Abstract
In many countries health system data remain too weak to accurately enumerate Plasmodium falciparum malaria cases. In response, cartographic approaches have been developed that link maps of infection prevalence with mathematical relationships to predict the incidence rate of clinical malaria. Microsimulation (or ‘agent-based') models represent a powerful new paradigm for defining such relationships; however, differences in model structure and calibration data mean that no consensus yet exists on the optimal form for use in disease-burden estimation. Here we develop a Bayesian statistical procedure combining functional regression-based model emulation with Markov Chain Monte Carlo sampling to calibrate three selected microsimulation models against a purpose-built data set of age-structured prevalence and incidence counts. This allows the generation of ensemble forecasts of the prevalence–incidence relationship stratified by age, transmission seasonality, treatment level and exposure history, from which we predict accelerating returns on investments in large-scale intervention campaigns as transmission and prevalence are progressively reduced.
 Mathematical models are used to predict malaria burden to inform disease control efforts. Here, Cameron et al. use Bayesian statistics to calibrate previous models against a data set of age-structured prevalence and incidence, generating stratified forecasts of the prevalence–incidence relationship.
Mathematical models are used to predict malaria burden to inform disease control efforts. Here, Cameron et al. use Bayesian statistics to calibrate previous models against a data set of age-structured prevalence and incidence, generating stratified forecasts of the prevalence–incidence relationship.
Despite encouraging recent progress, Plasmodium falciparum continues to impose an enormous burden of disease and death across sub-Saharan Africa1. In many countries with the most intense transmission, disease-reporting infrastructures are weak and precise enumeration of the burden on human health arising from malaria is challenging. This, in turn, limits evidence-based disease-control planning, implementation and evaluation. In response, cartographic approaches have been developed that use maps of infection prevalence (termed the P. falciparum parasite rate, PfPR)2,3 or other transmission metrics4 as a basis for estimating the incidence rate of clinical disease in different locations1,5. While maps of PfPR are becoming increasingly robust, in part because of the proliferation of high-quality data on infection prevalence from nation-wide household surveys, the relationship between PfPR and clinical incidence remains relatively poorly understood and informed by a much smaller and less standardized empirical evidence base.
Recent efforts to construct a suitable PfPR–incidence relationship for P. falciparum burden estimation include purely data-driven fits of varying degrees of sophistication from first-order stratification by endemicity class to hierarchical Gaussian process regression6,7, and projections based on the calibration of a steady-state compartmental transmission model8. Over the past decade, a number of sophisticated microsimulation models have been developed that aim to capture all important components of the malaria transmission system, providing a platform to investigate many aspects on the basic epidemiology of the disease and the likely effect of different control strategies8,9,10. Such models simulate infections at the level of distinct individuals within a population, each having experienced a unique history of past exposure and treatment11,12, and therefore allow inference of the community-level PfPR–incidence relationship. However, conflicts in their predictions arising from differences in the conceptual structures of these models cannot yet be distinguished from those simply because of differences in the data sets used in their calibration, nor indeed from any potential spatiotemporal or ethnic heterogeneity in the underlying relationship. Hence, no consensus yet exists on an appropriate form of the PfPR–incidence curve for use in disease-burden estimation and for addressing other important public-health questions.
The unique potential of microsimulation models for performing detailed epidemiological modelling under realistic conditions13 comes at the price of a much greater computational demand than for steady-state models. As a result, the calibration of microsimulation models against empirical data sets has proven a persistent difficulty for applications of these methods across the health sciences14, and in particular for malariology15,16: the common experience being that sophisticated statistical algorithms are required to achieve computational tractability whether the goal is maximum likelihood estimation of model parameters or full posterior inference. To overcome this challenge in the present study we introduce a novel model-emulation procedure on the basis of the technique of functional regression17,18—in which kernel-weighting methods are used to generate a map from the input space of entomological inoculation rate (EIR) seasonality profile plus model parameter vector to the output space of age-incidence curve plus age-PfPR curve on the basis of a pre-compiled library of noisy, small runtime simulation outputs. The emulator of each model allows fast approximate likelihood evaluations, thereby facilitating thorough posterior sampling under a Markov Chain Monte Carlo (MCMC) algorithm.
In this article we aim to apply the emulator approach to three P. falciparum microsimulation modelling frameworks and a standardized calibration data set to define an ensemble model for the PfPR–incidence relationship that incorporates both empirical uncertainty (driven by a limited and noisy calibration data set) and conceptual uncertainty (driven by structural differences between the models). These three frameworks were selected from among the wider family of contemporary mechanistic models on the basis of four criteria: (i) outputs are generated through microsimulation or stochastic transitions through a compartmental structure; (ii) immunity to clinical illness is explicitly modelled; (iii) either software was readily available or the algorithm was sufficiently transparent to replicate the model independently; and (iv) the modelling framework has been extensively documented in peer-reviewed publications. A number of models were identified as satisfying the first criterion with reference to the systematic review of Reiner et al.19 but were ultimately rejected on the second13,20, while the third and fourth criteria exclude a minority of codes in development or restricted to proprietary use (for example, the in-house GlaxoSmithKline model for roll-out of the RTS,S vaccine candidate). With the resulting ensemble model we are able to account during calibration for (i) the ‘observer effect' (or ‘Hawthorne effect') arising from ethical study designs in which the monitoring campaign itself introduces a treatment rate higher than that previously typical of the target community and (ii) site-to-site differences in the seasonality of malaria transmission. We then further account on prediction for (iii) age-, treatment- and seasonality dependence in the PfPR–incidence relationship and (iv) for the effects of recent declines from historically high levels of transmission (and hence exposure-based immunity). From an analysis of these end points, we predict accelerating returns on investments in large-scale intervention campaigns as transmission and prevalence are progressively reduced.
Results
Data
The data against which we calibrate each of the three transmission models explored in this study represent a subset of the compilation prepared by Battle et al.21 in their exhaustive literature review of studies reporting direct measurements of incidence for both P. falciparum and P. vivax malaria. Here we restricted our focus to those sub-Saharan African P. falciparum surveys with active case detection (ACD, where malaria cases are detected in the community) conducted no less frequently than monthly. In a number of these studies, passive case detection (where cases are detected after seeking care at health facilities) was additionally deployed to alleviate missingness from a fraction of febrile episodes occurring entirely between ACD visits. A further constraint imposed was that the incidence observations are available as raw counts with matched person–year observed tallies in at least four distinct age bins. Where incidence observations were presented under multiple case definitions, we select that with a parasite-density threshold, and where multiple thresholds are reported we select that closest to 5,000 parasites per μl. For continuity with previous work8 we also included a single passive case detection-only study22 that was not otherwise identified by the above criteria. Our final data set is thus composed of measurements from 24 separate studies reporting data for a total of 30 unique sites observed between 1981 and 2011 (Table 1). Contemporaneous, age-structured parasite prevalence data were extracted from the literature to supplement the incidence data for 28 of these sites. Eight of these studies report incidence under a case definition of fever with any detectable parasitaemia (that is, without application of a higher parasite-density threshold designed to improve specificity). In addition, worth noting is that 11 of the 24 studies included in our final data set were not utilized in the previous calibration of the Griffin et al. model8 (adding 10 unique sites to the 20 used previously).
Table 1. Overview of the prevalence–incidence data set used for model calibration.
