Abstract
Background
The whipworms Trichuris trichiura and Trichuris suis are two parasitic nematodes of humans and pigs, respectively. Although whipworms in human and non-human primates historically have been referred to as T. trichiura, recent reports suggest that several Trichuris spp. are found in primates.
Methods and Findings
We sequenced and annotated complete mitochondrial genomes of Trichuris recovered from a human in Uganda, an olive baboon in the US, a hamadryas baboon in Denmark, and two pigs from Denmark and Uganda. Comparative analyses using other published mitochondrial genomes of Trichuris recovered from a human and a porcine host in China and from a françois’ leaf-monkey (China) were performed, including phylogenetic analyses and pairwise genetic and amino acid distances. Genetic and protein distances between human Trichuris in Uganda and China were high (~19% and 15%, respectively) suggesting that they represented different species. Trichuris from the olive baboon in US was genetically related to human Trichuris in China, while the other from the hamadryas baboon in Denmark was nearly identical to human Trichuris from Uganda. Baboon-derived Trichuris was genetically distinct from Trichuris from françois’ leaf monkey, suggesting multiple whipworm species circulating among non-human primates. The genetic and protein distances between pig Trichuris from Denmark and other regions were roughly 9% and 6%, respectively, while Chinese and Ugandan whipworms were more closely related.
Conclusion and Significance
Our results indicate that Trichuris species infecting humans and pigs are phylogenetically distinct across geographical regions, which might have important implications for the implementation of suitable and effective control strategies in different regions. Moreover, we provide support for the hypothesis that Trichuris infecting primates represents a complex of cryptic species with some species being able to infect both humans and non-human primates.
Author Summary
Trichuris trichiura and Trichuris suis are whipworms found in humans and pigs, respectively, causing morbidity in humans and being associated with production losses in pigs. Although Trichuris from non-human primates is attributed to T. trichiura, hence considered the same species as the one infecting humans, several recent reports question this assumption. Morphologically similar parasites that have a wide global distribution and/or those capable of infecting several host species may comprise several ‘hidden’ species. In this study, we sequenced, annotated, and compared the mitochondrial genomes (including published genomes) of Trichuris obtained from different hosts in different geographical regions, including humans (Uganda and China), pigs (China, Uganda, and Denmark) and two types of non-human primates (baboons and françois’ leaf monkey). We found high genetic distinctiveness between human Trichuris from China and Uganda. Likewise, pig Trichuris from Denmark and other regions also showed considerable, although lower, genetic diversity. This suggests that both pig- and human-derived Trichuris may represent different species with potential differences in endemicity, which may have important implications for implementing effective control strategies. Our data also suggests that Trichuris infecting primates comprises several species and may be transmitted from non-human primates to humans.
Introduction
Neglected tropical diseases, including helminthiases, have a devastating effect on human health. It is estimated that about one billion people are infected with soil-transmitted helminths (STHs), including the common roundworm (Ascaris), hookworms (Necator and Ancylostoma spp.), and whipworm (Trichuris), mostly in underprivileged regions of the world [1]. Approximately 0.5 billion people are infected with T. trichiura, resulting in the loss of 0.64 million disability-adjusted life years [2]. Compared with adults, children are more prone to developing clinical symptoms such as dysentery, bloody diarrhea, rectal prolapse, and cognitive impairment in cases of chronic infection [3, 4].
Although whipworm infections in non-human primates are usually called T. trichiura, recent studies suggest that primates may host multiple species of Trichuris. Ravasi et al. [5] found evidence of two Trichuris species in both baboons and humans based on the sequences of the internal transcribed spacers (ITS) of nuclear ribosomal DNA. Another study by Hansen et al. [6] based on studies of the beta-tubulin gene and ITS-2 sequencing suggested that humans and baboons host shared Trichuris species. On the other hand, Liu et al. [7] identified a potentially novel species of Trichuris in a non-human primate (françois’ leaf monkey) based on complete mitochondrial genome analysis and the ITS-1 and -2 regions. Recently, Ghai et al. [8] suggested that Trichuris spp. in human and non-human primates represent several species that differ in host specificity. Therefore, there is a need to further explore which species of Trichuris that infect primates and investigate potential (zoonotic) routes of transmission between host species.
The whipworm of pigs, Trichuris suis is associated with production losses due to reduced growth rates and lower feed conversion efficiency [9]. Although morphologically indistinguishable from T. trichiura, several studies identified extensive genetic diversity between T. trichiura and T. suis based on nuclear and mitochondrial DNA analysis [10–12]. However, molecular characterization of Trichuris from sympatric pigs and humans indicated that T. suis can cause zoonotic infection in humans, emphasizing the public health importance of this pig parasite [12].
The circular mitochondrial (mt) genomes are relatively small in size (13–26 kb) and encode enzymes required for oxidative phosphorylation. Mitochondrial DNA has a number of advantages for delimiting closely related species due to its high substitution rate coupled with its low effective population size, which leads to rapid lineage sorting following speciation [13]. Comparative mitochondrial DNA analysis is therefore useful for identifying cryptic (“hidden”) species, i.e., those that cannot be differentiated by traditional methods, including morphological analysis. On the other hand, mitochondrial pseudogenes (numts) in the nuclear genome may lead to incorrect phylogenetic inferences, which is why caution is warranted whenever mt genes are used in phylogenetic analyses [14]. Moreover, sole dependence on mtDNA for delineating the taxonomic status might also lead to ambiguous phylogeny and misidentification of individuals due to incomplete lineage sorting or mitochondrial introgression [15].
Parasites with a wide geographical distribution or multiple host species may comprise cryptic species [13]. For instance, Hypodontus macropi, an intestinal parasite in macropodid marsupials, was found to consist of several cryptic species based on mt genome analysis [16]. In the study by Blouin [17], the genetic difference between sibling nematode species typically ranges between 10%–20% using cox1 and nad4 mt genes, whereas intra-species variation is usually below 2%.
In the present study, we logically extend previous investigations to investigate levels of genetic variation among specimens of Trichuris from a human from Uganda, two baboons and pigs from Denmark and Uganda. To do this, we (i) sequenced and characterized complete mt genomes from individual adult worms from these three host species and (ii) compared them (at the amino acid sequence level) with those of Trichuris spp. of human, françois’ leaf-monkey and pig determined in previous studies, in order to assess levels of genetic variation within Trichuris among host species and geographical regions.
