Abstract
A carbapenem resistant Salmonella enterica serovar Senftenberg isolate BCH 2406 was isolated from a diarrheal child attending an outpatient unit of B.C. Roy Hospital in Kolkata, India. This isolate was positive for the blaNDM-1 in the PCR assay, which was confirmed by amplicon sequencing. Except for tetracycline, this isolate was resistant to all the tested antimicrobials. The blaNDM-1 was found to be located on a 146.13-kb mega plasmid pNDM-SAL, which could be conjugally transferred into Escherichia coli and other enteric pathogens such as Vibrio cholerae O1 Ogawa and Shigella flexneri 2a. However, the expression of β-lactam resistance is not the same in different bacteria. The whole genome sequence of pNDM-SAL was determined and compared with other pNDM plasmids available in public domain. This plasmid is an IncA/C incompatibility type composed of 155 predicted coding sequences and shares homology with plasmids of E. coli pNDM-1_Dok01, Klebsiella pNDM-KN, and Citrobacter pNDM-CIT. In pNDM-SAL, gene cluster containing blaNDM-1 was located between IS26 and IS4321 elements. Between the IS26 element and the blaNDM-1, a truncated ISAba125 insertion sequence was identified. Downstream of the blaNDM-1, other genes, such as bleMBL, trpF, tat, and an ISCR1 element with class 1 integron containing aac(6′)-Ib were detected. Another β-lactacamase gene, blaCMY -4 was found to be inserted in IS1 element within the type IV conjugative transfer loci of the plasmid. This gene cluster had blc and sugE downstream of the blaCMY -4. From our findings, it appears that the strain S. Senftenberg could have acquired the NDM plasmid from the other members of Enterobacteriaceae. Transfer of NDM plasmids poses a danger in the management of infectious diseases.
Keywords: NDM, carbapenemase, S. Senftenberg, enteric pathogens, pNDM-SAL
Introduction
Emergence of carbapenem resistance among Gram-negative bacteria is a major public health problem as they are associated with critical infections. This drug has been considered as one of the vital drugs against pathogens which produce extended spectrum β-lactamases (ESBLs). New Delhi Metallo-β-lactamase-1(NDM-1), which is a recent addition to the carbapenemase has become a major concern worldwide due to its rapid spread across different members of Enterobacteriaceae and other Gram-negative bacteria. The NDM-1 encoding gene (blaNDM-1) was first detected in Klebsiella pneumoniae and Escherichia coli recovered from a Swedish patient who had undergone treatment in New Delhi, India (Yong et al., 2009; Ghosh et al., 2014). Thereafter, this gene was identified in different Gram-negative bacteria in several countries including the USA, Canada, France, Sweden, UK, Germany, Japan, Austria, Africa, and Australia (Rolain et al., 2010). Presence of blaNDM-1 was generally associated with resistance to most of the antimicrobials, including fluoroquinolones, aminoglycosides, and β-lactams. The blaNDM was found to be located of different large plasmids, which were readily transferable to other bacterial species (Kumarasamy et al., 2010; Rolain et al., 2010). The spread of the blaNDM-1 gene acquired by IncA/C MDR plasmids has drastically reduced the therapeutic options available to the physicians (Ghosh et al., 2014). In this study, we report the isolation of a Salmonella enterica serovar Senftenberg (S. Senftenberg) isolate carrying the blaNDM-1 gene on a large plasmid, which also possessed several genes responsible for extensively drug resistance (EDR).
Materials and Methods
Bacterial Strains
A carbapenem resistant Salmonella enterica isolate (BCH 2406) was isolated in 2012 from a five years old child who attended the outpatient department of B.C. Roy Memorial Hospital for Children, Kolkata for the treatment of diarrhea. This isolate was serotyped according to White-Kauffmann-Le Minor scheme with commercially available antisera (S&A Reagents Lab Ltd., Bangkok, Thailand). Tetracycline resistant XL1-Blue (TETR) and sodium azide resistant E. coli J53 (AzR) strains were used for conjugation experiments. In addition, ampicillin sensitive Vibrio cholerae O1 Ogawa (IDH 5313) and Shigella flexneri 2a (IDH 3077), isolated and identified from diarrheal patients were used as recipients. All the strains were preserved in Luria Bertani (LB) broth (Difco, Sparks, MD, USA) containing 15% glycerol at –80°C. Transconjugants were also maintained in nutrient agar (Difco) stab supplemented with 15 μg/ml ceftriaxone. E. coli ATCC 25922 was served as control in antimicrobial susceptibility testing.
Detection of Carbapenem Resistance Encoding Gene
Presence of blaNDM was identified by PCR with previously described primers (Chen et al., 2011) using Taq DNA polymerase (Roche, Mannheim, Germany). Amplicons were purified using a PCR product purification kit (Qiagen, Hilden, Germany) and sequenced using the ABI Big Dye terminator cycle sequencing ready reaction kit, version 3.1 (Applied Biosystems, Foster City, CA, USA) in an automated DNA sequencer (ABI 3730, Applied Biosystems). The sequences were assembled and analyzed using DNASTAR software (DNASTAR, Inc., Madison, WI, USA).
Conjugation
To test the mobility and promiscuity of the blaNDM-1 haboring plasmid, conjugation by broth mating technique was carried out using NDM-positive Salmonella isolate as donor with four different recipients, namely E. coli XL1-Blue (TETR), E. coli J53 (AzR), V. cholerae O1 Ogawa, and S. flexneri 2a. In brief, overnight cultures of the bacteria were diluted in LB broth and allowed to grow as late-exponential phase culture. Cell density was adjusted to 1.5 × 108cells/ml. Donor and recipient cells were mixed at 1:2 donor-to-recipient ratios in 1 ml of LB broth and allowed grow overnight at 37°C. In all cases, the donor and recipient suspensions were also diluted in phosphate buffer saline (PBS) with a dilution of 10-3 and 10-5 and plated on MacConkey agar (Difco) to confirm the purity and count the colonies.
