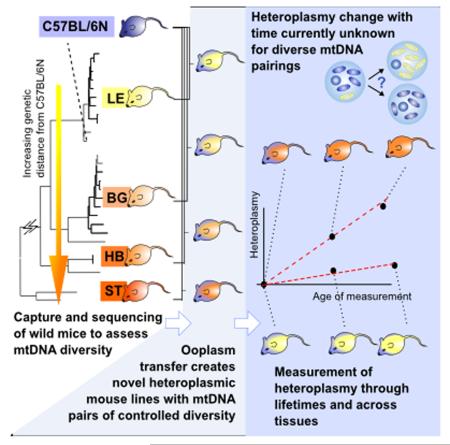
Summary
The dynamics by which mitochondrial DNA (mtDNA) evolves within organisms are still poorly understood, despite the fact that inheritance and proliferation of mutated mtDNA causes fatal and incurable diseases. When two mtDNA haplotypes are present in a cell, it is usually assumed that segregation (the proliferation of one haplotype over another) is negligible. We challenge this assumption showing that segregation depends on the genetic distance between haplotypes. We provide evidence by creating four new mouse models containing mtDNA haplotype pairs of varying diversity. We find tissue-specific segregation in all models over a wide range of tissues. Key findings are segregation in post mitotic tissues (important for disease models), and segregation covering all developmental stages from prenatal up to old age. We identify four new dynamic regimes of mtDNA segregation. We suggest “haplotype matching” as a means to avoid complications for therapies in human populations that our findings imply.
Graphical abstract
Introduction
Mitochondrial DNA (mtDNA) encodes functionally important elements of the electron transport chain, vital for providing the energy to fuel living processes in almost all eukaryotic cells. Pathological mtDNA mutations can lead to deficiencies in this energy supply and are responsible for several incurable inherited diseases (Poulton et al., 2010), and the question of how cellular mtDNA content evolves within organisms and between generations is of key importance in combatting these diseases.
Specific combinations of polymorphisms distinguish mtDNA into classes known as mitochondrial haplotypes and haplogroups (Mueller et al., 2012). Due to strict maternal inheritance, cells usually harbour only one mtDNA haplotype, with many identical mtDNA molecules present in a single cell, in a situation termed homoplasmy. During evolution, populations accumulate a high number of non-pathological mtDNA base substitutions that radiate along maternal lineages (Mueller et al., 2012). Cellular mtDNA populations may consist of a combination of different haplotypes: such a cellular population is termed heteroplasmic. Heteroplasmy can emerge naturally by inheritance or de novo mutations (Payne et al., 2013) and artificially by assisted reproductive techniques like cloning (reviewed in (St John et al., 2010)), ooplasm/ cytoplast transfer (Ferreira et al., 2010; Jenuth et al., 1996; St John, 2002) and gene therapies (reviewed in (St John and Campbell, 2010; Wallace and Chalkia, 2013)). The requirement of functional compatibility between two diverse mtDNA haplotypes in heteroplasmic organisms, as well as between mtDNA and nuclear DNA, are currently recognised not only concerning basic research (Sharpley et al., 2012), but also as unexplored concerns of gene therapy implementations (Reinhardt et al., 2013; St John and Campbell, 2010).
As mtDNA replicates and is degraded within cells quasi-independently of the cellular life cycle, heteroplasmic populations of mtDNA constitute an evolutionary system within cells. The dynamics governing the within- and between-generation evolution of heteroplasmic cellular mtDNA populations are largely unknown (Reinhardt et al., 2013; Sharpley et al., 2012; St John and Campbell, 2010), with current work mainly based on mouse-models with limited genetic diversity, usually utilising the New Zealand Black (NZB) laboratory mouse mtDNA haplotype mixed with mtDNAs of classical laboratory mouse strains (CIS) (Acton et al., 2007; Battersby and Shoubridge, 2001, 2007; Jenuth et al., 1997; Jokinen et al., 2010; Meirelles and Smith, 1997; Moreno-Loshuertos et al., 2006; Sharpley et al., 2012). In the absence of genetic engineering of mtDNA, the main sources of mtDNA variety in mouse models are randomly (pathologically) mutated mtDNAs (Farrar et al., 2013) or the very limited haplotypic variance between laboratory animals. The NZB mouse represents almost the only laboratory mouse that shows a haplotype with considerable genetic difference to the CIS and was therefore exclusively used in intra-subspecies heteroplasmic mouse models.
These experiments have illustrated that one haplotype can, for unknown reasons, proliferate faster and come to dominate over the other in a process termed mtDNA segregation bias: a potentially important mechanism modulating the balance of mtDNA populations. Such a segregation bias was shown to be specific for certain tissues, with the exception of post-mitotic tissues (Jenuth et al., 1997; Sharpley et al., 2012).
Despite intensive investigation of, and breakthroughs arising from, the NZB model, one key question remains unanswered: is the NZB model representative, or may other haplotypes display different segregation behaviours? This question is particularly pertinent when attempting to relate conclusions from studies on mouse lines to human systems, where mtDNA genetic diversity is pronounced and ubiquitous (Blanco et al., 2011) but in vivo experiments impossible. Moreover, the use of inbred laboratory animal strains was recently questioned, on the grounds that they do not mirror the natural (human) genetic variation (Hayden, 2013). Other currently unanswered questions regarding cellular mtDNA population dynamics include whether segregation occurs in post-mitotic tissues, of importance for the onset of inherited mitochondrial disease (St John and Campbell, 2010) (currently, segregation in post-mitotic tissues is viewed as neutral, with the exception of large scale deletions (Sato et al., 2007)), and the kinetics and influential factors that drive segregation in different tissues.
We address the questions of how genetic differences between mtDNA haplotypes influence segregation, and whether segregation effects could also be present in post-mitotic tissues, through a new experimental protocol. We create and use several new mouse models using mtDNA from wild-derived mouse populations gathered across Europe, thus incorporating a wide range of mtDNA variation, from very closely related haplotypes up to a cross-subspecies combination. These models allow us to control genetic distance and observe effects on segregation and mtDNA population dynamics. We analyse a large range of heteroplasmy data, sampled across tissues and over a range of organismal ages from prenatal to several years old, with a new and rigorous mathematical model for the evolution of mtDNA populations.
