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jiang C, Shi P, Li S, Dong R, Tian J, Wei J, et al. Gene expression profiling of skeletal muscle of nursing piglets. INT J BIOL SCI. 2010;6:627–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del PP, et al. FoxO3 controls autophagy in skeletal muscle in vivo. CELL METAB. 2007;6:458–71. [DOI] [PubMed] [Google Scholar]
- 13. Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. CELL METAB. 2007;6:472–83. [DOI] [PubMed] [Google Scholar]
- 14. Romer LH, Birukov KG, Garcia JG. Focal adhesions: paradigm for a signaling nexus. CIRC RES. 2006;98:606–16. [DOI] [PubMed] [Google Scholar]
- 15. Thomas K, Engler AJ, Meyer GA. Extracellular matrix regulation in the muscle satellite cell niche. CONNECT TISSUE RES. 2015;56:1–8. 10.3109/03008207.2014.947369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Serrano AL, Baeza-Raja B, Perdiguero E, Jardi M, Munoz-Canoves P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. CELL METAB. 2008;7:33–44. 10.1016/j.cmet.2007.11.011 [DOI] [PubMed] [Google Scholar]
- 17. Pedersen BK. Muscles and their myokines. J EXP BIOL. 2011;214:337–46. 10.1242/jeb.048074 [DOI] [PubMed] [Google Scholar]
- 18. Henriksen T, Green C, Pedersen BK. Myokines in myogenesis and health. Recent Pat Biotechnol. 2012;6:167–71. [DOI] [PubMed] [Google Scholar]
- 19. Cui TX, Lin G, LaPensee CR, Calinescu AA, Rathore M, Streeter C, et al. C/EBPbeta mediates growth hormone-regulated expression of multiple target genes. MOL ENDOCRINOL. 2011;25:681–93. 10.1210/me.2010-0232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marchildon F, Lala N, Li G, St-Louis C, Lamothe D, Keller C, et al. CCAAT/enhancer binding protein beta is expressed in satellite cells and controls myogenesis. STEM CELLS. 2012;30:2619–30. 10.1002/stem.1248 [DOI] [PubMed] [Google Scholar]
- 21. Wiper-Bergeron N, Salem HA, Tomlinson JJ, Wu D, Hache RJ. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5. Proc Natl Acad Sci U S A. 2007;104:2703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin H, Zhang S, Gilbert ER, Siegel PB, Zhu Q, Wong EA. Expression profiles of muscle genes in postnatal skeletal muscle in lines of chickens divergently selected for high and low body weight. Poult Sci. 2014;93:147–54. 10.3382/ps.2013-03612 [DOI] [PubMed] [Google Scholar]
- 23. Morosetti R, Mirabella M, Gliubizzi C, Broccolini A, De Angelis L, Tagliafico E, et al. MyoD expression restores defective myogenic differentiation of human mesoangioblasts from inclusion-body myositis muscle. Proc Natl Acad Sci U S A. 2006;103:16995–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. SEMIN CELL DEV BIOL. 2005;16:585–95. [DOI] [PubMed] [Google Scholar]
- 25. Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci U S A. 1990;87:7988–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blum R. Activation of muscle enhancers by MyoD and epigenetic modifiers. J CELL BIOCHEM. 2014;115:1855–67. 10.1002/jcb.24854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Blum R, Vethantham V, Bowman C, Rudnicki M, Dynlacht BD. Genome-wide identification of enhancers in skeletal muscle: the role of MyoD1. Genes Dev. 2012;26:2763–79. 10.1101/gad.200113.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lagirand-Cantaloube J, Cornille K, Csibi A, Batonnet-Pichon S, Leibovitch MP, Leibovitch SA. Inhibition of atrogin-1/MAFbx mediated MyoD proteolysis prevents skeletal muscle atrophy in vivo. PLOS ONE. 2009;4:e4973 10.1371/journal.pone.0004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. NATURE. 2000;404:782–7. [DOI] [PubMed] [Google Scholar]
- 30. Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, DiStefano PS, et al. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. SCIENCE. 2002;296:530–4. [DOI] [PubMed] [Google Scholar]
- 31. Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, et al. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. NATURE. 2002;419:316–21. [DOI] [PubMed] [Google Scholar]
- 32. Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. NATURE. 2013;494:323–7. 10.1038/nature11895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanchez AM, Candau RB, Bernardi H. FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. CELL MOL LIFE SCI. 2014;71:1657–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Vos KE, Coffer PJ. The extending network of FOXO transcriptional target genes. Antioxid Redox Signal. 2011;14:579–92. 10.1089/ars.2010.3419 [DOI] [PubMed] [Google Scholar]