To recover blaNDM-1 positive transconjugants, several selective media were used. Transconjugants (conjugally transferred, CT) of XL1-Blue (CT-E. coli XL1-Blue) and S. flexneri 2a (CT-S. flexneri) were selected on xylose lysine desoxycholate (XLD, Difco) agar supplemented with tetracycline (30 μg/ml) and ceftriaxone (5 μg/ml). Similarly, transconjugants of V. cholerae (CT-V. cholerae) were obtained with selection based on growth on ceftriaxone (5 μg/ml) containing thiosulphate citrate bile salts sucrose (TCBS, Eiken, Tokyo, Japan) agar and for transconjugants of E. coli J53 (CT-E. coli J53), MacConkey agar containing both ceftriaxone (5 μg/ml) and sodium azide (100 μg/ml) was used. Transconjugants were confirmed as blaNDM-1 positive by PCR analysis followed by PCR amplicon sequencing. The transfer frequencies were expressed as the number of transconjugants per donor cell.
Antimicrobial Susceptibility Testing
To confirm the transfer of resistance phenotype, antibiotic susceptibility patterns of the donor, recipients, and transconjugants were determined after their growth on Mueller–Hinton (MH, Difco) agar by disk diffusion method in accordance with Clinical and Laboratory Standards Institute [CLSI] (2014) using commercially available disks (Becton Dickinson Company, Sparks, MD) namely, ampicillin (AMP), cefuroxime (CXM), ceftriaxone (CRO), cefotaxime (CTX), cefotaxime/clavulanic acid (CTX-CLA), ceftazidime (CAZ), ceftazidime/clavulanic acid (CAZ-CLA), chloramphenicol (CHL), nalidixic acid (NA), ciprofloxacin (CIP), ofloxacin (OFX), norfloxacin (NOR), imipenem (IPM), streptomycin (STR), azithromycin (AZM) tetracycline (TET), trimethoprim/sulfamethoxazole (SXT). To measure the increase of resistance in the transconjugants, MICs of antibiotics (ceftriaxone, imipenem, tetracycline, and sulfamethoxazole/trimethoprim) were determined by E test (AB bioMérieux, Solna, Sweden) in comparison with the wild NDM-positive Salmonella isolate.
Plasmid analysis
Plasmid DNA was extracted from donor, recipients, and transconjugants by Kado and Liu’s (1981) method and analyzed by gel electrophoresis using 0.8% agarose. Presence of blaNDM-1 in the purified plasmid of the donor and transconjugants was determined by PCR. For size determination, plasmid analysis by S1 nuclease-pulsed-field gel electrophoresis (PFGE) (Barton et al., 1995) was performed with the donor and CT-E. coli J53. Total bacterial DNA prepared in agarose plugs was digested with S1 nuclease (Fermentas, Waltham, MA, USA) and separated using a CHEF-Mapper PFGE system (Bio-Rad, Hercules CA, USA), as reported previously (Kumarasamy et al., 2010). The PFGE conditions were run time 18 h gradient 6 V/cm, temperature 14°C, included angle 120° and initial and final pulses conducted for 2.16 s and 54.17 s, respectively. DNA of S. enterica serovar Braenderup H9812 digested with Xba1 (Roche) was included as control size marker. Incompatibility group of plasmid was determined for the Salmonella isolate and E. coli J53 transconjugant (as E. coli J53 AzR was devoid of plasmids) using the PCR-based replicon typing (PBRT) method as described by Carattoli et al. (2005).
Sequencing
Plasmid DNA was prepared from the E. coli J53 tranconjugant using Qiagen Maxiprep kit (Qiagen). Paired-end libraries (300–500 bp fragments) were constructed by using the Illumina H TruSeqTM DNA Sample preparation kit (Illumina). Each library was deposited onto a HiSeq Flow Cell and sequenced using an Illumina HiSeq-2000 next-generation DNA sequencer. The distributions of “base quality” “base composition” and %GC of the plasmid were checked. Based on these distributions, the first 15 bases and last one base were trimmed to avoid specific sequence bias and poor quality bases. Errors were corrected in the sequence data using the HiTEC tool. Contig assembly and predicted gaps were then confirmed and filled by PCR-based gap closure, confirmed by DNA sequencing of the amplicons (Applied Biosystems). To assemble the contigs, ABySS and Edena software were used by varying the parameter ‘k’ and ‘overlap cut-off’, respectively. This step produced several contigs for each parameter setting. The contigs were then integrated using contig integrator for sequence assembly (CISA). A blastN search was made against the ‘nt’ database for each contig and two contigs were retained for further downstream analysis. The open reading frames (ORFs) from the contigs were generated by CISA using Glimmer-MG program. For all these ORFs, the nucleotide sequence and amino acid sequences were obtained and translated in the appropriate frame. The predicted ORFs were annotated using an in-house pipeline (CANoPI-Contig Annotator Pipeline) that also includes blastX search for each ORF sequence against ‘nr’ database provided by NCBI. ORF search and gene prediction was performed for the complete plasmid sequence with Lasergene software (DNASTAR, Inc., Madison, WI, USA) and pairwise alignment was analyzed by blastN and blastP homology search using the NCBI database (http://www.ncbi.nlm.nih.gov/blast).
Nucleotide Sequence Accession Number
The complete sequence of plasmid pNDM-SAL was submitted to GenBank under accession number KP742988.1
Results
Identification and Characterization of blaNDM-1-Positive Isolate
The isolated Salmonella enterica was found to be positive for blaNDM-1, which was confirmed by amplicon sequencing. The sequence of the blaNDM-1 showed 100% homology with those reported previously (Chen et al., 2011; Ho et al., 2011; Sekizuka et al., 2011; Bonnin et al., 2012; Carattoli et al., 2012; Fu et al., 2012; Dolejska et al., 2013). By serology, this isolate was identified as S. Senftenberg presenting antigenic formula as 1,3,19 : g,s,t : -. The isolate was found to be resistant to almost all antibiotics, including nalidixic acid, ciprofloxacin, norfloxacin, ofloxacin, chloramphenicol, streptomycin, azithromycin, sulfamethoxazole/trimethoprim, ampicillin, cefuroxime, ceftriaxone, cefotaxime, ceftazidime, and also to β-lactamase inhibitor combinations (cefotaxime/clavulanate and ceftazidime/clavulanate). However, it was susceptible to tetracycline and showed reduced susceptibility towards imipenem.