| Study | Site, Country | Year (s) | PfPR2–10 | Treat. | τACD* | Threshold |
|---|---|---|---|---|---|---|
| Ba et al.54† | Ndiop, Senegal | 1993 | 0.26 | 0.639 | 1 | >3,600 p μl−1 |
| Bloland et al.55† | Asembo Bay, Kenya | 1992 | 0.79 | 0.870 | 14 | Age specific |
| Bonnet et al.37 | Koundou, Cameroon | 1997–1998 | 0.72 | 0.807 | 1 | >1,000 p μl−1 |
| Ebolakouno, Cameroon | 1997–1998 | 0.66 | 0.807 | 1 | >1,000 p μl−1 | |
| Bougouma et al.56,57† | Saponé, Burkina Faso | 2007 | 0.67 | 0.770 | 3 | >2,500 p μl−1 |
| Coulibaly et al.58† | Bandiagara, Mali | 1999 | 0.29 | 0.944 | 7 | Any patent |
| Diallo et al.59,60 | Dakar, Senegal | 1996–1997 | 0.014 | 0.495 | 7 | Any patent |
| S. Dakar, | 1994 | 0.003 | 0.495 | 7 | Any patent | |
| Dicko et al.61 | Donéguébougou, Mali | 1999–2000 | 0.403 | 0.944 | 7 | Any patent |
| Sotuba, Mali | 1999–2000 | 0.086 | 0.944 | 7 | Any patent | |
| Fillol et al.62† | Niakhar, Senegal | 2003 | 0.22 | 0.820 | 7 | >3,000 p μl−1 |
| Greenwood et al.63† | Farafenni, The Gambia | 1981–1982 | 0.32 | 0.682 | 30 | Age specific |
| Guinovart et al.22 | Manhiça, Mozambique | 2003–2005 | 0.20 | 0.820 | · | Any patent |
| Henry et al.64 | Katiola, Côte d'Ivoire | 1997–1998 | 0.91 | 0.686 | 1 | Age specific |
| Korhogo, Côte d'Ivoire | 1997–1998 | 0.88 | 0.686 | 1 | Age specific | |
| Korhogo, Côte d'Ivoire | 1997–1998 | 0.83 | 0.686 | 1 | Age specific | |
| Loha et al.65† | Chano Mille, Ethiopia | 2009–2011 | 0.044 | 0.812 | 7 | Any patent |
| Lusingu et al.66 | Mgome, Tanzania | 2001 | 0.91 | 0.850 | 30 | Age specific |
| Ubiri, Tanzania | 2001 | 0.27 | 0.850 | 30 | Age specific | |
| Magamba, Tanzania | 2001 | 0.067 | 0.850 | 30 | Age specific | |
| Molez et al.67† | Barkedji, Senegal | 1994–1995 | 0.098 | 0.443 | 10 | Age specific |
| Mwangi et al.68,69 | Ngerenya, Kenya | 1999–2001 | 0.25 | 0.870 | 7 | >2,500 p μl−1 |
| Chonyi, Kenya | 1999–2001 | 0.41 | 0.870 | 7 | >2,500 p μl−1 | |
| Nebie et al.70† | Balonghin, Burkina Faso | 2003 | 0.63 | 0.722 | 1 | >5,000 p μl−1 |
| Owusu-Agyei et al.71 | Kintampo, Ghana | 2004 | 0.72 | 0.922 | 2.33 | >5,000 p μl−1 |
| Rogier et al.72 | Dielmo, Senegal | 1990 | 0.89 | 0.639 | 1 | Sharp increase |
| Saute et al.73 | Manhiça, Mozambique | 1996–1999 | 0.26 | 0.384 | 7 | Any patient |
| Schellenberg et al.47 | Ifakara, Tanzania | 2000–2001 | 0.19 | 0.850 | 7 | Any patient |
| Thompson et al.74 | Matola, Mozambique | 1992–1995 | 0.38 | 0.526 | 1 | Age specific |
| Trape et al.75† | Linzolo, Republic of Congo | 1983–1984 | 0.79 | 0.788 | 1 | p/leu. >2 |
| Trape et al.40 | Dielmo, Senegal | 2007–2008 | 0.20 | 0.950 | 2.33 | p/leu. >3.5 |
| Velema et al.76† | Pahou, Benin | 1989 | 0.51 | 0.661 | 30 | >1,000 p μl−1 |
ACD, active case detection; PfPR, P. falciparum parasite rate.
*Here τACD denotes the period of ACD in days for each study. Note also that in the Threshold column p μl−1 stands for parasites per microlitre, and p/leu. the ratio of parasites to leucocytes.
†Highlights studies not included in the previous Griffin et al.8 model calibration.
·Symbol denotes the one study here that did not conduct ACD, but was included here for consistency with the previous analysis of Griffin et al.
Transmission models
The three contemporary transmission platforms employed in this study were OpenMalaria9,23,24,25 (run in a single baseline configuration, rather than as full ensemble itself), the EMOD DTK v1.6 (ref. 10, 12, 26, 27, 28) and the Griffin et al. model8. Here we employ the publicly available microsimulation codes for the former two and, for the latter, a bespoke code based on the compartmental model described therein (we will refer to this implementation as ‘the Griffin IS', that is, Individual Simulation). Each model was run with a 5-day time step under a forced EIR configuration, whereby a pre-determined transmission intensity is imposed as a direct model input, in contrast to ‘full vector' mode simulations in which the EIR is only indirectly controllable through adjustment of ancillary climate and mosquito model parameters. The use of forced EIR here thus is a pragmatic decision to facilitate model fitting at the expense of our ability to capture the dynamic response between host and vector populations (most important at low EIRs) with these simulations. The case management system in each model was configured to yield a 35% probability of effective treatment per febrile episode (formally, per 2-week period with illness in the case of OpenMalaria) during a 90-year period of warm-up simulation time to establish equilibrium levels of immunity under realistic conditions. Here we use the term ‘probability of effective treatment' to mean the direct probability of parasite clearance through drug-based intervention over the course of an illness: that is, the product of a series of steps in the health-care seeking and treatment chain not necessarily modelled explicitly in each code. A year of baseline observations was then sampled before reconfiguration to an 85% probability of effective treatment to simulate the potential ‘observer effect' of an ethical study design at this near-maximal treatment level. The simulation observables here are annual counts of clinical fevers, parasite positives and population size in age bins with end points spaced as {0, 1, 2, 3, 4, 5, 7.5, 10, 15, 25, 35, 45, 60 and 90 years old (y/o)}. Simulations with EIR declining after the warm-up period were effected by direct control, where allowed, by the EMOD DTK and Griffin IS, and through a generic intervention module providing a proportional reduction in the force of infection in the case of OpenMalaria.
Age dependence of the PfPR2–10–incidence relationship
Although fitted against a common data set, our posterior calibrations of the three microsimulation models exhibited a number of subtle differences in their predictions for the PfPR2–10–incidence relationship stratified by age. Figure 1 presents a direct comparison of their posterior envelopes from simulations under a low seasonality profile (here constant EIR) for three key age groups chosen for consistency with the reporting conventions (and prevalence/incidence-to-mortality modelling methodologies) of the Global Burden of Disease project29 and the World Malaria Report1: ‘infants and young children' (0–5 y/o exclusive: that is, up to the fifth birthday), ‘older children' (5–15 y/o) and ‘adults' (15+ y/o). Important to note when interpreting these plots is that the prevalence baseline is that for the 2–10 y/o age group (PfPR2–10) targeted by the spatiotemporal prevalence maps, to which these curves may be applied for burden estimation3. Moreover, the modelled relationships between prevalence and the force of infection are highly nonlinear, so neither should be interpreted naively as a proxy for the other. This is reflected by the convexity in the PfPR2–10–incidence curve for infants and young children in the Griffin IS: at low transmission, incidence scales linearly with both EIR and prevalence as each infected individual is unlikely to face an infectious challenge while currently infected; however, as transmission intensity increases, prevalence saturates, whereas super-infection can lead to episodes of clinical disease, hence, the appearance of a faster-than-linear scaling at 20–40% PfPR2–10. (This effect is also seen in the original Griffin et al. model8 fits as highlighted in Supplementary Fig. 9 of our Supplementary Information File.)
Figure 1. Calibrated posterior predictions of the P. falciparum prevalence–incidence relationship under conditions of low historical treatment and low transmission seasonality from the three microsimulation models comprising our ensemble, stratified by age.
(a,d,g) OpenMalaria; (b,e,h) EMOD DTK; (c,f,i) Griffin IS. In each panel the coloured curve and shaded zones illustrate the (pointwise) median and surrounding 68 and 95% credible intervals for incidence detectable with daily ACD supposing no change to treatment, while the dashed black lines illustrate the median prediction corresponding to a study year intervention increasing the effective treatment rate from 35 to 85% (that is, the ‘observer effect' of ethical study designs).
As each model implements a similar function for age dependence of the biting rate, the drivers of between-model differences observed here lie primarily in differences between exposure and the development of clinical immunity in each model8,24,27,30. In particular, the observation that neither OpenMalaria nor the EMOD DTK exhibits the above-noted convexity in their PfPR2–10–incidence curves for infants and young children can be traced to the operation of parts of their exposure-based immunity models on timescales much shorter than those of the Griffin IS. In the latter, the decay timescales of pre-erythrocytic and clinical disease immunity are fixed a priori to 10 and 30 years, respectively, which limits the fitting flexibility of these components since reductions to the predicted incidence at young ages are coupled strongly to reductions at older ages. In OpenMalaria, however, the dynamic parasite-density threshold model for clinical illness23 has a half-life of just 0.33 years, which enables it to regulate increases in the incidence in infants and young children without forcing the incidence in adults to zero. A similar effect is achieved in the EMOD DTK via the explicit mechanistic simulation of antigenic variation as a modulator of exposure-based immunity.