The high number of time points and analysed tissues makes it possible to elucidate several previously unknown segregation mechanisms. To the best of our knowledge we are the first to find segregation in post mitotic tissues within the same subspecies (important for disease models), and directly showing a prenatal segregation pattern. We characterize two new time-kinetics of mtDNA segregation. Moreover we correlate mtDNA segregation rates to genetic distance, mtDNA- and cell-turnover. We further propose and discuss applications of our findings in the context of recently proposed therapies in human assisted reproduction.
Results
Creation and phylogenetic analysis of wild-derived mouse models exhibiting a range of mtDNA differences
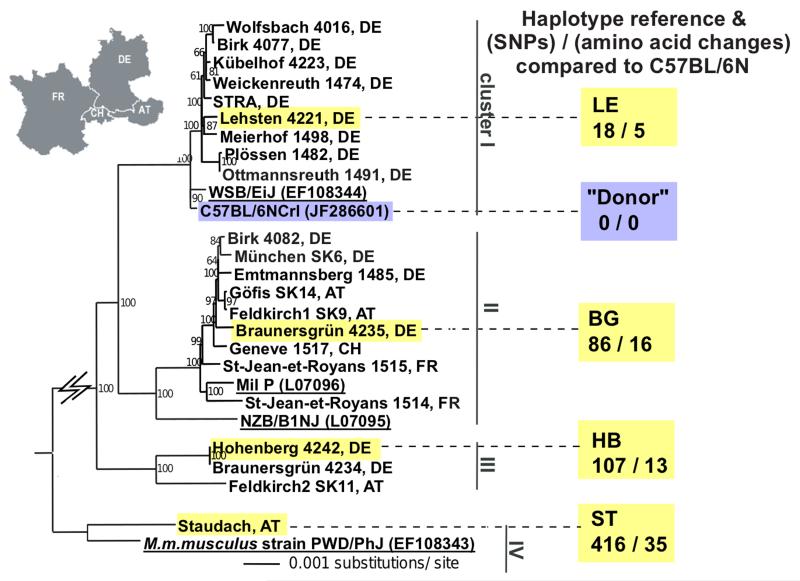
The mtDNA of 22 wild-derived house mouse lines (Mus musculus domesticus), captured in Western/ Central Europe were analysed. We used the total number of single nucleotide polymorphisms (SNPs), and the number of SNPs causing amino acid (AA) changes, as indicators of the genetic divergence between mtDNA haplotypes (Blanco et al., 2011; Green et al., 2008). By 454-sequencing and phylogenetic analysis we identified 3 clusters, with 18 to 107 SNPs and 5 to 16 AA changes compared to the C57BL/6N (B6N) mtDNA (cluster representatives: LE: 18 SNPs, 5 AA; BG: 86 SNPs, 16 AA; HB: 107 SNPs, 13 AA; Figure 1, Table S1). One mouse showed an mtDNA of another subspecies (Mus musculus musculus, eastern house mouse) although derived from western Austria (ST), “cluster IV”; ST: 416 SNPs, 35 AA changes.
Figure 1. Neighbour-joining phylogenetic analysis of 22 novel mitochondrial genomes of wild-derived house mice.
The mtDNA genomes of 22 wild-derived mice (M. m. domesticus) captured in four countries cluster into three groups, with the different subspecies M. m. musculus as an additional cluster (cluster IV).
Black: wild-derived mice; yellow highlighted: wild-derived mice used to generate the four heteroplasmic mouse models in this study; black and underlined: reference laboratory strains with GenBank accession number bracketed; blue highlighted: C57BL/6N (B6N) “donor” laboratory mouse. The country of origin of the mouse is designated by a two-letter code (AT: Austria, FR: France, DE: Germany, CH: Switzerland). Support values for the internal branches of the tree topology are shown as percentages. The number of SNPs and amino acid changes calculated relative to the B6N reference mtDNA are shown below the four lines (LE, BG, HB, ST). See also Table S1.
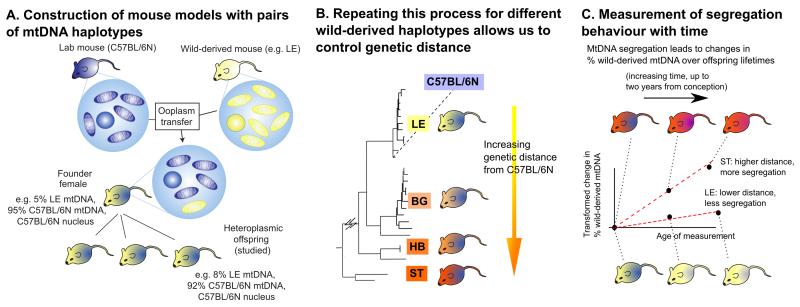
To mirror (and exceed) the variability range found for pairwise comparisons in human mtDNAs with up to 130 SNPs ((Blanco et al., 2011) with 29.3 and 78.3 SNPs average for the European and African population respectively (Lippold et al., 2014)) and 20 AA changes (Craven et al., 2011), we included one mouse of all four clusters in the subsequent study. Through ooplasm transfer four heteroplasmic mouse lines were created, one from each of the three M. m. domesticus clusters and the M. m. musculus (see Figure 2A and Experimental Procedures). The relative amount of wild-derived mtDNA in the founder females ranged from 5% to 7% (with an additional BG founder with 50% created by blastomere fusion, see Experimental Procedures). To avoid potential artefactual effects from the creation process, these founder females were not used in our analysis: instead, numerous offspring from the F1-F4 generations were used, having naturally inherited their levels of heteroplasmy.
Figure 2. Creation and measurement of wild-derived heteroplasmic mouse lines with diverse mtDNA haplotypes.
