Abstract
Genome-wide association studies have reported an association between NCAN rs1064395 genotype and bipolar disorder. This association was later extended to schizophrenia and major depression. However, the neurobiological underpinnings of these associations are poorly understood. NCAN is implicated in neuronal plasticity and expressed in subcortical brain areas, such as the amygdala and hippocampus, which are critically involved in dysfunctional emotion processing and regulation across diagnostic boundaries. We hypothesized that the NCAN risk variant is associated with reduced gray matter volumes in these areas. Gray matter structure was assessed by voxel-based morphometry on structural MRI data in two independent German samples (healthy subjects, n=512; depressed inpatients, n=171). All participants were genotyped for NCAN rs1064395. Hippocampal and amygdala region-of-interest analyses were performed within each sample. In addition, whole-brain data from the combined sample were analyzed. Risk (A)-allele carriers showed reduced amygdala and hippocampal gray matter volumes in both cohorts with a remarkable spatial overlap. In the combined sample, genotype effects observed for the amygdala and hippocampus survived correction for entire brain volume. Further effects were also observed in the left orbitofrontal cortex and the cerebellum/fusiform gyrus. We conclude that NCAN genotype is associated with limbic gray matter alterations in healthy and depressed subjects in brain areas implicated in emotion perception and regulation. The present data suggest that NCAN forms susceptibility to neurostructural deficits in the amygdala, hippocampus, and prefrontal areas independent of disease, which might lead to disorder onset in the presence of other genetic or environmental risk factors.
INTRODUCTION
Bipolar disorder (BD), major depressive disorder (MDD), and schizophrenia (SCZ) are highly heritable neuropsychiatric disorders. Genome-wide association studies have implicated several genes in these disorders, and have revealed a high degree of overlap in terms of common genetic factors (Lee et al, 2013). Although each of the identified genome-wide significant variants leads to only a marginal increase in individual disease risk, they may provide insights into the etiology and molecular pathways involved in these disorders. In recent years, imaging genetics has emerged as a powerful tool for exploring the involvement of previously identified genetic variants in the etiology of psychiatric disorders. This approach characterizes the influence of genetic variants on brain structure and function that are the consequence of normal and abnormal biological pathways (Dannlowski et al, 2010, 2011, 2014; Erk et al, 2010; Stein et al, 2012).
One of the most recently identified genome-wide significant candidates for BD is the single-nucleotide polymorphism (SNP) rs1064395 in Neurocan (NCAN) (Cichon et al, 2011). Subsequent candidate studies also found associations of this SNP with SCZ (p=2.28 × 10−3, Mühleisen et al, 2012) as well as with MDD in the overall PGC MDD sample (p=0.013, www.broadinstitute.org/mpg/ricopili) as well as in the German sample (p=0.009) suggesting that variation in NCAN is a susceptibility factor across psychiatric diagnostic categories.
NCAN is a brain-specific chrondoitin sulfate proteoglycan (CSPG), which is mainly expressed during early brain development (http://www.informatics.jax.org/assay/MGI:4826610, Schultz et al, 2014). It is widely expressed throughout the mammalian brain, including subcortical structures, such as the hippocampus and the amygdala (Cichon et al, 2011). NCAN is a component of the extracellular matrix, where it is involved in cell adhesion, cell migration, and neurite growth (Rauch et al, 2001). In gene expression studies of Brodman areas 8/9 and 46/10, as well as cerebellum, NCAN is differentially regulated in MDD patients compared with controls in Brodman area 46/10 (https://www.stanleygenomics.org/).
The subcortical distribution of NCAN expression shows remarkable parallels to regions implicated in neuroanatomical models of major psychoses on the basis of numerous studies reporting functional and structural aberrations in the amygdala and hippocampus, which have been linked to dysfunctions of emotion processing and regulation (Phillips and Swartz, 2014; Strakowski et al, 2012). Particularly, gray matter volume abnormalities in the amygdala and hippocampus appear to be a common feature of affective disorders and SCZ alike (Brown et al, 2011; Redlich et al, 2014; Stratmann et al, 2014). Therefore, it should be a plausible hypothesis that NCAN genotype is associated with subcortical gray matter structure.
In an early imaging genetics study, Schultz et al (2014) demonstrated that NCAN rs1064395 genotype influenced cortical folding in the occipital and prefrontal cortex. The authors hypothesized that this may establish disease susceptibility during neurodevelopment . However, the investigation was restricted to cortical structures only, and involved a relatively small sample. To our knowledge, no study to date has investigated NCAN genotype effects on either subcortical structures or gray matter volumes across the entire brain. The aim of the present study therefore was to investigate an association between NCAN rs1064395 and regional gray matter volume in a large sample of healthy subjects (N=512), and in a second, clinical sample of patients with MDD (N=171). Brain-wide local gray matter volume was measured using voxel-based morphometry (VBM). In each sample, hippocampal and amygdala region-of-interest (ROI) analyses were performed. In addition, analyses of whole-brain data were conducted for each sample separately and the combined sample.
As the minor allele A of rs1064395 has been shown to confer a risk effect that increases disease susceptibility, whereas the G-allele is likely to confer a protective effect (Cichon et al, 2011; Mühleisen et al, 2012), we hypothesized that carriers of the NCAN genotypes AA and AG would exhibit reduced gray matter volumes, particularly in the amygdala and hippocampus, as well as in other structures with an involvement in emotion perception and regulation, such as prefrontal areas.