Despite these subtle differences in the PfPR2–10–incidence relations predicted by each model at fixed age, there is a strong agreement as to the overall strength of exposure-based immunity in shaping the age dependence of clinical illness from P. falciparum malaria: namely, that at low transmission levels corresponding to PfPR2–10 levels below 10% the greatest burden is among the adult population, but at higher transmission levels the balance of morbidity quickly shifts towards children (cf. refs 8, 23). It is this general agreement we hope to capture in our ensemble predictions, which we produce from a weighted pool of each model's posterior predictive envelopes with weights chosen algorithmically (as described in the Methods under Ensemble Model) to favour two- and three-way agreements. Figure 2 presents our ensemble predictions for the age-structured PfPR2-–10–incidence relationships under low seasonality transmission (as shown separately for each model in Fig. 1), as well as for high seasonality transmission and one characterized by a recent decline in transmission intensity.
Figure 2. Ensemble model predictions of the P. falciparum prevalence–incidence relationship, stratified by age.
Predictions are given under conditions of low historical treatment and low transmission seasonality (a,d,g), high seasonality (b,e,h) and low seasonality after a 90% decline in EIR over the past 5 years (c,f,i). In each panel the coloured curve and shaded zones illustrate the (pointwise) median and surrounding 68 and 95% credible intervals for incidence detectable with daily ACD. These ensemble predictions represent a weighted average of the calibrated posteriors from each transmission model with weights assigned by the median of subset posteriors algorithm.
Effects of seasonality and a decline in transmission
A comparison between the age-structured PfPR2–10–incidence relationships of our ensemble model under conditions of low and high transmission seasonality (the left and middle columns of Fig. 2, respectively) reveals only a modest dependence; most notable is the re-emergence of convexity in the high seasonality curve for infants and young children not seen in the ensemble version at low seasonality. The same trend is observed in the high seasonality simulations presented in Griffin et al.8 and can be understood as a consequence of the definition of PfPR2–10 prevalence used here (and in Griffin et al.8) as the annualized average: with few parasite-positive cases expected during the long dry season, the relationship between prevalence and transmission intensity is steeper than for the benchmark low seasonality case. Nevertheless, the overall age dependence of the PfPR2–10–incidence relationship (that is, the shifting age burden with increasing intensity) is little affected by differences in the seasonality profile. However, in the case that EIR has declined from a historically higher level (illustrated in the right column of Fig. 2 for the scenario of a 90% decline over the past 5 years), the age dependence is notably exaggerated, such that infants and young children bear the majority of the burden at all ages. This effect was readily anticipated, given the presence of a long-lived component (>10 year decay timescale) to exposure-based immunity in all three models. Its quantification here is clearly important for an accurate assessment of burden in the context of recent declines in transmission intensity across much of the African continent3.
Discussion
Through a novel emulator-based approach we have been able to calibrate three contemporary microsimulation models against a common, purpose-built data set of age-structured prevalence and incidence counts across 30 unique sites in sub-Saharan Africa. These calibrations reveal subtle morphological differences between the age-structured PfPR–incidence relationships predicted by each model, but also a general agreement in the age dependence of the burden of clinical illness due to P. falciparum malaria at varying levels of transmission. As an ensemble, the combined predictive power of these three models allows the construction of consensus forecasts for the responses of these PfPR–incidence relationships to variations in the seasonality of transmission intensity and the effects of a recent decline in overall EIR. These curves represent a powerful new tool for improving the estimation of malaria disease burden and understanding the implications of changing transmission.
Important to note is that, despite the broad consensus with regard to the expected age distribution of incidence revealed in these ensemble predictions, a substantial degree of uncertainty remains in the overall normalization of the PfPR–incidence relationship owing to the great dispersion observed in total counts between field incidence surveys at different sites with comparable transmission levels. For policy makers considering cartographic burden estimates produced from these curves, it should therefore be emphasized that, although the resulting 95% credible intervals will typically indicate a margin of error of order 33% in the total number of incident cases, corresponding estimates of the proportional change in incidence relative to a given starting year can be made to higher precision, being largely robust against this normalization error. As such, the relative change may allow a more faithful assessment of progress towards elimination than absolute case tallies alone.
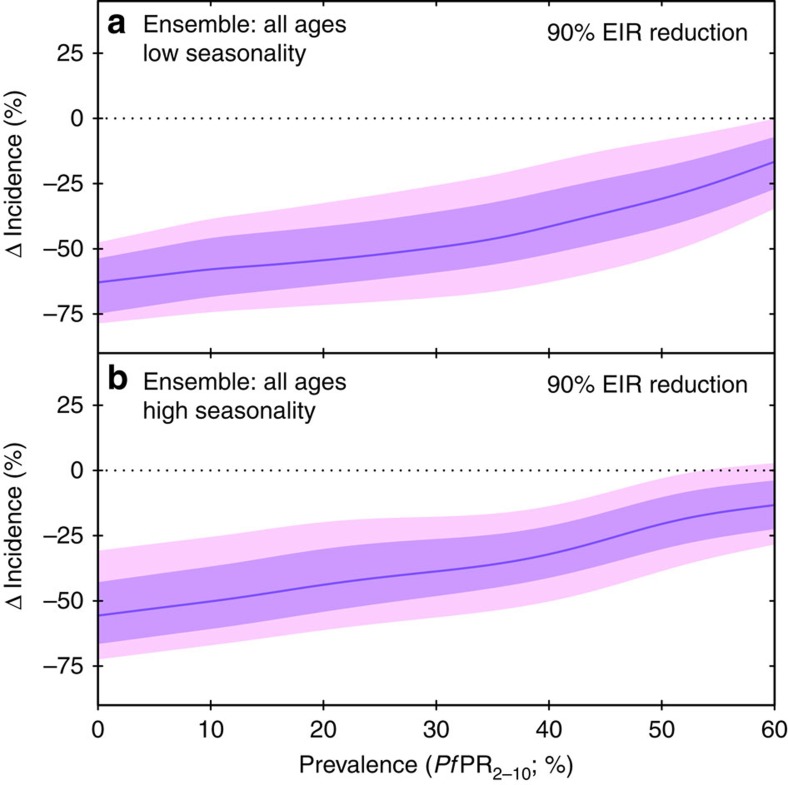
The form of the ensemble PfPR–incidence curves presented here also provides a simple insight with profoundly important implications for global malaria elimination and eradication efforts. Figure 3 shows the changes in clinical incidence that our ensemble model predicts for a given fixed reduction in transmission and how this varies depending on the pre-reduction prevalence level. Using the example of a 90% reduction in EIR over a 5-year period, we demonstrate how the proportional reductions in morbidity accelerate as the transmission reduction is applied to progressively lower prevalence settings. In practical terms, this means that a control programme beginning to successfully reduce PfPR in a highly endemic area may initially see only modest improvements in case incidence. However, as intervention coverage continues to scale up and new control measures are introduced, each successive drop in PfPR will yield progressively larger proportional reductions in cases. The origin of this effect lies in the importance of exposure-based immunity for P. falciparum malaria, as captured in the microsimulation models explored here, which in turn aim to reproduce the complex relationship between transmission intensity and the age dependence of clinical illness observed in field studies31,32. While purpose-built microsimulation studies remain essential to estimate the impacts of specific interventions with uncertain efficiency profiles15,33, our ensemble model demonstrates that for those interventions successful in effecting a general reduction in transmission intensity ever greater pay-offs can be expected as prevalence is brought down progressively across the African continent. This should serve as a rallying call to continue to intensify control efforts that have already yielded substantial declines in infection prevalence and now stand to make increasing impacts on disease burden.
Figure 3. Ensemble model predictions of changes to clinical incidence resulting from a 90% reduction in EIR over the past 5 years, as a function of starting prevalence.
In each panel the coloured curve and shaded zones illustrate the (pointwise) median and surrounding 68 and 95% credible intervals for the change in incidence detectable with daily ACD. Predictions are given for low (a) and high (b) seasonality settings.
Methods
Model emulation
To build a fast emulator for each of the three microsimulation transmission models comprising our ensemble, we adapted a technique from the field of functional data analysis known as functional regression. In this framework we aim to predict the noise-free age-prevalence, PfPR(a), and age-incidence curves, I(a), that would be returned by long runtime (that is, large population) simulations with each model for a given list of input parameters (including the effective treatment rate), θ, and annual EIR time series curve, E(t), using only the noisy age-prevalence and age-incidence curves returned by a reference library of short runtime (small population) simulations. That is, we sought a regression operator,
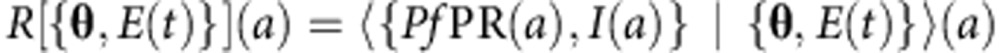 |
where 〈·|·〉 denotes conditional expectation with respect to the (hidden) stochastic process assumed to generate zero-mean noise in the short runtime output. The nonparametric functional regression solution to this problem17 is to construct a kernel-based estimator for R in which the output is a (pointwise) mean of functions from the reference library weighted by the ‘distance', d(·, ·), of their inputs from those of the target,
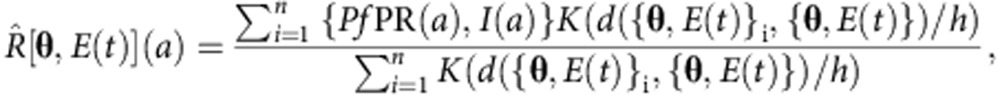 |
for kernel, K(·), and bandwidth parameter, h. The intuition here is that the long-run model output for a given target input can be estimated as a weighted mean of the ‘noisy' outputs, with greatest weight given to those of the latter simulated under inputs close to our target.