(A) Founder females were created by ooplasmic injection from a wild-derived mouse (yellow) into a zygote of a standard laboratory mouse (C57BL/6N (B6N), blue). Only offspring of the founder females were used in this study, to avoid artefacts associated with the creation process. (B) This process was repeated for each of four wild-derived mtDNA haplotypes (LE pictured) to achieve a range of genetic differences between haplotype pairs, allowing us to address for the first time the effect of mtDNA diversity on segregation. (C) To compute segregation rates, levels of the wild-derived mtDNA haplotype were measured in many mice from each lineage at different ages. Our inferential machinery allows us to compute the change in levels of the wild-derived haplotype since conception, and recording this heteroplasmy change as a function of the age of the measured mouse allows us to infer segregation rate in each tissue in each lineage.
We thus obtain a set of heteroplasmic mouse models, each exhibiting a different amount of genetic difference between their two mtDNA haplotypes (Figure 2B), allowing us to explore for the first time the influence of genetic difference on segregation behaviour.
Tissue-dependent segregation in all four mouse lines
The mtDNA heteroplasmy was measured with ARMS-qPCR in 13 tissues from heteroplasmic mice that were sacrificed at various ages (from prenatal up to 750 days, see Figure 2C). Additionally at the age of 21 days, tail biopsies were analysed to provide further information about heteroplasmy in the young mouse. In total, over 1600 tissue-samples of 154 mice were analysed (Tables S2-5). Table 1 gives exemplary percentage heteroplasmy values for each of the four heteroplasmic mouse lines. From each haplotype a heteroplasmic young mouse (1-3 days old) and an adult mouse (1-2 years old) are shown. While in the young mice the various organs showed similar heteroplasmy, in the adult animals the variation was much higher. Percentage heteroplasmy values of all mice analysed in this study are shown in Tables S2-5.
Table 1. mtDNA heteroplasmy changes between tissues over time; selected samples.
| % mtDNA heteroplasmyb |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mtDNA | mousea | age days | tail biopsy 21 days | brain | heart | muscle | kidney | liver | lung | blood | intestine | skin | spleen | tail | testis | uterus |
| LE | LE 729 p2 | 3 | - | 16.6 | 20.7 | 16.7 | 18.4 | 18.4 | 19.1 | 16.0 | 14.7 | 17.4 | 15.9 | 16.9 | 14.9 | - |
| LE 583 | 516 | 8.2 | 7.4 | 7.3 | 6.8 | 12.1 | 22.8 | 19.7 | 9.6 | 13.8 | 7.9 | 18.1 | 4.6 | n.d. | n.d. | |
|
| ||||||||||||||||
| HB | HB 172 p2 | 1 | - | 4.1 | 2.1 | 2.5 | 3.3 | 3.7 | 4.5 | 2.8 | 3.3 | 2.4 | 2.9 | 3.4 | n.d. | n.d. |
| HB 450 | 680 | 12.1 | 8.4 | 2.1 | 4.4 | 32.4 | 91.0 | 19.0 | 52.9 | 7.3 | 10.4 | 52.6 | 15.4 | n.d. | n.d. | |
|
| ||||||||||||||||
| BG | BG 667 p3 | 3 | - | 13.5 | 8.2 | 17.5 | 11.0 | 10.6 | 11.0 | 13.9 | 17.6 | 14.3 | 17.1 | 10.4 | - | 11.6 |
| BG 78 | 416 | 10.8 | 13.7 | 10.0 | 7.5 | 13.0 | 10.3 | 12.1 | 21.0 | 36.0 | 15.7 | 26.1 | 39.2 | - | 38.0 | |
|
| ||||||||||||||||
| ST | ST 344 p1 | 3 | - | 84.5 | 83.0 | 87.7 | 93.9 | 92.6 | 88.8 | 91.7 | 90.5 | 89.5 | 89.0 | 92.5 | n.d. | n.d. |
| ST 102 | 376 | 14.4 | 20.9 | 18.6 | 24.6 | 62.2 | 63.9 | 38.9 | 87.4 | 33.7 | 31.6 | 75.4 | 44.2 | - | 54.4 | |
Mice were exemplarily selected for similar initial heteroplasmy values, for ST only pups with high initial heteroplasmy were available
Tissues that were found to show a significant segregation bias in the subsequent statistical analysis of all mice used in this study are underlined (bold, increase of wild-derived mtDNA; italic, decrease of wild-derived mtDNA).
As the magnitude of mtDNA segregation depends on initial heteroplasmy levels, mice with different initial heteroplasmy values cannot be directly compared. For example, it is not immediately clear whether an observed change from 50% to 60% corresponds to more, less, or equally strong segregation compared to a change from 95% to 98%: slower absolute changes are expected as the limits of 0% and 100% are approached. To allow the comparison of mice with such different initial heteroplasmy values, we calculated the “proliferation rate” per day of the respective mtDNAs in each model (see Experimental Procedures and Supplemental Experimental Procedures for details, transformed heteroplasmy values of all mice analysed in this study are shown in Tables S6-9). These proliferation rates indicate how fast the proportion of a given mtDNA haplotype changes quantitatively over time, and are independent of initial heteroplasmy. This allows us to compare results across all our mice.
Positive proliferation rates indicate a proliferative advantage for the respective wild-derived mtDNA, negative rates indicate a proliferative advantage for the B6N mtDNA: e.g. changes in heteroplasmy from an inferred 15% at conception to 91% (liver) and 2.1% (heart) at 680 days of age (approximately the changes observed in mouse HB 450, Table 1) correspond to wild-derived mtDNA proliferation rates of 0.0060 (liver) and −0.0031 (heart) per day (Figure 3 shows the average proliferation rates of the respective tissues). Over the exemplary 680 days these rates result in a total transformed heteroplasmy change of 4.1 (liver) and −2.1 (heart) as shown in Figure 6.
