Abstract
Context:
Familial isolated pituitary adenoma (FIPA) due to aryl hydrocarbon receptor interacting protein (AIP) gene mutations is an autosomal dominant disease with incomplete penetrance. Clinical screening of apparently unaffected AIP mutation (AIPmut) carriers could identify previously unrecognized disease.
Objective:
To determine the AIP mutational status of FIPA and young pituitary adenoma patients, analyzing their clinical characteristics, and to perform clinical screening of apparently unaffected AIPmut carrier family members.
Design:
This was an observational, longitudinal study conducted over 7 years.
Setting:
International collaborative study conducted at referral centers for pituitary diseases.
Participants:
FIPA families (n = 216) and sporadic young-onset (≤30 y) pituitary adenoma patients (n = 404) participated in the study.
Interventions:
We performed genetic screening of patients for AIPmuts, clinical assessment of their family members, and genetic screening for somatic GNAS1 mutations and the germline FGFR4 p.G388R variant.
Main Outcome Measure(s):
We assessed clinical disease in mutation carriers, comparison of characteristics of AIPmut positive and negative patients, results of GNAS1, and FGFR4 analysis.
Results:
Thirty-seven FIPA families and 34 sporadic patients had AIPmuts. Patients with truncating AIPmuts had a younger age at disease onset and diagnosis, compared with patients with nontruncating AIPmuts. Somatic GNAS1 mutations were absent in tumors from AIPmut-positive patients, and the studied FGFR4 variant did not modify the disease behavior or penetrance in AIPmut-positive individuals. A total of 164 AIPmut-positive unaffected family members were identified; pituitary disease was detected in 18 of those who underwent clinical screening.
Conclusions:
A quarter of the AIPmut carriers screened were diagnosed with pituitary disease, justifying this screening and suggesting a variable clinical course for AIPmut-positive pituitary adenomas.
Familial isolated pituitary adenoma (FIPA) is characterized by the presence of pituitary adenomas in two or more members of the same family in the absence of other syndromic clinical features, such as those characteristic of multiple endocrine neoplasia (MEN) type 1 and MEN4, Carney complex or tumors related to mutations in the succinate dehydrogenase genes. FIPA is a heterogeneous condition, encompassing cases with unknown genetic cause and patients with mutations in the aryl-hydrocarbon receptor interacting protein gene (AIP), with distinctive clinical characteristics. Germline AIP mutations (AIPmuts) play a role not only in a subset of FIPA families (1–4) but also in sporadically diagnosed pituitary adenomas (5–9), and in the setting of somatostatin analog (SSA)-resistant acromegaly (10). Another form of FIPA, X-linked acrogigantism, due to microduplications in the Xq26.3 region, has been recently identified in patients with very young-onset gigantism and pituitary adenoma/hyperplasia (11).
The phenotype of AIPmut-associated pituitary adenomas has been described before (2–4, 12), but a systematic follow-up of cases and families is lacking due to the relative novelty of this pathogenic association (1), the variable disease penetrance (4, 12–14), and the rarity of this clinical entity. We present the clinical and genetic characteristics of a large cohort of FIPA and simplex (patients with germline mutation and no family history) AIPmut-positive patients, aiming for the following: 1) to perform a systematic follow-up of families to identify and characterize AIPmut-positive carriers, 2) to seek the role of disease-modifying genes on the variable phenotype and penetrance of the disease, and 3) to confirm and extend the description of the phenotype of AIPmut-positive patients, providing a comparison with AIPmut-negative cases. We establish that genetic screening followed by clinical assessment identifies a large percentage of family members with pituitary abnormalities, supporting the facilitation of genetic diagnosis and follow-up of these patients and their families.
Patients and Methods
Our study population (1725 subjects, Table 1) was recruited via the collaborative research network of the International FIPA Consortium (15). Pituitary adenoma patients were grouped into 11 clinical diagnostic categories (Supplemental Table 1). The diagnoses of acromegaly, acromegaly/prolactinoma, gigantism, gigantism/prolactinoma, and mild acromegaly (16) were grouped together under the category of GH excess for some analyses.
Table 1.
Study Population: Demographics and General Description
| Familial Cohort | Sporadic Cohort | Combined | |
|---|---|---|---|
| Total individuals, n, % | 1231 (71.4) | 494 (28.6) | 1725 (100) |
| Females, n, % | 668 (54.3) | 250 (50.6) | 918 (53.2) |
| Current age, median (range [IQR]) | 46.2 (2–97 [32–62]) | 35 (3–77 [26–42]) | 42.6 (2–97 [29–56]) |
| Clinical status, n, % | |||
| Affected | 502 (40.8) | 404 (81.8) | 906 (52.5) |
| Unaffected | 729 (59.2) | 90 (18.2) | 819 (47.5) |
| Affected males, n, % | 219 (43.6) | 203 (50.2) | 422 (46.6) |
| Affected females, n, % | 283 (56.4) | 201 (49.8) | 484 (53.4) |
| Diagnoses, n, % | |||
| Acromegaly | 170 (33.9) | 203 (50.2) | 373 (41.2) |
| Acromegaly/prolactinoma | 17 (3.4) | 12 (3) | 29 (3.2) |
| Cushing's disease | 24 (4.8) | 21 (5.2) | 45 (5) |
| FSHoma | 2 (0.4) | 1 (0.2) | 3 (0.3) |
| Gigantism | 44 (8.8) | 65 (16.1) | 109 (12) |
| Gigantism/prolactinoma | 1 (0.2) | 10 (2.5) | 11 (1.2) |
| Mild acromegaly | 2 (0.4) | — | 2 (0.2) |
| NFPA | 91 (18.1) | 21 (5.2) | 112 (12.4) |
| Pituitary tumor | 17 (3.4) | 2 (0.5) | 19 (2.1) |
| Prolactinoma | 134 (26.7) | 67 (16.6) | 201 (22.2) |
| TSHoma | — | 2 (0.5) | 2 (0.2) |
| GH excess patients, n, % | 234 (46.6) | 290 (71.8) | 524 (57.8) |
Abbreviations: FSHoma, FSH secreting adenoma. TSHoma, thyrotropinoma.
Dash indicates no patients in this category.
Between January 2007 and January 2014, we recruited patients from 35 countries from two different groups: either members of FIPA families, defined by the presence of pituitary adenomas in two or more members of a family without other associated clinical features (1–3, 17) (familial cohort), or sporadically diagnosed pituitary adenoma patients with disease onset at 30 years of age or younger (sporadic cohort). As an exception to these inclusion criteria, one AIPmut-positive sporadic patient older than 30 years was found thanks to AIP screening in the setting of a research study, and the screening of his relatives detected a second AIPmut-positive pituitary adenoma case; this family was included in the familial cohort. The first patient reported in each FIPA family and all the sporadic patients were considered probands. All the patients received treatment and were followed up in accordance with the guidelines and clinical criteria of their respective centers. Relevant clinical and family structure data were received from clinicians and/or patients, and genetic screening was performed in the families of all the AIPmut-positive probands, selecting individuals according to their risk of inheriting the mutation, based on their position in the family tree, and extending the screening to as many generations as possible. In both familial and sporadic cases, other causes of familial pituitary adenomas, such as MEN1 and MEN4, Carney complex, pheochromocytoma/paraganglioma and pituitary adenoma syndrome, and X-linked acrogigantism were ruled out by clinical, biochemical and, in some cases, genetic tests, as appropriate.
The study population included a great majority of new cases but also previously diagnosed patients being followed up by the participating centers and a few historical cases, corresponding to deceased members of FIPA families (further details in Supplemental Results). Four AIPmut-positive patients (two with diagnosis of acromegaly and two with gigantism) died in the postrecruitment period. Three of the deaths were due to cardiovascular causes (stroke, chronic heart failure, and acute coronary syndrome), whereas the exact cause of death is unknown in the fourth, a patient with long-standing untreated familial acromegaly.
All the patients and family members included agreed to take part by providing signed informed consent forms approved by the local ethics committee. Further details on the study population and the procedures for genetic/clinical screening and search for disease-modifying genes are described in the Supplemental Material.
Statistical analysis
The qualitative, categorical variables were expressed as percentages and compared using the χ2 test or the Fisher's exact test, as appropriate. The normal distribution of the quantitative variables was verified using the Shapiro-Wilk and the Kolmogorov-Smirnov tests for normality. Means and SDs were used to report parametric data, and nonparametric data were expressed as median and interquartile ranges. Parametric data were analyzed with the unpaired t test, with a 95% confidence interval (CI), whereas the Mann-Whitney U test was used for the nonparametric data. Statistical significance was considered when the P value was < .05. All the statistical analyses were carried out using the GraphPad Prism 6 (GraphPad Software Inc) and Stata 12 (StataCorp LP) statistical software.
Results
Study population
The familial cohort was composed of 216 FIPA families, including 156 new families (989 subjects: 337 patients and 652 unaffected family members) and 60 previously described families (3, 12), in which 46 new subjects (15 patients and 31 unaffected family members) were added to the previously reported 196 individuals (150 patients and 46 unaffected family members). The sporadic cohort originally included 409 pituitary adenoma patients 30 years old or younger at disease onset, with no known family history of pituitary adenoma, but we excluded five patients from further analysis due to harboring an Xq26.3 microduplication. Of the remaining 404 sporadic patients, six were reported previously (3). In addition to the AIPmut screening, a subset of AIPmut-negative FIPA (n = 55) and sporadic (n = 45) patients underwent genetic screening for other endocrine neoplasia-associated genes (Supplemental Table 2). All of these tests were negative for pathogenic variants. After the genetic screening and follow-up of the patients and carriers, 60 individuals in the familial cohort and seven in the sporadic cohort were classified as not at risk of inheriting an AIPmut and were excluded from further analysis. Twenty-three individuals initially thought to be unaffected were identified with pituitary abnormalities (see details in Prospective diagnosis).
