Abstract
Drug resistance is a global threat and one of the main contributing factors to tuberculosis (TB) outbreaks. The goal of this study was to analyse the molecular profile of multidrug-resistant TB (MDR-TB) in the state of Santa Catarina in southern Brazil. Fifty-three MDR Mycobacterium tuberculosisclinical isolates were analysed by spoligotyping and a partial region of therpoB gene, which is associated with rifampicin resistance (RMP-R), was sequenced. Some isolates were also distinguished by their mycobacterial interspersed repetitive units (MIRU). S531L was the most prevalent mutation found within rpoB in RMP-R isolates (58.5%), followed by S531W (20.8%). Only two MDR isolates showed no mutations withinrpoB. Isolates of the Latin American Mediterranean (LAM) family were the most prevalent (45.3%) found by spoligotyping, followed by Haarlem (9.4%) and T (7.5%) families. SIT106 was found in 26.4% of isolates and all SIT106 isolates typed by MIRU-12 (5 out of 14) belong to MIT251. There was a high correlation between the S531W mutation and the LAM family mainly because all SIT2263 (LAM9) isolates carry this mutation. Among isolates with the S531W mutation in rpoB MIRU demonstrates a cluster formed by four isolates (SIT2263 and MIT163) and very similar profiles were observed between eight of the nine isolates. Better characterisation of TB isolates may lead to new ways in which to control and treat TB in this region of Brazil.
Keywords: tuberculosis, MDR-TB, rpoB, spoligotyping, MIRU
Among the many infectious diseases, tuberculosis (TB) remains the world’s leading cause of death. Despite the advances in TB therapy over the past century, there were an estimated nine million new cases and 1.1 million deaths in 2013 (WHO 2014). Mycobacterial drug resistance is a global threat and one of the main problems associated with this illness, as evidenced by multidrug-resistant TB (MDR-TB). For MDR-TB, multidrug resistance is defined as resistance to isoniazid (INH) and rifampicin (RMP), the two key drugs used to treat TB. Thus, MDR-TB requires second-line drugs for treatment, which are more toxic, expensive and less efficient (Dye & Williams 2000, Espinal 2013).
The acquisition of resistance by Mycobacterium tuberculosis occurs spontaneously due to the natural mutation rate of genomic DNA and there are no known plasmids that confer resistance (Riska et al. 2000). The molecular basis of RMP resistance is mainly due to several mutations within the 81-bp hotspot region of the rpoB gene (corresponding to codons 507-533), most frequently at codons 531, 526 and 516. This gene encodes the β-subunit of the M. tuberculosis RNA polymerase (RNAP), the target of RMP (Telenti et al. 1993). Mutations in this hotspot region are found in almost 96% of RMP-resistant strains, makingrpoB the molecular marker of multidrug resistance and, in fact, the majority of the RMP-resistant isolates are MDR-TB (Somoskovi et al. 2001, Sam et al. 2006, Zhang & Yew 2009).
Genotyping tools are used to elucidate the distribution of TB and its transmission dynamics in the population (Demay et al. 2012). Spoligotyping is a genotyping technique based on the polymorphism of the direct repeat (DR) locus present within M. tuberculosis. The DR contains 36-bp well-conserved repeat sequences interspersed with 34-41-bp nonrepetitive spacers. The profile of the presence/absence of the 43 spacers is compared to more than 58,000 strains deposited in the SITVITWEB database (pasteur-guadaloupe.fr:8081/SITVITD) (Kamerbeek et al. 1997, Demay et al. 2012). Another important genotyping method is based on the copy number of the variable number tandem repeats of mycobacterial interspersed repeat units (MIRU-VNTR) (Supply et al. 2000). In fact, out of the 41 loci that contain MIRUs, the most prevalent VNTRs are the 12, 15 and 24-loci.