Conjugation and Transfer of Resistance
Plasmid harboring the blaNDM-1 was transferable to E. coli strains J53, XL1-Blue and to other enteric pathogens like V. cholerae O1 Ogawa and S. flexneri 2a isolated from the diarrheal patients. Conjugation frequencies were observed at higher rates (∼10-5 tranconjugants per donor cell) while using E. coli J53 and S. flexneri 2a as recipients than E. coli XL1-Blue (∼10-6). The transfer of NDM plasmid into V. cholerae O1 Ogawa was even less efficient (∼10-8) compared to the other strains (Table 1). In addition, the transconjugants acquired additional resistance against β-lactam antibiotics, namely ampicillin, cefuroxime, ceftriaxone, cefotaxime, ceftazidime, and also toward cephalosporin inhibitor combinations like cefotaxime/clavulanate and ceftazidime/clavulanate (Table 1). Interestingly, sulfamethoxazole resistance was noticed only in S. flexneri transconjugant. Though the CT-E. coli J53 was susceptible to sulfamethoxazole/trimethoprim, its MIC was found to increase by 2.7-fold. In CT-S. flexneri, the MIC of SXT rose by 31.6-fold, suggesting that sulfamethoxazole resistance was greatly expressed in CT-S. flexneri but not in CT-E. coli strains.
Table 1.
Antibiotic susceptibilities of donor, recipients, and transconjugants.
| Strain | Bacteria | Resistance profile | MIC value (μg/ml) | |||
|---|---|---|---|---|---|---|
| TET | CRO | IPM | SXT | |||
| BCH 2406 (Donor) | Salmonella Senftenberg | NA, NOR, CIP, OFX, SXT, CHL, AZM, STR, AMP, CXM, CRO, CTX, CTX-CLA, CAZ, CAZ-CLA, IPM (I) | 3 | >256 | 6 | >32 |
| XL1-Blue (Recipient) | Escherichia coli | NA, TET | 192 | 0.5 | 0.125 | |
| CT-E. coli XL1-Blue (Transconjugant) | E. coli Transfer frequency∗: 3.6 × 10-6 | NA, TET, AMP, CXM, CRO, CTX, CTX-CLA, CAZ, CAZ-CLA | 96 | >256 | 3 | |
| J53 (Recipient) | E. coli | AZD | 0.5 | 0.125 | 0.047 | |
| CT-E. coli J53 (Transconjugant) | E. coli Transfer frequency∗: 1.9 × 10-5 | AMP, CXM, CRO, CTX, CTX-CLA, CAZ, CAZ-CLA, AZD | >256 | 1.5 | 0.125 | |
| IDH 5313 (Recipient) | Vibrio cholerae O1 Ogawa | NA, SXT, STR | 0.5 | 1.5 | >32 | |
| CT-V. cholerae (Transconjugant) | V. cholerae O1 Ogawa Transfer frequency∗: 1.5 × 10-8 | NA, SXT, STR, AMP, CXM, CRO, CTX, CTX-CLA, CAZ, CAZ-CLA, IPM | >256 | >32 | >32 | |
| IDH 3077 (Recipient) | Shigella flexneri 2a | NA, STR, TET | 0.75 | 0.25 | 0.38 | |
| CT-S. flexneri (Transconjugant) | S. flexneri 2a Transfer frequency∗: 3.7 × 10-5 | NA, TET, STR, SXT, AMP, CXM, CRO, CTX, CTX-CLA, CAZ, CAZ-CLA, IPM | >256 | 32 | 12 | |
NA, nalidixic acid; CIP, ciprofloxacin; NOR, norfloxacin; OFX, ofloxacin; AZM, azithromycin; STR, streptomycin; AMP, ampicillin; CXM, cefuroxime; CRO, ceftriaxone; CTX, cefotaxime; CAZ, ceftazidime; CTX-CLA, cefotaxime/clavulanate; CAZ-CLA, ceftazidime/clavulanate; IPM, Imipenem; SXT, sulfamethoxazole/trimethoprim; CHL, chloramphenicol; TET, tetracycline; AZD, azide; (I) = intermediate resistance.
∗Frequency of transfer was calculated as number of transconjugants per donor cell.
The CT-E. coli showed reduced susceptibility to carbapenems (Table 1). On the contrary, the transconjugants of clinical strains (CT-V. cholerae and CT-S. flexneri) showed high level resistance to imipenem, far greater than the donor strain S. Senftenberg (>5-fold). The MIC values for imipenem were found to be higher in CT-S. flexneri (128-fold) than those in CT-V. cholerae (>21-fold), CT-E. coli XL1-Blue (24-fold), and CT-E. coli J53 (12-fold). For ceftriaxone, more than 300-fold rise in MIC was observed for all CTs compared to the respective recipients.