Genetic screening results
Thirty-seven of 216 FIPA families screened (17.1%) and 34 of 404 sporadic patients (8.4%) were positive for pathogenic or likely pathogenic AIPmuts, accounting for a total of 71 AIPmut-positive kindreds and 144 AIPmut-positive patients (76.4% familial and 23.6% simplex, Table 2). We also identified 164 AIPmut-positive apparently unaffected family members (see Follow-up and prospective diagnosis). Samples were not available from family members of 25 AIPmut-positive simplex cases to establish the presence or lack of de novo mutations. We identified three pituitary adenoma patients (two with clinically nonfunctioning pituitary adenoma [NFPA] and one with a microprolactinoma) belonging to AIPmut-positive FIPA families and being at risk of inheriting but not carrying an AIPmut; therefore, they were considered as phenocopies.
Table 2.
Screening for AIPmuts
| Familial Cohort |
Sporadic Cohort |
Combined |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| AIPmut-Positive Familial | AIPmut-Negative Familial | Total Familial | AIPmut-Positive Simplex | AIPmut-Negative Sporadic | Total Sporadic | AIPmut-Positive Familial and Simplex | AIPmut-Negative Familial and Sporadic | Total | |
| Total number of kindreds, n, % | 37 (17.1% of familial) | 179 (82.9% of familial) | 216 (34.8% of total) | 34 (8.4% of sporadic) | 370 (91.6% of sporadic) | 404 (65.2% of total) | 71 (11.5% of total) | 549 (88.5% of total) | 620 (100) |
| Total individuals, n, % | 475 (38.6% of familial) | 756 (61.4% of familial) | 1231 (71.4% of total) | 82 (16.6% of sporadic) | 412 (83.4% of sporadic) | 494 (28.6% of total) | 557 (32.3% of total) | 1168 (67.7% of total) | 1725 (100) |
| Genetic status, n, % | |||||||||
| AIPmut-negative patients | 3 (0.6) | 389 (51.5)a | 392 (31.8) | — | 370 (89.8) | 370 (74.9) | 3 (0.5) | 759 (65) | 762 (44.2) |
| AIPmut-positive tested patients | 95 (20) | — | 95 (7.7) | 34 (41.5) | — | 34 (6.9) | 129 (23.2) | — | 129 (7.5) |
| At risk but not tested | 33 (6.9) | — | 33 (2.7) | 8 (9.8) | — | 8 (1.6) | 41 (7.4) | — | 41 (2.4) |
| Not at risk | 48 (10.1) | 12 (1.6) | 60 (4.9) | 7 (8.5) | — | 7 (1.4) | 55 (9.9) | 12 (1) | 67 (3.9) |
| Obligate unaffected carriers, not tested | 8 (1.7) | — | 8 (0.6) | 2 (2.4) | — | 2 (0.4) | 10 (1.8) | — | 10 (0.6) |
| Predicted AIPmut-positive patients | 15 (3.2) | — | 15 (1.2) | — | — | — | 15 (2.7) | — | 15 (0.9) |
| Unaffected AIPmut tested carriers | 120 (25.3) | — | 120 (9.7) | 16 (19.5) | — | 16 (3.2) | 136 (24.4) | — | 136 (7.9) |
| Unaffected and AIPmut negative | 153 (32.2) | — | 153 (12.4) | 15 (18.3) | — | 15 (3) | 168 (30.2) | — | 168 (9.7) |
| Unaffected relatives of AIPmut-negative patients | — | 355 (47) | 355 (28.8) | — | 42 (10.2) | 42 (8.5) | — | 397 (34) | 397 (23) |
| Summary of AIPmut-positive individuals, n, % | |||||||||
| Total AIPmut-positive patientsb | 110 (23.2) | — | 110 (8.9) | 34 (41.5) | — | 34 (6.9) | 144 (25.9) | — | 144 (8.3) |
| Total unaffected AIPmut carriersc | 128 (26.9) | — | 128 (10.4) | 18 (22) | — | 18 (3.6) | 146 (26.2) | — | 146 (8.5) |
Dash indicates no individuals in this category.
In AIPmut-negative FIPA families, 199 patients were tested for AIPmuts; the rest (n = 190) were assumed to be negative.
This is equal to the sum of tested AIPmut-positive patients plus the predicted AIPmut-positive patients.
Sum of tested unaffected carriers plus obligate unaffected carriers.
Thirty-one different AIPmuts (10 not previously reported) were identified in our study population: 12 exclusively in familial cases, 12 in simplex cases only, and seven in both settings (Table 3 and Supplemental Figure 1). Of the total mutations, 70.8% (22/31) predicted a truncated or missing protein and were termed as truncating AIPmuts (Supplemental Figure 2). We also identified 11 apparently nonpathogenic AIP variants (three of them novel) in our population (Supplemental Table 3).
Table 3.
AIP Pathogenic or Likely Pathogenic Mutations in the Familial and Sporadic Cohorts
| Mutation (DNA Level [Protein Level]) | Mutation Type | Pathogenic | Location in Protein | Familial Cohort (n = 238)a | Simplex Cohort (n = 52)a | Combined (n = 290)a | References/SRb |
|---|---|---|---|---|---|---|---|
| g.4856_4857CG>AA | Promoter | Yesc | Not in protein (5′ UTR) | 3 (1.3) | — | 3 (1) | (3, 12)/(SR30) |
| c.3G>A (p.?) | Start codon | Likelyc | N terminus | 2 (0.8) | — | 2 (0.7) | This paper |
| c.40C>T (p.Q14*) | Nonsense | Yesc | N terminus | 2 (0.8) | — | 2 (0.7) | (1)/(SR31, 32) |
| c.70G>T (p.E24*) | Nonsense | Yesc | N terminus | 9 (3.8) | — | 9 (3.1) | (3)/(SR33) |
| c.74_81delins7 (p.L25Pfs*130) | Frameshift | Yesc | PPIase domain | 10 (4.2) | — | 10 (3.4) | (12)/(SR34) |
| c.100–1025_279 + 357del (ex2del) (p.A34_K93del) | Large genomic deletion | Yesc | PPIase domain | 12 (5) | 2 (4) | 14 (4.8) | (SR35) |
| c.100–18C>T | Intronic | Likely | Not in protein (intron 1) | — | 3 (6) | 3 (1) | (3, 7, 10)/(SR31) |
| c.241C>T (p.R81*) | Nonsense | Yesc | PPIase domain | 12 (5) | 4 (8) | 16 (5.5) | (3)/(SR30, 36–38) |
| c.249G>T (p.G83Afs*15) | Splice site (cryptic splice site) | Yesc | PPIase domain | 4 (1.7) | — | 4 (1.4) | (12) |
| c.338_341dup (p.L115Pfs*16) | Frameshift | Yesc | PPIase domain | — | 2 (4) | 2 (0.7) | (6) |
| c.427C>T (p.Q143*) | Nonsense | Yesc | Between PPIase and TPR1 domains | — | 1 (2) | 1 (0.3) | This paper |
| c.469–2A>G (p.E158_Q184del) | Splice site | Likely | TPR1 domain | — | 1 (2) | 1 (0.3) | (5)/(SR39, 40) |
| c.490C>T (p.Q164*) | Nonsense | Yesc | Between PPIase and TPR1 domains | 3 (1.3) | — | 3 (1) | (12) |
| c.570C>G (p.Y190*) | Nonsense | Yesc | TPR1 domain | 9 (3.8) | — | 9 (3.1) | This paper |
| c.662dupC (p.E222*) | Nonsense | Yesc | Between TPR1 and 2 domains | 3 (1.3) | — | 3 (1) | (12) |
| c.713G>A (p.C238Y) | Missense | Yes | TPR2 domain | 4 (1.7) | — | 4 (1.4) | (3)/(SR33) |
| c.783C>G (p.Y261*) | Nonsense | Yesc | TPR2 domain | 4 (1.7) | — | 4 (1.4) | (9)/(SR39, 41, 42) |
| c.787 + 9C>T | Intronic | Uncertain | Not in protein (intron 5) | — | 1 (2) | 1 (0.3) | This paper |
| c.804C>A (p.Y268*) | Nonsense | Yesc | TPR3 domain | 19 (8) | 3 (6) | 22 (7.6) | (SR43, 44) |
| c.805_825dup (p.F269_H275dup) | In-frame insertion | Yes | TPR3 domain | 16 (6.7) | 2 (4) | 18 (6.2) | (3)/(SR30, 39, 45) |
| c.807C>T (p.(=)) | Splice site (reduced transcript level) | Yes | TPR3 domain | 7 (2.9) | 4 (8) | 11 (3.8) | (3, 5, 7, 10, 12)/(SR46, 47) |
| c.811C>T (p.R271W) | Missense | Yes | TPR3 domain | — | 1 (2) | 1 (0.3) | (2, 7, 12)/(SR48) |
| c.816delC (p.K273Rfs*30) | Frameshift | Yesc | TPR3 domain | — | 1 (2) | 1 (0.3) | This paper |
| c.868A>T (p.K290*) | Nonsense | Yesc | TPR3 domain | — | 1 (2) | 1 (0.3) | This paper |
| c.872_877delTGCTGG (p.V291_L292del) | In-frame deletion | Yes | TPR3 domain | — | 1 (2) | 1 (0.3) | This paper |
| c.910C>T (p.R304*) | Nonsense | Yesc | C-terminal α-helix | 88 (37) | 16 (31) | 104 (35.9) | (1–3, 5, 7, 9, 12, 14)/ (SR39, 49–51) |
| c.911G>A (p.R304Q) | Missense | Yes | C-terminal α-helix | 20 (8.4) | 3 (6) | 23 (7.9) | (3, 5, 7, 9, 12)/(SR31, 39, 52, 53) |
| c.967delC (p.R323Gfs*39) | Frameshift | Yesc | C-terminal α-helix | — | 4 (8) | 4 (1.4) | This paper |
| c.976_977insC (p.G326Afs*?) | Frameshift | Yesc | C-terminal α-helix | — | 1 (2) | 1 (0.3) | This paper |
| c.978dupG (p.I327Dfs*?) | Frameshift | Yesc | C-terminal α-helix | — | 1 (2) | 1 (0.3) | This paper |
| c.1-?_993+?del− (whole gene deletion) | Large genomic deletion | Yesc | Absence of the whole protein | 11 (4.6) | — | 11 (3.8) | (12) |
Abbreviations: PPIase, peptidylprolyl isomerase; SR, supplemental references; TPR, tetratricopeptide repeat; UTR, untranslated region.