Despite the relevance of drug resistance to the treatment and control of TB and MDR-TB, few studies have been published about circulating resistant strains in Brazil. Thus, the aim of this study was to analyse the molecular profile of MDR-TB strains in the state of Santa Catarina (SC), Brazil.
SUBJECTS, MATERIALS AND METHODS
Bacterial isolates and DNA extraction - From March 2008-March 2012, 53 MDR M. tuberculosis isolates were obtained from clinical specimens (only 1 isolate per patient) during routine work at the Central Laboratory of the State of Santa Catarina, the reference laboratory for TB diagnosis in SC. SC has an estimated population of 6,727,148 inhabitants and a demographic density of 65.27 inhabitants per km2 (IBGE 2015). These strains represent all strains detected by the reference laboratory in that period. Susceptibility testing to INH, RMP, ethambutol (EMB) and streptomycin (SM) was performed using BACTEC-960™-MGIT™ according to the manufacturer’s instructions. The H37Rv strain was used as a RMP-susceptible control.
Liquid cultures from BACTEC-960™-MGIT™ were transferred to Ogawa-Kudoh medium and, after four weeks at 36ºC, DNA was extracted using the CTAB method (van Soolingen et al. 1994).
PCR and DNA sequencing - A partial region of rpoB(411 bp) was sequenced using primers described by Telenti et al. (1993). A 50 µL polymerase chain reaction (PCR) was assembled with 1U Taq DNA polymerase, 25 pmol of each primer, 200 µM dNTPs, 1.5 mM MgCl, 20 mM Tris-HCl (pH 8.4) and 50 mM KCl. The following cycling protocol was applied: 94°C for 1 min, 55°C for 1 min and 72°C for 1 min for 40 cycles.
The rpoB PCR fragments were purified by PureLink®Invitrogen PCR purification kit. Sequencing was performed in an Applied Biosystems ABI model 3730 A automated DNA Sequencer (ThermoFisher Scientific, USA). Sequences were aligned and compared to the H37Rv strain sequence (GenBank accession U12205 and our H37Rv sequencing) using BioEdit 7.1.11 software.
Spoligotyping - Spoligotyping was carried out as described by Kamerbeek et al. (1997) using commercially available membranes (Ocimum Biosolutions, India). Hybridising fragments were detected by chemiluminescence using peroxidase-labelled streptavidin and ECL detection reagents.
Binary formats of spoligotypes were compared to those stored in the SITVITWEB database. Within the SITVITWEB database, spoligotype international type (SIT) designates patterns shared by two or more patient isolates, whereas “orphan” designates patterns reported for a single isolate (Demay et al. 2012).
MIRU - MIRU-12 was performed using agarose gel electrophoresis as referenced by Supply et al. (2000). The results were compared with those stored in the SITVITWEB database. MIRU international type (MIT) designates patterns shared by two or more patient isolates in the SITVITWEB database (Demay et al. 2012).
Data analysis - Nominal data were expressed as absolute or relative numbers. A cluster is defined as two or more isolates with identical spoligotypes and MIRU patterns. Comparisons between categorical variables were performed using the Fisher exact test. p values < 0.05 were considered statistically significance.
Ethics - Samples were collected with approval by the Research Ethical Committee of the Federal University of Santa Catarina (process 204.297).
RESULTS
Characterisation of samples - The median age of patients at the time of diagnosis was 36 ± 10 years with a male:female ratio of 2.3. Approximately 60% of cases were either new or relapsed; the remaining cases consisted of retreatments after dropout. Among the 53 MDR-TB isolates, 41 were resistant only to INH and RMP, eight to INH, RMP and EMB, three to INH, RMP and SM and only one to INH, RMP, EMB and SM.
Mutations associated with RMP resistance - The evaluation of 53 MDR isolates identified eight different mutations within rpoB. S531L was the most prevalent mutation, which was found in 31 isolates. The second most frequent mutation was S531W, which was detected in 11 isolates. Other single mutations were found in codons 526, 516 and 572. No mutations were found in two MDR isolates (Table I).