MATERIALS AND METHODS
Sample Characteristics
Two independent samples from separate studies have been collected at the University of Münster, at the same MRI scanner with identical sequences. Both samples were of Central European ancestry with an age range of 18–65 years. None of the subjects from either study have been included in any previous report regarding NCAN genotype.
Sample 1 comprised N=520 healthy subjects. All subjects were thoroughly investigated by experienced psychologists and were free from any lifetime history of psychiatric disorders according to DSM-IV criteria, assessed by means of the SCID interview. The data of eight subjects had to be excluded from the analyses due to the presence of anatomical abnormalities (abnormally enlarged ventricles) or artifacts discovered in the structural MRI Images checked by visual inspection and identification as extreme outliers in the check data quality function of the VBM8-toolbox (>3 SD from mean homogeneity). The remaining scans of N=512 subjects (mean age 33.3±11.6; N=223 male, N=289 female) were clear of such problems.
Sample 2 (clinical sample) comprised N=176 hospitalized inpatients with MDD and a current depressive episode, as diagnosed with the SCID interview, all under current antidepressant treatment. Comorbid diagnoses of substance-related disorders or psychotic disorders were exclusion criteria. Five data sets were removed after the quality check described above, leaving N=171 patients (mean age 38.6±11.7; N=66 male, N=105 female) in sample 2.
Common exclusion criteria for both samples were any neurological abnormalities including stroke, epilepsy, multiple sclerosis, Parkinson's disease or dementia, head trauma or unconsciousness, and the usual MRI contraindications (metal implants, cardiac pacers, and so on). The study was approved by the Ethics Committee of the University of Münster. After complete description of the study to the participants, written informed consent was obtained.
Verbal intelligence was estimated by the Mehrfachwahl-Wortschatz-Intelligenztest (multiple-choice vocabulary intelligence test; Lehrl, 1995). Depression level and trait anxiety were assessed by means of the BDI (Beck and Steer, 1987) and STAI-trait (Laux et al, 1981) questionnaire.
Compared with sample 1, participants in sample 2 (i) were significantly older (38.6±11.7 vs 33.3±11.6 years, t(681)=5.16, p<0.001); (ii) reported fewer years of education (14.2±2.2 vs 14.8±2.1 years, t(681)=3.25, p=0.001); and (iii) displayed lower verbal IQ scores (111.4±13.4 vs 116.0±12.5, t(594)=3.84, p<0.001). As expected, compared with sample 1, participants in sample 2 showed higher scores on both the BDI (23.3±11.1 vs 2.4±3.1, t(662)=37.9, p<0.001), and the STAI (58.8±9.3 vs 32.2±6.9, t(615)=37.0, p<0.001). The two samples did not differ in terms of gender, χ2(1)=1.29, p=0.26, or NCAN genotype distribution, χ2(2)=1.33, p=0.51.
DNA Extraction and Genotyping
Genomic DNA was prepared from whole blood using standard procedures. Genotyping was performed using a TaqMan 7900HT Fast Real-Time PCR System (Life Technolgies), and the SNP Genotyping Assay ID C_8931759_20 for the NCAN polymorphism rs1064395. Accuracy was assessed by duplicating 15% of the sample. Reproducibility was 100%.
Structural MRI Acquisition and Morphometry
A detailed description of both the MRI methodology and statistical approach is provided elsewhere (Baune et al, 2012a, 2012b; Dannlowski et al, 2012, 2014; Opel et al, 2014; Stacey et al, 2014). Briefly, T1-weighted high-resolution anatomical images were acquired using a 3 T-MRI scanner (Gyroscan Intera 3 T, Philips Medical Systems, Best, NL), and a 3D fast gradient echo sequence (‘Turbo Field Echo'). TR=7.4 ms, TE=3.4 ms, FA=9°, 2 signal averages, inversion prepulse every 814.5 ms, acquired over a field of view of 256 mm (FH) × 204 mm (AP) × 160 mm (RL, nominal slice selection direction) with 1-mm resolution in all directions, frequency encoding in FH direction, phase encoding in AP and RL direction, reconstructed to cubic voxels of 0.5 × 0.5 × 0.5 mm.
The VBM8-toolbox (http://dbm.neuro.uni-jena.de/vbm) was used for pre-processing all structural images using default parameters. Images were bias-corrected, tissue classified, and normalized to MNI space using linear (12-parameter affine) and nonlinear transformations, within a unified model including high-dimensional DARTEL normalization. Gray matter segments were modulated by the nonlinear components only in order to preserve actual gray matter values locally (modulated GM volumes). Use of these procedures precludes the need for further correction for total brain volume.
As mentioned above, homogeneity of gray matter images was checked using the covariance structure of each image with all other images, as implemented in the check data quality function. The modulated gray matter images were smoothed with a Gaussian kernel of 8 mm FWHW.
Group statistics were calculated using SPM8 in each sample separately. To investigate the effect of rs1064395, t-tests were conducted using genotype group (A-carriers vs GG homozygotes) as a between-subjects factor. Age and gender were added to the models as nuisance regressors. The scanner gradient system was upgraded in the course of recruitment (‘Master' Gradient System to ‘Quasar Dual' Gradient System). Although the MRI sequence used before and after the gradient system upgrade was identical, the scanner upgrade was modeled as a regressor of no interest.