Dash indicates no individuals in this category.
Number of positive individuals for each mutation, considering the AIPmut-positive tested individuals, the obligate carriers, and the predicted AIPmut patients.
For supplemental references, see Supplemental Material.
Truncating mutation.
A multiple regression analysis was performed to determine which clinical features could more accurately predict the likelihood of a patient to carry an AIPmut. An age at diagnosis of 10 years or older and younger than 20 years conferred an odds ratio (OR) of 5.8 (P = .000, 95% CI 3.1–10.8) of having an AIPmut, whereas the OR was 2.8 if the age at diagnosis was 20 years or older and younger than 30 years (P= .000, 95% CI 1.3–5.7); thus, an age at diagnosis between 10 and 30 years is the best predictor of AIPmuts. Inversely, a diagnosis of prolactinoma resulted in an OR of 0.2 (P= .000, 95% CI 0.1–0.5).
Genotype-phenotype correlation within the AIPmut-positive cohort
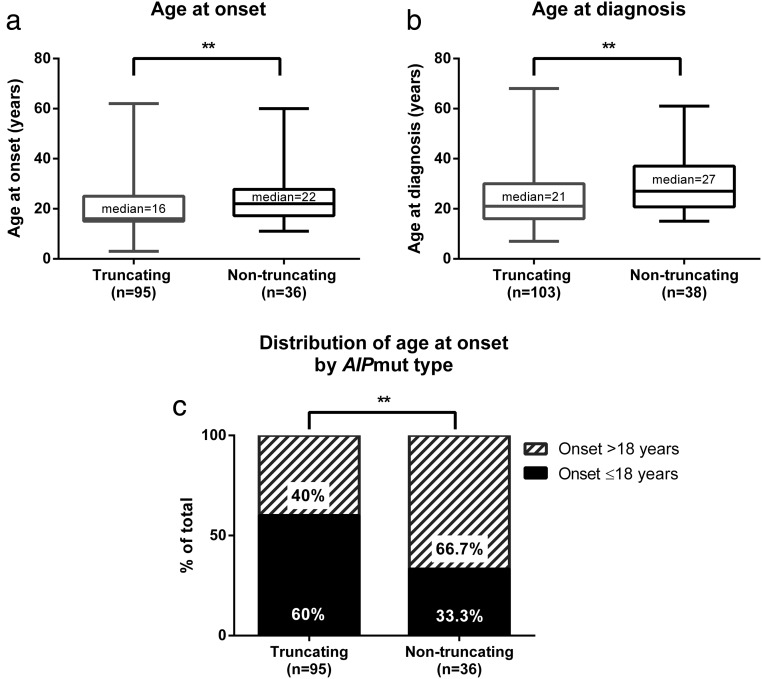
Truncating mutations accounted for 78.9% of the AIPmuts found in the familial cohort (15 of 19) and for 57.9% of those detected in the sporadic cohort (11 of 19). To study a possible difference in disease penetrance between truncating and nontruncating mutations, we compared the number of affected individuals with truncating AIPmuts in the familial (85 of 110 [77.3%]) and simplex cohorts (21 of 34 [61.8%]), finding no significant difference, although a trend was observed (P = .0729, analysis included prospectively diagnosed patients). No significant differences were found regarding the proportion of GH excess cases, number of patients per family, maximum tumoral diameter, frequency of macroadenomas, extrasellar invasion, or number of treatments received between the patients with truncating and nontruncating mutations. However, patients with truncating mutations were significantly younger at disease onset (median 16 [interquartile range (IQR) 15–25] vs 22 [IQR 17.3–27.8] y, P = .0046, Figure 1A) and at diagnosis (median 21 [IQR 16–30] vs 27 [IQR 20.8–37] y, P = .0028, Figure 1B), and the occurrence of pediatric cases was more common in this group (60% [57 of 95], Figure 1C), compared with the patients with nontruncating AIPmuts (33.3% [12 of 36], P = .0064). In concordance with these differences, gigantism accounted for a significantly higher proportion of the GH excess cases in the patients with truncating AIPmuts (54.7% [47 of 86]), compared with those with nontruncating AIPmuts (30% [9 of 30], P = .0200). Because p.R304* was the most common AIPmut in our study population (20 kindreds), we analyzed whether these patients behaved differently from other patients with truncating mutations. We found more affected individuals per family (median 4 [IQR 2.5–5]) among families carrying the p.R304* AIPmut, compared with families with other AIPmuts (median 2 [IQR 2–3], P = .0133). When considering all the AIPmut-positive patients together (familial and sporadic), we found a higher proportion of pediatric patients among those with the AIP p.R304* mutation (65.8% [25 of 38] vs 46.5% [40 of 86], P = .0475).
Figure 1.
Patients with truncating vs nontruncating AIPmuts. Patients with truncating AIPmuts present with a more aggressive phenotype, characterized by an earlier age at onset (A) (P = .005) and (B) at diagnosis (P = .003). C, This earlier disease onset results in a higher frequency of pediatric cases (n [total] = 131); in fact, most of the patients with truncating mutations present in childhood and adolescence. **, P < .01.
Clinical and histopathological features
Findings regarding gender distribution, age at onset/diagnosis, distribution of clinical diagnoses, tumor size/extension, pituitary apoplexy, histopathological features, extrapituitary tumors, and specific analyses of patients with GH excess and with gigantism are detailed in the Supplemental Material and depicted in Supplemental Tables 4 and 5 and Supplemental Figures 3–7.
Disease penetrance
To calculate the penetrance of pituitary adenomas among AIPmut positive families, complete data are needed both for phenotype and genotype. Therefore, we have selected three families (two with p.R304* and one with p.A34_K39del mutations) in which complete data were available in three or more generations for consenting at-risk individuals. The AIP genotype was known in 76.6% (range 68.4%–94.7%) of the individuals at risk; of them, 16.8% were patients and 83.2% were unaffected carriers. The gender distribution was similar between patients and unaffected carriers. The mean penetrance in these three families was 28.6% (19%–38.1%), and it decreased to 22.7% (18.2%–26.7%) when 50% of the individuals at risk with unknown genotype were considered as unaffected carriers. When the prospectively diagnosed patients were omitted from this calculation, the total penetrance of pituitary adenomas was 12.5%, highlighting the importance of the follow-up of apparently unaffected carriers for the correct calculation of the disease penetrance.
Because penetrance cannot be appropriately calculated for AIPmut-negative families, we assessed the number of affected family members. The AIPmut-positive families had more affected individuals per family than the AIPmut-negative families (P < .0001, Supplemental Figure 7E). Whereas 84.9% of the AIPmut-negative families (152 of 179) had only two affected members, 48.6% of the AIPmut-positive families (18 of 37) had three or more pituitary adenoma patients per family. The maximum number of affected individuals within the same family was eight (six of them prospectively diagnosed) in a family carrying the p.R304* AIPmut, and the maximum number of cases of gigantism in the same family was five, in a FIPA family with the p.E24* AIPmut.
Follow-up and prospective diagnosis
Of the 164 originally identified AIPmut carriers, 160 were available and advised to undergo biochemical and clinical screening. Prospective diagnosis of a pituitary adenoma was established in 11.3% (18 subjects, 11 males) of the individuals originally considered as unaffected AIPmut carriers.
Six of the prospectively diagnosed patients had acromegaly (one of them with prolactin [PRL] cosecretion), one patient had gigantism, two patients were diagnosed with mild acromegaly (16), and nine patients harbored NFPAs. Of the 142 individuals remaining as apparently unaffected AIPmut carriers, 79 (55.6%) underwent clinical assessment and one or more biochemical or imaging tests, whereas 63 subjects (44.4%) had only clinical evaluation.
The prospective cases were diagnosed at an older age than the rest of the patients (median 30 [IQR 22.8–39.5] vs 23 [IQR 16–33] y, P = .025). At diagnosis, seven of the prospectively diagnosed patients were symptomatic (headaches, arthralgias, acral growth, facial changes, weight gain, or hyperhidrosis). Five of the 18 prospectively diagnosed tumors were macroadenomas, in contrast with a predominance of macroadenomas (89.9%, 71 of 79) in the rest of the AIPmut-positive FIPA patients (P < .0001). The maximum diameter was significantly smaller for prospective cases (median 5.8 [IQR 4.7–14.4] vs 16.5 [IQR 10–29] mm, P = .0002). Four of the patients with macroadenomas had surgery, and the histopathological study confirmed GH- or GH/PRL-positive adenomas. The fifth macroadenoma was identified in a 68-year-old male patient with high IGF-1, well-controlled hypertension and diabetes mellitus and no other comorbidities or symptoms, who did not want to receive any treatment. In addition, one AIPmut-negative pituitary adenoma patient, harboring a 25-mm NFPA, was prospectively diagnosed as part of an AIPmut-positive family (brother of the AIPmut positive proband).