TABLE I. Frequencies of mutated rpoB gene found in 53 multidrug-resistant isolates of Mycobacterium tuberculosis .
| Amino acid change | Nucleotide change | Strains (n) |
|---|---|---|
| S531L | TCG → TTG | 31 |
| S531W | TCG → TGG | 11 |
| H526Y | CAC → TAC | 3 |
| H526D | CAC → GAC | 2 |
| D516V | GAC → GTC | 1 |
| F505V/D516F | TTC → GTC/GAC → TTC | 1 |
| S512G/H526N | AGC → GGC/CAC → AAC | 1 |
| I572V | ATC → GTC | 1 |
| No mutation | - | 2 |
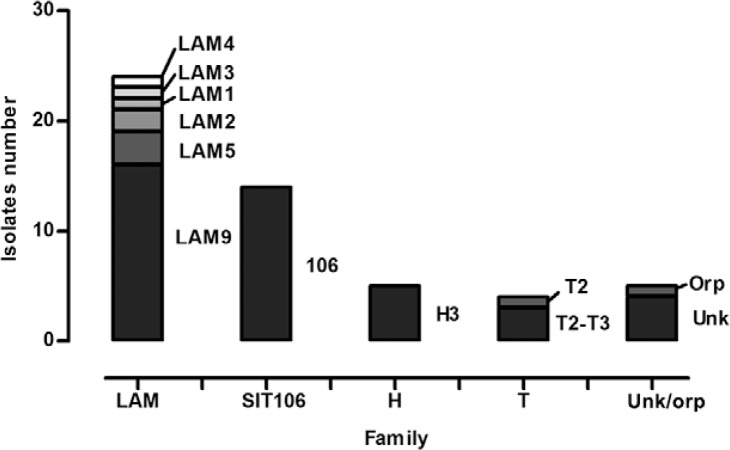
Spoligotyping patterns - Spoligotyping showed 22 different profiles, including 17 shared types, four unknowns and one orphan type (Table II). Between shared types, eight subfamilies and three families were found. As expected, Latin American Mediterranean (LAM) (47.2%) was the most prevalent family, followed by Haarlem (H) (9.4%) and T (7.5%) families. SIT106 represented 26.4% of the isolates and was considered a different group because it is not related to a specific family. The five main shared types were SIT106 (26.4%), SIT42 (13.2%), SIT2263 (13.2%), SIT50 (5.7%) and SIT73 (5.7%) (Figure).
Spoligotyping profiles of 53 multidrug-resistant tuberculosis isolates grouped by families. H: Haarlem; LAM: Latin American Mediterranean; SIT: spoligotype international type; Unk/orp: unknown and orphan.
TABLE II. Spoligotyping profiles found in 53 multidrug-resistant tuberculosis isolates from the state of Santa Catarina, Brazil.

H: Haarlem; LAM: Latin American Mediterranean; SIT: spoligotype international type.
MIRU-12 and associations between mutations and molecular typing methods -Isolates with enough DNA from two groups were subjected to MIRU-12; these isolates were those with the S531W mutation (9/12) and isolates from SIT106 (5/14). Among 14 SIT106 isolates, it was possible to type five isolates by MIRU-12 analysis. These five isolates belonged to the MIT251 (224326133323). Interestingly, SIT106 showed six different mutations in rpoB: S531L (10 isolates), S531W (1 isolate), D516V (1 isolate) and H526D (1 isolate), with D516F and F505V mutations being located in the same isolate.
The S531W mutation showed significant association with the LAM family (p < 0.001) as shown in Table III. This mutation was present in 40% of LAM isolates and 7.2% of SIT106 isolates and was not found in the other families identified in this study. Among the isolates with the S531W mutation, seven belong to SIT2263, two to SIT42, one to SIT50 and one to SIT106. These isolates, excluding SIT106, belong to the LAM9 subfamily. MIRU-12 analysis defined a cluster formed by four isolates (SIT2263 and MIRU 224126152321; isolates 5, 6, 8 and 9) (Table IV) and very similar profiles between eight of the nine isolates that were possible apply MIRU (isolates 1-9) (Table IV).