Methods
Ethics statement
The human Trichuris was recovered from the feces of a child after anthelmintic treatment as part of an efficacy study as described previously [12]. Permission was obtained from the Ministry of Health and the National Council of Science and Technology in Uganda, and the Danish Central Medical Ethics Committee approved the study. The parents and children were informed about the study and received a consent form in both English and the local language. Written informed consent was received for each individual participating in the study. Worms from baboons in the Southwest National Primate Research Center, Texas, USA and the Copenhagen Zoo, Denmark were recovered during post mortem examination, which is performed both places on all animals culled on a routine basis. T. suis was obtained from an experimentally infected pig in Denmark. The Animal Experiments Inspectorate, Ministry of Justice, Denmark, approved the animal study protocol, which was carried out according to stipulated guidelines (License no. 2005/561-1060). T. suis was obtained from a naturally infected pig in Uganda raised on a private farm, slaughtered, and used for local consumption. Permission to recover worms from the animal was obtained from the owner.
Parasites, DNA extraction, and genotyping of worms
Adult Trichuris worms were recovered from an olive baboon, Papio anubis, at Southwest National Primate Research Center, Texas, USA, and a hamadryas baboon, Papio hamadryas, in Copenhagen Zoo, Denmark, both during post mortem examination. Adult Trichuris were collected from domesticated pigs post mortem from Denmark and Uganda and recovered from a human stool sample from Uganda upon anthelmintic treatment as described [12]. Worms were rinsed with tap water and stored in 70% ethanol at 5°C until DNA extraction.
The MasterPure DNA Purification Kit (Epicenter Biotechnologies) was used to extract total genomic DNA from the anterior thin part of the worms according to manufacturer's protocol. Worm material was homogenized in lysis solution in an Eppendorf tube using a matching plastic pestle followed by incubation at 56°C for at least six hours.
PCR-linked restriction fragment length polymorphism analysis (PCR-RFLP) of the internal transcribed spacer-2 (ITS-2) region was used to genotype worms, since Trichuris from primates, including humans, and pigs are morphologically indistinguishable [8, 12]. PCR products and digested fragments were resolved using 1.5% agarose gels, stained with GelRedTM (Biotium), and detected using UV light. Worms from humans and baboons showed banding patterns characteristic of T. trichiura (~130, 220 and 340 bp) and worms from pigs showed banding patterns characteristic of T. suis (~130 and 490 bp) [12].
Mitochondrial genome amplification and sequencing
Different primate- and pig-derived Trichuris were chosen for long-range PCR amplification and next generation sequencing (NGS). Two baboon worms, P. hamadryas (TTB1) and P. anubis (TTB2), one Uganda human worm (TTHUG), and two pig worms (TSDK and TSUG) from Denmark and Uganda, respectively were chosen based on their distinct haplotypes identified as part of another study when sequencing the rrnL gene of 140 Trichuris worms. The two mt genomes of T. trichiura and T. suis (Accession nos. GU385218 and GU070737, respectively) were aligned to identify conserved regions relevant to primer design. However, no suitable conserved regions were identified, which precluded the design of general primers applicable to all worm samples. Hence, different sets of primers were designed for each genome. Primers were designed based on the genome of T. trichiura (GU385218) to amplify the mt genomes of the baboon- and human-derived Trichuris in three overlapping fragments (~5 kbp each) and for pig-derived Trichuris TSDK and TSUG in three overlapping fragments (~6, 5, and 3 kbp) based on the genome of T. suis (GU070737) (Table 1). However, several obstacles were encountered in the amplification and sequencing processes. First, only TTB2 was amplified, and other sets of primers were therefore designed to amplify the TTB1 and TTHUG genomes in two overlapping fragments (~8 and ~6 kbp) (Table 1). However, due to the presence of non-specific bands, amplified DNA from the band representing the fragment nad1–rrnL was extracted from the agarose gel using spin columns (Millipore) as stipulated by the manufacturer’s protocol. Second, the library construction (see below) of the TTHUG genome failed, and the genome was amplified in 15 fragments of ~1,000 bp each, using 15 overlapping primer pairs designed based on the TTB1 mt genome (S1 Table) and sequenced by Sanger dideoxy-sequencing (Macrogen Inc., Seoul, South Korea).
Table 1. Primers used for complete mitochondrial genome amplification of Trichuris from baboons (TTB1 and TTB2) and pigs (TSDK and TSUG).
| Forward (5’—3’) | Reverse (5’—3’) | ||
|---|---|---|---|
| TTB2 | |||
| TTB2cox1F | CAGGAAATCACAAGAAAATTGG | TTB2nad5R | AGTGGTTGCAGGAACAATTC |
| TTB2nad5F | AGCAATCTGCGATATTGTTG | TTB2rrnLR | TCGCAACGGTTTAAACTCAA |
| TTB2rrnLF | CGCAGTAATCTGACTGTGC | TTB2cox1R | AAATTTTCCTGCTATGAATATGA |
| TTB1 | |||
| TTB1nad1F | ACAGCCCATCCTAGACGGTA | TTB1rrnL | ACCTGTCTCGCAACGGTTTA |
| TTB1rrnLF | TCTGACTGTGCAAAGGTAGCA | TTB1nad1R | TTGCGGACCAAAAGGTTATGAAT |
| TSDK & TSUG | |||
| TSrrnLF | TTAAATGGCCGCAGTAACCT | TSnad1R | AGCTCACCCTGTAATAATGATGT |
| TSnad1F | TCTGATCTGTGCTACCCTACAC | TSnad5R | CCAACACCCGTGAGTTCTT |
| TSnad5F | CTTTTGCAAGGGCATGATTA | TSrrnLR | TCACGTAATGTAGAATCGTCGA |
Long-range PCR was conducted in a total volume of 20 μL containing 2 μL 10X PCR buffer, 0.2 mM of each dNTP, 0.4 mM of each primer pair, 2.0 mM MgCl2, and 2.5 U of Long PCR Enzyme Mix (Thermo Scientific). PCR cycling conditions included initial denaturation at 92°C for 4 min, followed by 35 cycles of denaturation at 92°C (20 s), annealing at 50°C (30 s), extension at 62–67°C (7 min), and a final extension at 60–67°C for 10 min. PCR gradient and MgCl2 titration was used to optimize the PCR for each primer pair. PCR products were stained using GelRedTM (Biotium) and visualized after gel electrophoresis (0.8% agarose) under UV light. PCR products were cleaned enzymatically using 1 μL Exonuclease I (Fermentas) and 2 μL FastAP Thermosensitive Alkaline Phosphatase (1 U/μL) (Fermentas) for each 5μL of amplicons and incubated for 15 min at 37°C, followed by 15 min at 85°C. Finally, DNA concentration was measured using a NanoDrop 1000 spectrophotometer (Thermo Fischer Scientific), and equal amounts of fragments of each genome were pooled. Library construction, including tagging (indexing) of samples and NGS using the Illumina HiSeq 2000 platform, was performed by Macrogen Inc. (Seoul, South Korea).