Following ref. 18 we chose a locally adaptive kth nearest neighbours bandwidth, whereby for each input {θ, E(t)} the corresponding h was identified such that the distance of the kth nearest reference library member was scaled to unity and K(·) was set to have unit interval support (here we used the truncated standard Normal). Our distance metric was formed from a weighted combination of two separate metrics: one on the function space of EIR time series and the other on the p-dimensional space of input model parameters,
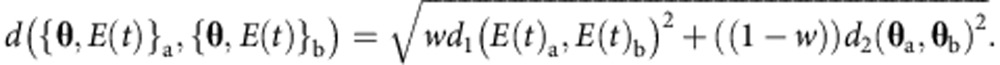 |
For d1(·, ·) we chose the logarithmic L2 distance and for d2(·, ·) the Mahalanobis distance with diagonal covariance matrix, Ξ, after θ was prior integral mass transformed to the unit hypercube. The optimal k,w and Ξ for each emulator were identified via a downhill gradient search through the space {2i, [0, 1], [1, m]mI} (where I denotes the identity matrix and m the dimension of the input parameter vector) for the combination maximizing mean predictive accuracy against a training sample of long runtime simulations.
Almost every choice made in the implementation of a functional regression procedure (or, more generally, a kernel-based regression) can potentially have an impact on its predictive performance: (i) the choice of kernel and bandwidth selection procedure34, (ii) the choice of distance metric imposed on the input space17 and (where relevant, as present) (iii) the design of the reference library. As described above, our approach to the former was to fix the kernel to Normal (Gaussian) and the bandwidth selection to an adaptive k nearest-neighbour strategy a priori, and to impose a strict form for the distance metric with just a handful of free parameters that we choose iteratively so as to optimize the predictive accuracy of the emulator against a library of long runtime benchmark simulations. However, this procedure can only operate after construction of the reference library of ‘noisy' short runtime simulation output, the design of which we describe next.
Reference library
The fundamental trade-off in construction of the reference library is between the accuracy of the simulations on which it is built (determined by the simulated population size) and the coverage of input parameter space (determined by the total number of simulations conducted). As a rule of thumb, given that each code is substantially different in its computational overheads: with each microsimulation model requiring up to 90 years of ‘warm-up' time to ensure equilibrium levels of acquired immunity, even simulations with a population of just 200 people can have runtimes in the tens of seconds, while runs with 10,000 people typically take minutes, and runs with 100,000 people tend towards hours. Hence, although larger populations give more stable outputs, it is clearly infeasible to thoroughly populate a library at such runtimes with inputs drawn from a >13-dimensional parameter space. However, indeed, since unbiased estimation is impossible with Nadaraya–Watson-type estimators in the noise-free limit, the reduction of simulation noise to zero would remain undesirable for the purposes of our model emulation, were the computational burdens any less. Through a process of trial and error we eventually settled on population sizes of 5,000 as a suitable basis for building our microsimulation emulators as with this choice it was possible to build libraries of 100,000 realizations spanning densely the input parameter space of each model. For bandwidth optimization and validation (see Supplementary Fig. 1) we produced a further 100-long runtime simulations with a population size of 100,000.
Transmission code details for openmalaria and EMOD DTK
Numerous aspects of the computational implementation and model structure in both the OpenMalaria and EMOD DTK v1.6 codes have been made extensively customizable to facilitate their application across a diverse range of modelling goals. To ensure the reproducibility of our analysis, we describe here the precise settings used in construction of the reference libraries of simulated age-prevalence and age-incidence curves serving as the reference library in our model emulator.
Our model settings for OpenMalaria (schema version 32) were chosen to follow closely the specification of the ‘base model' described in ref. 15: namely, no decay of immunity (that is, both the ‘IMMUNE_EFFECTOR_DECAY' and ‘ASEXUAL_IMMUNITY_DECAY' parameters set to zero), no mass action effect35 of EIR heterogeneity (that is, ‘LOGNORMAL_MASS_ACTION' set to ‘false'), no heterogeneity in treatment seeking or comorbidities and fixed parameters for the age dependence of the biting rate (S∞=0.049 and E*=0.032). As an exploratory analysis we ran OpenMalaria over a range of EIR levels with zero seasonality for each of the 14 model variants in the best-fit parameterizations described in ref. 15 to trace out approximate PfPR–incidence relationships for each. As only four of these variants (those with the fastest fixed immune decay) exhibited any appreciable difference in this regard, we would broadly expect the results presented herein to be robust against our decision to proceed with the ‘base model' only. To improve the flexibility of the model in representing the diversity of observed age-incidence and age-prevalence counts, we allow the threshold for microscopy-based parasite detection to vary between 20 and 200 parasites per μl in building the reference library.
Where relevant, our model settings for EMOD DTK were then largely chosen in sympathy with those described above for OpenMalaria. In particular, transmission heterogeneity is confirmed zero for MALARIA_SIM mode and we select ‘SURFACE_AREA_DEPENDENT' for the ‘Age_Dependent_Biting_Risk_Type' as the functional form described for this risk profile matches that used in ref. 16. Other key EMOD DTK control option choices here were ‘Enable_Disesase_Mortality' set to zero, ‘Enable_Maternal_Transmission' set to one and ‘Enable_Superinfection' set to one. Again, the parameter controlling the threshold of microscopy-based diagnosis (‘Parasite_Smear_Sensitivity') was allowed to vary over a range equivalent to 20–200 parasites per μl.
For both OpenMalaria and EMOD DTK, the health system settings were simplified to represent administration of a generic antimalarial with the effective treatment rate specified directly as the efficacy (compliance being set to 100%). Simulation of the ‘observer effect' of introducing an enhanced treatment level to a site after years of historically low treatment was implemented via the ‘changeHS' and ‘SimpleHealthTriggeredIntervention' modules, respectively. Specification of EIR seasonality in OpenMalaria was implemented via the ‘fourierSeries' parameterization (discussed further below under Transmission Code Details for EIR Time Series) with a later decline in the mean EIR affected in simulations via the introduction of a generic pre-erythrocytic vaccine intervention (‘vaccineType' set to ‘PEV') blocking a certain fraction of infectious challenges, while in EMOD DTK both these aspects of EIR were specified via the ‘Monthly_EIR' intervention. Finally, it is important to note that each code was run in ‘forced EIR' mode in which the dynamical feedback of transmission intensity between human and vector hosts is turned off. Although potentially less ‘realistic' (being unable to capture the effects of stochastically driven feedback loops between vector and host disease reservoirs), this mode of operation allows for much shorter simulation times, and exploratory analyses with both codes revealed minimal differences in the outputs of interest for EIRs above 0.1 bites per person per year.
Transmission code details for the Griffin IS
In contrast to the general OpenMalaria and EMOD DTK malaria simulation frameworks, the compartmental model described in ref. 8 features only a single structural form in which (i) immunity decays (with a half-life of dClog(2)=20.8 years for acquired immunity), (ii) transmission is strictly heterogeneous but treatment seeking is not and (iii) the age dependence of the biting rate takes a fixed form (parameters ρ=0.85 and a0=8 years). In the ‘Griffin IS' microsimulation code we developed for this model, the disease states of individuals in a mock population are simulated stochastically using a 5-day time step given the out-of-state transition matrix defined by the equations of Griffin et al.8 conditioned by their age, past exposure history, and transmission heterogeneity level. After every month of simulated time the population balance is compared with the input template and bins exhibiting a significant discrepancy are resampled and new births added as necessary to maintain a stable demography despite ageing. EIR seasonality and long-term mean declines are imposed directly at each 5-day time step in a manner equivalent to the ‘forced EIR' modes of OpenMalaria and EMOD DTK. The structure of the Griffin IS model so described was found to be well suited to the object-oriented programming paradigm of the c++ language in which we chose to code it, and satisfactory runtimes were easily achieved with help from the GNU scientific library for simulation from parametric probability densities. Extensive comparisons of the output from our microsimulation code against that of the steady-state compartmental version under zero seasonality were performed to validate its behaviour in at least this classical regime.