TABLE III. Association between rpoB S531W mutation and spoligotyping families.
| Spoligotyping family |
rpoBS531W |
||
|---|---|---|---|
| Mutated (n) | Nonmutate (n) | p | |
| SIT106 | 1 | 13 | 0.11 |
| Non-SIT106 | 10 | 29 | |
| H | 0 | 5 | 0.30 |
| Non-H | 11 | 37 | |
| LAM | 10 | 14 | < 0.001 |
| Non-LAM | 1 | 28 | |
| T | 0 | 4 | 0.43 |
| Non-T | 11 | 38 | |
| Unk/orp | 0 | 6 | 0.23 |
| Non-unk/orp | 11 | 36 | |
H: Haarlem; LAM: Latin American Mediterranean; SIT: spoligotype international type; unk/orp: unknown/ohphan.
TABLE IV. Characteristics of rpoB S531W isolates.
| Isolate ID | SIT | Subfamily | MIRU | MIT |
|---|---|---|---|---|
| 1 | 106 | - | 224326133323 | 251 |
| 2 | 42 | LAM9 | 224126152321 | 163 |
| 3 | 42 | LAM9 | 225126152321 | Orphan |
| 4 | 150 | LAM9 | 224126152321 | 163 |
| 5 | 2263 | LAM9 | 224126152321 | 163 |
| 6 | 2263 | LAM9 | 224126152321 | 163 |
| 7 | 2263 | LAM9 | 224226112321 | Unknown |
| 8 | 2263 | LAM9 | 224126152321 | 163 |
| 9 | 2263 | LAM9 | 224126152321 | 163 |
| 10 | 2263 | LAM9 | Not performed MIRU | - |
| 11 | 2263 | LAM9 | Not performed MIRU | - |
bold indicate different number of copy of majority. LAM: Latin American Mediterranean; MIT: mycobacterial interspersed repeat unit (MIRU) international type; SIT: spoligotype international type.
DISCUSSION
RMP represents a great advance in TB treatment because the drug enables short-course chemotherapy via fast bactericidal effects on TB bacilli. However, treatment and control of RMP-resistant TB, mainly MDR-TB, remain a challenge in the XXI century.
Analysis of mutations in the rpoB hotspot region revealed 10 different mutations in 96.2% of resistant strains included in our study. Single nucleotide polymorphisms in codon 531 were responsible for 79.2% of allrpoB mutations. S531L was the most prevalent mutation found in 58.5% of RMP-R isolates, which is in agreement with several studies from Brazil (Hofling et al. 2005, Perizzolo et al. 2012) and other countries (Hilleman et al. 2005, Prammananan et al. 2008, Bolotin et al. 2009, Imperiale et al. 2012, Tang et al. 2013) where prevalence ranged from 40-70%. S531W was the second most common mutation in the rpoB, as verified in our study and was detected in 20.8% of our RMP-R isolates. Araya et al. (2011) found S531W mutations in 28% of RMP-R strains from Chile. Clemente et al. (2008) described the same mutation in 14.3% of RMP-R isolates from Brazil. In contrast, several studies from other Brazilian regions and other countries observed this mutation in less than 10% of RMP-R isolates (Bodmer et al. 1995, Spíndola de Miranda et al. 2001, Cavusoglu et al. 2002, Hofling et al. 2005, Lee et al. 2005, Alves et al. 2011, Tang et al. 2013). The high prevalence of S531W mutations is likely due to a clonal expansion of the LAM9 subfamily (discussed below).