Assembly, annotation, and genome sequence analysis
Reads (~100 bp) of each genome were assembled using CLC Genomics Workbench v.6.0.4 (CLC Inc, Aarhus, Denmark) de novo except for sample TSDK that was assembled using TSUG and GenBank entry GU070737 (TSCH). The files of the NGS raw data can be provided upon request. For TTHUG, sequences were manually checked, edited, and trimmed using Vector NTI [18] and BioEdit [19] and aligned to TTB1. After assembly, genome annotation was performed using the pipeline MITOS [20] and BLAST search tools available through NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Secondary structures for all tRNAs were predicted using tRNAScan-SE [21] and ARWEN [22].
The genomes were compared with T. trichiura from a human in China (TTHCH) (GU385218); Trichuris sp. GHL from francois' leaf monkey (T.GHL) from China (KC461179), and T. suis from China (TSCH) (GU070737). Protein-coding genes (PCGs) and ribosomal DNA genes were individually extracted and aligned by ClustalW using default settings. Another data set was generated by concatenating all PCG and rDNA sequences. Genetic distances were estimated for these data sets using MEGA v.6.1 [23]. Nucleotide diversity (π) was calculated across the genomes of Trichuris from humans and non-human primates and Trichuris from pigs using a sliding window of 100 bp with 25 bp steps implemented in DnaSP v.5 [24].
Phylogenetic analysis
Three different methods were used for phylogenetic analysis, namely Neighbor Joining (NJ), Maximum Likelihood (ML), and Bayesian Inferences (BI). Two different data sets (DNA and amino acid sequences) were generated for the phylogenetic analyses. Amino acid sequences for the 13 PCGs were aligned using ClustalW for 10 Trichuris spp., namely Trichuris from baboons (TTB1, TTB2), from humans (TTHUG and TTHCH), from francois' leaf monkey (KC461179), from pigs (TSUG, TSDK and TSCH), and from T. ovis and T. discolor (JQ996232 and JQ996231, respectively). Similarly, the DNA sequences representing the PCGs and rDNA genes were aligned using ClustalW. Trichinella spiralis (AF293969) was used as an outgroup in the phylogenetic analyses. ML and NJ trees were generated using MEGA v.6.1 [23]. The best-to-fit substitution model was identified using jModelTest0.1.1 [25] under Akaike information criterion (AIC) [26] for each dataset. BEAST v.1.6.1 [27] was used for the BI on the two data sets. Uncorrelated log normal was used as prior for the mutation rate with mtRev as the substitution model for protein sequences and the General Time Reversible (GTR) model for DNA sequences, with gamma distribution and invariant sites assumed in both substitution models. A random starting tree with Yule prior was assumed as well. Three independent runs with 10 million steps each with a burn-in of 10,000 steps were carried out. Tracer v.1.6 [27] was used to analyze log files of the MCMC chains, and the reliability of parameters was checked by recording effective sample size values above 200. Tree Annotater v.1.6.1 [27] was used to summarize the tree data with a posterior probability limit of 0.5.
Cox1 phylogeny
In order to investigate the phylogenetic relationship between the mt genome haplotypes identified in this study with other Trichuris haplotypes from primates and pigs in different geographical regions, partial (372bp) cox1 sequences from GenBank were obtained (Table 2) for phylogenetic analyses. ML and NJ trees were generated using MEGA v.6.1 [23], and the best-to-fit model was identified using jModelTest0.1.1 [25] under Akaik information criterion (AIC) [26]. Ascaris lumbricoides (AB591799) was used as an outgroup.
Table 2. Partial cox1 sequences retrieved from the GenBank database with accession numbers, host, and the country from which the worms were sampled.
All the non-human primates represented in the table were held in captivity.
| Host | Country | Accession No. in GenBank |
|---|---|---|
| Colobus guereza kikuyensis (Black-and-white colobus) | Spain | HE653116, HE653117, HE653118, HE653119. |
| Papio anubis (Olive baboon) | Czech Republic | JF690964 |
| Theropithecus gelada (Gelada baboon) | Czech Republic | JF690965 |
| Papio hamadryas (Hamadryas baboon) | Czech Republic | JF690963 |
| Macaca fascicularis (Longtailed macaque) | Czech Republic | JF690967 |
| Human | Czech Republic | JF690962 |
| Sus scrofa domestica (Domestic pig) | China | HQ204208, HQ204209, HQ183740, HQ183741 |
| Sus scrofa domestica (Domestic pig) | Spain | HE653124, HE653125, HE653126 |
| Sus scrofa scrofa (Wild boar) | Spain | HE653127, HE653128, HE653129 |
Results
Annotation and features of mitochondrial genomes
The complete mt genomes of the primate worms TTB1, TTB2, and TTHUG comprised 13,984, 14,009, and 14,079 bp, respectively, whereas the two pig worms, TSDK and TSUG comprised 14,521 and 14,410 bp (GenBank accession nos. KT449822-KT449826). The genomes contain 13 PCGs, 22 tRNAs, and two ribosomal RNA genes (Tables 3 and 4). The general mt features, including gene synteny, is the same as previously described for Trichuris spp. [7, 11,28] as all genes are transcribed from the heavy strand, except 4 PCGs (nad2, nad5, nad4, and nad4L) and 10 tRNA motifs (tRNA-Met, tRNA-Phe, tRNA-His, tRNA-Arg, tRNA-Pro, tRNA-Trp, tRNA-Ile, tRNA-Gly, tRNA-Cys, and tRNA-Tyr), which are transcribed from the light strand. The starting and termination codons for some PCGs differed between Trichuris spp. recovered from identical host species. For instance, the starting codon for TSDK is ATA for the nad4 gene, while it reads ATG in the TSUG genome; for the atp6 gene, the starting codon is GTA in TSDK, while being GTG in TSUG. Moreover, the termination codon in the cox1 gene is TAG for TSDK and TAA for TSUG, and in the nad4 gene, TAA is the termination codon in TSDK, while it reads TAG in TSUG.
Table 3. Mitochondrial genomes of baboon Trichuris (TTB1 and TTB2) and human Trichuris (TTHUG).