In a first step, a ROI approach was applied, restricting the analyses to the bilateral amygdala and hippocampus. An anatomical mask was generated using the Anatomy Toolbox (http://www.fz-juelich.de/SharedDocs/Downloads/INM/INM-1/DE/Toolbox/Toolbox_20.html). The mask encompassed all parts of the amygdala and the hippocampus, as defined on the basis of probabilistic cytoarchitectonic maps (Amunts et al, 2005). To control for multiple statistical testing within the ROI, a cluster-level false-positive detection rate was maintained at p<0.05 using a voxel-level threshold of p<0.01 and a cluster extent (k) empirically determined by Monte Carlo simulations (n=5000 iterations), as reported previously (Baune et al, 2012a, 2012b; Dannlowski et al, 2012). This was performed using the AlphaSim procedure, which accounts for spatial smoothness (estimated from the residuals), as implemented in the REST toolbox (http://restfmri.net/forum/index.php). The empirically determined cluster threshold for the bilateral hippocampus/amygdala mask was k=160 voxels.
In a second step, exploratory whole-brain analyses were conducted in order to detect brain areas for which no a priori hypothesis existed, but which were potentially affected by NCAN genotype. The statistical threshold for whole brain analyses was p<0.001, k=10. The overlap of genotype effects in the two samples was assessed at the same threshold by calculating all voxels surviving p<0.01 in both sample 1 and sample 2, thereby yielding a combined threshold of p<0.001 (cluster threshold k=10 was used as well).
To explore whether genotype effects were influenced by age or gender, two supplementary models were conducted in the healthy sample, modeling the interaction term of genotype × age and genotype × sex. These analyses were restricted to all areas showing significant genotype effects in this sample (as listed in Table 2).
Finally, genotype effects were additionally calculated for the combined sample (N=683). The previously used nuisance regressors were included (age, gender, and gradient upgrade). In the combined sample, sample (healthy vs MDD) was modeled as an additional regressor, also including the interaction term of sample and genotype group. From this model, also main effects of sample (healthy vs MDD) were calculated.
RESULTS
Genotyping Results
Sample 1 comprised N=123 A-allele carriers (12 AA homozygotes, 111 AG heterozygotes) vs N=389 GG homozygotes. Given the small number of AA homozygotes, these subjects were grouped together with the AG-carriers for the purposes of the present analyses. Genotype frequencies did not deviate from Hardy–Weinberg equilibrium (HWE), (exact test: p=0.24). Sample 2 comprised N=124 GG homozygotes vs N=47 A-carriers (N=3 AA, N=44 AG). Again, allele frequencies did not deviate from HWE (exact test: p=1). Table 1 shows the sociodemographic and questionnaire data of the two samples according to rs1064395 genotype.
Table 1. Sample Characteristics According to NCAN rs1064395 Status.
|
Sample 1 (N=512 healthy subjects) | |||
|---|---|---|---|
| AA/AG (N=123) | GG (N=389) | p-valuea | |
| Age | 33.9±12.8 | 33.1±11.3 | 0.50 |
| Sex (m/f) | 48/75 | 175/214 | 0.25 |
| Verbal IQb | 115.6±13.0 | 116.2±12.4 | 0.68 |
| Years of education | 14.5±2.0 | 14.9±2.1 | 0.09 |
| STAI-traitc | 32.1±6.5 | 32.3±7.0 | 0.90 |
| BDId | 2.7±3.8 | 2.3±2.9 | 0.20 |
|
Sample 2 (N=171 MDD patients) | |||
|---|---|---|---|
| AG/AA (N=47) | GG (N=124) | p-value | |
| Age | 38.6±12.5 | 38.7±11.5 | 0.96 |
| Sex (m/f) | 15/32 | 51/73 | 0.27 |
| Verbal IQb | 110.5±13.3 | 111.7±13.5 | 0.62 |
| Years of education | 14.1±2.3 | 14.2±2.2 | 0.68 |
| STAI-traitc | 60.7±7.5 | 58.0±9.8 | 0.12 |
| BDId | 24.3±10.2 | 22.9±11.4 | 0.48 |
According to t-tests or χ2-tests.
Assessed with the German language Mehrfachwahl-Wortschatz-Test (multiple-choice vocabulary test (Lehrl, 1995)). Data were available for N=449 (sample 1) and N=147 (sample 2) subjects.
State–Trait Anxiety Inventory. Data were available for N=476 (sample 1) and N=141 (sample 2) subjects.
Beck Depression Inventory. Data were available for N=501 (sample 1) and N=163 (sample 2) subjects.
Region-of-Interest Analysis of the Bilateral Amygdala and Hippocampus
In healthy subjects (sample 1), a significant rs1064395 genotype effect on bilateral amygdala, and hippocampus gray matter volumes was observed. Carriers of one or two risk (A) alleles had significantly decreased gray matter volumes compared with subjects with GG status (left: x=−26, y=0, z=−23, t(507)=3.05, k=323, p=0.0012; right: x=18, y=−12, z=−9, t(507)=3.31, k=436, p=0.0005). There were no significant differences between AA homozygotes and AG-carriers.
In depressed patients (sample 2), very similar effects in the same direction were observed (left: x=−16, y=0, z=−20, t(166)=2.87, k=182, p=0.0023; right: x=30, y=−13, z=−20, t(166)=3.43, k=305, p=0.0003).
Whole-Brain Data
In healthy subjects, several areas were found showing lower gray matter volume in A-carriers compared with GG homozygotes, including the amygdala and hippocampus, cerebellum, prefrontal, particularly orbitofrontal areas, and in the temporal and occipital cortex (see Table 2). There was no area showing larger gray matter volumes in A-carriers compared with GG homozygotes.