All strains with rpoB mutations showed changes in one of 12 amino acids that surround the RMP binding pocket, which contains the RNAP active site. These mutations cause a modification in the active site of RMP because they lead to the replacement of an amino acid with a compact side chain with an amino acid containing a large side chain (Campbell et al. 2001). Thus, this exchange inactivates the RNAP site. Interestingly, in this study, the double mutations observed in two of the isolates include one amino acid within the group that surrounds the RMP binding pocket and another amino acid that does not belong to this group. We believe that these mutations could be compensatory mutations, improving the fitness cost imposed by the primary mutation that is responsible for RMP resistance.
Molecular tools reduce the time in which it takes to detect drug resistance and, consequently, improve treatment and prevent transmission of MDR-TB (Jacobson et al. 2013). In this study, only two isolates did not have mutations in rpoB. Additionally, although two mutations (F505V and I572V) outside of the 81-bp hotspot of therpoB gene were identified in two of the isolates, one of these isolates had a second mutation in rpoB (F505V/D516F). As reported in previous studies from other Brazilian regions and other countries (Spíndola de Miranda et al. 2001, Hofling et al. 2005, Santos et al. 2010, Alves et al. 2011, Perizzolo et al. 2012), these results confirm that the 81-bp hotspot region can be useful for molecular diagnosis of RMP resistance in TB isolates from SC.
Spoligotyping patterns revealed that the LAM family was the most prevalent (47.2% of isolates), followed by H (9.5%) and T (7.5%). Together, these three families cover 64.2% of the isolates included in this study, which is less than that expected for South America (more than 90% expected) (Demay et al. 2012). This finding could be explained by the high number of SIT106 isolates, corresponding to 26.4% of all MDR isolates included in this study. SIT106 is related to the subfamily U (LAM3) in SpolDB4 (Brudey et al. 2006), but recently, in the SITVITWEB database, was reclassified as “unknown” because it did not belong strictly to any lineage (Demay et al. 2012). This SIT was not reported in a study containing 1,991 isolates from 11 states within Brazil (not including SC) (Gomes et al. 2012). In our study, all isolates typed by MIRU-12 (5) from SIT106 belonged to MIT251 (224326133323). Although it was not possible to analyse nine of the remaining isolates, this group probably represents a big large cluster formed by 14 isolates. Indeed, 10 out of the 14 isolates had the S531L mutation, which represented 18.9% of all isolates in this study. Furthermore, five other different mutations were found in SIT106 isolates. Thus, resistant bacteria from this SIT may generate strains with different mutations due to antibiotic pressure. Isolates within SIT106 and MIT251 were only reported in the SITVITWEB database between 2003-2004 in Spain, Belgium and the United States of America, but they were not related to any spoligotype family (Demay et al. 2012). Interestingly, Gavin et al. (2012), in a study conducted in Spain, found a cluster formed by 14 SIT106 isolates with the S531L mutation that were classified as belonging to the LAM family. Although they performed MIRU-15, overlapping repetitive regions (MIRU04, MIRU10, MIRU16, MIRU26, MIRU31, MIRU39 and MIRU40) had the same number of copies as our SIT106 isolates typed by MIRU-12.
In this study, we demonstrate an association between the rpoB S531W mutation and the LAM family because all SIT2263 isolates (7) had this mutation. Because spoligotyping alone is not sufficient to confirm an epidemiological link, a second method with higher discriminatory power was utilised to highlight potential epidemiological clusters (Jagielski et al. 2014). MIRU-12 was performed on nine isolates with the S531W mutation (isolates 1-9) (Table IV) and a cluster formed by four isolates with SIT2263/MIT163 was confirmed (isolates 5, 6, 8 and 9) (Table IV). Two isolates were not analysed by MIRU-12 (isolates 10 and 11) (Table IV) because there was not enough DNA for analysis. Interestingly, among the four isolates with S531W mutations (those that were not SIT2263; isolates 1-4) (Table IV), two had the same profile as the four initial isolates and only one had a single different allele. The final isolate with the S531W mutation belonged to SIT106 and was not related to the other isolates (isolate 1) (Table IV). Only a single spacer, as determined by spoligotyping of 43 spacers, distinguished SIT42 from SIT150, as well as SIT42 from SIT2263. Our MIRU-12 results indicate a recent spread by infectious MDR-TB containing the S531W mutation. Mariam el al. (2004), in a study with Harlingen strains (T family), demonstrated that the S531W mutation reduces the fitness of M. tuberculosis in vitro both in single cultures and in macrophage cultures, imposing a biological cost to tubercle bacillus. However, fitness cost may be dependent not only on the rpoB mutation, but also on a strain’s genotype (Gagneaux et al. 2006). Therefore, it is possible that strains from the LAM9 subfamily may be better adapted than T family strains.