Protein coding, transfer RNA (tRNA), and ribosomal DNA (rDNA) genes with lengths in nucleotides (nt) are given. The lengths of TTB1 and TTHUG are identical, and differences are given in parentheses for (TTB2/TTHUG); likewise for the initiation and termination codons.
| Genes | Positions | Lengths | Codons | Strand | |||
|---|---|---|---|---|---|---|---|
| TTB2 | TTB1 | TTHUG | nt | Initiation | Termination | ||
| cox1 | 1–1545 | 1–1545 | 1–1545 | 1545 | ATG | TAA | + |
| cox2 | 1560–2234 | 1558–2232 | 1558–2232 | 675 | ATG | TAG(TAA) | + |
| tRNA-leu | 2248–2308 | 2255–2317 | 2255–2317 | 63(-3) | + | ||
| tRNA-glu | 2318–2374 | 2324–2384 | 2324–2384 | 61(-4) | + | ||
| nad1 | 2397–3296 | 2406–3305 | 2406–3305 | 900 | ATA | TAG(TAA) | + |
| tRNA-lys | 3424–3484 | 3334–3399 | 3425–3490 | 66(-5) | + | ||
| nad2 | 3487–4383 | 3397–4293 | 3488–4384 | 897(-12) | ATA(GTA) | TAA | - |
| tRNA-met | 4384–4444 | 4294–4354 | 4385–4445 | 61 | - | ||
| tRNA-phe | 4441–4496 | 4349–4405 | 4440–4496 | 57(-1) | - | ||
| nad5 | 4496–6043 | 4397–5953 | 4488–6044 | 1557(-9) | ATA | TAG(TAA) | - |
| tRNA-his | 6041–6094 | 5947–6004 | 6038–6095 | 58(-4) | - | ||
| tRNA-arg | 6096–6158 | 6006–6069 | 6097–6160 | 64 | - | ||
| nad4 | 6160–7371 | 6074–7294 | 6165–7382 | 1221(-9/-3) | ATG | TAA | - |
| nad4L | 7402–7650 | 7317–7529 | 7405–7617 | 213(+36) | ATA | TAA | - |
| tRNA-thr | 7655–7710 | 7570–7627 | 7658–7715 | 58(-2) | + | ||
| tRNA-pro | 7712–7770 | 7627–7685 | 7715–7773 | 59 | - | ||
| nad6 | 7763–8239 | 7678–8154 | 7766–8242 | 477 | ATT | TAA | + |
| cytb | 8246–9352 | 8161–9267 | 8249–9355 | 1107 | ATG | TAG | + |
| tRNA-ser | 9351–9400 | 9266–9318 | 9354–9406 | 53(-3) | + | ||
| rrnS | 9393–10086 | 9311–10009 | 9399–10102 | 699(-5) | + | ||
| tRNA-val | 10044–10144 | 10011–10067 | 10104–10160 | 57 | + | ||
| rrnL | 10144–11153 | 10069–11077 | 10162–11170 | 1009(+2) | + | ||
| atp6 | 11124–11963 | 11048–11860 | 11141–11953 | 813(+27) | ATG(GTG) | TAA | + |
| cox3 | 11938–12711 | 11866–12639 | 11959–12732 | 774 | ATG | TAA | + |
| tRNA-trp | 12718–12780 | 12652–12714 | 12745–12807 | 63 | - | ||
| tRNA-gln | 12784–12836 | 12718–12773 | 12811–12866 | 56(-3) | + | ||
| tRNA-Ile | 12838–12898 | 12776–12836 | 12869–12929 | 61 | - | ||
| tRNA-gly | 12908–12964 | 12850–12906 | 12943–12999 | 57 | - | ||
| tRNA-asp | 12970–13034 | 12913–12970 | 13006–13063 | 58(+8) | + | ||
| atp8 | 13016–13180 | 12959–13126 | 13052–13219 | 168(-3) | ATT(ATA) | TAG | + |
| nad3 | 13190–13531 | 13136–13477 | 13229–13570 | 342 | ATT | TAA | + |
| tRNA-ser | 13626–13675 | 13571–13620 | 13664–13714 | 50 | + | ||
| tRNA-asn | 13676–13729 | 13621–13675 | 13715–13769 | 55(-1) | + | ||
| tRNA-leu | 13737–13799 | 13683–13742 | 13777–13836 | 60(+3) | + | ||
| tRNA-ala | 13806–13858 | 13754–13811 | 13848–13905 | 58(-4) | + | ||
| tRNA-cys | 13888–13940 | 13854–13907 | 13948–14001 | 54(-1) | - | ||
| tRNA-tyr | 13949–14009 | 13908–13968 | 14002–14062 | 61 | - | ||
Table 4. Mitochondrial genomes of pig Trichuris TSDK and TSUG from Denmark and Uganda, respectively.
Protein-coding, transfer RNA (tRNA), and ribosomal DNA (rDNA) genes are indicated with lengths in nucleotides (nt) for the respective genes. Gene lengths are given for TSDK, and the differences from TSUG are given in parentheses; likewise for the initiation and termination codons.
| Regions | Positions | Lengths | Codons | Strand | ||
|---|---|---|---|---|---|---|
| TSDK | TSUG | nt | Initiation | Termination | ||
| cox1 | 1–1542 | 1–1542 | 1542 | ATG | TAG(TAA) | + |
| cox2 | 1579–2259 | 1577–2257 | 681 | ATG | TAA | + |
| tRNA-leu | 2271–2330 | 2269–2328 | 60 | + | ||
| tRNA-glu | 2338–2394 | 2336–2392 | 57 | + | ||
| nad1 | 2416–3315 | 2414–3313 | 900 | ATT | TAA | + |
| tRNA-lys | 3447–3506 | 3454–3513 | 60 | + | ||
| nad2 | 3521–4402 | 3528–4409 | 882 | ATA | TAG | - |
| tRNA-met | 4412–4473 | 4419–4480 | 62 | - | ||
| tRNA-phe | 4477–4537 | 4484–4542 | 61(-2) | - | ||
| nad5 | 4528–6084 | 4533–6089 | 1557 | ATA | TAG | - |
| tRNA-his | 6085–6140 | 6090–6145 | 56 | - | ||
| tRNA-arg | 6144–6207 | 6149–6212 | 64 | - | ||
| nad4 | 6213–7616 | 6218–7606 | 1404(-15) | ATA(ATG) | TAA(TAG) | - |
| nad4L | 7726–7968 | 7628–7891 | 240(-21) | ATA | TAG | - |
| tRNA-thr | 7970–8024 | 7884–7938 | 55 | + | ||
| tRNA-pro | 8017–8087 | 7944–8001 | 71(-14) | - | ||
| nad6 | 8080–8550 | 7994–8464 | 471 | ATT | TAA | + |
| cytb | 8563–9675 | 8476–9588 | 1113 | ATG | TAG | + |
| tRNA-ser | 9674–9728 | 9587–9641 | 55 | + | ||
| rrnS | 9726–10435 | 9639–10349 | 710 | + | ||
| tRNA-val | 10435–10491 | 10349–10405 | 57 | + | ||
| rrnL | 10500–11510 | 10414–11420 | 1011 | + | ||
| atp6 | 11506–12309 | 11410–12219 | 804(+6) | ATT(GTG) | TAA | + |
| cox3 | 12318–13094 | 12229–13005 | 777 | ATG | TAA | + |
| tRNA-trp | 13099–13165 | 13010–13076 | 67 | - | ||
| tRNA-gln | 13169–13225 | 13080–13136 | 57 | + | ||
| tRNA-ile | 13228–13293 | 13139–13204 | 66 | - | ||
| tRNA-gly | 13312–13367 | 13223–13278 | 56 | - | ||
| tRNA-asp | 13382–13441 | 13290–13350 | 60 | + | ||
| atp8 | 13421–13591 | 13331–13501 | 171 | TTG | TAA | + |
| nad3 | 13616–13957 | 13526–13867 | 342 | ATA | TAA | + |
| tRNA-ser | 14065–14118 | 13975–14026 | 54(-2) | + | ||
| tRNA-asn | 14118–14177 | 14026–14084 | 60(-1) | + | ||
| tRNA-leu | 14193–14252 | 14102–14164 | 60(+3) | + | ||
| tRNA-ala | 14258–14312 | 14167–14221 | 55 | + | ||
| tRNA-cys | 14344–14401 | 14245–14299 | 58(-3) | - | ||
| tRNA-tyr | 14401–14457 | 14300–14356 | 57 | - | ||
Similar observations were found in the Trichuris genomes from baboons. For nad2 and atp6, the starting codons were ATA and ATG, respectively, in TTB1, while they read GTA and GTG, respectively, in TTB2. Likewise, the termination codons read TAG for the cox2, nad1, and nad5 genes in TTB1, while they read TAA for the same genes in the TTB2 genome. Finally, the length of the open reading frame (ORF) for some of the genes differed between the genomes. nad4 and nad4L showed different ORF lengths between TSDK and TSUG. For the TTB1 and TTB2 genomes, nad1, nad2, nad5, nad4, nad4L, atp6, and atp8 also varied in terms of respective ORF lengths. However, TTB1 and TTHUG were identical in terms of all initiation and termination codons and gene lengths, except for the nad4 gene, which was one amino acid (3 nucleotides) shorter in TTHUG.