Transmission code details for EIR time series
Following the approach of Stuckey et al.36 we model transmission seasonality as a sinusoidal time series in the logarithm of daily EIR, that is,
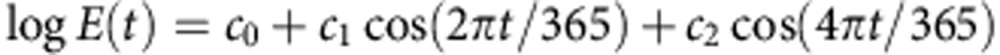 |
where c1=c2=0 corresponds to a constant EIR (no seasonality), c1≠0, c2=0 a single peak of seasonal transmission and c1≠0, c2≠0a double-peaked profile with half a year between peaks (as seen, for instance, in the monthly EIR time series for Ebolakounou reported in ref. 37). By way of reference we note that for a single-peak seasonal profile (that is, c2=0) a value of c1=1 concentrates roughly 75% of transmission within a 6-month period, while a value of c1=2.5 concentrates the same percentage into just 3 months, equivalent to the definitions of high seasonality previously advocated by Roca-Feltrer et al.38 and Cairns et al.39, respectively. We therefore employ the latter (c1=2.5) as our benchmark for high seasonality posterior prediction and use c1=0 as our low seasonality benchmark.
When fitting the age-incidence and (where available) age-prevalence data for each site we treat the EIR and its seasonality profile as nuisance parameters, which we integrate out (stochastically) via our MCMC algorithm. For this purpose we suppose the following priors:
 |
that is, we restrict c2 to be no more than two-thirds the value of c1, and given the resulting seasonality profile we draw its mean EIR from a broad log-Normal distribution centred on an EIR of 100.
An important question for the modelling of seasonality in this context is whether or not the seasonal profile is indeed identifiable, given only age-structured incidence counts as data and no site-specific prior information concerning the annual EIR time series. An analysis of our posterior inferences for Ndiop and Dielmo (1990–1993) suggests the affirmative as our estimates of EIR=63 (15–120 (95% CrI)) and c1=0.9 (0.2–2.6), and EIR=313 (100–670) and c1=0.5 (0–1.5), respectively, are comparable to their contemporary estimates of EIR=20, with transmission restricted to the brief rainy season in Ndiop and EIR=200 with year-round transmission owing to the presence of a nearby river in Dielmo40,41. In principle, one might also seek to allow for a diversity of long-term historic changes in transmission during fitting; however, aside from identifiability concerns, the computational requirements to build a well-sampled emulator in this case could become excessive. It is also worth noting, as a caveat to our simulations of the ‘observer effect' of treatment, that by running these microsimulation codes in forced EIR mode we cannot capture any follow-on effect of treatment itself reducing transmission (through reduced human infectiousness to mosquitoes). This may lead to a slight overcompensation for the ‘observer effect' in our fits; however, we judged this preferable to neglect the issue.
Model calibration
With our model emulator able to provide rapidly a near approximation to the long runtime limit {PfPR(a), I(a)} for each transmission model belonging to any given {θ, E(t)} pairing, the remaining requirements for posterior exploration were specification of a likelihood function for the observed data given the model, and specification of priors on the input parameters. For the former we introduced a hierarchical Bayesian structure allowing for both site-level random effects and overdispersion in the data, with these and the annual EIR time series treated as nuisance parameters. With yijk denoting the observed incidence in the kth incidence age bin of the jth site in the ith study, and pijm the observed prevalence in the mth prevalence age bin of the same (where available; for all but two sites),
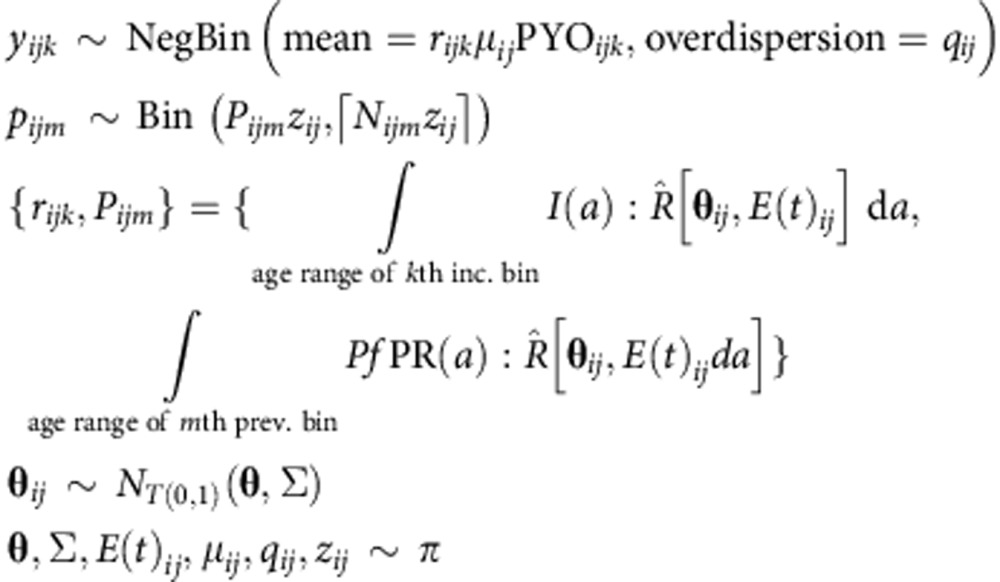 |
where rijk represents the expected (long runtime) incidence rate in the given incidence age bin, and Pijm the expected prevalence in the given prevalence age bin, approximated via the emulator for the transmission model being fit. The likelihood was completed through prior densities (represented here by the place holder, π) on the expected model parameters and site-specific EIR time series, along with the site-specific random effects, μij, and overdispersion terms, qij and zij, described below under random effect and overdispersion priors.
Although an investigation of the posterior predictives for the age-incidence and age-prevalence data on a site-by-site basis under each emulator confirms that the adopted noise model is sufficiently flexible to account for the observational variance here, visual inspection of the discrepancy between model and data in the age-incidence (Supplementary Figs 3, 5 and 7) and age-prevalence (Supplementary Figs 4, 6 and 8) plots for some of these sites (for example, Ngerenya and Kenya) reveals a degree of ‘structural noise' (that is, a limitation of the models to reproduce the observed age dependencies). This issue, also noted in previous studies8,23, likely follows primarily from discrepancies in the levels of heterogeneity in exposure and case management between model and site42, although one might hypothesize that unmodelled spatial variation in the underlying transmission dynamics (for example, variation in vector species43 or parasite genetic diversity44) could also play a role. Both issues warrant future investigation when further ACD-based incidence studies increasing the coverage of countries in Central and Southern Africa (currently under-represented in our data set) become available.
Rather than performing joint MCMC sampling of the complete space of this statistical model—which is of dimensionality >100 as it comprises the full set of nuisance parameters for each study, location and year observed in addition to the global model parameters of interest (θ) and their site-specific random realizations (θij)—we instead performed MCMC (with simulated tempering45 to improve mixing) on each site separately and combined these to approximate the full posterior with importance sample reweighting46. The feasibility of this approach is in part facilitated by the weakness of the parameter constraints imposed by the data from each site individually, although en masse the full data set is ultimately strongly informative with regard to particular parameters from each model. With the magnitude of the observed incidence rate commonly held as a far less reliable indicator of the ground truth than the shape of its age dependence47, and the magnitude of the predicted incidence in each model similarly sensitive to assumptions regarding the maximum duration of a single illness event, our prior on the site-specific random effect allowing for incidence scaling, μij, was deliberately made broad and minimally informative. As a result, a secondary phase of normalization is required to achieve concordance between our model parameter posteriors and the average magnitude of observed counts at each site, which we implement via a linear regression of the expected-to-observed all-age incidence ratios against the frequency of ACD for each draw from our full posterior (as described further under Normalization and Calibration to Daily ACD). All statistical computations were performed in the R environment48.
Parameter priors for transmission model parameters
Each of the three microsimulation codes comprising our ensemble requires the user to specify values for numerous controlling parameters. Previous studies with OpenMalaria15,23,24,25,30 and EMOD DTK10,12,27,28 have sought to constrain these in an essentially stepwise manner through comparisons against disjoint data sets targeting specific segments of each model, whereas the Griffin et al. model has been calibrated only in its steady-state (non-microsimulation) form8 against a homogeneous age-incidence data set including a significant fraction of the studies used here (Table 1). To allow each model a comparable degree of flexibility in the present analysis, we adopt deliberately broad priors on the parameters of all three codes with respect to the constraints suggested by previous analyses. In the Supplementary Discussion (see Supplementary Information) we examine the impact on the posterior predictive PfPR2–10–incidence curves of returning to some of the previously fit or ‘default' values of the key OpenMalaria and EMOD DTK parameters.