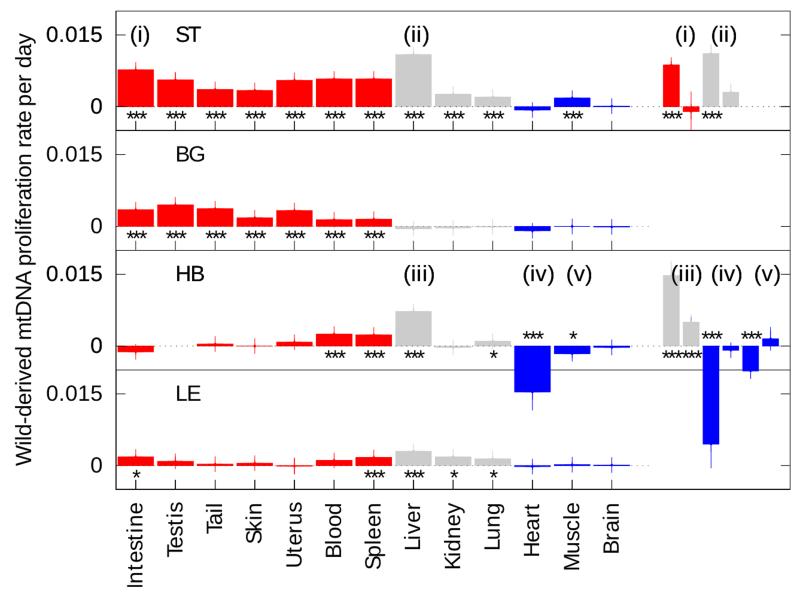
Figure 3. mtDNA segregation rates across tissues, in correlation to cell-turnover.
The mean proliferation rate for each wild-derived mtDNA haplotype (see Figure 1 for their definition, Tables S2-9 for raw data) is reported across tissues, with the standard deviation of the associated bootstrapped distribution (vertical lines). Positive values indicate relative increase of the respective wild-derived mtDNA over time, negative values relative increase of C57BL/6N (B6N) mtDNA (y-axis). Tissues are coloured according to mitotic rate: (left, red) highly mitotic; (centre, grey) mitotic; (right, blue) post-mitotic (details in Table S10). Cases where the segregation rate significantly differs from zero are reported (bootstrapping with the percentile method; * p < 0.05, ** p < 0.01, *** p < 0.001 after Bonferroni correction). n.d., not determined. While in ST almost all tissues show increase of wild-derived mtDNA, in BG this happens only in self-renewing tissues with high cell-turnover (red). HB shows complex segregation pattern that includes a decrease of wild-derived mtDNA in heart and muscle. LE is the haplotype most closely related to B6N, and nevertheless shows significant segregation in several tissues, possibly resembling ST haplotype at a slower segregation rate.
(i) to (v) denote tissues where the magnitude of the proliferation rate was significantly higher in younger than older animals. Each of (i)-(v) is re-plotted as an inset, early segregation rate on the left, late on the right (likelihood ratio test, p < 0.01 after Bonferroni correction). See also Figure 6. (n= 31; 34; 56; 33 for LE; BG; HB; ST respectively)
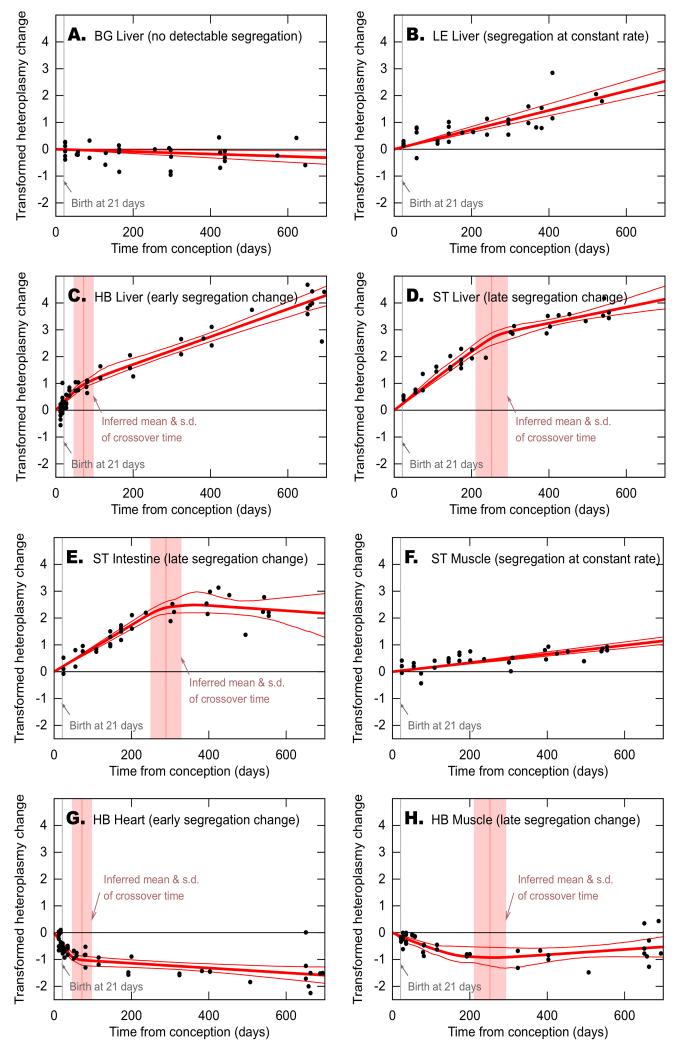
Figure 6. Segregation regimes vary based on haplotype and tissue.