A further seven subjects had abnormalities in their screening tests, but a pituitary disease was not confirmed: five individuals had slightly elevated IGF-1 levels for their age/gender, one patient displayed acromegaloid features but normal pituitary magnetic resonance imaging (MRI) and biochemistry, and a 17-year-old female had repeatedly borderline high IGF-1 and incompletely suppressed GH on oral glucose tolerance test, but her bulky pituitary gland (11 mm in height), normal at this age group, is not changing during follow-up and her biochemical results are now within the normal range, after 3 years of follow-up.
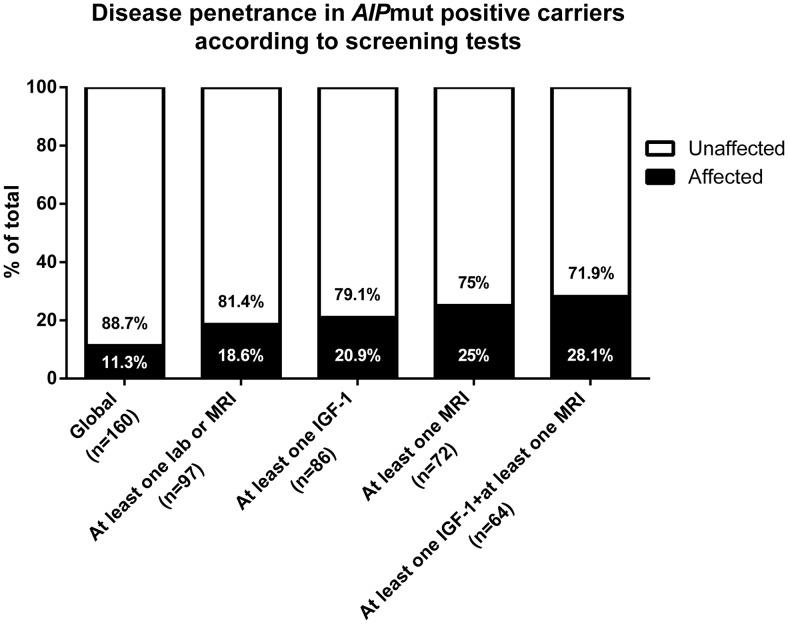
The global penetrance of pituitary adenomas among the individuals initially considered as unaffected AIPmut carriers was 11.3% (18 of 160). However, the penetrance was higher in the group of carriers who underwent biochemical and imaging investigations, varying between 18.6% and 28.1%, depending on the depth of screening (Figure 2). Overall, these data suggest that approximately 20%–25% of the apparently unaffected AIPmut carriers screened with biochemical or imaging tests will be identified with a pituitary adenoma at some point in their lives.
Figure 2.
Penetrance in screened AIPmut-positive carriers (n [total] = 160). The probability of detecting new cases of pituitary adenomas within apparently unaffected AIPmut carriers depends on the clinical assessment and the type of complementary biochemical/imaging studies included in the screening protocol (see text).
Clinical screening was not systematically performed in the AIPmut-negative FIPA unaffected family members. Nevertheless, due to the increased disease awareness given by the existence of previous pituitary adenoma cases within their families, four individuals (three females and one male) from three different AIPmut-negative FIPA families were prospectively diagnosed. Three of them harbored NFPAs, but we lack complete information about the fourth patient. The mean age at diagnosis in the three NFPA cases was 37 years, and only one patient referred symptoms at diagnosis (galactorrhea, not clearly associated to stalk compression, and lethargy). All of them had microadenomas, with a mean diameter of 6.5 mm and did not require any therapeutic intervention other than hormonal replacement in one case. The characteristics of these cases resemble those of incidentalomas; however, the occurrence of two prospective cases in the same family supports an apparent inherited component.
Disease-modifying genes
We have studied the role of two possible disease-modifying genes: GNAS1 (somatic) (18) and FGFR4 (germline) (19). GNAS1 mutations were absent in all the studied AIPmut-positive somatotropinomas (n = 23) but were detected in 50% of the AIPmut-negative familial somatotropinomas (5 of 10), 16.7% of the AIPmut-negative, young-onset cases (1 of 6), and 26.3% of the unselected acromegaly cases studied (5 of 19). The distribution of the FGFR4 p.G388R single-nucleotide polymorphism (SNP) conserved the Hardy-Weinberg equilibrium (20), and the genotype distribution was similar between patients (n = 98) and AIPmut carriers (n = 108) (P = .523). The age at onset and at diagnosis, tumor size, and frequency of extrasellar invasion were not significantly different between the GG (wild type) and GR/RR patients.
Discussion
AIPmuts are prevalent in young-onset, GH excess patients (24%) and FIPA (17.1%), with more than double frequency in patients with gigantism (46.7%) in our cohort, in concordance with other studies (7, 9, 21, 22). However, in contrast to previous reports, in this large and extensively studied cohort, there was no predominance of male patients among the AIPmut-positive familial cases, and equal numbers of male and female unaffected carriers were identified. Earlier studies (3, 4, 12, 23) may have had an ascertainment bias for families with cases of gigantism, a disease that is more prevalent in males, at least partly due to the physiologically later puberty and therefore later cessation of growth in boys.
We have demonstrated that approximately a quarter of the individuals initially identified as unaffected AIPmut carriers who underwent clinical screening tests were diagnosed with pituitary abnormalities. Full clinical screening identified 28.1% of the carriers, with fewer tests understandably resulting in fewer positive cases. Our data suggest that not all the AIPmut-associated pituitary adenomas have a rapidly growing, aggressive phenotype. The follow-up of these patients allowed us to observe some probably very early cases of acromegaly, in which the current clinical scenario had not indicated intervention at data closure. We cannot rule out that some of the small NFPAs are indeed incidentalomas, similar to those frequently observed in AIPmut-negative subjects of the general population.
This frequency of prospective diagnosis may justify the clinical screening and, possibly, follow-up of all the AIPmut-positive unaffected carriers. Our data would support the assessment of all the newly identified AIPmut carriers (clinical examination/history, PRL, and IGF-1, as a minimum, up to a full screening, also including an oral glucose tolerance test and contrast enhanced pituitary MRI). Follow-up of the younger family members should continue until at least 30 years of age, preferably annually, with clinical assessment and basal pituitary hormonal levels, leaving stimulation tests for cases with suspicion of pituitary disease and a follow-up MRI if necessary (24, 25). The cost-effectiveness and the possible psychological burden of this approach will need future study. Stopping the follow-up should be considered in older patients, given the low possibility of detecting new pituitary adenoma patients in these individuals after the fifth decade of life (24, 25). Once a case has been prospectively diagnosed, the treatment and follow-up should proceed as for the general population of pituitary adenoma patients because there are no data to suggest a different type of treatment in AIPmut-positive patients (26) although reduced SSA responsiveness has been described.
The genetic and clinical screening of AIPmut-negative FIPA families is uncertain at this point. Baseline screening and follow-up of obligate carriers could be considered, keeping in mind that the age of onset is considerably older in these families. Education on possible signs and symptoms of family members is a viable option in the routine setting. We expect that the identification of further genes implicated in the pathogenesis of FIPA in the next years will allow us to tailor these recommendations in accordance with the clinical behavior of each genetic entity.
Patients with GH excess starting before the age of 5 years should be tested for the recently identified Xq26.3 chromosomal microduplications (11). The genetic screening of other sporadic, young-onset pituitary adenoma patients with no evidence of other endocrine tumors should be focused on AIPmuts in first instance in cases of GH excess (with or without PRL cosecretion) and on MEN1 mutations, especially in cases of prolactinoma (9) because this can be the first manifestation of MEN1 (27). Whether it would be advisable to continue screening young patients with other diagnoses for AIPmuts out of the setting of research studies needs longer follow-up.
To explain the variable clinical phenotype in our AIPmut-positive patients, we evaluated the possible influence of two disease-modifying genes, GNAS1 and FGFR4. Whereas somatic GNAS1 mutations are common in unselected somatotropinomas (4.4%–59% of the cases) (28–35), we have not identified any in adenomas from AIPmut-positive patients, suggesting that germline AIPmuts and somatic GNAS1 mutations are mutually exclusive in somatotropinomas. GNAS1 mutations have rarely been studied in pediatric patients with acromegaly and gigantism, and they seem to be an extremely infrequent finding in this age group (36, 37). A recent study has shown no change in the AIP immunostaining in sporadic somatotropinomas in the presence of GNAS1 mutations (38). The characteristic phenotype of adenomas containing the GNAS1 mutations (small [32, 39], highly responsive to the treatment with SSAs, and more often densely granulated according to some [40], but not all studies [41]), seems to be in contrast with the typical AIPmut-positive tumor phenotype. On the other hand, in somatotroph adenomas of AIPmut-negative FIPA patients, half of the tested samples had GNAS1 mutations. This suggests that in AIPmut-negative FIPA, the somatic GNAS1 mutations could exist in a similar frequency as in unselected somatotropinomas and possibly, in addition to a germline predisposing mutation, may play a role in their pathogenesis.
The FGFR4 gene SNP rs351855 (c.1162G>A, p.G388R), with a minor allele frequency of 0.3, is a predictor of progression and poor prognosis in a variety of human neoplasms (42). A role for rs351855 as a facilitator of somatotroph cell tumorigenesis has been recently proposed (19), and we hypothesized that this variant could increase the penetrance and/or size and extension of AIPmut-positive pituitary adenomas. The screening for this SNP in our AIPmut-positive patients failed to show increase in size, extension, or apoplexy, even though this association had previously been suggested in sporadic acromegaly patients (19), and no earlier onset or higher penetrance was observed. The lack of association with these two potentially disease-modifying genes suggests that AIPmut-related pituitary adenomas are regulated by different pathogenic mechanisms than unselected somatotropinomas.