Comparative sequence analysis
Genetic distances between each PCG and rDNA gene of the different genomes of Trichuris spp. in primates and pigs are listed in Table 5, together with differences in amino acid sequences, based on all encoded proteins. The genetic distances between worms for individual PCGs and rDNA genes are given in S2 Table. The highest genetic variation was found in the atp8 gene, and the most conserved gene was rrnS. Among all the PCGs, cox1 and atp8 were found to be the most and least conserved gene, respectively. The overall differences in nucleotide and amino acid sequences between the genomes of TTHCH and TTHUG were high (18.8% and 14.6%, respectively), whereas the baboon Trichuris TTB1 was genetically nearly identical to the human TTHUG. TTB2 was most closely related to TTHCH with an overall nucleotide difference of 6.5%. Among the primate-derived Trichuris, T.GHL from francois' leaf-monkey was most distinct, with a nucleotide difference of 27%–28% compared with worms from humans and baboons. Nucleotide differences between TSUG and TSCH (3.1%) were much lower compared with TSDK (~9%).
Table 5. Overall genetic and protein distances between the Trichuris spp. genomes derived from baboons (TTB1 and TTB2), humans (TTHCH and TTHUG from China and Uganda, respectively), pigs (TSCH, TSUG and TSDK from China, Uganda and Denmark, respectively), and francois' leaf monkey (T.GHL).
The amino acid sequence distances are given above the diagonal and genetic distances below the diagonal.
| Trichuris genomes | TTHCH | TTHUG | TTB1 | TTB2 | T.GHL | TSDK | TSUG | TSCH |
|---|---|---|---|---|---|---|---|---|
| TTHCH | 14.6 | 14.8 | 4.7 | 27.6 | 39.2 | 33.6 | 35.3 | |
| TTHUG | 18.8 | 0.8 | 14.1 | 26.7 | 39.1 | 33.3 | 35.0 | |
| TTB1 | 19.9 | 1.0 | 14.4 | 26.7 | 39.1 | 33.2 | 35.0 | |
| TTB2 | 6.5 | 19.0 | 19.4 | 27.9 | 38.9 | 33.4 | 35.0 | |
| T.GHL | 28.1 | 27.6 | 27.9 | 28.8 | 40.9 | 35.7 | 37.2 | |
| TSDK | 31.2 | 30.3 | 30.6 | 31.6 | 32.0 | 5.5 | 6.1 | |
| TSUG | 31.2 | 30.2 | 30.5 | 31.6 | 32.2 | 9.1 | 2.2 | |
| TSCH | 30.9 | 30.4 | 30.7 | 31.1 | 32.2 | 8.8 | 3.1 |
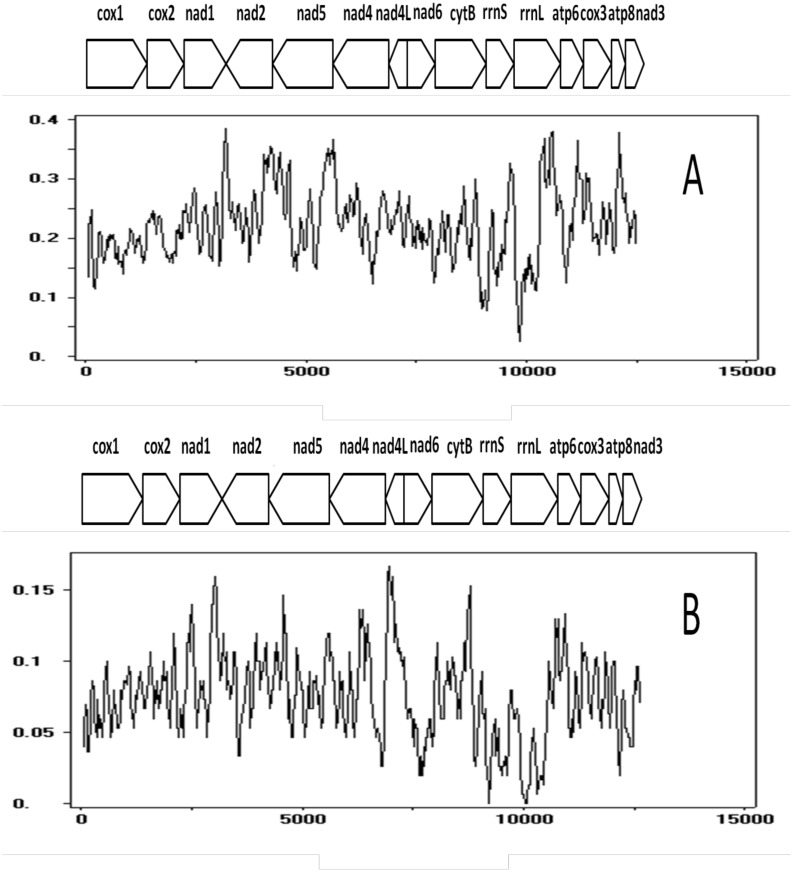
Nucleotide diversity among the Trichuris genomes was analyzed using the sliding window approach for all the PCGs and rDNA genes. The variation estimated for all primate- and pig-derived Trichuris is given in two separate windows (Fig 1). The overall variation within the primate-derived Trichuris was higher compared with that of pig-derived Trichuris. The rDNA and cox1 genes were found to have the lowest nucleotide diversity among pig- and primate-derived Trichuris.