Table 2. Results of Whole-Brain Analyses of NCAN rs1064395 Genotype Effects on Local Gray Matter Volume in the Healthy Sample (N=512) and the Patient Sample (N=171).
| Anatomical region | Side | Cluster size | x | y | z | Z-score | |
|---|---|---|---|---|---|---|---|
| Sample 1 (healthy sample) | |||||||
| Cerebellum, FFG | L | 2131 | −27 | −57 | −29 | 4.19 | |
| ITG, MTG, TP | R | 154 | 52 | 2 | −39 | 3.88 | |
| Orbital parts of MidFG, IFG, SFG; SFG, MidFG | L | 427 | −52 | 51 | 12 | 3.84 | |
| IPG, postcentral gyrus | R | 25 | 44 | −37 | 54 | 3.69 | |
| MTG | R | 162 | 69 | −30 | 9 | 3.64 | |
| HC, PHG, amygdala | R | 36 | 15 | −3 | −18 | 3.29 | |
| STG | L | 18 | −52 | −42 | 18 | 3.27 | |
| SFG, MidFG | R | 10 | 22 | 60 | 24 | 3.25 | |
| Calcarine gyrus | L | 25 | −2 | −94 | −9 | 3.24 | |
| ITG, IOG | L | 11 | −56 | −66 | 12 | 3.20 | |
| ITG, FFG | R | 10 | 38 | 3 | −38 | 3.16 | |
| Sample 2 (patient sample) | |||||||
| HC | R | 45 | 16 | −9 | −9 | 3.66 | |
| SFG | R | 11 | 15 | 51 | 19 | 3.32 | |
| Cerebellum | L | 23 | −8 | −39 | −11 | 3.26 | |
Abbreviations: FFG, fusiform gyrus; HC, hippocampus; IFG, inferior frontal gyrus; IOG, inferior occipital gyrus; ITG, inferior temporal gyrus; MidFG, middle frontal gyrus; MTG, middle temporal gyrus; PHC, parahippocampal gyrus; SFG, superior frontal gyrus; STG, superior temporal gyrus; TP, temporal pole.
Coordinates are given in MNI space.
In the depressed sample, clusters showing effects in the same direction were found in the hippocampus, superior frontal gyrus and cerebellum (Table 2).
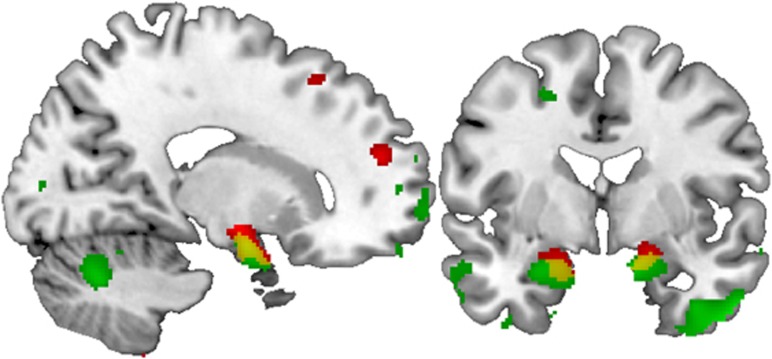
The clusters within the two samples showed spatial overlap (k=334 voxels surviving p<0.01 in both groups, see Figure 1). The two largest overlapping clusters were located in the bilateral amygdala/hippocampus head (left: x=−21, y=−1, z=−21, k=92; right: x=15, y=−3, z=−18, k=189). Checking this overlapping area regarding the affected hippocampal and amygdala substructures (Amunts et al, 2005) by means of the Anatomy-Toolbox yielded a major involvement of the basolateral amygdala (right: 25.3%, left: 84.3% of the overlapping area) and superficial amygdala (right: 62.7%, left: 13.2%), with additional minor involvements of the entorhinal cortex, subiculum, and the hippocampus–amygdala transition area. Other overlapping areas were found in the insula/temporal pole (x=−39, y=9, z=−15, k=19) and cerebellum (x=−6, y=−42, z=−12, k=17).
Figure 1.
Coronal (y=−2) and sagittal (x=17) views of the amygdala and the hippocampus region depicting the effect of NCAN rs1064395 genotype on gray matter volumes in the two samples, as corrected for gender and age and rendered on a standard brain template in MNI space. Green, results from sample 1 (healthy subjects) showing N=389 GG homozygotes >N=123 A-carriers with a threshold of p<0.01. Red, results from sample 2 (MDD) showing N=124 GG>N=47 A-carriers at the same statistical threshold. Yellow, overlap between the clusters of each sample with a combined threshold of p<0.001.
There were no genotype × age or genotype × gender interactions within the areas showing main effects of genotype.
In our whole-brain approach within the entire sample (N=683), we detected five clusters influenced by genotype surviving a whole-brain correction. These were located in the bilateral amygdala/hippocampus/parahippocampal gyrus, lateral orbitofrontal cortex, and cerebellum/fusiform gyrus (see Supplementary Table 1 and Figure 1).
Matching several previous studies, effects of diagnosis (healthy subjects vs controls) were found in the hippocampus, amygdala, insula, thalamus, and orbitofrontal cortex, among others (see Supplementary Table 2). However, there was no area in the entire brain showing a significant genotype × sample interaction, indicating that genotype effects were comparable in healthy subjects and in depressed patients.