We recognize the numerous limitations of our study. We chose an arbitrary age cutoff (≤30 y), in concordance with previous AIP-related publications, but our data show that only 13.2% of the AIPmut-positive patients had disease onset after the age of 30 years. Our patients were recruited from different genetic backgrounds, and this could have influenced the disease penetrance and presentation. On the other hand, 19.7% of the AIPmut-positive kindreds (24.3% of the AIPmut positive patients) belong to a cohort with a founder AIPmut (p.R304*), originally from Northern Ireland (14). The larger number of subjects screened in these families provided a higher number of carriers and chance for detection of affected individuals. Additional genetic traits possibly cosegregating with this founder mutation could modify the phenotype and thus introduce a bias into our results. Full genotype and phenotype data were not available for all the families; therefore, we limited our penetrance calculations to three large, well-characterized families. A better assessment of the prevalence of pituitary apoplexy and extrapituitary adenomas in AIPmut-positive patients would require a large control group, screened ad hoc, which was beyond the scope of this study. Finally, the data about therapeutic modalities were limited, hampering the analysis of the response to different treatments.
Conclusions
The analysis of this large cohort of FIPA patients allowed us to establish a number of novel aspects of FIPA. A phenotype-genotype correlation was found with younger onset of disease in patients with truncating AIPmuts. We identified a surprisingly high percentage of somatic GNAS1 mutations in the AIPmut-negative somatotropinomas and their absence in AIPmut-positive tumors. The lack of influence of the germline FGFR4 p.G388R variant on disease penetrance/severity suggests that currently unknown factors drive penetrance and variable phenotype in AIPmut-positive pituitary adenomas. The presence of milder, more indolent disease in some AIPmut-positive subjects has been established. Genetic and clinical screening leads to the prospective identification of an unexpectedly high proportion of affected patients in the originally apparently unaffected carrier group, resulting in earlier diagnosis and treatment and, possibly, better long-term outcome (25). The recruitment of a large study population with this uncommon disease has only been possible thanks to worldwide collaboration.
Acknowledgments
We are very grateful for patients and their family members and the numerous health care professionals supporting the study. We acknowledge Jonathan P. Bestwick (Wolfson Institute of Preventive Medicine, Barts and The London School of Medicine, Queen Mary University of London) for his valuable help with the statistical analysis and Dr Richard J. M. Ross (Department of Human Metabolism, School of Medicine and Biomedical Science, University of Sheffield, Sheffield, United Kingdom) for his collaboration in the recruitment of patients and collection of clinical data.
The International FIPA Consortium includes the following: Amar Agha, MD (Beaumont Hospital, Dublin, Republic of Ireland); Scott A. Akker, MD (St Bartholomew's Hospital, London, United Kingdom); Elena D. Aflorei, MD (Barts and The London School of Medicine, Queen Mary University of London, London, United Kingdom); Sándor Alföldi, MD (Szent Imre Egyeteni Oktatókórház, Budapest, Hungary); Professor Wiebke Arlt, MD (University of Birmingham, Birmingham, United Kingdom); Professor Brew Atkinson (Royal Victoria Hospital, Belfast, Northern Ireland, United Kingdom); Anna Aulinas-Masó, MD (Hospital de Sant Pau, Universitat Autònoma de Barcelona, Barcelona, Spain); Simon J. Aylwin, MD (Kings College Hospital National Health Service Foundation Trust, London, United Kingdom); Professor Philippe F. Backeljauw (Cincinnati Children's Hospital Medical Center, Cincinnati, Ohio); Corin Badiu, MD (Carol Davila University of Medicine and Pharmacy, Bucharest, Romania); Stephanie Baldeweg, MD (University College London Hospital, London, United Kingdom); Gul Bano, MD (St George's University of London, London, United Kingdom); Professor Ariel Barkan (University of Michigan Medical Center, Ann Arbor, Michigan); Julian Barwell, MD (Leicester Royal Infirmary, Leicester, United Kingdom); Carmen Bernal-González, MD (Hospital Universitario “12 de Octubre,” Madrid, Spain); Professor G. Michael Besser (St Bartholomew's Hospital, London, United Kingdom); Professor John S. Bevan (Aberdeen Royal Infirmary, Foresterhill, Aberdeen, United Kingdom); Jo Blair, MD (Alder Hey Children's National Health Service Foundation Trust, Liverpool, United Kingdom); Pierre Bouloux, MD (Royal Free and University College School of Medicine, London, United Kingdom); Lisa Bradley, MD (St George's Healthcare National Health Service Trust, London, United Kingdom); Michael Buchfelder, MD (University of Erlangen, Erlangen, Germany); Professor Mehtap Cakir (Meram School of Medicine, Konya Necmettin Erbakan University, Konya, Turkey); Natalie Canham, MD (North West Thames Regional Genetics Service, Northwick Park Hospital, London, United Kingdom); Paul Carroll, MD (Guy's and St Thomas' National Health Service Foundation Trust, London, United Kingdom); Harvinder S. Chahal, MD, PhD (Imperial College Healthcare National Health Service Trust, London, United Kingdom); Tim Cheetham, MD (University of Newcastle, Newcastle, United Kingdom); Farida Chentli, MD (Bab El Oued Teching Hospital, Algiers, Algeria); Richard N. Clayton, MD (University of Keele, Stoke-on-Trent, United Kingdom); Mark Cohen, MD (Royal Free National Health Service Foundation Trust, Barnet Hospital, Barnet, United Kingdom); Trevor Cole, MD (Birmingham Women's Hospital, Birmingham, United Kingdom); Hamish Courtney, MD (Royal Victoria Hospital, Belfast, Northern Ireland, United Kingdom); Elizabeth Crowne, MD (University Hospitals Bristol Foundation Trust, Bristol, United Kingdom); Daniel Cuthbertson, MD (University of Liverpool, Liverpool Merseyside, United Kingdom); Jacob Dal, MD (Aarhus University Hospital, Aarhus, Denmark); Nadezhda Dalantaeva, MD (Endocrinology Research Centre, Moscow, Russia); Christina Daousi, MD (University Hospital Aintree, Clinical Sciences Centre, University of Liverpool, Liverpool, United Kingdom); Ken Darzy, MD (Lister Hospital, Stevenage, United Kingdom); Professor Mehul Dattani, MD (University College London Institute of Child Health, London, United Kingdom); Justin H. Davies, MD (University Hospital Southampton, Southampton, United Kingdom); Professor Julian Davis, MD (Faculty of Medical and Human Sciences, University of Manchester and Central Manchester University Hospitals National Health Service Foundation Trust, Manchester, United Kingdom); Margaret De Castro, MD (Ribeirao Preto School of Medicine, University of Sao Paulo, Sao Paulo, Brazil); Laura De Marinis, MD (Università Cattolica del Sacro Cuore, Policlinico Universitario A. Gemelli, Rome, Italy); Professor William Drake, MD (St Bartholomew's Hospital, London, United Kingdom); Pinaki Dutta, MD (Postgraduate Institute of Medical Education and Research, Chandigarh, India); Larisa Dzeranova, MD (Endocrinology Research Centre, Moscow, Russia); Britt Edén-Engström, MD (Uppsala University Hospital, Uppsala, Sweden); Professor Rosalind Eeles, MD (Sutton Hospital, Sutton, United Kingdom); Maria Elfving, MD (Lund University Hospital, Lund, Sweden); Marianne Elston, MD (Waikato Hospital, Hamilton, New Zealand and Waikato Clinical School, University of Auckland, Hamilton, New Zealand); Louise Emmerson, MD (All Wales Medical Genetics Service, Glan Clwyd Hospital, Rhyl, United Kingdom); Naomi Fersht, MD (Department of Oncology, University College London Hospitals, London, United Kingdom); Professor Simona Fica, MD (Elias Hospital, Carol Davila University of Medicine and Pharmacy, Bucharest, Romania); Stefan Fischli, MD (Luzerner Kantonsspital, Luzern, Switzerland); Daniel Flanagan, MD (Derriford Hospital, Plymouth, United Kingdom); Maria Fleseriu, MD (Northwest Pituitary Center, Oregon Health and Science University, Portland, Oregon); Pamela U. Freda, MD (Columbia University College of Physicians and Surgeons, New York, New York); Professor Theodore Friedman, MD (Charles R. Drew University of Medicine and Science, Los Angeles, California); Professor Lawrence A. Frohman, MD (University of Illinois at Chicago, Chicago, Illinois); Patricia Gallego, MD (Western University, Children's Hospital, London Health Science Centre, London, Ontario, Canada); Evelien Gevers, MD (Barts and The London School of Medicine, Queen Mary University of London, London, United Kingdom); Edit Gláz, MD (Semmelweis University, Budapest, Hungary); James A. Goldman, MD (Harvard Vanguard Medical Associates, Boston Massachusetts); Anthony P. Goldstone (Imperial College Healthcare National Health Service Trust, Hammersmith Hospital, London, United Kingdom); Miklos Goth, MD (Health Center, Hungarian Defense Forces, Budapest, Hungary); Lynn Greenhalgh, MD (Alder Hey Children's Hospital Eaton Road, Liverpool, United Kingdom); Joan Grieve, MD (National Hospital for Neurology and Neurosurgery, Queen Square, London, United Kingdom); Mirtha Guitelman, MD (Hospital Durand, Buenos Aires, Argentina); Alper Gürlek, MD (Faculty of Medicine, Hacettepe University, Ankara, Sihhiye, Turkey); Mark Gurnell, MD (University of Cambridge and Cambridge Biomedical Research Centre, Addenbrooke's Hospital, Cambridge, United Kingdom); Katalin Horvath, MD (Gyor Hospital, Gyor, Hungary); Trevor A. Howlett, MD (University Hospitals of