Fig 1. Nucleotide diversity (π) for protein coding regions and ribosomal DNA (rrnS and rrnL) measured every 25 bp over 100 bp windows.
The aligned dataset for Trichuris in primates (humans, baboons, francois’ leaf monkey) is given in (A), while that of the pig-derived Trichuris is given in (B).
Phylogenetic analysis
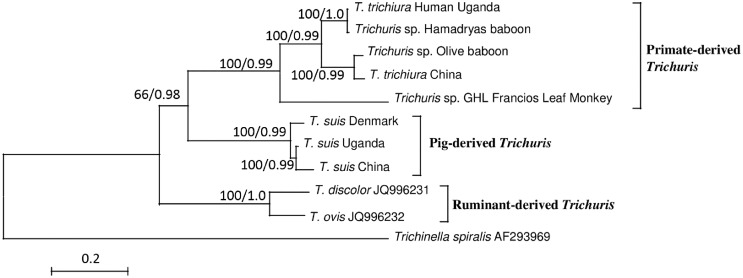
Amino acid and nucleotide data sets gave similar tree topologies by all the methods applied (NJ, ML and BI). The best-to-fit model was mtREV+G+I+F for the amino acid sequences and General Time Reversible with gamma distribution and invariant sites (GTR+G+I) for the nucleotide sequences. Three major groups were identified in the phylogeny based on the mt genomes (amino acid sequences), namely primate-, pig-, and ruminant-derived Trichuris (Fig 2). The nucleotide sequence-based phylogeny is provided in S1 Fig and depicts similar tree topology.
Fig 2. Inferred phylogenetic relationship among Trichuris spp. using concatenated amino acid sequences and Maximum Likelihood (ML) and Bayesian Inferences (BI).
The three major groups identified by the phylogenetic tree include primate-, pig- and ruminant- derived Trichuris. Bootstrap frequencies (BF) and posterior probabilities (PP) are indicated on the branches (BF/PP). Scale bar represents the number of substitutions per site.
Cox1 phylogeny
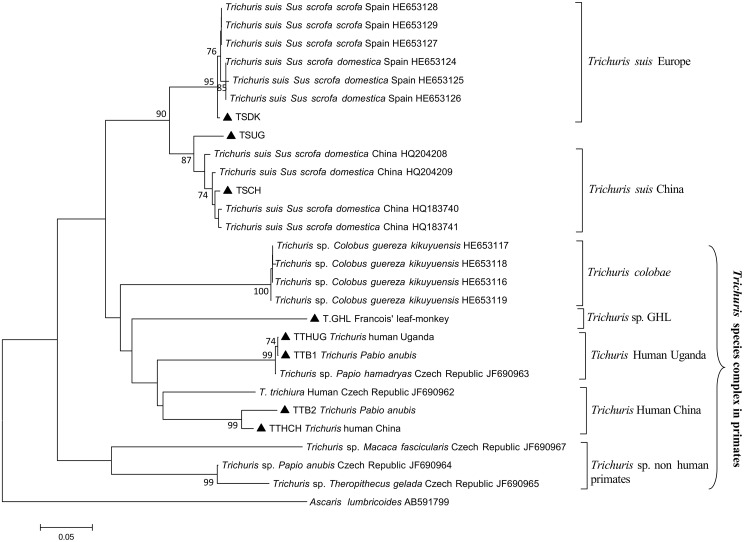
For the cox1 sequences, the best data fit was obtained with the Tamura 3-parameter model with gamma distribution. Both NJ and ML depicted similar tree topologies; hence, the NJ tree is depicted in Fig 3. TSDK clustered with Spanish T. suis and forms the 'T. suis Europe' clade, and TSCH is found in the 'T. suis China' clade. TSUG is most closely related to T. suis from China, which is concordant with the mt genome phylogeny. The cox1 phylogeny also supports the presence of a Trichuris species complex infecting primates, identifying five distinct clades, which were named after the Trichuris spp. recovered from these hosts, namely T. colobae recovered from Colobus guereza kikuyensis [29] and Trichuris sp. GHL from francois' leaf monkey [7]. Human-derived Trichuris TTHUG and TTHCH were found in two separate clades named after the country of origin, 'Human Trichuris Uganda' and 'Human Trichuris China', respectively, while the last clade comprises Trichuris sp. from different non-human primates (Theropithecus gelada, Macaca fascicularis, and P. anubis) and here named 'Trichuris sp. non-human primates'. The baboon Trichuris TTB1 clusters with the Trichuris from baboon (P. hamadryas) in the 'Human Trichuris Uganda' clade, while TTB2 clusters with the 'Human Trichuris China' according to the mt genome phylogeny (Fig 2). The human Trichuris from Czech Republic is found clustering with the human Trichuris from China. Remarkably, Trichuris sp. from P. anubis from the Czech Republic is genetically very distinct and clusters in a clade (Trichuris sp. non-human primates) distant to that of TTB2, although both are Trichuris isolated from the same host species.
Fig 3. Inferred phylogeny among Trichuris spp. recovered from pigs and primates based on partial cox1 sequences and NJ clustering.
Samples for which the full mitochondrial genome is sequenced are indicated by a solid triangle. The phylogeny identified two major clades for Trichuris in pigs, namely ‘T. suis Europe’ and ‘T. suis China’. Five distinct clades for Trichuris spp. recovered from primates (including humans) were identified, namely ‘T. colobae’, ‘Trichuris sp. GHL’, ‘Trichuris Human Uganda’, ‘Trichuris Human China’, and ‘Trichuris sp. non-human primates’. Scale bar indicates number nucleotide substitutions per site. Only bootstrap values > 70 are given.
Discussion
We sequenced the complete mt genomes of Trichuris spp. recovered from a human, baboons, and pigs and evaluated their genetic and evolutionary relationships. Several major haplotypes with clear genetic distinctiveness were observed, suggesting that multiple Trichuris species infect these host species and supporting the hypothesis that whipworms in primates comprise a species complex, which may also be the case for whipworms in pigs (S3 Fig).