Plasmid Analysis
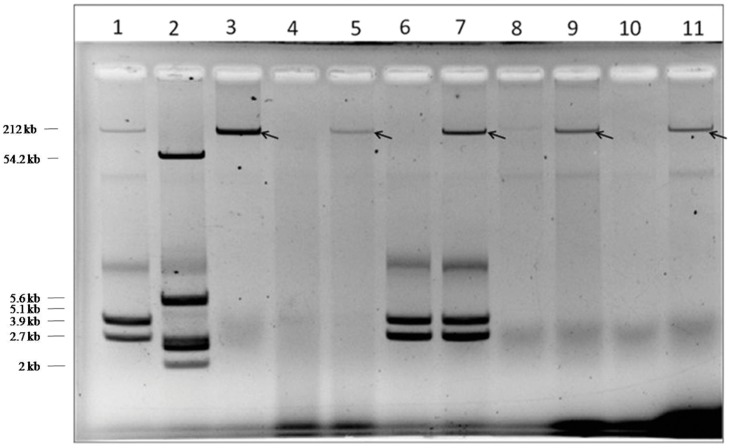
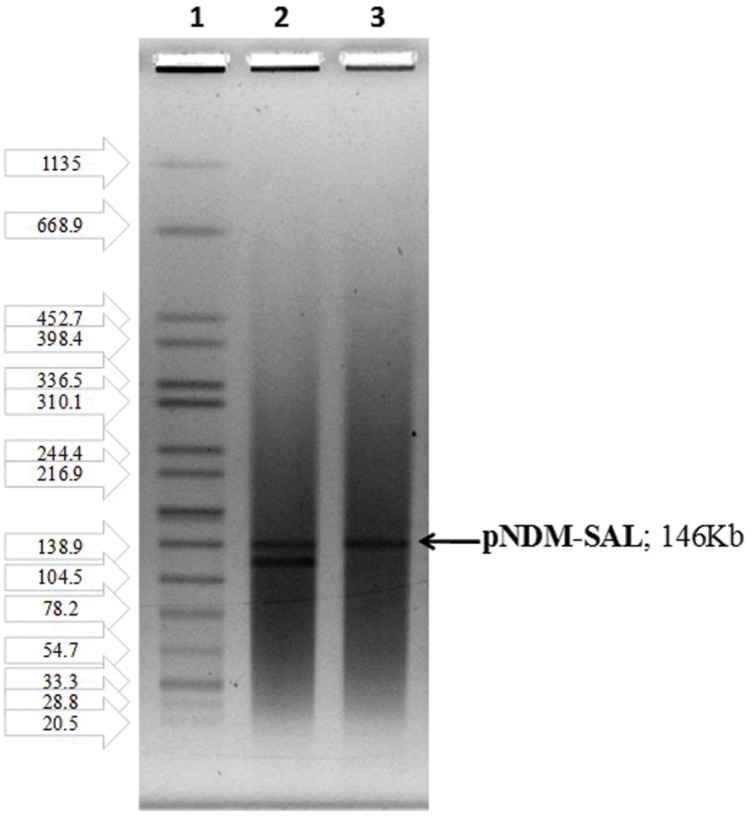
Plasmid profiles of the donor and transconjugants (Figure 1) revealed the transfer of a mega plasmid from S. Senftenberg which conferred β-lactam resistance to the recipient strains of E. coli, V. cholerae, and S. flexneri. NDM-1 PCR assay confirmed the presence of blaNDM-1 in all the transconjugants. S1-PFGE clearly showed that the resistance phenotype was associated with the transfer of single plasmid of about 146-kb (Figure 2) and the NDM-encoding gene was detected by PCR on the plasmid isolated from the transconjugants. Plasmid pNDM-SAL was assigned to the IncA/C incompatibility group using the PCR-based PBRT method.
FIGURE 1.
Plasmid profile of donor, recipient, and transconjugants. Lanes 1 and 2, Shigella flexneri YSH 6000 and Escherichia coli K12 V517 were used as reference standards, respectively; Lane 3, BCH 2406: Salmonella Senftenberg (donor); Lane 4, IDH 5313: Vibrio cholerae O1 Ogawa (recipient); Lane 5, CT-V. cholerae (transconjugant); Lane 6, IDH 3077: S. flexneri 2a (recipient); Lane 7, CT-S. flexneri (transconjugant); Lane 8, XL1-Blue, E. coli (recipient); Lane 9, CT-E. coli XL1-Blue (transconjugant); Lane 10, E. coli J53 (recipient); Lane 11, CT-E. coli J53 (transconjugant).
FIGURE 2.
S1-PFGE indicating the transfer of single plasmid harboring blaNDM-1: Lane1, H-9812 (PFGE marker strain, S. Braenderup H9812); Lane 2, BCH 2406 S. Senftenberg (donor); Lane 3, CT-E. coli J53 (transconjugant).
Sequence Analysis of Plasmid pNDM-SAL
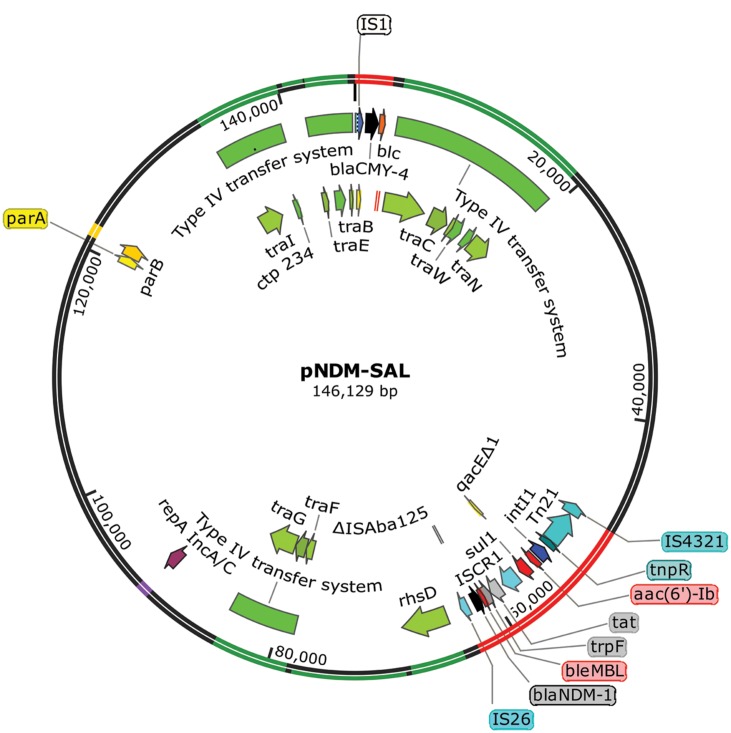
Whole plasmid showed the plasmid pNDM-SAL to consist of 146.13-kb with an average %GC content of 51.7. In addition to its own replication machinery with partitioning system, it has two separate gene clusters for blaNDM-1 and blaCMY -4 (Figure 3). It contained 155 predicted coding sequences (CDSs). This plasmid shared extensive homology (99% identity with 98% query coverage) with an IncA/C plasmid, pNDM-1_Dok01 (195.56-kb, described for E. coli, accession no. AP012208) including the complete array of genes for replication, type IV conjugative transfer machinery, partition and stabilization, except for the flanking region around blaNDM-1 and blaCMY -4. In addition, it had sequence similarity with Klebsiella plasmid pNDM-KN (JN157804) and Citrobacter plasmid pNDM-CIT (JX182975) showing 99% identity with 87% query coverage and 99% identity with 87% query coverage, respectively.
FIGURE 3.
Salient features of blaNDM-1 and blaCMY -4 encoding plasmid pNDM-SAL with transposons and type IV conjugative transfer system. Starting from the outside, the first circle indicates the coordinate of the complete plasmid circle. Transfer machinery and variable region are showed in green and red, respectively. rep and par loci are showed in purple and yellow, respectively. The open reading frames (ORFs) were annotated in the inner circle with arrows representing the direction of transcription. β-lactam resistance genes are denoted by black color and other resistance genes as red color. All transposons and IS elements are indicated in shades of blue. ORFs responsible for conjugation are denoted by shades of green.