This study sought to define the molecular basis behind RMP resistance in clinical TB isolates from SC. Molecular characterisation of resistant strains is important to suggest nucleic acid amplification methods that can reduce the time required to detect resistance. We found an association between the rpoB S531W mutation and LAM family strains, which highlights a successful strategy by S531W-containing strains to resist antibiotics. These results may lead to a way in which to improve the control and treatment of MDR-TB within SC.
ACKNOWLEDGEMENTS
To Dr Silvana Spíndola de Miranda, from UFMG, in MIRU technique training, and to PDTIS sequencing platform from Fiocruz.
Funding Statement
Financial support: CNPq (473402/2011-4)
Footnotes
Financial support: CNPq (473402/2011-4)
REFERENCES
- Alves SLA, Metzker FS, Araújo JA, Filho, Junqueira-Kipnis AP, Kipnis A. Clinical data and molecular analysis of Mycobacterium tuberculosis isolates from drug-resistant tuberculosis patients in Goiás, Brazil. Mem Inst Oswaldo Cruz. 2011;106:655–661. doi: 10.1590/s0074-02762011000600003. [DOI] [PubMed] [Google Scholar]
- Araya P, Velasco M, Tognarelli J, Arias F, Leiva T, Sccapatticio A, Alviz P, Fernández P. Detection of genes associated with drug resistance in Mycobacterium tuberculosis in Chile. Rev Med Chile. 2011;139:467–473. [PubMed] [Google Scholar]
- Bodmer T, Zürcher G, Imboden P, Telenti A. Mutation position and type of substitution in the β-subunit of the DNA polymerase influence in vitro activity of rifamycins in rifampin-resistant Mycobacterium tuberculosis. J Antimicrob Chemother. 1995;35:345–348. doi: 10.1093/jac/35.2.345. [DOI] [PubMed] [Google Scholar]
- Bolotin S, Alexander DC, Chedore P, Drews SJ, Jamieson F. Molecular characterization of drug-resistant Mycobacterium tuberculosis isolates from Ontario, Canada. J Antimicrob Chemother. 2009;64:263–266. doi: 10.1093/jac/dkp183. [DOI] [PubMed] [Google Scholar]
- Brudey K, Driscoll JR, Rigouts L, Prodiger WM, Gori A, Al-Hajoj S, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Viedma DG, Garzelli C, Gazzola L, Gomes HM, Gutierrez MC, Hawkey PM, Van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kilkarni SP, Liens B, Lillebaek T, Ly HM, Martin C, Martin C, Makrousov I, Narvskaia O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolfo-Razanamparany V, Rasalonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, Singh UB, Somoskovi A, Skuce RA, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren RM, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. 23BMC Microbiol. 2006;6 doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- Cavusoglu C, Hilmioglu S, Guneri S, Bilgic A. Characterization of rpoB mutations in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Turkay by DNA sequencing and line probe assay. J Clin Microbiol. 2002;40:4435–4438. doi: 10.1128/JCM.40.12.4435-4438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente WT, Lima SSS, Palaci M, Silva MSN, Rodrigues VFS, Costa ERD, Possuelo L, Cafrune PI, Ribeiro FK, Gomes HM, Serufo JC. Phenotypic and genotypic characterization of drug-resistant Mycobacterium tuberculosis strains. Diagn Microbiol Infect Dis. 2008;62:199–204. doi: 10.1016/j.diagmicrobio.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N. SITVITWEB - A publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12:755–766. doi: 10.1016/j.meegid.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Dye C, Williams BG. Criteria for the control of drug-resistant tuberculosis. Proc Natl Acad Sci. 2000;97:8180–8185. doi: 10.1073/pnas.140102797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinal MA. The global situation of MDR-TB. Tuberculosis. 2003;83:44–51. doi: 10.1016/s1472-9792(02)00058-6. [DOI] [PubMed] [Google Scholar]