Leicester National Health Service Trust, Leicester Royal Infirmary, Leicester, United Kingdom); Charlotte Höybye, MD (Karolinska University Hospital, Stockholm, Sweden); Steven Hunter, MD (Royal Victoria Hospital, Belfast, Northern Ireland, United Kingdom); Donato Iacovazzo, MD (Barts and The London School of Medicine, Queen Mary University of London, London, United Kingdom, and Università Cattolica del Sacro Cuore, Policlinico Universitario A. Gemelli, Rome, Italy); Peter Igaz, MD (Faculty of Medicine, Semmelweis University, Budapest, Hungary); Warrick J. Inder, MD (School of Medicine, The University of Queensland, Brisbane, Queensland, Australia); Takeo Iwata, MD (Institute of Health Biosciences, The University of Tokushima Graduate School, Toskushima City, Japan); Louise Izatt (Guy's and St Thomas' Foundation Trust, Guy's Hospital, London, United Kingdom); Sujatha Jagadeesh, MD (Mediscan, Chennai, India); Gregory Kaltsas, MD (Laiko General Hospital, School of Medicine, National and Kapodistrian University of Athens, Athens, Greece); Felicity Kaplan, MD (Lister Hospital, Stevenage, United Kingdom); Niki Karavitaki, MD (Oxford Centre for Diabetes, Endocrinology, and Metabolism, Churchill Hospital, Oxford, United Kingdom); Darko Kastelan, MD (University Hospital Zagreb, and School of Medicine University of Zagreb, Zagreb, Croatia); Michelle Katz, MD (Massachusetts General Hospital and Harvard Medical School, Boston, Massachusetts); Tara Kearney, MD (Greater Manchester Neurosciences Centre, Salford Royal Foundation Trust, Manchester, United Kingdom); Bernard Khoo, MD (University College London, London, United Kingdom); Cathy Kiraly-Borri, MD (King Edward Memorial Hospital for Women, Subiaco, Australia); Robertas Knispelis, MD (Medical Academy, Lithuanian University of Health Sciences, Kaunas, Lithuania); Gábor László Kovács, MD (Flór Ferenc Hospital, Kistarcsa, Hungary); Ajith V. Kumar, MD (Great Ormond Street Hospital, London, United Kingdom); Edward R. Laws Jr, MD (Brigham and Women's Hospital, Boston, Massachusetts); Ronald M. Lechan, MD (Tupper Research Institute, Tufts Medical Center, Tufts University School of Medicine, Boston, Massachusetts); Miles J. Levy, MD (University Hospitals of Leicester National Health Service Trust, Leicester Royal Infirmary, Leicester, United Kingdom); Krzysztof Lewandowski, MD (Polish Mother's Memorial Hospital-Research Institute, and Medical University, Lodz, Poland); Janet Lo, MD (Harvard Medical School, Massachusetts General Hospital, Boston, Massachusetts); Niki Maartens, MD (University of Brisbaine, Brisbaine, Australia); Professor Akira Matsuno (Teikyo University, Tokyo, Japan); Barbara McGowan, MD (Guy's and St Thomas' Foundation Trust, St Thomas' Hospital, London, United Kingdom); Siobhán E. McQuaid, MD (Mater Misericordiae University Hospital, Dublin, Republic of Ireland); Milica Medic-Stojanoska, MD (Clinical Center of Vojvodina and Medical Faculty, University of Novi Sad, Novi Sad, Serbia); Professor Moisés Mercado-Atri, MD (Hospital de Especialidades Centro Médico Nacional Siglo XXI, Instituto Mexicano del Seguro Social, Mexico City, Mexico); Emese Mezősi, MD (Faculty of Medicine, University of Pécs, Pécs, Hungary); Dragana Miljic, MD (Clinical Center of Serbia and Medical Faculty, University of Belgrade, Belgrade, Serbia); Karen K. Miller, MD (Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts); Silvia Modenesi, MD (Hospital das Clinicas, Minas Gerais Federal University, Belo Horizonte, Brazil); Mark E. Molitch, MD (Northwestern University Feinberg School of Medicine, Chicago, Illinois); Professor John Monson, MD (St Bartholomew's Hospital, London, United Kingdom); Damian G. Morris, MD (The Ipswich Hospital, Ipswich, United Kingdom); Patrick J. Morrison, MD (Belfast Health and Social Care Trust, Belfast, Northern Ireland, United Kingdom); Alia Munir, MD (Pinderfields Hospital, Yorkshire, United Kingdom, and Royal Hallamshire Hospital, Sheffield, United Kingdom); Professor Robert D. Murray, MD (Leeds Teaching Hospitals National Health Service Trust, St James's University Hospital, Leeds, United Kingdom); Madalina Musat, MD (Carol Davila University of Medicine and Pharmacy, Bucharest, Romania); Nina Musolino, MD (Universidade de São Paulo, São Paulo, Brazil); Lisa Nachtigall, MD (Harvard Medical School, Massachusetts General Hospital, Boston, Massachusetts); Professor John Newell-Price (School of Medicine and Biomedical Science, University of Sheffield, Sheffield, United Kingdom); Arla Ogilvie, MD (Watford Hospital, Watford, United Kingdom); Steve M. Orme, MD (Leeds General Infirmary, Leeds, United Kingdom); Ionela, Paşcanu MD (University of Medicine and Pharmacy, Tirgu-Mures, Romania); Attila Patócs, MD (Semmelweis University, Budapest, and Hungarian Academy of Sciences, Budapest, Hungary); Catherine Patterson, MD (Queen Margaret Hospital, Fife, United Kingdom); Simon H. Pearce, MD (Newcastle University, Newcastle-upon-Tyne, United Kingdom); Francesca Pecori Giraldi, MD (University of Milan, and Istituto Auxologico Italiano Istituto di Ricovero e Cura a Carattere Scientifico, Milan, Italy); Professor Marija Pfeifer, MD (University Medical Center Ljubljana, Ljubljana, Slovenia); Professor Vera Popovic (Clinical Center of Serbia and Medical Faculty, University of Belgrade, Belgrade, Serbia); Nicola Poplawski, MD (South Australia Pathology at the Women's and Children's Hospital, North Adelaide, South Australia, Australia); Michael Powell, MD (The National Hospital for Neurology and Neurosurgery, Queen Square, London, United Kingdom); Peter Pullan, MD (Sir Charles Gairdner Hospital, Nedlands, West Australia, Australia); Richard Quinton, MD (Institute of Genetic Medicine, University of Newcastle on Tyne, Royal Victoria Infirmary, Newcastle, United Kingdom); Serban Radian, MD, PhD (Barts and The London School of Medicine, Queen Mary University of London, London, United Kingdom); Harpal Randeva, MD (University of Warwick, Warwick, United Kingdom); Antônio Ribeiro-Oliveira Jr, MD (Hospital das Clinicas, Minas Gerais Federal University, Belo Horizonte, Brazil); Celia Rodd, MD (Wiinipeg University, Winnipeg, Canada); Fiona Ryan, MD (The John Radcliffe Hospital, Oxford, United Kingdom); Roberto Salvatori, MD (Johns Hopkins University School of Medicine, Baltimore, Maryland); Professor Christof Schöfl (Universitätsklinikum Erlangen, Friedrich-Alexander-Universität, Erlangen-Nürnberg, Germany); Debbie Shears, MD (Churchill Hospital, Oxford University Hospitals National Health Service Trust, Oxford, United Kingdom); Kevin Shotliff, MD (Chelsea and Westminster Hospital National Health Service Foundation Trust, London, United Kingdom); Beatriz S. Soares, MD (Hospital das Clinicas, Minas Gerais Federal University, Belo Horizonte, Brazil); Noel Somasundaram (National Hospital of Sri Lanka, Colombo, Sri Lanka); Professor Anna Spada, MD (Fondazione Cà Granda Istituto di Ricovero e Cura a Carattere Scientifico Ospedale Maggiore, University of Milan, Milan, Italy); James Sperber, MD (Endocrine Clinic, San Clemente, California); Helen A. Spoudeas, MD (The London Centre for Pediatric Endocrinology and Diabetes, Great Ormond Street Hospital for Children National Health Service Foundation Trust, London, United Kingdom); Susan Stewart, MD (University Hospital Birmingham, and Birmingham Women's Hospital, Birmingham, United Kingdom); Helen Storr, MD (Barts and The London School of Medicine, Queen Mary University of London, London, United Kingdom); Christian Strasburger, MD (Charite Campus Mitte, Berlin, Germany); Maria Elisabeth Street, MD (Santa Maria Nuova Hospital and Research Institute, Reggio-Emilia, Italy); Francesca Swords, MD (Norfolk and Norwich University Hospital, Norwich, United Kingdom); Professor Rajesh V. Thakker, MD (University of Oxford, Oxford Centre for Diabetes, Endocrinology, and Metabolism, Churchill Hospital, Oxford, United Kingdom); Elaine Tham, MD (Women's and Children's Hospital, Adelaide, Australia); Chris Thompson, MD (Beaumont Hospital, Dublin, Republic of Ireland); Dr Michael O. Thorner (University of Virginia, Charlottesville, Virginia); Miklós Tóth, MD (Faculty of Medicine, Semmelweis University, Budapest, Hungary); Professor Peter J. Trainer, MD (The Christie National Health Service Foundation Trust, Manchester, United Kingdom); Stylianos Tsagarakis, MD (Evangelismos Hospital, Athens, Greece); Marinella Tzanela, MD (Evangelismos Hospital, Athens, Greece); János Vadász, MD (Szolnok Hospital, Szolnok, Hungary); Vladimir Vaks, MD (Great Western Hospitals National Health Service Foundation Trust, Swindon, United Kingdom); Rasa Verkauskiene, MD (Institute of Endocrinology, Medical Academy, Lithuanian University of Health Sciences, Kaunas, Lithuania); Professor John A. Wass, MD (Oxford Centre for Diabetes, Endocrinology, and Metabolism, Churchill Hospital, Oxford, United Kingdom); Susan M. Webb, MD (Hospital Sant Pau, Centre for Biomedical Research on Rare Diseases (Centro de Investigación Biomédica en Red de Enfermedades Raras Unit 747); Universitat Autònoma de Barcelona, Barcelona, Spain); Astrid Weber, MD (Liverpool Women's National Health Service Foundation Trust, Liverpool, United Kingdom); Shozo Yamada, MD (Toranomon Hospital, Tokyo, Japan); Sema Yarman, MD (Istanbul University, Istanbul Faculty of Medicine, Istanbul, Turkey); Philip Yeoh, MD (The London Clinic, London, United Kingdom); Katsuhiko Yoshimoto, MD (Institute of Health Biosciences, The University of Tokushima Graduate School, Tokushima City, Japan); and Nicola N. Zammitt, MD (Royal Infirmary of Edinburgh, Edinburgh, Scotland, United Kingdom).