DISCUSSION
The present study investigated the impact of a single-nucleotide variant in NCAN, which has previously associated with BD, SCZ, and MDD, on gray matter volume. The investigations were performed in a large sample of healthy controls, which is a first-time finding in the literature, and were repeated in a second sample of hospitalized depressed patients. The two samples showed comparable associations of genotype and brain structure with an overlap of genotype effects mapping predominantly to the bilateral amygdala and hippocampal subfields. In the combined sample, the amygdala/hippocampus clusters survived correction for entire brain volume, as did areas in the left orbitofrontal cortex and cerebellum/fusiform gyrus. As these findings were obtained in healthy subjects and a clinical sample alike, they might indicate a disease-independent genetic effect on corticolimbic brain circuitries.
Previous neuroimaging studies have highlighted the role of the amygdala and the hippocampal formation in the etiology of BD, SCZ, and MDD. For example, several functional neuroimaging studies have shown that amygdala dysfunction is a key feature of BD (Almeida and Phillips, 2012; Blond et al, 2012; Fournier et al, 2013; Grotegerd et al, 2013, 2014; Lim et al, 2013), SCZ (Pankow et al, 2013; Rauch et al, 2010), and MDD (Dannlowski et al, 2007; Groenewold et al, 2012; Siegle et al, 2007; Stuhrmann et al, 2011, 2013; Suslow et al, 2010). Also structural imaging studies have demonstrated decreased amygdala volumes in BD (Blumberg et al, 2003; Foland-Ross et al, 2013; Redlich et al, 2014) and gray matter volume reductions have been described in the hippocampus of BD patients (Hajek et al, 2012a, 2012b; Redlich et al, 2014), although the respective literature is less consistent than that for the amygdala (Blumberg et al, 2003; Hartberg et al, 2011).
Further research has also demonstrated structural alterations in the amygdala, hippocampus, and prefrontal areas in MDD and SCZ. Gray matter deficits of the hippocampus, for example, are one the most frequently described imaging finding in MDD (MacQueen and Frodl, 2010; Stratmann et al, 2014), which was also found in the present study, and gray matter volume reductions in the amygdala and hippocampus have also been confirmed in meta-analyses of SCZ data (Shepherd et al, 2012). As the amygdala and the hippocampus have a key role in the generation and regulation of affective states, gray matter structural deficits might correspond to the genetic association findings for NCAN, which cross the diagnostic boundaries for the major psychoses (Miró et al, 2012).
NCAN is a CSPG. CSPGs are crucial for ECM organization, and participate in cell adhesion and migration. They are thus crucial for neuroplasticity, neuronal growth, and axon guidance during development, as well as for regenerative processes in neurons following central nervous system injury (Avram et al, 2014). The synergistic effects of cell proliferation and differentiation are the basis for the development of gray matter structure (Zilles et al, 2013), and the lack of CSPGs results in improved axonal regeneration and recovery (Galtrey and Fawcett, 2007). A plausible hypothesis therefore is that rs1064395 A-allele status leads to an upregulation of NCAN, which results in turn in growth inhibition. One possible explanation for the observed effects of NCAN variation on grey matter structure in the amygdala and hippocampus could lie in the interplay of the protein with binding partners, which are only present in the regions in question. However, to date, only few binding partners of NCAN have been described, among them PTPRS (Shen et al, 2009) and NCAM1, L1CAM, and contactin2 (Rauch et al, 2001) as well as other proteoglycans (www.string-db.org). None of these proteins are exclusively expressed in the amygdala and/or hippocampus.
However, the precise molecular mechanisms through which NCAN is involved in the etiology of psychiatric disorders, and impacts on gray matter volume, still await elucidation. This process will require molecular genetic studies, including, for example, the search for tissue-specific binding partners by screening amygdala- and hippocampus-specific cDNA libraries using an adapted yeast two-hybrid system.
The effects of NCAN genotype appear to be disease independent as they have been found in healthy subjects and a patient sample alike and interaction effects of genotype and disease status were missing. It seems that these disease-independent effects form susceptibility to structural brain deficits that may be in turn associated with mental illness. Other genetic or environmental risk factors in patients (or protective factors in healthy subjects) probably need to be present to exhibit psychopathology (or prevent disease onset).
The results agree well with a recent study (Raum et al, 2015), showing that NCAN rs1064395 A-carriers have significant deficits in memory tasks associated with hippocampal function compared with GG homozygotes, and show differences in brain activation in temporal areas, which were also affected by genotype in our healthy sample. Conversely, different results were obtained in a study by Schultz et al, who reported NCAN genotype effects on cortical folding only in schizophrenia patients but not in healthy controls. However, sample sizes were weak (N=65 healthy subjects), and the morphometric methods and the patient sample differed much compared with the present study.
The present study had limitations. First, the current lack of data concerning the functional properties of the NCAN SNP rs1064395 renders our statements regarding potential molecular mechanisms speculative. Furthermore, behavioral and clinical correlates of reduced amygdala, hippocampal, and PFC gray matter volume remain to be investigated in more detail. The inclusion of a second healthy sample would have represented a truly independent second sample compared with a patient sample. As patients and controls were not matched for age and other sociodemographic data, the effects of sample on gray matter volumes must be taken with care. Finally, we cannot exclude the possibility that the observed effect was driven by another genetic variant that is in linkage disequilibrium with rs1064395.