- Gagneaux S, Long CD, Small PM, Van T, Schoolnik GK, Bohannnan JM. The competitive cost of antibiotic resistance in Mycobacterium tuberculosis. Science. 2006;312:1944–1946. doi: 10.1126/science.1124410. [DOI] [PubMed] [Google Scholar]
- Gavin P, Iglesias MJ, Jiménez MS, Rodríguez-Valín E, Ibarz D, Lezcano MA, Revillo MJ, Martín C, Samper S. Long-term molecular surveillance of multidrug-resistat tuberculosis in Spain. Infect Genet Evol. 2012;12:701–710. doi: 10.1016/j.meegid.2011.05.016. on behalf of The Spanish Working Group on MDR-TB. [DOI] [PubMed] [Google Scholar]
- Gomes HM, Elias AR, Oelemann MA, Pereira MA, Montes FF, Marsico AG, Kritski AL, Filho LD, Caldas PC, Possuelo LG, Cafrune P, Rossetti ML, Lucena N, Saad MH, Cavalcanti HR, Leite CQ, Brito RC, Lopes ML, Lima K, Souza M, Trindade RD, Zozio T, Sola C, Rastogi N, Suffys PN. Spoligotypes of Mycobacterium tuberculosis complex isolates from patients residents of 11 states of Brazil. Infect Genet Evol. 2012;12:649–656. doi: 10.1016/j.meegid.2011.08.027. [DOI] [PubMed] [Google Scholar]
- Hilleman D, Kubica T, Rüsch-Gerdes S, Niemann S. Disequilibrium in distribution of resistance mutations among Mycobacterium tuberculosis Beijing and non-Beijing strains isolated from patients in Germany. Antimicrob Agents Chemother. 2005;49:1229–1231. doi: 10.1128/AAC.49.3.1229-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofling CC, Pavan EM, Giampaglia CMS, Ferrazoli L, Aily DCG, Albuquerque DM, Ramos MC. Prevalence of the katG Ser315 substitution and rpoB mutations in isoniazid-resistant Mycobacterium tuberculosis isolates from Brazil. Inter J Tuberc Lung D. 2005;9:87–93. [PubMed] [Google Scholar]
- IBGE . Estados @. Brasília: 2015. Instituto Brasileiro de Geografia Estatística. http://ibge.gov.br/estadosat/perfil.php?sigla=sc. [Google Scholar]
- Imperiale BR, Zamárraga MJ, Weltman G, Zudiker R, Cataldi, Morcillo NS. First evaluation in Argentina of the GenoType® MTBDRplus assay for multidrug-resistant Mycobacterium tuberculosis detection from clinical isolates and specimens. Rev Argent Microbiol. 2012;44:283–289. [PubMed] [Google Scholar]
- Jacobson KR, Theron D, Kendall EA, Franke ML, Barnard M, Van Herlden PD, Victor TC, Streicher EM, Murray MB, Warren RM. Implementation of GenoType MTBDRplus reduces time to multidrug-resistant tuberculosis therapy initiation in South Africa. Clin Infect Dis. 2013;56:503–508. doi: 10.1093/cid/cis920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagielski T, Van Ingen T, Rastogi N, Dziadek J, Mazur PK, Bielocki J. Current methods in the molecular typing of Mycobacterium tuberculosis and other mycobacteria. Biomed Res Int. 2014;2014 doi: 10.1155/2014/645802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamerbeek J, Shouls L, Kolk A, Van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw H, Goyal M, Van Embden J. Simultaneous detection and stain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ASG, Lim IHK, Tang LLH, Wong SY. High frequency of mutations in the rpoB gene in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Singapore. J Clin Microbiol. 