Analysis and Comparison of Gene Organization Around blaNDM-1 and between Other Plasmids
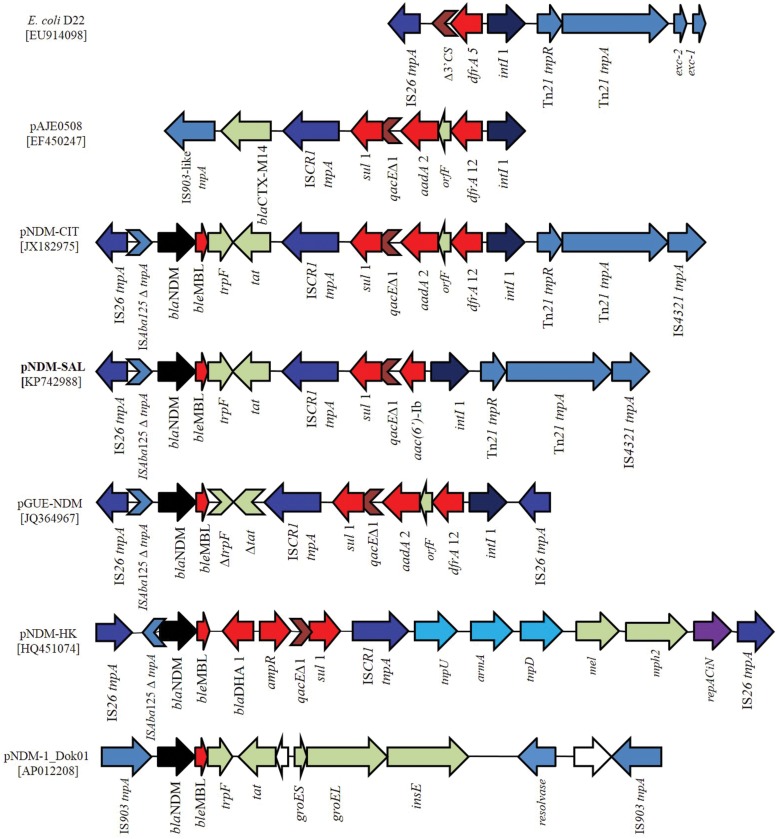
The blaNDM-1 gene was localized in a multidrug resistance region of 15.3-kb (Figure 4). This region was bracketed by two different copies of IS elements in inverted orientation, suggesting that the gene was acquired as a composite transposon. Unlike, pNDM-1_Dok01 and pGUE-NDM (87-kb, described in E. coli, accession no JQ36496) where blaNDM-1 gene was flanked by IS903 and IS26, respectively, pNDM-SAL possessed IS26 and IS4321 upstream and downstream of the blaNDM-1. This orientation is much similar to that found in the pNDM-CIT. In between the IS26 element and the blaNDM-1 gene, a remnant of ISAba125 insertion sequence was identified which comprised –35 promoter sequences leading to the high expression of NDM.
FIGURE 4.
Schematic representation of the DNA sequences surrounding the blaNDM-1 genes in pNDM-SAL compared to the sequences in E. coli D22 (EU914098), pAJE0508 (EF450247), pNDM-CIT (JX182975), pGUE-NDM (JQ36496), pNDM-HK (HQ451074), and pNDM-1_Dok01 (AP012208). Arrows showed the direction of transcription. Gaps between the ORFs are indicated by straight line. The same color and label are used to represent homologous genes. blaNDM-1 are denoted by black color and other resistance genes as red color. All transposons and IS elements are indicated in shades of blue. Other ORFs are denoted by light gray and white. Violet arrow indicates the repAciN gene.
Downstream of the blaNDM-1, the bleMBL gene encoding resistance to bleomycin followed by a truncated phosphorybosilanthranilate isomerase gene (trpF), and a twin-arginine translocation pathway signal protein gene (tat) was identified. Such an arrangement has been reported previously in other plasmid scaffolds (Sekizuka et al., 2011). Following this, a ISCR1 element was identified trailed by a class 1 integron containing aac(6′)-Ib as its variable resistance gene cassette. Such genetic arrangement has been found in a complex class 1 integron containing ISCR1 element of ESBL positive E. coli (EF450247) and V. cholerae (DQ310703). ISCR1 and IS26 are known to mediate transposition and/or expression of multiple resistance genes in their close proximity (Bae et al., 2007; Kiiru et al., 2013). Similarly, class 1 integron bearing ISCR1 was detected in the previously described NDM plasmids, namely pGUE-NDM (JQ364967) and pNDM-CIT (JX182975). The only difference being that the other plasmids harbored dfrA12-ofrF-aadA2 as their integron resistance gene cassettes (Figure 4). However, in all the cases qacEΔ1 and sul1 gene are present in the integron structure. The intI1 gene was followed by the tnpR and tnpA, the genes of transposon Tn21 and then by a copy of insertion element IS4321, as previously identified in pNDM-CIT (JX182975). Like pNDM-SAL, linkage between Tn21 transposon bearing class 1 integron and IS26 element has been seen in NDM negative E. coli of human and animal origin (Dawes et al., 2010) and in E. coli D22 (EU914098). From the in silico analysis, it was not possible to determine the genetic events that led to the formation of this heterogeneous genetic structure. However, it is likely that multiple genetic events contributed to the acquisition of the blaNDM-1 containing locus by the plasmid.