Our calibration of OpenMalaria includes 14 free parameters assigned priors of the following forms typically matching the mean but at least doubling the 95% CI ranges quoted in previous papers9,23,24,25,30: Simm (Beta), γp (log-Normal), 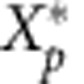 (Gamma),
(Gamma), 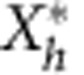 (log-Normal),
(log-Normal), 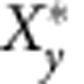 (Gamma),
(Gamma), 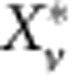 (log-Normal), am (Beta),
(log-Normal), am (Beta), 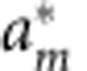 (Gamma),
(Gamma), 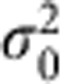 (Normal), α (log-Normal),
(Normal), α (log-Normal), 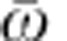 (log-Normal),
(log-Normal), 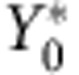 (log-Normal),
(log-Normal), 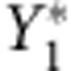 (log-Normal) and
(log-Normal) and  (log-Normal). Our calibration of EMOD DTK includes 13 free parameters assigned priors to the following forms chosen primarily to concentrate mass near the ‘default' values suggested in the EMOD documentation: ‘Antigen_Switch_Rate' (log-Normal), ‘Clinical_Fever_ Threshold_High' (uniform), ‘Clinical_Fever_Threshold_Low' (uniform), ‘Falciparum_MSP_Variants' (Poisson), ‘Falciparum_PfEMP1_variants' (Poisson), ‘Maternal_Antibody_Protection' (Beta), ‘Maternal_Antibody_Decay_Rate' (log-Normal), ‘MSP1_Merozoite_Kill_Fraction' (Beta), ‘MSP2_Merozoite_Kill_Fraction' (Beta), ‘Pyrogenic_Threshold' (log-Normal), ‘Falciparum_Nonspecific_Types' (Poisson), ‘Max_Individual_Infections' (Uniform) and ‘Nonspecific_Antigenicity_Factor' (log-Uniform). Finally, our calibration of the Griffin IS includes 19 free parameters assigned priors of the following forms, roughly matching the 95% prior credible intervals of Griffin et al.8: dU (log-Normal), ID0 (log-Normal), κD (Normal), uD (log-Normal), b0 (Beta), IB0 (log-Normal), κB (log-Normal), uB (log-Normal), φ0 (Beta), φ1 (Beta), IC0 (log-Normal), κC (log-Normal), uC (log-Normal), PM (Beta), dM (log-Normal), d1 (Beta), fD0 (Beta), aD (log-Normal) and γD (log-Normal).
(log-Normal). Our calibration of EMOD DTK includes 13 free parameters assigned priors to the following forms chosen primarily to concentrate mass near the ‘default' values suggested in the EMOD documentation: ‘Antigen_Switch_Rate' (log-Normal), ‘Clinical_Fever_ Threshold_High' (uniform), ‘Clinical_Fever_Threshold_Low' (uniform), ‘Falciparum_MSP_Variants' (Poisson), ‘Falciparum_PfEMP1_variants' (Poisson), ‘Maternal_Antibody_Protection' (Beta), ‘Maternal_Antibody_Decay_Rate' (log-Normal), ‘MSP1_Merozoite_Kill_Fraction' (Beta), ‘MSP2_Merozoite_Kill_Fraction' (Beta), ‘Pyrogenic_Threshold' (log-Normal), ‘Falciparum_Nonspecific_Types' (Poisson), ‘Max_Individual_Infections' (Uniform) and ‘Nonspecific_Antigenicity_Factor' (log-Uniform). Finally, our calibration of the Griffin IS includes 19 free parameters assigned priors of the following forms, roughly matching the 95% prior credible intervals of Griffin et al.8: dU (log-Normal), ID0 (log-Normal), κD (Normal), uD (log-Normal), b0 (Beta), IB0 (log-Normal), κB (log-Normal), uB (log-Normal), φ0 (Beta), φ1 (Beta), IC0 (log-Normal), κC (log-Normal), uC (log-Normal), PM (Beta), dM (log-Normal), d1 (Beta), fD0 (Beta), aD (log-Normal) and γD (log-Normal).
Random effect and overdispersion priors
To complete our hierarchical Bayesian model for the observed data set of age-structured incidence and prevalence counts described above (under Model Calibration), we must add priors for the distributions of site-specific random effects and overdispersions to those on the EIR time series parameters given earlier. Inspection of the fits presented in previous work8 to calibrate the steady-state version of their transmission model led us to expect both marked variations in normalizations of the observed age-incidence and age-prevalence relations at a similar EIR (that is, a potentially large random effects term) and (occasionally) marked structural departures from the model (that is, potentially large overdispersion terms). Hence, we allowed broad priors on each of the form:
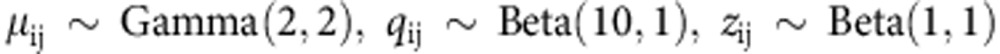 |
where the Gamma distribution takes the shape–rate parameterization. These specifications give an expectation of one for the random effect term in a given survey (corresponding to no rescaling of incidence) and a weak overdispersion of 0.91 (close to the Poissonian limit of one) in the case of incidence, but both allow wide variation about these means if the data so demands; the overdispersion term acting on the observed prevalence, zij, is given greater freedom as the sample sizes of prevalence surveys are typically much larger than those of incidence, increasing the potential for structural conflict between the model and data. Finally, to ensure that our posterior inferences regarding the mean parameterization, θ, are not unnecessarily weakened by the assumed variance on the site-specific realizations, θij, unless the data strongly favour this outcome for a certain parameter, we place an Exponential prior on Σ of mean 0.01. Important to note is that we have not at this stage adopted a specific model for the influence of ACD frequency on the observed incidence counts. Instead, we allow any contribution of this form to be absorbed into our fitted random effect terms, only re-calibrating our predictions to daily ACD during our subsequent normalization step (as described below).
Normalization and calibration to daily ACD
To facilitate exploration of the parameter posterior for each model we specified above only a weak prior on the random effects term, μij, scaling the overall incidence of each site, nor do we explicitly include a covariate representing the frequency of ACD in our likelihood function, although one might expect this to be an important contributor to between-study variance. Hence, we instead perform a further stage of analysis to normalize our emulator posterior the prediction of incidence observable via daily ACD. We implement this calibration through a simple regression model to infer the mean ratio between total incidence observed and model predicted at daily ACD via a simple linear regression against the logarithm of ACD period for those studies applying a parasite threshold in their case definition (assumed to improve specificity). This normalization is performed on each posterior draw; a useful illustration is that shown in Supplementary Fig. 2: for each model we have extracted the mean and s.d. of μij at each site and performed a joint linear regression against the logarithm of ACD period in which the slope is shared across models, while the intercept is not. The resulting plot highlights the relatively small fraction of the observational variance explained by differences in the ACD period, although (as expected) there is a general trend towards surveys conducted at longer ACD follow-up periods to yield lower incidence estimates (by a factor of order 2).
Posterior prediction
By drawing from the MCMC output representing the parameter posterior for each transmission model, and re-running our model emulator across a range of mean EIR levels, we are able to construct the posterior predictive curves for the PfPR–incidence relationship in each age group under a given seasonality profile and treatment history. We therefore chose a representative template for low seasonality (constant EIR) and high seasonality (sinusoidal in log EIR with a factor of 7.3 variation between maximum and minimum; cf. ref. 38) and generated posterior predictive curves for each seasonality and treatment history. A further library of posterior predictive curves was then generated using the emulator approach with an additional set of noisy input simulations from each transmission model under the scenario of a 90% decline in the mean EIR over the past 5 years. This accounts for immunity acquired at historically higher transmission levels when forecasting incidence at sites with recent success in scaling up interventions.
Ensemble model
The canonical Bayesian approach to ensemble prediction under multiple competing models in the Bayesian paradigm is that of model averaging49, in which a weighted average of the posterior predictives is formed with weights proportional to the marginal likelihoods of the models under consideration. However, although attractive for their perceived ‘Occam's Razor'-like penalization of model complexity, marginal likelihoods are potentially highly sensitive to the parameter priors assigned to each model46,50, and in this case we have limited prior information to inform our choices (as discussed above under Parameter Priors). As such, we did not attempt Bayesian model averaging in this case. Nevertheless, rather than defaulting simply to an equal weighting for each model, we would prefer to reward consistency between the predictions of these competing models (following the paradigm of ‘weighting by agreement' identified in a recent review51), such that where two agree with similar credible intervals but the third does not, we up-weight the former two relative to the latter. To achieve this effect with a quantitative, reproducible algorithm we adopted the ‘M-posteriors' routine52; although originally described for combining posterior samples under equal partitions of a single observed data set fit with a single model, the version described therein of Weiszfeld's algorithm for constructing a median of point measures with kernel-based discrepancy distance provides exactly the functionality we required here. Worth noting is that in this setting (that is, location of the empirical posterior median distribution) the problem is one of convex optimization to which the Weiszfeld algorithm ensures a stable solution53. A caveat to this general scheme for ensemble construction is that one may over-reward ‘group-think' where model structures have not been arrived at completely independently—although we note that substantial differences are evident in both the conceptualization and actualization of each microsimulation model considered in this study. Unlike in Bayesian model averaging, the weights assigned to each model here are different for each separate set of posterior predictive curves (low/high seasonality, low/high treatment and so on). Testament to the overall consistency of these three models, it is to be noted that no model is ever assigned less than an 18% contribution to the ensemble by the M-posteriors algorithm.