Black dots are experimental data of single mice; dark red lines show mean and 95% confidence intervals of inferred segregation trajectories. In the models where there appears to be a bi-phasic timecourse vertical light pink lines show mean and standard deviation of inferred crossover time between two different segregation speeds. The y-axis shows transformed heteroplasmy change, giving the segregation rate of wild-derived mtDNA. Positive values indicate relative increase of the respective wild-derived mtDNA, negative values relative increase of the C57BL/6N (B6N) mtDNA. In BG liver (A), no significant segregation is observed (zero heteroplasmy change lies within the confidence intervals). In LE liver (B) and ST muscle (F) segregation occurs at constant rates, leading to single gradients of heteroplasmy changes with time. In other tissues pictured, there is statistical support (likelihood ratio test, p < 0.01 after Bonferroni) for a dynamic regime involving fast segregation before a certain crossover time and slower segregation after this time. In HB liver (C) and heart (G), this crossover time is inferred to be during the growth phase of the mice (day 72 ±26 and 50 ±12 respectively); in ST liver (D) and intestine (E), and HB muscle (H), the crossover time is much later in life (day 252 ±42, 289 ±40 and 227 ±57 respectively), possibly due to physiological and metabolic changes at these times.
(n= 31; 34; 56; 33 for LE; BG; HB; ST respectively)
Figure 3 shows the segregation rates for all analyzed tissues of the respective haplotypes. Tissues are color coded according to their average mitotic activity. All four tested wild-derived mtDNAs showed significant segregation bias across most tissues compared to B6N mtDNA (see also heatmap colours in raw-data, Tables S2-5). The BG haplotype showed segregation bias exclusively in tissues with high mitotic activity (red). The ST haplotype (M. m. musculus), relatively increased in all analysed tissues, with the exception of brain and heart. The LE haplotype showed a highly significant increase in spleen and liver, and the effects generally resembled the ST haplotype with lower segregation magnitudes.
The HB haplotype followed a different pattern, with proliferative advantages detected in blood, spleen, liver and lung. Importantly, a decrease of the HB mtDNA in heart and skeletal muscle also occurred.
Correlation between genetic distance and segregation strength
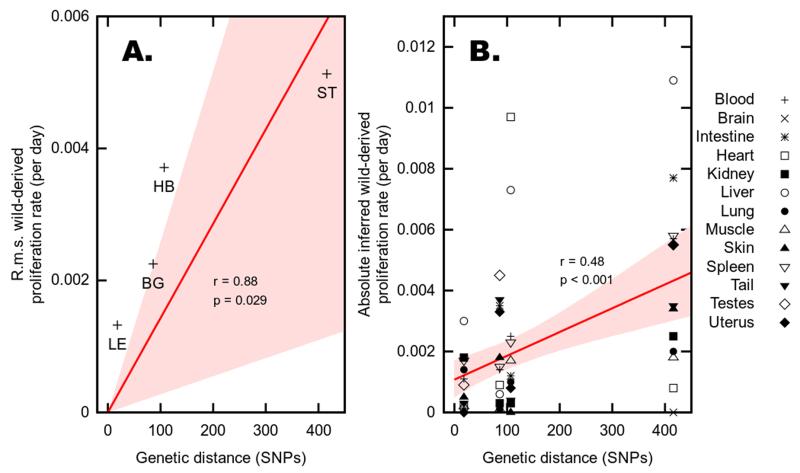
The mtDNA sequences of the four mice used for ooplasm transfer (LE, BG, HB, and ST) show 18, 86, 107 and 416 SNPs compared to the B6N mtDNA. We find a fit with significant statistical support (r = 0.88, p = 0.029) to a linear model in which root mean square proliferation rate across all tissues is directly proportional to genetic distance between haplotype pairs measured as number of SNPs (0.00133, 18 SNPs; 0.00225, 86 SNPs; 0.00372, 107 SNPs; 0.00513, 416 SNPs for LE, BG, HB and ST respectively, Figure 4A). We also found a highly significant correlation between the magnitudes of individual tissue-specific proliferation rates and the magnitude of mtDNA genetic difference in the mouse model in which each measurement was taken (r = 0.48, p < 0.001; Figure 4B). This result is intuitively visible in Figure 3, where, for example, the proliferation rates observed in ST (the most diverse haplotype pairing) are of much higher magnitudes than those in LE (the least diverse pairing) across a broad range of tissues. These results strongly support our key hypothesis: that segregation is more pronounced between mtDNA haplotypes with large genetic differences (Figure 4B). We now discuss the more fine-grained mechanisms by which segregation occurs.
Figure 4. Correlation between genetic distance and segregation.
The proliferation rate of wild derived haplotypes correlates with the genetic distance between haplotypes both as root-mean-square (r.m.s) over all tissues (A) and between tissues (B). (A) Increasing genetic distance leads to increase of the wild-derived mtDNA, measured over all tissues. Genetic distance between haplotypes (in number of SNPs) is shown against r.m.s. proliferation rates of wild-derived mtDNA across all tissues. Regression line and shaded region (red) show a linear model fit with zero intercept (as identical haplotypes experience no proliferative difference) and 95% confidence intervals. p-value is reported against the null hypothesis of zero gradient (i.e. a horizontal regression line). (B) Increasing genetic distance leads to increase of the wild-derived mtDNA, measured as individual tissues. Genetic distance between haplotypes (in number of SNPs) is shown against wild-derived proliferation rate in each tissue. Regression line and shaded region (red) show linear model fit and 95% confidence intervals; p-value is reported against the null hypothesis of no correlation (i.e. a horizontal regression line).