The blaCMY Module Region
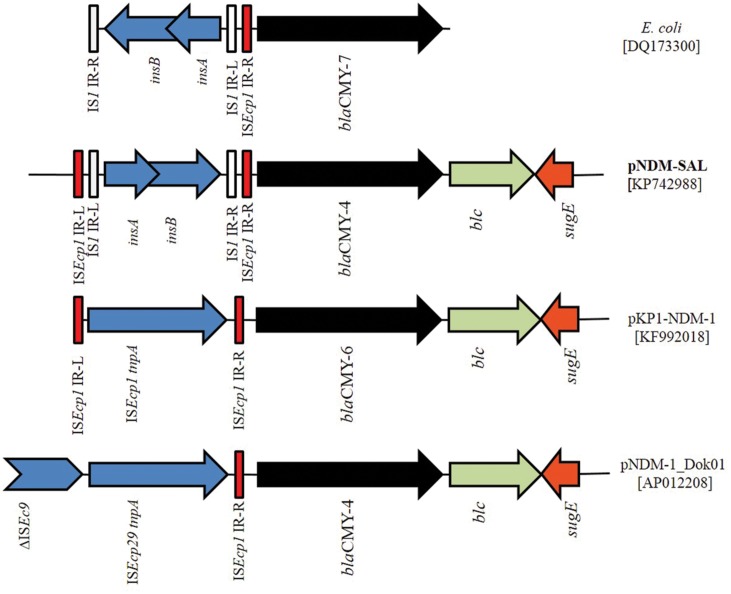
In addition to the blaNDM-1, pNDM-SAL carried an additional β-lactam resistance gene, blaCMY -4, distantly located from the NDM harboring composite transposon (Figure 5). The blaCMY gene was preceded by the IS1 instead of ISEcp1, as reported previously for other NDM plasmids, pNDM-1_Dok01 and pKP1-NDM-1 (KF992018). Remarkably, blaCMY gene associated with IS1 element was detected before in NDM-negative E. coli (DQ173300) with inverted repeats (IRs) of ISEcp1 (Hopkins et al., 2006). This makes it tempting to speculate that an intact copy of ISEcP1 was responsible for the early transposition and mobility of blaCMY -4 followed by insertion of the IS1 element; a process similar to what has been reported previously for E. coli (DQ173300). Downstream of the blaCMY -4 gene, blc gene encoding outer membrane lipoprotein was detected followed by the sugE encoding quaternary ammonium compound resistance protein (Figure 5), a feature common in many of the blaCMY regions in other NDM-positive plasmids (Kang et al., 2006; Sekizuka et al., 2011). This IS1-blaCMY module was located within the tra locus. Similar insertion with ISEcp1 has been demonstrated before (Poole et al., 2009).
FIGURE 5.
Schematic representation of the DNA sequences surrounding the blaCMY -4 genes in pNDM-SAL and comparison with the sequences in E. coli (DQ173300), pKP1-NDM-1 (KF992018), and pNDM-1_Dok01 (AP012208). Arrows indicating blaCMY -4 (black), blc (gray) and sugE (orange) are showed according the direction of transcription. Gaps between the ORFs are indicated by straight line. The same color and label are used to represent homologous genes. IS elements are indicated in shades of blue. Red and white color indicates the inverted repeat (IR) of ISEcp1 and IS1, respectively.
Discussion
The present study revealed EDR features of the pNDM-SAL isolated from S. Senftenberg. This finding highlights the ease with which the resistance determinants can move to other enteric pathogens. Firstly, the blaNDM-1 is located on a broad host range IncA/C plasmid, providing a possible explanation why the pNDM-SAL could be transferred by conjugation to E. coli (E. coli J53 and XL1-Blue) as well as to wild-type strains of V. cholerae O1 Ogawa and S. flexneri 2a easily. In repeated experiments, CT-V. cholerae and CT-S. flexneri showed a higher level of imipenem resistance compared to the donor strains. This could be hypothesized to be due to “possibly” a higher copy number of the plasmid carrying the gene in the recipient than in the donor strains. In other words due to “gene dosage” effect. There could be other possible explanations as well. Examples of such phenomenon, transconjugants showing higher drug resistance are documented in the literature (Ouellette et al., 1988; Petroni et al., 2002).
Since IncA/C plasmids possess highly mobile nature, it has commonly been found in NDM harboring bacteria, especially among the members of Enterobacteriaceae (Ghosh et al., 2014). The size of plasmid varies considerably, ranging from 35.9 to 400-kb (Ghosh et al., 2014) with certain shared genetic background. A recent study has shown that the NDM carrying plasmids from Enterobacteriaceae at New Delhi, shared the replicon type IncA/C with140-kb in size (Walsh et al., 2011). Large plasmids belonging to the IncA/C group have received increased attention, primarily due to their ability to confer resistance to a diverse group of antimicrobial agents (Fernandez-Alarcon et al., 2011). Transmission of pNDM-SAL was also found to be closely associated with transfer of multi drug resistance. Since clinical strains of Vibrio or Shigella are typically resistant to some antimicrobials, transfer of pNDM-SAL carrying blaNDM-1 and other genes could make them multi resistance. This situation is of major concern in the clinical management of infections. Since the pNDM-SAL harbored antibiotic resistance genes are clustered in a few integration hotspots within complex genetic structures, they may be able to acquire novel antibiotic resistance genes through homologous recombination (Doublet et al., 2012). In the pNDM-SAL, rhs locus remains near blaNDM-1 with a phage-integrase and hence they are likely to be the hotspots for integration of accessory genes within the IncA/C plasmids (Carattoli et al., 2012).
In pNDM-SAL, blaNDM-1 was located between IS26 and IS4321. Similar genetic arrangements have previously been observed in other blaNDM-1 bearing plasmids (Sekizuka et al., 2011; Bonnin et al., 2012; Dolejska et al., 2013). Comparison of the flanking regions of blaNDM-1 present in different plasmids suggests that different genetic events may have supported acquisition of this gene in different plasmids (Ghosh et al., 2014). In pNDM-SAL, the linkage of blaNDM-1 with IS26 creates a condition favorable for the mobilization of blaNDM-1. Interestingly, in many NDM negative E. coli strains, arrangement of class 1 integron between Tn21 transposon and IS26 was a characteristic feature (Dawes et al., 2010) as noticed in the pNDM-SAL. The IS26 element, a member of the IS6 family, is widespread among Enterobacteriaceae and exists adjacent to the β-lactamases region, which is a part of transposon-like structure in many plasmids (Yu et al., 2006; Ho et al., 2011). Similar to the other plasmids such as E. coli DVR22 (JF922606), pNDM-HK (HQ451074), pKpANDM-1 (FN396877) and p271A (JF785549), a truncated ISAba125 has been identified upstream of blaNDM-1 in pNDM-SAL. Interestingly, in Acinetobacter baumanii isolates of African origin, the NDM encoding gene was found within the Tn125- like or ISAba125 (Bonnin et al., 2013). This suggests that the ISAba125 insertion sequence has possibly played an important role in the mobilization of blaNDM-1 from a common progenitor and that event was followed by subsequent transfer events mediated by the other insertion elements like IS26 as seen in Acinetobacter spp. and Enterobacteriaceae (Ghosh et al., 2014). In pGUE-NDM (E. coli, JQ364967) and pNDM-MAR (Klebseilla, JN420336) NDM regions are fully bracketed by IS26. But pNDM-SAL possesses IS4321 and Tn21 tranposases in one end similar to what is found in the pNDM-CIT (Citrobacter, JX182975). Instead of blaDHA-1 gene commonly found downstream of many NDM plasmids, eg. pNDM-HK (E. coli, HQ451074) the pNDM-SAL ISCR1 bearing class 1 integron has been inversely located (Figure 4). The association between bleMBL and blaNDM appears to be strong as they are expressed by a common promoter. This may be the reason why both the genes transfer en bloc (Fu et al., 2012; Ghosh et al., 2014).