In summary, the present study is the first to demonstrate an influence of a cross-disorder risk variant in NCAN on subcortical brain structure in healthy and depressed subjects. In our imaging genetics approach, the risk allele showed strong impact on gray matter volume in brain areas critically involved in emotion processing, which also show functional and structural abnormalities in BD, MDD, and SCZ. The present data suggest that the increased disease susceptibility in risk allele carriers might be mediated by neurostructural deficits in the amygdala, hippocampus, and prefrontal areas, leading to dysfunctional emotion processing and regulation.
FUNDING AND DISCLOSURE
The study was supported by grants from the German Research Foundation (DFG) grant FOR 2107 (DA1151/5-1 to UD, KO4291/3-1 to CK, KI588/14-1 to TK, and JA1890/7-1 to AJ); DFG KR3822/2-1 to AK, TK, and MR; DFG NO246/10-1 to MMN; Innovative Medizinische Forschung (IMF) of the Medical Faculty of Münster (DA120903 to UD, DA111107 to UD, and DA211012 to UD); the Rolf-Dierichs-Stiftung (ZUW80037 to UD); and the Interdisziplinäres Zentrum für Klinische Forschung (IZKF) of the Medical Faculty of Münster (research group 4 to CK). The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7) under grant agreement no. 279227 to MR. This study was further supported by the German Federal Ministry of Education and Research (BMBF) through the Integrated Network IntegraMent (Integrated Understanding of Causes and Mechanisms in Mental Disorders), under the auspices of the e:Med Programme (grant 01ZX1314A to MMN, grant 01ZX1314G to MR). MMN is a member of the DFG-funded Excellence-Cluster ImmunoSensation. MMN also received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. V. Arolt is a member of the advisory board of, or has given presentations on behalf of, the following companies: Astra-Zeneca, Janssen-Organon, Lilly, Lundbeck, Servier, Pfizer, Otsuka, and Trommsdorff. C. Konrad received fees for an educational program from esparma GmbH/Aristo Pharma GmbH, Lilly Deutschland GmbH, Servier Deutschland GmbH, and MagVenture GmbH, as well as travel support and speakers honoraria from Lundbeck and Servier. K. Domschke has received speaker fees from Pfizer, Lilly and Bristol-Myers Squibb, she was a consultant for Johnson & Johnson and has received funding by Astra Zeneca. H. Kugel has received consultation fees from MR:comp GmbH, Testing Services for MR Safety. These affiliations are of no relevance to the work described in the manuscript. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Almeida JRC, Phillips ML. Distinguishing between unipolar depression and bipolar depression: current and future clinical and neuroimaging perspectives. Biol Psychiatry. 2012;73:111–118. doi: 10.1016/j.biopsych.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts K, Kedo O, Kindler M, Pieperhoff P, Mohlberg H, Shah NJ, et al. Cytoarchitectonic mapping of the human amygdala, hippocampal region and entorhinal cortex: intersubject variability and probability maps. Anat Embryol (Berl) 2005;210:343–352. doi: 10.1007/s00429-005-0025-5. [DOI] [PubMed] [Google Scholar]
- Avram S, Shaposhnikov S, Buiu C, Mernea M. Chondroitin sulfate proteoglycans: structure-function relationship with implication in neural development and brain disorders. Biomed Res Int. 2014;2014:642798. doi: 10.1155/2014/642798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baune BT, Konrad C, Grotegerd D, Suslow T, Birosova E, Ohrmann P, et al. Interleukin-6 gene (IL-6): a possible role in brain morphology in the healthy adult brain. J Neuroinflammation. 2012;9:125. doi: 10.1186/1742-2094-9-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baune BT, Konrad C, Grotegerd D, Suslow T, Ohrmann P, Bauer J, et al. Tumor necrosis factor gene variation predicts hippocampus volume in healthy individuals. Biol Psychiatry. 2012;72:655–662. doi: 10.1016/j.biopsych.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Beck AT, Steer RA. Beck Depression Inventory: Manual. Psychological Corporation Harcourt Brace Jovanovich: San Antonio, TX, USA; 1987. [Google Scholar]
- Blond BN, Fredericks CA, Blumberg HP. Functional neuroanatomy of bipolar disorder: structure, function, and connectivity in an amygdala-anterior paralimbic neural system. Bipolar Disord. 2012;14:340–355. doi: 10.1111/j.1399-5618.2012.01015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg HP, Kaufman J, Martin AA, Whiteman R, Zhang JH, Gore JC, et al. Amygdala and hippocampal volumes in adolescents and adults with bipolar disorder. Arch Gen Psychiatry. 2003;60:1201–1208. doi: 10.1001/archpsyc.60.12.1201. [DOI] [PubMed] [Google Scholar]
- Brown GG, Lee J-S, Strigo IA, Caligiuri MP, Meloy MJ, Lohr J. Voxel-based morphometry of patients with schizophrenia or bipolar I disorder: a matched control study. Psychiatry Res. 2011;194:149–156. doi: 10.1016/j.pscychresns.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannlowski U, Grabe HJ, Wittfeld K, Klaus J, Konrad C, Grotegerd D, et al. Multimodal imaging of a tescalcin (TESC)-regulating polymorphism (rs7294919)-specific effects on hippocampal gray matter structure. Mol Psychiatry. 2014;20:398–404. doi: 10.1038/mp.2014.39. [DOI] [PubMed] [Google Scholar]