2005;43:2026–2027. doi: 10.1128/JCM.43.4.2026-2027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2004;48:1289–1294. doi: 10.1128/AAC.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perizzolo PF, Costa ERD, Ribeiro AW, Spies FS, Ribeiro MO, Spies FS, Ribeiro MO, Dias CF, Unis G, Silva PA, Gomes HM, Suffys PN. Characteristics of multidrug-resistant Mycobacterium tuberculosis in southern Brazil. Tuberculosis. 2012;92:56–59. doi: 10.1016/j.tube.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Prammananan T, Cheunoy W, Taechamahapun D, Yorsangsukkamol J, Phumpruch S, Phdarat P, Leechawengwong M, Chaiprasert A. Distribution of rpoB mutations among multidrug-resistant Mycobacterium tuberculosis (MDRTB) strains from Thailand and development of a rapid method for mutation detection. Clin Microbiol Infect. 2008;14:446–453. doi: 10.1111/j.1469-0691.2008.01951.x. [DOI] [PubMed] [Google Scholar]
- Riska PF, Jacobs WR, Alland D. Molecular determinants of drug resistance in tuberculosis. Int J Tuberc Lung Dis. 2000;4:54–60. [PubMed] [Google Scholar]
- Sam I, Drobniewski F, More P, Kemp M, Brown T. Mycobacterium tuberculosis and rifampin resistance. Emerg Infect Dis. 2006;12:752–759. doi: 10.3201/eid1205.041339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos LC, Bousquet HM, Pereira AM, Junqueira-Kipnis AP, Kipnis A. A high prevalence of resistance in new tuberculosis cases of midwestern Brazil. Infect Genet Evol. 2010;10:1052–1057. doi: 10.1016/j.meegid.2010.06.018. [DOI] [PubMed] [Google Scholar]
- Somoskovi A, Parsons LM, Salfinger M. The molecular basis of resistance to isoniazid, rifampin and pyrazinamide in Mycobacterium tuberculosis. Respir Res. 2001;2:164–168. doi: 10.1186/rr54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spíndola de Miranda S, Kritski AL, Filliol I, Mabilat C, Panteix G, Drouet E. Mutations in the rpoB gene of rifampicin-resistant Mycobacterium tuberculosis strains isolated in Brazil and France. Mem Inst Oswaldo Cruz. 2001;96:247–250. doi: 10.1590/s0074-02762001000200019. [DOI] [PubMed] [Google Scholar]
- Supply P, Mazars E, Lesjean S, Vincent V, Gicquel B, Locht C. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol Microbiol. 2000;36:762–771. doi: 10.1046/j.1365-2958.2000.01905.x. [DOI] [PubMed] [Google Scholar]
- Tang K, Sun H, Zhao Y, Guo J, Zhang C, Feng Q, He Y, Luo M, Li Y, Sun Q. Characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Sichuan in China. Tuberculosis. 2013;93:89–95. doi: 10.1016/j.tube.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Telenti A, Imboden P, Marchesi P, Cole S, Colston M, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- van Soolingen D, Hass PEW, Hermans PWM, van Embden JDA. DNA firgerprinting of Mycobacterium tuberculosis. Meth Enzym. 1994;235:196–204. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- WHO World Health Organization . Global tuberculosis report 2014. WHO; Geneva: 2014. 289 [Google Scholar]
- Zhang Y, Yew WW. Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuber C Lung Dis. 2009;13:1320–1330. [PubMed] [Google Scholar]