Based on the genetic environment of blaNDM-1 in pNDM-SAL, we hypothesize that (i) a common transposon structure with ISAba125 could be responsible for the early acquisition of blaNDM-1, (ii) by extensive genetic rearrangement; it was captured in a Tn21 linked complex class 1 integron bearing ISCR1, (iii) later on it was bracketed by two IS elements, namely IS4321 preceding the Tn21 transposase and IS26 truncating the ISAba125.
The plasmid pNDM-SAL has similarity to other NDM-plasmids, which harbor blaCMY -4 and the complex class 1 integron carrying several antibiotic resistance-conferring genes (Toleman et al., 2006a,b). AmpC-like cephalosporinase (blaCMY) genes that have been frequently mobilized by IncA/C-type plasmids, could be identified in E. coli and Salmonella isolates not only of human but also of animal origins in the United States, Canada, and Europe (Winokur et al., 2001; Carattoli et al., 2012). The evolutionary relationship between NDM-1-bearing plasmids and the IncA/C plasmids carrying blaCMY suggests that IncA/C blaCMY-carrying plasmid could have acquired the blaNDM-1 within its scaffold due to a secondary event (Carattoli et al., 2012).
From the data presented in this paper it can be seen that plasmid pNDM-SAL possesses many interesting features as it contain gene encoding the metallo-β-lactamase NDM-1, which is positively associated with multidrug resistance (Figure 3). Besides, pNDM-SAL harbors a large arsenal of genetic elements (integrons, transposons, and ISCRs), giving it the ability to acquire and disseminate antibiotic resistance genes. However, the extent of carbapenem resistance due to the presence of NDM plasmid varied in different hosts. The trend of antimicrobial susceptibility is shifting toward old generation antibiotics, which are less commonly used in recent years (Meziane-Cherif and Courvalin, 2014). In many studies, it was shown that NDM producers were susceptible to old generation antibiotics such as chloramphenicol, tetracycline etc. (Abdul Rahim et al., 2015; Uppu et al., 2015). In our finding, excerpt for tetracycline, the NDM-positive S. Senftenberg were resistant to most of the old generation antibiotics such as ampicillin, trimethoprim, sulfamethoxazole, streptomycin, nalidixic acid, and chloramphenicol. It appears that the maintenance of resistance to any given antibiotic may vary from species to species.
From our data it can be inferred that the strain S. Senftenberg probably was not the natural host for this NDM plasmid but once acquired, the plasmid it had the ability to transfer it to a broad range of pathogenic and non-pathogenic Gram-negative bacteria, an observation which merits serious and important attention to our findings.
Author Contributions
AS, GP, and GC isolated and identified the pathogens, performed phenotypic and genetic analysis. AG and TR analyzed the data, conceived the idea, and wrote the manuscript. All authors were involved in the compilation of the report and approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank George A. Jacoby, Lehey Hospital and Medical Center, Burlington, MA, USA for kindly providing the Escherichia coli J53 (AzR) strain.
Footnotes
Funding. Our work was funded in part by the Indian Council of Medical Research and Indian National Science Academy, New Delhi.
References
- Abdul Rahim N., Cheah S. E., Johnson M. D., Yu H., Sidjabat H. E., Boyce J., et al. (2015). Synergistic killing of NDM-producing MDR Klebsiella pneumoniae by two ‘old’ antibiotics-polymyxin B and chloramphenicol. J. Antimicrob. Chemother. 70 2589–2597. 10.1093/jac/dkv135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae I. K., Lee Y. N., Lee W. G., Lee S. H., Jeong S. H. (2007). Novel complex class 1 integron bearing an ISCR1 element in an Escherichia coli isolate carrying the blaCTX-M-14 gene. Antimicrob. Agents Chemother. 51 3017–3019. 10.1128/AAC.00279-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton B. M., Harding G. P., Zuccarelli A. J. (1995). A general method for detecting and sizing large plasmids. Anal. Biochem. 226 235–240. 10.1006/abio.1995.1220 [DOI] [PubMed] [Google Scholar]
- Bonnin R. A., Cuzon G., Poirel L., Nordmann P. (2013). Multidrug-resistant Acinetobacter baumannii clone, France. Emerg. Infect. Dis. 19 822–823. 10.3201/eid1905.121618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnin R. A., Poirel L., Carattoli A., Nordmann P. (2012). Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS ONE 7:e34752 10.1371/journal.pone.0034752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Villa L., Poirel L., Bonnin R. A., Nordmann P. (2012). Evolution of IncA/C blaCMY-2 carrying plasmids by acquisition of the blaNDM-1 carbapenemase gene. Antimicrob. Agents Chemother. 56 783–786. 10.1128/AAC.05116-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhou Z., Jiang Y., Yu Y. (2011). Emergence of NDM-1 producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66 1255–1259. 10.1093/jac/dkr082 [DOI] [PubMed] [Google Scholar]
- CLSI [Clinical and Laboratory Standards Institute]. (2014). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Fourth Informational Supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Dawes F. E., Kuzevski A., Bettelheim K. A., Hornitzky M. A., Djordjevic S. P., Walker M. J. (2010). Distribution of class 1 integrons with IS26-mediated deletions in their 3’-conserved segments in Escherichia coli of human and animal origin. PLoS ONE 5:e12754 10.1371/journal.pone.0012754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolejska M., Villa L., Poirel L., Nordmann P., Carattoli A. (2013). Complete sequencing of an IncHI1 plasmid encoding the carbapenemase NDM-1, the ArmA 16S RNA methylase and a resistance-nodulation-cell division/multidrug efflux pump. J. Antimicrob. Chemother. 68 34–39. 10.1093/jac/dks357 [DOI] [PubMed] [Google Scholar]