- Dannlowski U, Konrad C, Kugel H, Zwitserlood P, Domschke K, Schöning S, et al. Emotion specific modulation of automatic amygdala responses by 5-HTTLPR genotype. Neuroimage. 2010;53:893–898. doi: 10.1016/j.neuroimage.2009.11.073. [DOI] [PubMed] [Google Scholar]
- Dannlowski U, Kugel H, Franke F, Stuhrmann A, Hohoff C, Zwanzger P, et al. Neuropeptide-S (NPS) receptor genotype modulates basolateral amygdala responsiveness to aversive stimuli. Neuropsychopharmacology. 2011;36:1–7. doi: 10.1038/npp.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannlowski U, Ohrmann P, Bauer J, Kugel H, Arolt V, Heindel W, et al. Amygdala reactivity to masked negative faces is associated with automatic judgmental bias in major depression: a 3 T fMRI study. J Psychiatry Neurosci. 2007;32:423–429. [PMC free article] [PubMed] [Google Scholar]
- Dannlowski U, Stuhrmann A, Beutelmann V, Zwanzger P, Lenzen T, Grotegerd D, et al. Limbic scars: long-term consequences of childhood maltreatment revealed by functional and structural MRI. Biol Psychiatry. 2012;71:286–293. doi: 10.1016/j.biopsych.2011.10.021. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Schnell K, Opitz von Boberfeld C, Esslinger C, Kirsch P, et al. Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch Gen Psychiatry. 2010;67:803–811. doi: 10.1001/archgenpsychiatry.2010.94. [DOI] [PubMed] [Google Scholar]
- Foland-Ross LC, Thompson PM, Sugar Ca, Narr KL, Penfold C, Vasquez RE, et al. Three-dimensional mapping of hippocampal and amygdalar structure in euthymic adults with bipolar disorder not treated with lithium. Psychiatry Res. 2013;211:195–201. doi: 10.1016/j.pscychresns.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier JC, Keener MT, Almeida JRC, Kronhaus DM, Phillips ML. Amygdala and whole-brain activity to emotional faces distinguishes major depressive disorder and bipolar disorder. Bipolar Disord. 2013;15:741–752. doi: 10.1111/bdi.12106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtrey CM, Fawcett JW. The role of chondroitin sulfate proteoglycans in regeneration and plasticity in the central nervous system. Brain Res Rev. 2007;54:1–18. doi: 10.1016/j.brainresrev.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Groenewold NA, Opmeer EM, de Jonge P, Aleman A, Costafreda SG. Emotional valence modulates brain functional abnormalities in depression: evidence from a meta-analysis of fMRI studies. Neurosci Biobehav Rev. 2012;37:152–163. doi: 10.1016/j.neubiorev.2012.11.015. [DOI] [PubMed] [Google Scholar]
- Grotegerd D, Stuhrmann A, Kugel H, Schmidt S, Redlich R, Zwanzger P, et al. Amygdala excitability to subliminally presented emotional faces distinguishes unipolar and bipolar depression: an fMRI and pattern classification study. Hum Brain Mapp. 2014;35:2995–3007. doi: 10.1002/hbm.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotegerd D, Suslow T, Bauer J, Ohrmann P, Arolt V, Stuhrmann A, et al. Discriminating unipolar and bipolar depression by means of fMRI and pattern classification: a pilot study. Eur Arch Psychiatry Clin Neurosci. 2013;263:119–131. doi: 10.1007/s00406-012-0329-4. [DOI] [PubMed] [Google Scholar]
- Hajek T, Cullis J, Novak T, Kopecek M, Höschl C, Blagdon R, et al. Hippocampal volumes in bipolar disorders: opposing effects of illness burden and lithium treatment. Bipolar Disord. 2012;14:261–270. doi: 10.1111/j.1399-5618.2012.01013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajek T, Kopecek M, Höschl C, Alda M. Smaller hippocampal volumes in patients with bipolar disorder are masked by exposure to lithium: a meta-analysis. J Psychiatry Neurosci. 2012;37:333–343. doi: 10.1503/jpn.110143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartberg CB, Sundet K, Rimol LM, Haukvik UK, Lange EH, Nesvåg R, et al. Subcortical brain volumes relate to neurocognition in schizophrenia and bipolar disorder and healthy controls. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1122–1130. doi: 10.1016/j.pnpbp.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Laux L, Glanzmann P, Schaffner P, Spielberger CD. Das State-Trait Angstinventar. Beltz: Weinheim, Germany; 1981. [Google Scholar]
- Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, Perlis RH, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrl S. Mehrfachwahl-Wortschatz-Intelligenztest MWT-B. Hogrefe: Göttingen, Germany; 1995. [Google Scholar]
- Lim CS, Baldessarini RJ, Vieta E, Yücel M, Bora E, Sim K. Longitudinal neuroimaging and neuropsychological changes in bipolar disorder patients: review of the evidence. Neurosci Biobehav Rev. 2013;37:418–435. doi: 10.1016/j.neubiorev.2013.01.003. [DOI] [PubMed] [Google Scholar]
- MacQueen GM, Frodl T. The hippocampus in major depression: evidence for the convergence of the bench and bedside in psychiatric research. Mol Psychiatry. 2010;16:252–264. doi: 10.1038/mp.2010.80. [DOI] [PubMed] [Google Scholar]
- Miró X, Meier S, Dreisow MLM, Frank J, Strohmaier J, Breuer R, et al. Studies in humans and mice implicate neurocan in the etiology of mania. Am J Psychiatry. 2012;169:982–990. doi: 10.1176/appi.ajp.2012.11101585. [DOI] [PubMed] [Google Scholar]