Additional information
How to cite this article: Cameron, E. et al. Defining the relationship between infection prevalence and clinical incidence of Plasmodium falciparum malaria. Nat. Commun. 6:8170 doi: 10.1038/ncomms9170 (2015).
Supplementary Material
Supplementary Figures 1-9, Supplementary Discussion and Supplementary References
Acknowledgments
P.W.G. is a Career Development Fellow (#K00669X) jointly funded by the UK Medical Research Council (MRC) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement and receives support from the Bill and Melinda Gates Foundation (#OPP1068048 and #OPP1106023). These grants also support E.C., S.B., B.M., U.D., D.J.W. and D.B. The Swiss TPH component was supported through the project #OPP1032350 funded by the Bill and Melinda Gates Foundation (BMGF). S.I.H. is funded by a Senior Research Fellowship from the Wellcome Trust (#095066), which also supports K.E.B., and a grant from the Bill & Melinda Gates Foundation (#OPP1119467). He also acknowledges funding support from the RAPIDD program of the Science & Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health. J.T.G. is funded by an MRC Fellowship (#G1002284). E.A.W. and P.A.E. are funded by the Global Good Fund. We also appreciate the support and interactions facilitated by the Bill & Melinda Gates Foundation-funded Malaria Modeling Consortium (OPP1119467).
Footnotes
Author contributions P.W.G. conceived and designed the research. E.C. drafted the manuscript and Supplementary Information with support from P.W.G. K.E.B. assembled the incidence survey data. E.C. ran each transmission model with advice from E.A.W., P.A.E., J.T.G., M.A.P. and T.A.S. E.C. designed and performed all statistical analyses used in the calibration process. All authors discussed the results and contributed to the revision of the final manuscript.
References
- World Health Organisation. World Malaria Report 1-227 WHO, Switzerland (2014). [Google Scholar]
- Hay S. I. et al. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 6, e1000048 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething P. W. et al. A new world malaria map: Plasmodium falciparum endemicity in 2010. Malar. J. 10, 378 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M. H., Snow R. W. & le Sueur D. A climate-based distribution model of malaria transmission in sub-Saharan Africa. Parasitol. Today 15, 105–111 (1999). [DOI] [PubMed] [Google Scholar]
- Hay S. I. et al. Estimating the global clinical burden of Plasmodium falciparum malaria in 2007. PLoS Med. 7, e1000290 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A. P. et al. Defining the relationship between Plasmodium falciparum parasite rate and clinical disease: statistical models for disease burden estimation. Malar. J. 8, 186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis R. E., Aregawi M., Williams R., Otten M. & Dye C. Worldwide incidence of malaria in 2009: estimates, time trends, and a critique of methods. PLoS Med. 8, e1001142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. T., Ferguson N. M. & Ghani A. C. Estimates of the changing age-burden of Plasmodium falciparum malaria disease in sub-Saharan Africa. Nat. Commun. 5, 3136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. et al. Mathematical modeling of the impact of malaria vaccines on the clinical epidemiology and natural history of Plasmodium falciparum malaria: Overview. Am. J. Trop. Med. Hyg. 75, 1–10 (2006). [DOI] [PubMed] [Google Scholar]
- Eckhoff P. A. A malaria transmission-directed model of mosquito life cycle and ecology. Malar. J. 10, 303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. et al. Towards a comprehensive simulation model of malaria epidemiology and control. Parasitology 135, 1507–1516 (2008). [DOI] [PubMed] [Google Scholar]
- Eckhoff P. Mathematical models of within-host and transmission dynamics to determine effects of malaria interventions in a variety of transmission settings. Am. J. Trop. Med. Hyg. 88, 817–827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W. et al. An individual-based model of Plasmodium falciparum malaria transmission on the coast of Kenya. Trans. R Soc. Trop. Med. Hyg. 97, 43–50 (2003). [DOI] [PubMed] [Google Scholar]
- Rutter C. M., Zaslavsky A. M. & Feuer E. J. Dynamic microsimulation models for health outcomes: a review. Med. Decis. Making 31, 10–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. et al. Ensemble modeling of the likely public health impact of a pre-erythrocytic malaria vaccine. PLoS Med. 9, e1001157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K. A., Wenger E. A., Huynh G. H. & Eckhoff P. A. Calibration of an intrahost malaria model and parameter ensemble evaluation of a pre-erythrocytic vaccine. Malar. J. 14, 6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraty F., Van Keilegom I. & Vieu P. Regression when both response and predictor are functions. J. Multivar. Anal. 109, 10–28 (2012). [Google Scholar]
- Ciollaro M. et al. Nonparametric functional prediction of the unabsorbed flux continuum in the Lyman-α forest of quasar spectra. Contributions in infinite-dimensional statistics and related topics 91–96 (2014). [Google Scholar]
- Reiner R. C. Jr. et al. A systematic review of mathematical models of mosquito-borne pathogen transmission: 1970-2010. J. R. Soc. Interface 10, 20120921 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. L., Smith D. L. & Fidock D. A. Malaria's missing number: calculating the human component of R0 by a within-host mechanistic model of Plasmodium falciparum infection and transmission. PLoS Comput. Biol. 9, e1003025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle K. E. et al. Global database of matched Plasmodium falciparum and P. vivax incidence and prevalence records from 1985–2013. Sci. Data 2, 150012 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinovart C. et al. Malaria in rural Mozambique. Part I: children attending the outpatient clinic. Malar. J. 7, 36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. et al. An epidemiologic model of the incidence of acute illness in Plasmodium falciparum malaria. Am. J. Trop. Med. Hyg. 75, 56–62 (2006). [DOI] [PubMed] [Google Scholar]
- Maire N. et al. A model for natural immunity to asexual blood stages of Plasmodium falciparum malaria in endemic areas. Am. J. Trop. Med. Hyg. 75, 19–31 (2006). [DOI] [PubMed] [Google Scholar]
- Ross A., Maire N., Molineaux L. & Smith T. An epidemiologic model of severe morbidity and mortality caused by Plasmodium falciparum. Am. J. Trop. Med. Hyg. 75, 63–73 (2006). [DOI] [PubMed] [Google Scholar]
- Eckhoff P. A. Malaria parasite diversity and transmission intensity affect development of parasitological immunity in a mathematical model. Malar. J. 11, 419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhoff P. Plasmodium falciparum infection durations and infectiousness are shaped by antigenic variation and innate and adaptive host immunity in a mathematical model. PLoS ONE 7, 0044950 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger E. A. & Eckhoff P. A. A mathematical model of the impact of present and future malaria vaccines. Malar. J. 12, 126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. J. & Lopez A. D. Alternative projections of mortality and disability by cause 1990-2020: Global Burden of Disease Study. Lancet 349, 1498–1504 (1997). [DOI] [PubMed] [Google Scholar]
- Smith T. et al. Relationship between the entomologic inoculation rate and the force of infection for Plasmodium falciparum malaria. Am. J. Trop. Med. Hyg. 75, 11–18 (2006). [DOI] [PubMed] [Google Scholar]
- Carneiro I. et al. Age-patterns of malaria vary with severity, transmission intensity and seasonality in sub-Saharan Africa: a systematic review and pooled analysis. PLoS ONE 5, 0008988 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca-Feltrer A. et al. The age patterns of severe malaria syndromes in sub-Saharan Africa across a range of transmission intensities and seasonality settings. Malar. J. 9, 282 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire N., Shillcutt S. D., Walker D. G., Tediosi F. & Smith T. A. Cost-effectiveness of the introduction of a pre-erythrocytic malaria vaccine into the expanded program on immunization in sub-Saharan Africa: analysis of uncertainties using a stochastic individual-based simulation model of Plasmodium falciparum malaria. Value Health 14, 1028–1038 (2011). [DOI] [PubMed] [Google Scholar]