Haplotype-specific correlation of segregation with mtDNA turnover
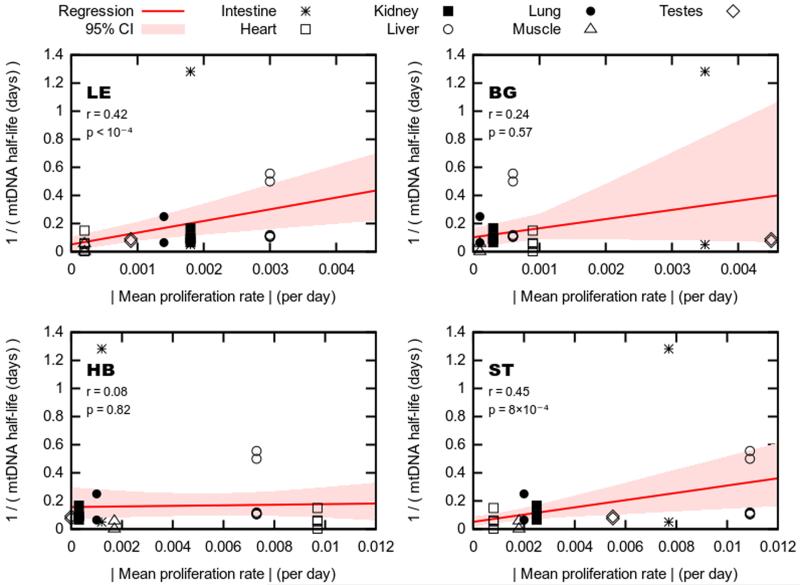
mtDNA replicates quasi-independently of the cell cycle in all tissues, at tissue-dependent rates. As the rate of mtDNA turnover may be expected to strongly influence the dynamics of haplotype segregation, we explored the connection between mtDNA turnover rates and segregation rate. The literature on mtDNA turnover rates is relatively sparse and spans several decades, with methods utilized and tissues examined differing among studies (Poovathingal et al., 2009). To explore this phenomenology as broadly as possible we amalgamated all relevant measurements into a single dataset and sought correlations using this broad set of values, reasoning that if such a correlation existed despite the high variance, it would likely reflect a true underlying trend. Two of the four tested haplotypes showed a highly significant correlation between the segregation rates of eight target tissues detected in this study and mtDNA turnover times reported across the literature (LE, ST; Figure 5).
Figure 5. Correlation between mtDNA turnover and mtDNA segregation rate.
Measurements on mtDNA half-life in different tissues amalgamated from across the literature (y-axis; for raw data see Table S10) against wild-derived proliferation rate in each tissue (x-axis), for each haplotype pair. Regression lines (red) show a linear model fit relating proliferation and turnover measurements; shaded regions (red) give 95% confidence intervals on this fit. p-values are reported against the null hypothesis of no relationship (i.e. corresponding to a horizontal line).
Two mtDNA haplotypes (ST, LE) show significant correlation between mtDNA turnover (half-life) and speed of segregation in the respective tissues. Only tissues for which mtDNA turnover data was available in the literature were included in the analysis.
See also Table S 10.
Four different segregation regimes depending on developmental stage
In most tissues analysed in this study a single segregation rate was sufficient to explain the data, i.e. heteroplasmy changed constantly over time at the same rate between birth and senescence at about two years (Figures 6B, F; S1 A-C).
In contrast to this continuous segregation behaviour, in several tissue/ mtDNA combinations the data is not adequately explained by this simple model: instead, there is significantly greater support for a model in which segregation occurs at an increased rate during early development, before slowing to a lower rate thereafter. The time of change is defined as crossover time (Figure 6C, D, E, G, H, likelihood ratio test, p < 0.01).
Two regimes are discernible, defined by the crossover time:
(i) Early segregation change: the wild-mouse derived mtDNA increased or decreased relative to B6N mtDNA at a constant fast rate, which changed to a slower constant rate during early adulthood. This regime was only observed in the HB haplotype. In HB liver the HB mtDNA increased at a fast rate until postnatal day (P) 51 (± 26), slowing to a lower rate after this crossover-time. In HB heart, the HB mtDNA decreased at a fast rate until P29 (±12), again slowing to a lower rate thereafter (Figure 6C, G; note that in Figure 6 times are shown from conception rather than from birth).
(ii) Late segregation change: the wild-mouse derived mtDNA increased or decreased relative to B6N mtDNA at a constant rate until later in life (around 8 months), and changed to a slower constant rate (or stops) thereafter. This regime could be seen in HB muscle and ST intestine and liver with crossover-times at days P206 (±57), P268 (±40) and P231 (±42) respectively (Figure 6D, E, H). Our mathematical model excludes the possibility that this observation is simply due to saturation of heteroplasmy at 0% or 100%; moreover, in tissues where this effect is seen, few samples approach 99% (e.g. ST liver), while in other tissues one haplotype increases at a constant rate close to 99% (e.g. ST blood; Table S5).
Prenatal segregation
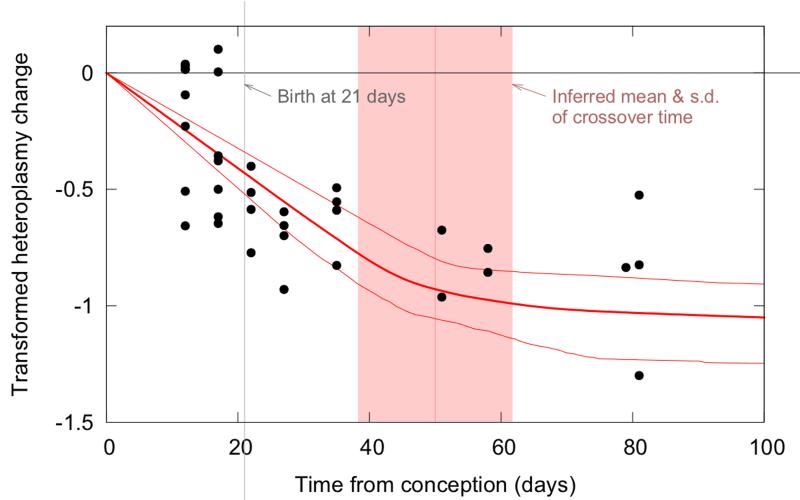
New-born and juvenile HB mice already showed very low levels of HB mtDNA in heart compared to other tissues (Table S4). To find out whether the above described decrease of HB mtDNA in heart starts prenatally, 15 fetuses were additionally sampled (see Table S4, heart tissue compared to other tissues). The proliferation rate in HB heart over the prenatal period was nonzero with high significance (p < 10−4), showing that that tissue-specific segregation bias occurred between conception and birth (Figure 7).
Figure 7. Tissue-specific mtDNA segregation starts prenatally in HB heart.
New-born and juvenile HB mice already show very low levels of HB mtDNA in heart compared to other tissues. Additional analysis of 15 fetuses (left of the grey vertical line indicating birth at 21 days after conception) confirms that reduction of HB mtDNA (and increase of C57BL/6N (B6N) mtDNA) starts already prenatally.