- 35. Hu P, Geles KG, Paik JH, DePinho RA, Tjian R. Codependent activators direct myoblast-specific MyoD transcription. DEV CELL. 2008;15:534–46. 10.1016/j.devcel.2008.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. CELL. 2004;117:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–601. [DOI] [PubMed] [Google Scholar]
- 38. Calnan DR, Brunet A. The FoxO code. ONCOGENE. 2008;27:2276–88. 10.1038/onc.2008.21 [DOI] [PubMed] [Google Scholar]
- 39. Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. SCIENCE. 2001;294:1704–8. [DOI] [PubMed] [Google Scholar]
- 40. Cleveland BM, Evenhuis JP. Molecular characterization of atrogin-1/F-box protein-32 (FBXO32) and F-box protein-25 (FBXO25) in rainbow trout (Oncorhynchus mykiss): Expression across tissues in response to feed deprivation. Comp Biochem Physiol B Biochem Mol Biol. 2010;157:248–57. 10.1016/j.cbpb.2010.06.010 [DOI] [PubMed] [Google Scholar]
- 41. Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. CELL. 2004;117:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murphy BA, Wagner AL, McGlynn OF, Kharazyan F, Browne JA, Elliott JA. Exercise influences circadian gene expression in equine skeletal muscle. VET J. 2014;201:39–45. 10.1016/j.tvjl.2014.03.028 [DOI] [PubMed] [Google Scholar]
- 43. Mallinson JE, Constantin-Teodosiu D, Glaves PD, Martin EA, Davies WJ, Westwood FR, et al. Pharmacological activation of the pyruvate dehydrogenase complex reduces statin-mediated upregulation of FOXO gene targets and protects against statin myopathy in rodents. J Physiol. 2012;590:6389–402. 10.1113/jphysiol.2012.238022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim JR, Jung HS, Bae SW, Kim JH, Park BL, Choi YH, et al. Polymorphisms in FOXO gene family and association analysis with BMI. Obesity (Silver Spring). 2006;14:188–93. [DOI] [PubMed] [Google Scholar]
- 45. Wang M, Wang Q, Pan Y. From QTL to QTN: candidate gene set approach and a case study in porcine IGF1-FoxO pathway. PLOS ONE. 2013;8:e53452 10.1371/journal.pone.0053452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14-3-3 proteins. Biochim Biophys Acta. 2011;1813:1938–45. 10.1016/j.bbamcr.2011.06.002 [DOI] [PubMed] [Google Scholar]
- 47. Hunt R, Sauna ZE, Ambudkar SV, Gottesman MM, Kimchi-Sarfaty C. Silent (synonymous) SNPs: should we care about them? Methods Mol Biol. 2009;578:23–39. 10.1007/978-1-60327-411-1_2 [DOI] [PubMed] [Google Scholar]
- 48. Teng S, Michonova-Alexova E, Alexov E. Approaches and resources for prediction of the effects of non-synonymous single nucleotide polymorphism on protein function and interactions. Curr Pharm Biotechnol. 2008;9:123–33. [DOI] [PubMed] [Google Scholar]
- 49. Yates CM, Sternberg MJ. The effects of non-synonymous single nucleotide polymorphisms (nsSNPs) on protein-protein interactions. J MOL BIOL. 2013;425:3949–63. 10.1016/j.jmb.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 50. Yu L, Zhou H, Hu F, Xu Y. Two novel mutations of the GTP cyclohydrolase 1 gene and genotype-phenotype correlation in Chinese Dopa-responsive dystonia patients. EUR J HUM GENET. 2013;21:731–5. 10.1038/ejhg.2012.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lei MM, Nie QH, Peng X, Zhang DX, Zhang XQ. Single nucleotide polymorphisms of the chicken insulin-like factor binding protein 2 gene associated with chicken growth and carcass traits. Poult Sci. 2005;84:1191–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
WRRh vs. WRRl, XHh vs. XHl, WRRh vs. XHh and WRRl vs. XHl indicate the comparisons between WRRh and WRRl, between XHh and XHl, between WRRh and XHh and between WRRl and XHl, respectively. In each comparisons, up-regulated indicates that the expression in the second group was higher than that in the first group, while down-regulated indicates that the expression in the first group was higher than that in the second group.
(DOC)
Sequences in the black square frame indicated the sequences absent in the second transcript. Sequences in the CDS are presented in uppercase, while those in the 3’ UTR are presented in lowercase.
(DOC)
A: DF-1 cells. B: Skeletal muscle cells.
(TIF)
(XLS)
(XLS)
(XLS)
(DOC)
(XLS)
(DOC)
Data Availability Statement
Relevant data are within the paper and its Supporting Information files. The raw RNA-seq data are available on Gene Expression Omnibus (GEO) at accession number GSE72424 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72424).