The two human Trichuris from Uganda and China were genetically distinct, and the difference in amino acid and nucleotide sequences was found to be around 14.6% and 18.8%, which is in the range of previously reported differences between different parasitic nematode species, suggesting the presence of at least two Trichuris species infecting humans. For instance, the difference in amino acids for mt protein sequences between T. ovis and T. discolor adds up to 15.4% [28], 11.7% between Wuchereria bancrofti and Brugia malayi [30], 10.3% between Chabertia ovina and C. erschowi [31], and ranges from 4% to 18% between different species of Trichinella [32]. The baboon Trichuris, TTB1, was nearly identical to the human Trichuris from Uganda, which is in accordance with a previous study analyzing beta tubulin genes and the ITS-2 region [6], while the other baboon Trichuris, TTB2, was genetically more related to the human Trichuris from China, suggesting that baboons—similar to humans—may also host at least two Trichuris spp. In accordance with our study, but based on ITS-1 and -2 sequence analyses, Ravasi et al. [5] identified two different Trichuris species in humans from Cameroon and China, which were also found in chacma baboons in South Africa. On the other hand, Trichuris from the leaf monkey was very distinct from baboon worms, suggesting different Trichuris species in non-human primates as proposed by Liu et al. [7]. Indeed, Ghai et al. [8] recently suggested that primates may be infected with several Trichuris species, with some species only found in humans and others only found in non-human primates, while others again are shared, suggesting various degrees of host specificity of the different Trichuris spp. in primates.
Amino acid sequence distances between TSDK compared with TSCH and TSUG were considerable (around 6.1% and 5.5%, respectively), while TSCH and TSUG were genetically more closely related (2.2%). Although the distances between TSDK, TSUG, and TSCH were not notably high, similar amino acid sequence distances between different parasitic nematode species have been reported, such as bewteen Ancylostoma duodenale and A. caninum (4%) [33,34] and different Toxocara spp. (5.6%–7.2%) [35], suggesting that pigs may also harbor different Trichuris species.
The cox1 phylogeny also supports that whipworms in primates and pigs make up a cryptic species complex. The human Trichuris TTHCH and TTHUG cluster in two distinct clades, here designated 'Human Trichuris China' and 'Human Trichuris Uganda'. The previously described Trichuris species from different non-human primates (T. colobae and Trichuris sp. GHL in black-and-white colobus and francois' leaf monkey, respectively) [7,29] were also found in distinct clades (Fig 2). Moreover, one of the clades included whipworms from other non-human primates (olive, gelada baboons, and long-tailed macaque), which could represent a different Trichuris species in non-human primates (‘Trichuris sp. non-human primates’ clade). Hence, the cox1 phylogeny suggested at least five potential Trichuris spp. infecting primates. Likewise, Ghai et al. [8] identified a distinct group of worms found only in non-human primates, and these might be related to the ‘Trichuris sp. non-human primates’ clade in our study. However, the Trichuris from a black-and-white colobus was not identified as a separate species by Ghai et al. [8], suggesting that this host can also be infected with different Trichuris spp., or it may reflect the use of different genetic markers between studies [36]. For pig Trichuris, the cox1 phylogeny identified the T. suis from Spain to be genetically closely related to TSDK (‘T. suis Europe’ clade) but distinct from T. suis from China, supporting the possibility that different T. suis species can be found in various geographical regions.
In addition to obvious transmission issues for Trichuris species that are shared between humans and non-human primates, the presence of different cryptic species might also be very important for implementation of appropriate control strategies. For instance, different cryptic species of the human trematode Opisthorchis viverrini in different localities (Laos and Thailand) were found to have significantly different fecundity as measured by eggs/g/worm [37]. Moreover, benzimidazole resistance has been associated with single nucleotide polymorphisms (SNPs) in the beta tubulin gene and has been detected in T. trichiura [38], but the presence and frequencies of these SNPs may vary with geography [6, 38] and between whipworms within the species complex. Hence, control and treatment in different areas may not be equally effective, and therefore, there is a need to further explore the species diversity and compare the pathology, epidemiology, and drug susceptibility of different Trichuris species [13].
In conclusion, based on complete mt genome analyses, we suggest the existence of a Trichuris species complex in primates and pigs. Moreover, a rich source of genetic markers is provided that can be used to inform further investigation into the genetic variation among Trichuris spp. infecting these hosts. There is an urgent need to further elucidate the Trichuris species infecting primates in order to illuminate transmission routes and to identify and implement appropriate control measures. Consequently, differences in pathology and treatment efficacy between species should be investigated. This study also suggests that Trichuris in pigs may consist of a cryptic species complex with similar implications. However, this hypothesis needs further testing including samples from various geographical regions and including nuclear DNA markers as well.
Supporting Information
(XLSX)
(XLSX)
Bayesian Inferences revealed a similar tree topology. Bootstrap frequencies (BF) and posterior probabilities (PP) are indicated on the branches (BF/PP). Scale bar represents the number of nucleotide substitutions per site.
(JPG)
Acknowledgments
We gratefully acknowledge the following for providing worm specimens: T. J. C. Anderson and the Southwest National Primate Research Center at Texas Biomedical Research Institute, San Antonio, Texas; M. Bertelsen, Copenhagen Zoo, Denmark; H. Namwanje, Vector Control Division, Ministry of Health, Kampala, Uganda; S. Nissen, I-H. Poulsen, A. Andreassen, H.H. Petersen University of Copenhagen, Denmark
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1. Hotez PJ, Fenwick A, Savioli L, Molyneux DH (2009) Rescuing the bottom billion through control of neglected tropical diseases. Lancet 373: 1570–1575. 10.1016/S0140-6736(09)60233-6 [DOI] [PubMed] [Google Scholar]
- 2. Pullan RL, Smith JL, Jasrasaria R, Brooker SJ (2014) Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors 7:37 10.1186/1756-3305-7-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sternberg RJ, Powell C, McGrane P, Grantham-McGregor S 81997) Effects of a parasitic infection on cognitive functioning. J Exp Psychol 3: 67–76. [Google Scholar]
- 4. Bethony J, Brooker S, Albonico M, Geiger SM, Loukas A, Diemert D, et al. (2006) Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367: 1521–1532. [DOI] [PubMed] [Google Scholar]