Black dots are experimental data of single mice; dark red lines show mean and 95% confidence intervals of inferred segregation trajectories. Vertical light pink lines show mean and standard deviation of inferred crossover time between two different segregation speeds. The y-axis shows transformed heteroplasmy change, giving the proliferation rate of HB mtDNA. Positive values indicate relative increase of HB mtDNA, negative values relative increase of the B6N mtDNA.
Discussion
To date, the NZB-CIS mtDNA based heteroplasmic mouse models have been viewed as an exception to the paradigm that non-pathological mtDNAs segregate neutrally (exceptions are cross-subspecies models (Ferreira et al., 2010; Takeda et al., 2000)). A more systematic analysis of this phenomenon was hampered by the lack of mtDNA variation in laboratory mice.
We overcame this problem by creating and analysing four new heteroplasmic mouse models, using mtDNA from wild-derived mice. This approach produced a previously unavailable range of genetic diversity, with differences in mtDNA between 18 and 416 SNPs compared to the B6N mtDNA. To the best of our knowledge this makes our study the first to describe the segregation effects of several admixtures of physiological mtDNA haplotypes, covering a broad genetic spectrum.
Across the models tested, we find significant statistical support (r = 0.88, p = 0.029) for a model in which root mean square proliferation rate is directly proportional to genetic distance between haplotypes. We provide the first evidence that segregation bias is higher in genetically more distant haplotypes, also within the same subspecies. This key finding suggests that in genetically diverse populations, segregation is likely to be common and pronounced. As all four tested haplotypes show segregation, we conclude that non-neutral segregation seems to be the rule rather than the exception between distinct mtDNA haplotypes under artificial heteroplasmic conditions.
The segregation mechanisms that are arguably the most relevant for clinical and basic research are those located in post-mitotic tissues (e.g. brain, heart- and skeletal muscle), as they are most often affected by mitochondrial diseases. However, the mechanisms of mtDNA segregation in these tissues are still largely unknown. Animal models with a single population of variant mtDNA exist only for highly pathogenic mutants, such as large scale deletions (Sato et al., 2007). The NZB model is neutral in post-mitotic tissues (Jenuth et al., 1997; Sharpley et al., 2012). In another heteroplasmic model utilising mtDNA of two different subspecies (C57BL/6, M. m. domesticus; RR, M. m. molossinus) a general increase of RR mtDNA was found in several tissues, but only relative to brain and heart (Takeda et al., 2000). However, these authors did not publish a time course for heteroplasmy in brain and heart (Takeda et al., 2000). In contrast to these previous studies, we find tissue-specific segregation in HB heart and skeletal muscle (and ST muscle), finally proving that tissue specific segregation in post-mitotic tissues may occur, even between two closely related, non-pathological mtDNA haplotypes. This segregation reaches a maximum during development, when there is still cell division in muscle, and slows down after adulthood in heart or stops altogether in muscle. This could be based on the slow mtDNA turnover in post-mitotic tissues (Collins et al., 2003), that in consequence also slows down segregation.
To find out more about a possible correlation between the rate of cell/ mtDNA-turnover and mtDNA segregation, we correlated the segregation speeds in our models with published mtDNA and turnover rates of the respective tissues. In fact, two haplotypes, ST and LE, show strong correlation between mtDNA turnover and segregation strength. Another, BG, only shows segregation in organs that include self-renewing tissues. While a similar selective advantage in mitotic tissues is known for the human A3243G mtDNA mutation (associated with Mitochondrial encephalopathy, lactic acidosis, and strokelike episodes (Frederiksen et al., 2006)), a correlation between mtDNA turnover and haplotype segregation constitutes a novel finding, pointing to an independent segregation mechanism. This mechanism could explain part of the quantitatively varying tissue-specific segregation found both in pathological and non-pathological mtDNA (Frederiksen et al., 2006; Sharpley et al., 2012).
However, the developmental stages of an organism like growth-phase, adulthood and senescence, and the accompanying metabolic changes could potentially influence cell- and mtDNA turnover, and in consequence the segregation rate. Such long-time scale dynamics of mtDNA in different cell populations of the body are also a key issue in understanding- and genetic counselling- of heteroplasmic mtDNA diseases (Poulton et al., 2010; Rajasimha et al., 2008). Currently little is known about the dynamics of mtDNA heteroplasmy over time, and knowledge is mostly derived from the NZB mouse model (Battersby et al., 2003) and various clinical studies (Rajasimha et al., 2008). In the NZB mouse model, the rate of selection is currently held to be independent of initial heteroplasmy, and constant with time (Battersby et al., 2003). The human A3243G mtDNA mutation is lost continually from the blood of patients (Rahman et al., 2001; Rajasimha et al., 2008). To find out whether mtDNA dynamics changes during organismal development or stays constant at all times we applied an appropriate population genetic model for mtDNA segregation on our data. This model allows us to discern finer-level details regarding the evolution of mtDNA populations. Besides neutral segregation, where both mtDNA types do not show any significant changes in their distribution over time, constant segregation bias as published (Frederiksen et al., 2006; Jenuth et al., 1997) was indeed the most abundant segregation regime: however, two new segregation regimes were found, each showing separate segregation speeds that change at certain cross-over times.
In the first, segregation bias starts high and slows down after two months of age, with the crossover-time closely matching the change between juvenile organ-development and adulthood. For example, cell numbers in the liver increase until postnatal day (P) 30, reaching a plateau at P60 (Epstein, 1967; Sakata et al., 1996), and heart muscle cells reach their final number at P4, but increase rapidly in volume until P14, again reaching a plateau around day P60 (Leu et al., 2001). These developmental changes correspond with the cross-over times between fast and slow mtDNA segregation with P51 (± 26) and P29 (±12) for liver and heart, respectively.