- Doublet B., Boyd D., Douard G., Praud K., Cloeckaert A., Mulvey M. R. (2012). Complete nucleotide sequence of the multidrug resistance IncA/C plasmid pR55 from Klebsiella pneumoniae isolated in 1969. J. Antimicrob. Chemother. 67 2354–2360. 10.1093/jac/dks251 [DOI] [PubMed] [Google Scholar]
- Fernandez-Alarcon C., Singer R. S., Johnson T. J. (2011). Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS ONE 6:e23415 10.1371/journal.pone.0023415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. L., Du X., Ji J., Chen Y., Jiang Y., Yu Y. (2012). Epidemiological characteristics and genetic structure of blaNDM-1 in non-baumannii Acinetobacter spp. in China. J. Antimicrob. Chemother. 67 2114–2122. 10.1093/jac/dks192 [DOI] [PubMed] [Google Scholar]
- Ghosh A., Sarkar A., Chowdhury G., Pazhani G. P., Ramamurthy T. (2014). An overview on the New Delhi Mettalo β-lactamase (NDM)-producers. Proc. Indian Natl. Sci. Acad. 80 547–563. 10.16943/ptinsa/2014/v80i3/55132 [DOI] [Google Scholar]
- Ho P. L., Lo W. U., Yeung M. K., Lin C. H., Chow K. H., Ang I., et al. (2011). Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS ONE 6:e17989 10.1371/journal.pone.0017989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins K. L., Batchelor M. J., Liebana E., Deheer-Graham A. P., Threlfall E. J. (2006). Characterisation of CTX-M and AmpC genes in human isolates of Escherichia coli identified between 1995 and 2003 in England and Wales. Int. J. Antimicrob. Agents 28 180–192. 10.1016/j.ijantimicag.2006.03.027 [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. (1981). Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. S., Besser T. E., Call D. R. (2006). Variability in the region downstream of the blaCMY-2-lactamase gene in Escherichia coli and Salmonella enterica plasmids. Antimicrob. Agents Chemother. 50 1590–1593. 10.1128/AAC.50.4.1590-1593.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiiru J., Butaye P., Goddeeris B. M., Kariuki S. (2013). Analysis for prevalence and physical linkages amongst integrons, ISEcp1, ISCR1, Tn21 and Tn7 encountered in Escherichia coli strains from hospitalized and non-hospitalized patients in Kenya during a 19-year period (1992-2011). BMC Microbiol. 13:109 10.1186/1471-2180-13-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumarasamy K. K., Toleman M. A., Walsh T. R., Bagaria J., Butt F., Balakrishnan R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10 597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meziane-Cherif D., Courvalin P. (2014). Antibiotic resistance: to the rescue of old drugs. Nature 510 477–478. 10.1038/510477a [DOI] [PubMed] [Google Scholar]
- Ouellette M., Gerbaud G., Courvalin P. (1988). Genetic, biochemical and molecular characterization of strains of Vibrio cholerae multiresistant to antibiotics. Ann. Inst. Pasteur Microbiol. 139 105–113. [PubMed] [Google Scholar]
- Petroni A., Corso A., Melano R., Cacace M. L., Bru A. M., Rossi A., et al. (2002). Plasmidic extended-spectrum beta-lactamases in Vibrio cholerae O1 El Tor isolates in Argentina. Antimicrob. Agents Chemother. 46 1462–1468. 10.1128/AAC.46.5.1462-1468.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole T. L., Edrington T. S., Brichta-Harhay D. M., Carattoli A., Anderson R. C., Nisbet D. J. (2009). Conjugative transferability of the A/C plasmids from Salmonella enterica isolates that possess or lack blaCMY in the A/C plasmid backbone. Foodborne Pathog. Dis. 6 1185–1194. 10.1089/fpd.2009.0316 [DOI] [PubMed] [Google Scholar]
- Rolain J. M., Parola P., Cornaglia G. (2010). New Delhi metallo-β-lactamase (NDM-1): towards a new pandemia? Clin. Microbiol. Infect. 16 1699–1701. 10.1111/j.1469-0691.2010.03385.x [DOI] [PubMed] [Google Scholar]
- Sekizuka T., Matsui M., Yamane K., Takeuchi F., Ohnishi M., Hishinuma A., et al. (2011). Complete sequencing of the blaNDM-1-positive IncA/C plasmid from Escherichia coli ST38 isolate suggests a possible origin from plant pathogens. PLoS ONE 6:e25334 10.1371/journal.pone.0025334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Walsh T. R. (2006a). Common regions e.g. orf513 and antibiotic resistance: IS91-like elements evolving complex class 1 integrons. J. Antimicrob. Chemother. 58 1–6. 10.1093/jac/dkl204 [DOI] [PubMed] [Google Scholar]
- Toleman M. A., Bennett P. M., Walsh T. R. (2006b). ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev 70 296–316. 10.1128/MMBR.00048-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppu D. S., Manjunath G. B., Yarlagadda V., Kaviyil J. E., Ravikumar R., Paramanandham K., et al. (2015). Membrane-active macromolecules resensitize NDM-1 gram-negative clinical isolates to chloramphenicol antibiotics. PLoS ONE 10:e0119422 10.1371/journal.pone.0119422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R., Weeks J., Livermore D. M., Toleman M. A. (2011). Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect. Dis. 11 355–362. 10.1016/S1473-3099(11)70059-7 [DOI] [PubMed] [Google Scholar]
- Winokur P. L., Vonstein D. L., Hoffman L. J., Uhlenhopp E. K., Doern G. V. (2001). Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45 2716–2722. 10.1128/AAC.45.10.2716-2722.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D., Toleman M. A., Giske C. G., Cho H. S., Sundman K., Lee K., et al. (2009). Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53 5046–5054. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W. L., Chen S. C., Hung S. W., Chuang Y. C., Chung J. G., Chen I. C., et al. (2006). Genetic association of blaSHV-5 with transposable elements IS26 and IS5 in Klebsiella pneumoniae from Taiwan. Clin. Microbiol. Infect. 12 806–809. 10.1111/j.1469-0691.2006.01488.x [DOI] [PubMed] [Google Scholar]