- 5. Ravasi DF, O'Riain MJ, Davids F, Illing N (2012) Phylogenetic evidence that two distinct Trichuris genotypes infect both humans and non-human primates. PloS ONE 7:e44187 10.1371/journal.pone.0044187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hansen TV, Thamsborg SM, Olsen A, Prichard RK, Nejsum P (2013) Genetic variations in the beta-tubulin gene and the internal transcribed spacer 2 region of Trichuris species from man and baboons. Parasit Vectors 6: 236 10.1186/1756-3305-6-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu GH, Gasser RB, Nejsum P, Wang Y, Chen Q, Song HQ, et al. (2013) Mitochondrial and nuclear ribosomal DNA evidence supports the existence of a new Trichuris species in the endangered francois' leaf-monkey. PloS ONE 8: e66249 10.1371/journal.pone.0066249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ghai RR, Simons ND, Chapman CA, Omeja PA, Davies TJ, Ting N, et al. (2014) Hidden population structure and cross-species transmission of whipworms (Trichuris sp.) in humans and non-human primates in Uganda. PLoS Negl Trop Dis 8: e3256 10.1371/journal.pntd.0003256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roepstorff A, Mejer H, Nejsum P, Thamsborg SM (2011) Helminth parasites in pigs: new challenges in pig production and current research highlights. Vet Par 180: 72–81. [DOI] [PubMed] [Google Scholar]
- 10. Cutillas C, Callejon R, de Rojas M, Tewes B, Ubeda JM, Ariza C, et al. (2009) Trichuris suis and Trichuris trichiura are different nematode species. Acta Trop. 111: 299–307. 10.1016/j.actatropica.2009.05.011 [DOI] [PubMed] [Google Scholar]
- 11. Liu GH, Gasser RB, Su A, Nejsum P, Peng L, Lin RQ, et al. (2012) Clear genetic distinctiveness between human- and pig-derived Trichuris based on analyses of mitochondrial datasets. PLoS Negl Trop Dis 6: e1539 10.1371/journal.pntd.0001539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nissen S, Al-Jubury A, Hansen TV, Olsen A, Christensen H, Thamsborg SM, et al. (2012) Genetic analysis of Trichuris suis and Trichuris trichiura recovered from humans and pigs in a sympatric setting in Uganda. Vet Par 188: 68–77. [DOI] [PubMed] [Google Scholar]
- 13. Nadler SA, DE Leon GP (2011) Integrating molecular and morphological approaches for characterizing parasite cryptic species: implications for parasitology. Parasitol 138: 1688–1709. [DOI] [PubMed] [Google Scholar]
- 14. Bensasson D, Zhang D, Hartl DL, Hewitt GM (2001) Mitochondrial pseudogenes: evolution's misplaced witnesses. Trends Ecol Evol 16: 314–321. [DOI] [PubMed] [Google Scholar]
- 15. McGuire JA, Linkem CW, Koo MS, Hutchison DW, Lappin AK, Orange DI, et al. (2007) Mitochondrial introgression and incomplete lineage sorting through space and time: phylogenetics of crotaphytid lizards. Evolution 61: 2879–2897. [DOI] [PubMed] [Google Scholar]
- 16. Jabbar A, Beveridge I, Mohandas N, Chilton NB, Littlewood DT, Jex AR, et al. (2013) Analyses of mitochondrial amino acid sequence datasets support the proposal that specimens of Hypodontus macropi from three species of macropodid hosts represent distinct species. BMC Evol Biol 13:259 10.1186/1471-2148-13-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blouin MS (2002) Molecular prospecting for cryptic species of nematodes: mitochondrial DNA versus internal transcribed spacer. Int J Pararasitol 32: 527–531. [DOI] [PubMed] [Google Scholar]
- 18. Lu G, Moriyama EN (2004) Vector NTI, a balanced all-in-one sequence analysis suite. Briefings in bioinformatics 5: 378–388. [DOI] [PubMed] [Google Scholar]
- 19. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 20. Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, et al. (2013) MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol 69: 313–319. 10.1016/j.ympev.2012.08.023 [DOI] [PubMed] [Google Scholar]
- 21. Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33: W686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laslett D, Canback B (2008) ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24: 172–175. [DOI] [PubMed] [Google Scholar]
- 23. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 25. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 26. Akaike H (1974) A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- 27. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu GH, Wang Y, Xu MJ, Zhou DH, Ye YG, Li JY, et al. (2012) Characterization of the complete mitochondrial genomes of two whipworms Trichuris ovis and Trichuris discolor (Nematoda: Trichuridae). Infec Genet Evol12: 1635–1641. [DOI] [PubMed] [Google Scholar]
- 29. Cutillas C, de Rojas M, Zurita A, Oliveros R, Callejon R (2014) Trichuris colobae n. sp. (Nematoda: Trichuridae), a new species of Trichuris from Colobus guereza kikuyensis . Parasitol Res 113: 2725–2732. 10.1007/s00436-014-3933-6 [DOI] [PubMed] [Google Scholar]
- 30. Ramesh A, Small ST, Kloos ZA, Kazura JW, Nutman TB, Serre D, et al. (2012) The complete mitochondrial genome sequence of the filarial nematode Wuchereria bancrofti from three geographic isolates provides evidence of complex demographic history. Mol Biochem Parasitol 183: 32–41. 10.1016/j.molbiopara.2012.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu GH, Zhao L, Song HQ, Zhao GH, Cai JZ, Zhao Q, et al. (2014) Chabertia erschowi (Nematoda) is a distinct species based on nuclear ribosomal DNA sequences and mitochondrial DNA sequences. Parasit Vectors 7: 44 10.1186/1756-3305-7-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mohandas N, Pozio E, La Rosa G, Korhonen PK, Young ND, Koehler AV, et al. (2014) Mitochondrial genomes of Trichinella species and genotypes—a basis for diagnosis, and systematic and epidemiological explorations. Int J Parasitol 44: 1073–1080. 10.1016/j.ijpara.2014.08.010 [DOI] [PubMed] [Google Scholar]
- 33. Hu M, Chilton NB, Gasser RB (2002) The mitochondrial genomes of the human hookworms, Ancylostoma duodenale and Necator americanus (Nematoda: Secernentea). Int J Parasitol 32: 145–158. [DOI] [PubMed] [Google Scholar]
- 34. Jex AR, Waeschenbach A, Hu M, van Wyk JA, Beveridge I, Littlewood DT, et al. (2009) The mitochondrial genomes of Ancylostoma caninum and Bunostomum phlebotomum—two hookworms of animal health and zoonotic importance. BMC Genomics 10: 79 10.1186/1471-2164-10-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li MW, Lin RQ, Song HQ, Wu XY, Zhu XQ (2008) The complete mitochondrial genomes for three Toxocara species of human and animal health significance. BMC Genomics 9: 224 10.1186/1471-2164-9-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Callejon R, Nadler S, De Rojas M, Zurita A, Petrasova J, Cutillas C (2013) Molecular characterization and phylogeny of whipworm nematodes inferred from DNA sequences of cox1 mtDNA and 18S rDNA. Parasitolo Res 112: 3933–3949. [DOI] [PubMed] [Google Scholar]
- 37. Saijuntha W, Sithithaworn P, Wongkham S, Laha T, Pipitgool V, Tesana S, et al. (2007) Evidence of a species complex within the food-borne trematode Opisthorchis viverrini and possible co-evolution with their first intermediate hosts. Int J Parasitol 37: 695–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DA, Scott ME, et al. (2009) Assays to detect beta-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides . PLoS Negl Trop Dis 3: e397 10.1371/journal.pntd.0000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Bayesian Inferences revealed a similar tree topology. Bootstrap frequencies (BF) and posterior probabilities (PP) are indicated on the branches (BF/PP). Scale bar represents the number of nucleotide substitutions per site.
(JPG)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.