Authors' contributions include the following: LC.H.-R. collected and entered the clinical and genetic data in the database, performed the statistical analysis and GNAS1 and FGFR4 genotyping, and prepared the manuscript; P.G. managed the ethics, recruited the patients, managed the samples and patient's data, extracted the DNA, collected and entered the clinical and genetic data in the database, and contacted the collaborators; J.D. recruited the patients, managed the samples and patient's data, extracted the DNA, and collected and entered the clinical and genetic data in the database; K.S. performed the DNA sequencing and in silico analysis of AIPmuts; G.T. collected the genetic data and performed the in silico analysis of AIPmuts; D.T. performed the FGFR4 genotyping; F.F. performed the DNA extraction and FGFR4 genotyping; J.E. analyzed the MRI studies of the patients; S.E. supervised the DNA sequencing and in silico analysis of AIPmuts; A.B.G. recruited the patients, collected the clinical and genetic data, and reviewed the manuscript; F.R. reviewed and completed the histopathological diagnoses; M.R.G. recruited the patients, collected the clinical and genetic data, and reviewed the manuscript; M.K. designed and coordinated the study, recruited the patients, collected and entered the clinical and genetic data in the database, reviewed the in silico analyses, and prepared and reviewed the manuscript; and The International FIPA Consortium members recruited the patients and provided the clinical data.
The trial of Genetics of Endocrine Tumors-Familial Isolated Pituitary Adenoma-FIPA is registered with clinicaltrials.gov with the identifier of NCT00461188.
This work was supported by the Medical Research Council of the United Kingdom (G0701307), the Wellcome Trust (097970/Z/11/Z), the National Institute of Health Research, the Barts and The London Charity, the Royal Society, and Pfizer. L.C.H.-R. is supported by grants from the National Council of Science and Technology and the Secretariat of Public Education from the Mexican Government.
The funding bodies had no role in the study design, collection, analysis, and interpretation of the data or in the manuscript preparation.
Disclosure Summary: M.G. serves on the Medical Advisory Board of Novartis. M.K. has received research grants from Pfizer and Novartis and serves on the Medical Advisory Board of Pfizer Inc. S.E. is a Wellcome Trust Senior Investigator. The other authors have nothing to disclose.
Footnotes
- AIP
- aryl-hydrocarbon receptor interacting protein
- AIPmut
- AIP mutation
- CI
- confidence interval
- FIPA
- familial isolated pituitary adenoma
- IQR
- interquartile range
- MEN
- multiple endocrine neoplasia
- MRI
- magnetic resonance imaging
- NFPA
- nonfunctioning pituitary adenoma
- OR
- odds ratio
- PRL
- prolactin
- SNP
- single-nucleotide polymorphism
- SSA
- somatostatin analog.
Contributor Information
Collaborators: The International FIPA Consortium, Amar Agha, Scott A. Akker, Elena D. Aflorei, Sándor Alföldi, Professor Wiebke Arlt, Professor Brew Atkinson, Anna Aulinas-Masó, Simon J. Aylwin, Professor Philippe F. Backeljauw, Corin Badiu, Stephanie Baldeweg, Gul Bano, Professor Ariel Barkan, Julian Barwell, Carmen Bernal-González, Professor G. Michael Besser, Professor John S. Bevan, Jo Blair, Pierre Bouloux, Lisa Bradley, Michael Buchfelder, Professor Mehtap Cakir, Natalie Canham, Paul Carroll, Harvinder S. Chahal, Tim Cheetham, Farida Chentli, Richard N. Clayton, Mark Cohen, Trevor Cole, Hamish Courtney, Elizabeth Crowne, Daniel Cuthbertson, Jacob Dal, Nadezhda Dalantaeva, Christina Daousi, Ken Darzy, Professor Mehul Dattani, Justin H. Davies, Professor Julian Davis, Margaret De Castro, Laura De Marinis, Professor William Drake, Pinaki Dutta, Larisa Dzeranova, Britt Edén-Engström, Professor Rosalind Eeles, Maria Elfving, Marianne Elston, Louise Emmerson, Naomi Fersht, Professor Simona Fica, Stefan Fischli, Daniel Flanagan, Maria Fleseriu, Pamela U. Freda, Professor Theodore Friedman, Professor Lawrence A. Frohman, Patricia Gallego, Evelien Gevers, Edit Gláz, James A. Goldman, Anthony P. Goldstone, Miklos Goth, Lynn Greenhalgh, Joan Grieve, Mirtha Guitelman, Alper Gürlek, Mark Gurnell, Katalin Horvath, Trevor A. Howlett, Charlotte Höybye, Steven Hunter, Donato Iacovazzo, Peter Igaz, Warrick J. Inder, Takeo Iwata, Louise Izatt, Sujatha Jagadeesh, Gregory Kaltsas, Felicity Kaplan, Niki Karavitaki, Darko Kastelan, Michelle Katz, Tara Kearney, Bernard Khoo, Cathy Kiraly-Borri, Robertas Knispelis, Gábor László Kovács, Ajith V. Kumar, Edward R. Laws, Jr, Ronald M. Lechan, Miles J. Levy, Krzysztof Lewandowski, Janet Lo, Niki Maartens, Professor Akira Matsuno, Barbara McGowan, Siobhán E. McQuaid, Milica Medic-Stojanoska, Professor Moisés Mercado-Atri, Emese Mezősi, Dragana Miljic, Karen K. Miller, Silvia Modenesi, Mark E. Molitch, Professor John Monson, Damian G. Morris, Patrick J. Morrison, Alia Munir, Professor Robert D. Murray, Madalina Musat, Nina Musolino, Lisa Nachtigall, Professor John Newell-Price, Arla Ogilvie, Steve M. Orme, Ionela Paşcanu, Attila Patócs, Catherine Patterson, Simon H. Pearce, Francesca Pecori Giraldi, Professor Marija Pfeifer, Professor Vera Popovic, Nicola Poplawski, Michael Powell, Peter Pullan, Richard Quinton, Serban Radian, Harpal Randeva, Antônio Ribeiro-Oliveira, Jr, Celia Rodd, Fiona Ryan, Roberto Salvatori, Professor Christof Schöfl, Debbie Shears, Kevin Shotliff, Beatriz S. Soares, Noel Somasundaram, Professor Anna Spada, James Sperber, Helen A. Spoudeas, Susan Stewart, Helen Storr, Christian Strasburger, Maria Elisabeth Street, Francesca Swords, Professor Rajesh V. Thakker, Elaine Tham, Chris Thompson, Dr Michael O. Thorner, Miklós Tóth, Professor Peter J. Trainer, Stylianos Tsagarakis, Marinella Tzanela, János Vadász, Vladimir Vaks, Rasa Verkauskiene, Professor John A. Wass, Susan M. Webb, Astrid Weber, Shozo Yamada, Sema Yarman, Philip Yeoh, Katsuhiko Yoshimoto, and Nicola N. Zammitt
References
- 1. Vierimaa O, Georgitsi M, Lehtonen R, et al. Pituitary adenoma predisposition caused by germline mutations in the AIP gene. Science. 2006;312:1228–1230. [DOI] [PubMed] [Google Scholar]
- 2. Daly AF, Vanbellinghen JF, Khoo SK, et al. Aryl hydrocarbon receptor-interacting protein gene mutations in familial isolated pituitary adenomas: analysis in 73 families. J Clin Endocrinol Metab. 2007;92:1891–1896. [DOI] [PubMed] [Google Scholar]
- 3. Leontiou CA, Gueorguiev M, van der Spuy J, et al. The role of the aryl hydrocarbon receptor-interacting protein gene in familial and sporadic pituitary adenomas. J Clin Endocrinol Metab. 2008;93:2390–2401. [DOI] [PubMed] [Google Scholar]
- 4. Daly AF, Tichomirowa MA, Petrossians P, et al. Clinical characteristics and therapeutic responses in patients with germ-line AIP mutations and pituitary adenomas: an international collaborative study. J Clin Endocrinol Metab. 2010;95:E373–E383. [DOI] [PubMed] [Google Scholar]
- 5. Cazabat L, Libe R, Perlemoine K, et al. Germline inactivating mutations of the aryl hydrocarbon receptor-interacting protein gene in a large cohort of sporadic acromegaly: mutations are found in a subset of young patients with macroadenomas. Eur J Endocrinol. 2007;157:1–8. [DOI] [PubMed] [Google Scholar]
- 6. Stratakis CA, Tichomirowa MA, Boikos S, et al. The role of germline AIP, MEN1, PRKAR1A, CDKN1B and CDKN2C mutations in causing pituitary adenomas in a large cohort of children, adolescents, and patients with genetic syndromes. Clin Genet. 2010;78:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tichomirowa MA, Barlier A, Daly AF, et al. High prevalence of AIP gene mutations following focused screening in young patients with sporadic pituitary macroadenomas. Eur J Endocrinol. 2011;165:509–515. [DOI] [PubMed] [Google Scholar]
- 8. Cazabat L, Bouligand J, Salenave S, et al. Germline AIP mutations in apparently sporadic pituitary adenomas: prevalence in a prospective single-center cohort of 443 patients. J Clin Endocrinol Metab. 2012;97:E663–E670. [DOI] [PubMed] [Google Scholar]
- 9. Cuny T, Pertuit M, Sahnoun-Fathallah M, et al. Genetic analysis in young patients with sporadic pituitary macroadenomas: beside AIP don't forget MEN1 genetic analysis. Eur J Endocrinol. 2013;168:533–541. [DOI] [PubMed] [Google Scholar]
- 10. Oriola J, Lucas T, Halperin I, et al. Germline mutations of AIP gene in somatotropinomas resistant to somatostatin analogues. Eur J Endocrinol. 2013;168:9–13. [DOI] [PubMed] [Google Scholar]
- 11. Trivellin G, Daly AF, Faucz FR, et al. Gigantism and acromegaly due to Xq26 microduplications and GPR101 mutation. N Engl J Med. 2014;371:2363–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Igreja S, Chahal HS, King P, et al. Characterization of aryl hydrocarbon receptor interacting protein (AIP) mutations in familial isolated pituitary adenoma families. Hum Mutat. 2010;31:950–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Naves LA, Daly AF, Vanbellinghen JF, et al. Variable pathological and clinical features of a large Brazilian family harboring a mutation in the aryl hydrocarbon receptor-interacting protein gene. Eur J Endocrinol. 2007;157:383–391. [DOI] [PubMed] [Google Scholar]
- 14. Chahal HS, Stals K, Unterlander M, et al. AIP mutation in pituitary adenomas in the 18th century and today. N Engl J Med. 2011;364:43–50. [DOI] [PubMed] [Google Scholar]
- 15. The International FIPA Consortium. http://www.fipapatients.org/fipaconsortium/ Accessed November 17, 2014.