- Mühleisen TW, Mattheisen M, Strohmaier J, Degenhardt FA, Priebe L, Schultz CC, et al. Association between schizophrenia and common variation in neurocan (NCAN), a genetic risk factor for bipolar disorder. Schizophr Res. 2012;138:69–73. doi: 10.1016/j.schres.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Opel N, Redlich R, Zwanzger P, Grotegerd D, Arolt V, Heindel W, et al. Hippocampal atrophy in major depression: a function of childhood maltreatment rather than diagnosis. Neuropsychopharmacology. 2014;39:2723–2731. doi: 10.1038/npp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankow A, Friedel E, Sterzer P, Seiferth NY, Walter H, Heinz A, et al. Altered amygdala activation in schizophrenia patients during emotion processing. Schizophr Res. 2013;150:101–106. doi: 10.1016/j.schres.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Phillips ML, Swartz HA. A critical appraisal of neuroimaging studies of bipolar disorder: toward a new conceptualization of underlying neural circuitry and a road map for future research. Am J Psychiatry. 2014;171:829–843. doi: 10.1176/appi.ajp.2014.13081008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch AV, Reker M, Ohrmann P, Pedersen A, Bauer J, Dannlowski U, et al. Increased amygdala activation during automatic processing of facial emotion in schizophrenia. Psychiatry Res. 2010;182:200–206. doi: 10.1016/j.pscychresns.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Rauch U, Feng K, Zhou XH. Neurocan: a brain chondroitin sulfate proteoglycan. Cell Mol Life Sci. 2001;58:1842–1856. doi: 10.1007/PL00000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raum H, Dietsche B, Nagels A, Witt SH, Rietschel M, Kircher T, et al. A genome-wide supported psychiatric risk variant in NCAN influences brain function and cognitive performance in healthy subjects. Hum Brain Mapp. 2015;36:378–390. doi: 10.1002/hbm.22635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redlich R, Almeida JRC, Grotegerd D, Opel N, Kugel H, Heindel W, et al. Brain morphometric biomarkers distinguishing unipolar and bipolar depression: a VBM—pattern classification approach. JAMA Psychiatry. 2014;71:1222–1230. doi: 10.1001/jamapsychiatry.2014.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CC, Mühleisen TW, Nenadic I, Koch K, Wagner G, Schachtzabel C, et al. Common variation in NCAN, a risk factor for bipolar disorder and schizophrenia, influences local cortical folding in schizophrenia. Psychol Med. 2014;44:811–820. doi: 10.1017/S0033291713001414. [DOI] [PubMed] [Google Scholar]
- Shen Y, Tenney AP, Busch SA, Horn KP, Cuascut FX, Liu K, et al. PTPsigma is a receptor for chondroitin sulfate proteoglycan, an inhibitor of neural regeneration. Science. 2009;326:592–596. doi: 10.1126/science.1178310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd AM, Laurens KR, Matheson SL, Carr VJ, Green MJ. Systematic meta-review and quality assessment of the structural brain alterations in schizophrenia. Neurosci Biobehav Rev. 2012;36:1342–1356. doi: 10.1016/j.neubiorev.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Siegle GJ, Thompson WK, Carter CS, Steinhauer SR, Thase ME. Increased amygdala and decreased dorsolateral prefrontal BOLD responses in unipolar depression: related and independent features. Biol Psychiatry. 2007;61:198–209. doi: 10.1016/j.biopsych.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Stacey D, Redlich R, Opel N, Grotegerd D, Arolt V, Kugel H, et al. No evidence of DISC1-associated morphological changes in the hippocampus, anterior cingulate cortex, or striatum in major depressive disorder cases and healthy controls. J Affect Disord. 2014;166:103–107. doi: 10.1016/j.jad.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strakowski SM, Adler CM, Almeida JRC, Altshuler LL, Blumberg HP, Chang KD, et al. The functional neuroanatomy of bipolar disorder: a consensus model. Bipolar Disord. 2012;14:313–325. doi: 10.1111/j.1399-5618.2012.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann M, Konrad C, Kugel H, Krug A, Schöning S, Ohrmann P, et al. Insular and hippocampal gray matter volume reductions in patients with major depressive disorder. PLoS One. 2014;9:e102692. doi: 10.1371/journal.pone.0102692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhrmann A, Dohm K, Kugel H, Zwanzger P, Redlich R, Grotegerd D, et al. Mood-congruent amygdala responses to subliminally presented facial expressions in Major Depression: associations with anhedonia. J Psychiatry Neurosci. 2013;38:249–258. doi: 10.1503/jpn.120060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhrmann A, Suslow T, Dannlowski U. Facial emotion processing in major depression: a systematic review of neuroimaging findings. Biol Mood Anxiety Disord. 2011;1:10. doi: 10.1186/2045-5380-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suslow T, Konrad C, Kugel H, Rumstadt D, Zwitserlood P, Schöning S, et al. Automatic mood-congruent amygdala responses to masked facial expressions in major depression. Biol Psychiatry. 2010;67:155–160. doi: 10.1016/j.biopsych.2009.07.023. [DOI] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Amunts K. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 2013;36:275–284. doi: 10.1016/j.tins.2013.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.