- Silverman B. W. Weak and strong uniform consistency of kernel estimate of a density and its derivatives. Ann. Stat. 6, 177–184 (1978). [Google Scholar]
- Smith T. Estimation of heterogeneity in malaria transmission by stochastic modelling of apparent deviations from mass action kinetics. Malar. J. 7, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuckey E. M., Smith T. & Chitnis N. Seasonally dependent relationships between indicators of malaria transmission and disease provided by mathematical model simulations. PLoS Comput. Biol 10, e1003812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S. et al. Level and dynamics of malaria transmission and morbidity in an equatorial area of South Cameroon. Trop. Med. Int. Health 7, 249–256 (2002). [DOI] [PubMed] [Google Scholar]
- Roca-Feltrer A., Schellenberg J. R. M. A., Smith L. & Carneiro I. A simple method for defining malaria seasonality. Malar. J. 8, 276 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns M. et al. Estimating the potential public health impact of seasonal malaria chemoprevention in African children. Nat. Commun. 3, 881 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trape J. F. & Rogier C. Combating malaria morbidity and mortality by reducing transmission. Parasitol. Today 12, 236–240 (1996). [DOI] [PubMed] [Google Scholar]
- Fontenille D. et al. Four years' entomological study of the transmission of seasonal malaria in Senegal and the bionomics of Anopheles gambiae and A. arabiensis. Trans. R. Soc. Trop. Med. Hyg. 91, 647–652 (1997). [DOI] [PubMed] [Google Scholar]
- Ross A. & Smith T. Interpreting malaria age-prevalence and incidence curves: a simulation study of the effects of different types of heterogeneity. Malar. J. 9, 132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinka M. E. et al. A global map of dominant malaria vectors. Parasit. Vectors 5, 69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasey A. et al. Genetic diversity of Plasmodium falciparum shows geographical variation. Am. J. Trop. Med. Hyg. 42, 403–413 (1990). [DOI] [PubMed] [Google Scholar]
- Geyer C. Handbook of Markov Chain Monte Carlo 295–311 (2011). [Google Scholar]
- Cameron E. & Pettitt A. Handbook of Markov Chain Monte Carlo (eds. Brooks S., Gelman A., Jones G. L., Meng X.-L. 397–419Chapman & Hall/CRC, Boca Raton, FL (2014). [Google Scholar]
- Schellenberg D. M. et al. The incidence of clinical malaria detected by active case detection in children in Ifakara, southern Tanzania. Trans. R. Soc. Trop. Med. Hyg. 97, 647–654 (2003). [DOI] [PubMed] [Google Scholar]
- R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing (2015). [Google Scholar]
- Hoeting J. A., Madigan D., Raftery A. E. & Volinsky C. T. Bayesian model averaging: a tutorial. Stat. Sci. 14, 382–401 (1999). [Google Scholar]
- Kass R. E. & Raftery A. E. Bayes factors. J. Am. Stat. Assoc. 90, 773–795 (1995). [Google Scholar]
- Lindstrom T., Tildesley M. & Webb C. A bayesian ensemble approach for epidemiological projections. PLoS Comput. Biol. 11, e1004187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsker S., Srivastava S., Lin L. & Dunson D. B. Robust and scalable Bayes via a median of subset posterior measures. Preprint at http://arxiv.org/abs/1403.2660 (2014).
- Vygen J. Approximation Algorithms Facility Location Problems Forschungsinstitut für Diskrete Mathematik, Rheinische Friedrich-Wilhelms-Universität (2005). [Google Scholar]
- Ba F. THÈSE DE TROISIÈME CYCLE DE BIOLOGIE ANIMALE FATOU BA épouse FALL Université Cheikh Anta Diop de Dakar (2000). [Google Scholar]
- Bloland P. B. et al. Longitudinal cohort study of the epidemiology of malaria infections in an area of intense malaria transmission II. Descriptive epidemiology of malaria infection and disease among children. Am. J. Trop. Med. Hyg. 60, 641–648 (1999). [DOI] [PubMed] [Google Scholar]
- Bougouma E. C. et al. Haemoglobin variants and Plasmodium falciparum malaria in children under five years of age living in a high and seasonal malaria transmission area of Burkina Faso. Malar. J. 11, 154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouedraogo A. et al. Malaria morbidity in high and seasonal malaria transmission area of Burkina Faso. PLoS ONE 8, e50036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly D. et al. Impact of preseason treatment on incidence of falciparum malaria and parasite density at a site for testing malaria vaccines in Bandiagara, Mali. Am. J. Trop. Med. Hyg. 67, 604–610 (2002). [DOI] [PubMed] [Google Scholar]
- Diallo S. et al. Malaria in the central health district of Dakar (Senegal). Entomological, parasitological and clinical data. Sante 10, 221–229 (2000). [PubMed] [Google Scholar]
- Diallo S. et al. Malaria in the southern sanitary district of Dakar (Senegal). 1. Parasitemia and malarial attacks. Bull. Soc. Pathol. Exot. 91, 208–213 (1998). [PubMed] [Google Scholar]
- Dicko A. et al. Year-to-year variation in the age-specific incidence of clinical malaria in two potential vaccine testing sites in Mali with different levels of malaria transmission intensity. Am. J. Trop. Med. Hyg. 77, 1028–1033 (2007). [PubMed] [Google Scholar]
- Fillol F. et al. Influence of wasting and stunting at the onset of the rainy season on subsequent malaria morbidity among rural preschool children in Senegal. Am. J. Trop. Med. Hyg. 80, 202–208 (2009). [PubMed] [Google Scholar]
- Greenwood B. M. et al. Mortality and morbidity from malaria among children in a rural area of The Gambia, West Africa. T. R. Soc. Trop. Med. H 81, 478–486 (1987). [DOI] [PubMed] [Google Scholar]
- Henry M. C. et al. Inland valley rice production systems and malaria infection and disease in the savannah of Cote d'Ivoire. Trop. Med. Int. Health 8, 449–458 (2003). [DOI] [PubMed] [Google Scholar]
- Loha E., Lunde T. M. & Lindtjorn B. Effect of bednets and indoor residual spraying on spatio-temporal clustering of malaria in a village in south Ethiopia: a longitudinal study. PLoS ONE 7, e47354 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusingu J. P. et al. Malaria morbidity and immunity among residents of villages with different Plasmodium falciparum transmission intensity in North-Eastern Tanzania. Malar. J. 3, 26 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molez J. F., Diop A., Gaye O., Lemasson J. J. & Fontenille D. Malaria morbidity in Barkedji, village of Ferlo, in Senegal Sahelian area. Bull. Soc. Pathol. Exot. 99, 187–190 (2006). [PubMed] [Google Scholar]
- Mwangi T. W., Ross A., Marsh K. & Snow R. W. The effects of untreated bednets on malaria infection and morbidity on the Kenyan coast. Trans. R. Soc. Trop. Med. Hyg. 97, 369–372 (2003). [DOI] [PubMed] [Google Scholar]
- Mwangi T. W., Ross A., Snow R. W. & Marsh K. Case definitions of clinical malaria under different transmission conditions in Kilifi District, Kenya. J. Infect. Dis. 191, 1932–1939 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebie I. et al. Humoral responses to Plasmodium falciparum blood-stage antigens and association with incidence of clinical malaria in children living in an area of seasonal malaria transmission in Burkina Faso, West Africa. Infect. Immun. 76, 759–766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Agyei S. et al. Epidemiology of malaria in the forest-savanna transitional zone of Ghana. Malar. J. 8, 220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogier C. & Trape J. F. Malaria attacks in children exposed to high transmission: who is protected? Trans. R. Soc. Trop. Med. Hyg. 87, 245–246 (1993). [DOI] [PubMed] [Google Scholar]
- Saute F. et al. Malaria in southern Mozambique: incidence of clinical malaria in children living in a rural community in Manhica district. Trans. R. Soc. Trop. Med. Hyg. 97, 655–660 (2003). [DOI] [PubMed] [Google Scholar]
- Thompson R. et al. The Matola malaria project: a temporal and spatial study of malaria transmission and disease in a suburban area of Maputo, Mozambique. Am. J. Trop. Med. Hyg. 57, 550–559 (1997). [DOI] [PubMed] [Google Scholar]
- Trape J. F., Zoulani A. & Quinet M. C. Assessment of the incidence and prevalence of clinical malaria in semi-immune children exposed to intense and perennial transmission. Am. J. Epidemiol. 126, 193–201 (1987). [DOI] [PubMed] [Google Scholar]
- Velema J. P. et al. Malaria morbidity and mortality in children under three years of age on the coast of Benin, West Africa. Trans. R. Soc. Trop. Med. Hyg. 85, 430–435 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-9, Supplementary Discussion and Supplementary References