- 16. Dimaraki EV, Jaffe CA, DeMott-Friberg R, Chandler WF, Barkan AL. Acromegaly with apparently normal GH secretion: implications for diagnosis and follow-up. J Clin Endocrinol Metab. 2002;87:3537–3542. [DOI] [PubMed] [Google Scholar]
- 17. Daly AF, Jaffrain-Rea ML, Ciccarelli A, et al. Clinical characterization of familial isolated pituitary adenomas. J Clin Endocrinol Metab. 2006;91:3316–3323. [DOI] [PubMed] [Google Scholar]
- 18. Landis CA, Masters SB, Spada A, Pace AM, Bourne HR, Vallar L. GTPase inhibiting mutations activate the alpha chain of Gs and stimulate adenylyl cyclase in human pituitary tumours. Nature. 1989;340:692–696. [DOI] [PubMed] [Google Scholar]
- 19. Tateno T, Asa SL, Zheng L, Mayr T, Ullrich A, Ezzat S. The FGFR4–G388R polymorphism promotes mitochondrial STAT3 serine phosphorylation to facilitate pituitary growth hormone cell tumorigenesis. PLoS Genet. 2011;7:e1002400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hardy-Weinberg Equilibrium Calculator. http://www.koonec.com/k-blog/2010/06/20/hardy-weinberg-equilibrium-calculator/ Accessed January 3, 2014.
- 21. Rostomyan L, Daly AF, Lila A, et al. Gigantism: results of an international clinical and genetic study. Endocr Rev. 2013;34(03_MeetingAbstracts):OR20–6. [Google Scholar]
- 22. Schofl C, Honegger J, Droste M, et al. Frequency of AIP gene mutations in young patients with acromegaly: a registry-based study. J Clin Endocrinol Metab. 2014;99(12):E2789–E2793. [DOI] [PubMed] [Google Scholar]
- 23. Beckers A, Daly AF. The clinical, pathological, and genetic features of familial isolated pituitary adenomas. Eur J Endocrinol. 2007;157:371–382. [DOI] [PubMed] [Google Scholar]
- 24. Korbonits M, Storr H, Kumar AV. Familial pituitary adenomas—who should be tested for AIP mutations? Clin Endocrinol (Oxf). 2012;77:351–356. [DOI] [PubMed] [Google Scholar]
- 25. Williams F, Hunter S, Bradley L, et al. Clinical experience in the screening and management of a large kindred with familial isolated pituitary adenoma due to an aryl hydrocarbon receptor interacting protein (AIP) mutation. J Clin Endocrinol Metab. 2014;99:1122–1131. [DOI] [PubMed] [Google Scholar]
- 26. Beckers A, Aaltonen LA, Daly AF, Karhu A. Familial isolated pituitary adenomas (FIPA) and the pituitary adenoma predisposition due to mutations in the aryl hydrocarbon receptor interacting protein (AIP) gene. Endocr Rev. 2013;34:239–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thakker RV, Newey PJ, Walls GV, et al. Clinical practice guidelines for multiple endocrine neoplasia type 1 (MEN1). J Clin Endocrinol Metab. 2012;97:2990–3011. [DOI] [PubMed] [Google Scholar]
- 28. Landis CA, Harsh G, Lyons J, Davis RL, McCormick F, Bourne HR. Clinical characteristics of acromegalic patients whose pituitary tumors contain mutant Gs protein. J Clin Endocrinol Metab. 1990;71:1416–1420. [DOI] [PubMed] [Google Scholar]
- 29. Hosoi E, Yokogoshi Y, Hosoi E, et al. Analysis of the Gsα gene in growth hormone-secreting pituitary adenomas by the polymerase chain reaction-direct sequencing method using paraffin-embedded tissues. Acta Endocrinol (Copenh). 1993;129:301–306. [DOI] [PubMed] [Google Scholar]
- 30. Yoshimoto K, Iwahana H, Fukuda A, Sano T, Itakura M. Rare mutations of the Gsα subunit gene in human endocrine tumors. Mutation detection by polymerase chain reaction-primer-introduced restriction analysis. Cancer. 1993;72:1386–1393. [DOI] [PubMed] [Google Scholar]
- 31. Shi Y, Tang D, Deng J, Su C. Detection of gsp oncogene in growth hormone-secreting pituitary adenomas and the study of clinical characteristics of acromegalic patients with gsp-positive pituitary tumors. Chin Med J (Engl). 1998;111:891–894. [PubMed] [Google Scholar]
- 32. Buchfelder M, Fahlbusch R, Merz T, Symowski H, Adams EF. Clinical correlates in acromegalic patients with pituitary tumors expressing GSP oncogenes. Pituitary. 1999;1:181–185. [DOI] [PubMed] [Google Scholar]
- 33. Park C, Yang I, Woo J, et al. Somatostatin (SRIF) receptor subtype 2 and 5 gene expression in growth hormone-secreting pituitary adenomas: the relationship with endogenous srif activity and response to octreotide. Endocr J. 2004;51:227–236. [DOI] [PubMed] [Google Scholar]
- 34. Mendoza V, Sosa E, Espinosa-de-los-Monteros AL, et al. GSPα mutations in Mexican patients with acromegaly: potential impact on long term prognosis. Growth Horm IGF Res. 2005;15:28–32. [DOI] [PubMed] [Google Scholar]
- 35. Freda PU, Chung WK, Matsuoka N, et al. Analysis of GNAS mutations in 60 growth hormone secreting pituitary tumors: correlation with clinical and pathological characteristics and surgical outcome based on highly sensitive GH and IGF-I criteria for remission. Pituitary. 2007;10:275–282. [DOI] [PubMed] [Google Scholar]
- 36. Johnson MC, Codner E, Eggers M, Mosso L, Rodriguez JA, Cassorla F. Gps mutations in Chilean patients harboring growth hormone-secreting pituitary tumors. J Pediatr Endocrinol Metab. 1999;12:381–387. [DOI] [PubMed] [Google Scholar]
- 37. Metzler M, Luedecke DK, Saeger W, et al. Low prevalence of Gsα mutations in somatotroph adenomas of children and adolescents. Cancer Genet Cytogenet. 2006;166:146–151. [DOI] [PubMed] [Google Scholar]
- 38. Jaffrain-Rea ML, Rotondi S, Turchi A, et al. Somatostatin analogues increase AIP expression in somatotropinomas, irrespective of Gsp mutations. Endocr Relat Cancer. 2013;20:753–766. [DOI] [PubMed] [Google Scholar]
- 39. Larkin S, Reddy R, Karavitaki N, Cudlip S, Wass J, Ansorge O. Granulation pattern, but not GSP or GHR mutation, is associated with clinical characteristics in somatostatin-naive patients with somatotroph adenomas. Eur J Endocrinol. 2013;168:491–499. [DOI] [PubMed] [Google Scholar]
- 40. Spada A, Arosio M, Bochicchio D, et al. Clinical, biochemical, and morphological correlates in patients bearing growth hormone-secreting pituitary tumors with or without constitutively active adenylyl cyclase. J Clin Endocrinol Metab. 1990;71:1421–1426. [DOI] [PubMed] [Google Scholar]
- 41. Mayr B, Buslei R, Theodoropoulou M, Stalla GK, Buchfelder M, Schofl C. Molecular and functional properties of densely and sparsely granulated GH-producing pituitary adenomas. Eur J Endocrinol. 2013;169:391–400. [DOI] [PubMed] [Google Scholar]
- 42. Frullanti E, Berking C, Harbeck N, et al. Meta and pooled analyses of FGFR4 Gly388Arg polymorphism as a cancer prognostic factor. Eur J Cancer Prev. 2011;20:340–347. [DOI] [PubMed] [Google Scholar]