The second novel segregation regime is similar to the previous one, but the change between fast and slow segregation happens as late as around eight months of age, with no apparent influence of the growing phase. A change so late in life could be based on various age-related metabolic changes. For example, in liver mtDNA copy number per nuclear genome decreases constantly over time starting between 2-5 months (Masuyama et al., 2005).
These segregation regimes are based on separated mechanisms that are not necessarily tissue-specific: all four segregation regimes could be observed in liver, with each model showing a different segregation regime. While constant segregation seems to be the main segregation regime, the two new segregation kinetics of our models should be taken into account when long-term dynamics of novel heteroplasmic populations (e.g.in patients) have to be evaluated.
Interestingly, the above mentioned segregation regimes already start prenatally. In a recently created mouse model utilising a pathogenic tRNA mutation (one of a total of 35 mtDNA mutations), purifying selection during gestation was proposed as explanation for differences in heteroplasmy between mother and offspring (Freyer et al., 2012). This could not be shown directly, but was deduced from differing heteroplasmy levels between mutant levels in the mothers and offspring. In contrast, we now found in one mouse model (HB heart) a strong tissue-specific prenatal segregation bias. Moreover, we directly show this segregation bias in fetal tissues. This not only finally proves that prenatal segregation can occur at a measurable rate even in the short gestation of the mouse, but importantly does so in a tissue-specific way.
In this study we compare four mtDNAs of wild-derived mice against a standardized background of C57BL/6N mtDNA. While the mtDNA derived from wild-derived mice is presumably shaped by natural selection, mtDNA in laboratory mouse strains is likely to accumulate slightly deleterious mutations due to relaxed selection (Yonezawa and Hasegawa, 2014). Can the segregation bias towards wild-derived strains be due to the re-introduction of fitter wild-type mitochondria, which then receive a proliferative advantage? Several pieces of evidence suggest this is not the case. First, if this bias is due to the replacement of less fit mtDNA of the laboratory strain by fitter wild-type mtDNAs, we would see unidirectional bias in favour of wild haplotypes. However, in the energy-demanding tissues of heart and skeletal muscle of the HB strain the laboratory mouse mtDNA increases. Second, the proliferation advantage was not uniform in the models, with varying effects and segregation speeds between models or tissues, arguing against a simple basic mechanism. Third, also natural mouse populations were found to harbour comparable slightly deleterious mutations as do laboratory mice, with a higher incidence in intra- than inter- species comparisons, putatively due to longer operating natural selection in the latter (Nachman et al., 1994). Therefore the effects observed in this study must be intrinsic to the various wild-derived mtDNAs and the results suggest the differences are linked to partitioning of genetic variation. Which mutations are responsible for these effects and their underlying genesis has to be the analysed in further studies.
While the focus of this study is the segregation patterns of heteroplasmic mice, the results may help to evaluate a currently intensively discussed risk in human assisted reproductive medicine. To help female carriers of mitochondrial disease to conceive healthy children, the nuclear genome of a parent ‘donor’ oocyte may be transferred to a ‘recipient’ oocyte with no nucleus and healthy mtDNA (karyoplast transfer). Current implementations of this protocol lead to an inevitable low-level heteroplasmy in the resulting embryo (St John and Campbell, 2010; Wallace and Chalkia, 2013). While this low-level heteroplasmy is insufficient to cause disease, if the donor mtDNA experiences a proliferative advantage due to haplotypic differences between ‘donor’ and ‘recipient’ mtDNA as in the mouse models, the disease could later become manifest despite the treatment, as the initially low-level donor mtDNA comes to dominate the cellular population. Our study does not exactly model the transfer process itself as implemented in human systems. However, it does provide an adequate representation both of the post-transfer dynamics of a mixed mtDNA population, and model the mtDNA dynamics that may be expected in subsequent heteroplasmic generations. We show in these cases that the aforementioned (potentially pathological) proliferative advantage of donor mtDNA cannot be discounted in pairings from a diverse population. That mtDNA segregation of presumably non-pathological mtDNA is generally possible in humans was demonstrated by the detection of tissue-specific positive selection for recurrent mutations in unrelated individuals (Samuels et al., 2013).
We therefore suggest that a careful choice, or exact matching, of mtDNA haplotypes between donor and recipient (discounting the pathological mutation) is required to guarantee the benefits of karyoplast transfer (see Supplemental discussion for details).
To conclude, we have shown using newly constructed wild derived mouse models that mtDNA segregation bias between two haplotypes is common and strongly depends on the genetic details of those haplotypes. MtDNA segregation occurs at all developmental stages from prenatal up to old age. We considerably expand previous studies by showing that segregation occurs not only in specific tissues, but in almost all tissues. Most importantly we find segregation in post mitotic tissues. The underlying mechanisms are however more complicated than expected. Nevertheless, they can be grouped into four distinct segregation kinetics, depending on the respective tissue and haplotype. They can be further correlated to cell- and mtDNA turnover rates of the respective tissues.
These results represent the first systematic study on mtDNA segregation behaviour controlling the genetic distance between the haplotypes in question.
Experimental Procedures
Ethics Statement
The study was discussed and approved by the institutional ethics committee and performed in accordance with Austrian, Czech and European laws. FELASA recommendations for the health monitoring of SPF mice were followed. Animal experiment licenses: BMWF 68.205/0215-C/GT/2007, BMWF-68.205/0230-II/10b/2009, and 27/2007.
Wild derived mouse lines
Wild-derived Mus musculus mice were obtained from the Research Facility Studenec, Institute of Vertebrate Biology, Academy of Sciences of the Czech Republic, Brno, Czech Republic.
DNA extraction from tissues
DNA was extracted using the NucleoSpin® Tissue Kit (Macherey-Nagel, Germany) according to the protocol for animal tissue (no RNAse treatment). Details in Supplemental Experimental Procedures.
454 sequencing of the mitochondrial genome
mtDNA sequencing was performed on a Roche 454 2nd generation sequencing system; LE, BG, HB, ST mtDNA was additionally Sanger resequenced (LGC Genomics, Germany). Details in Supplemental Experimental Procedures.
Generation of heteroplasmic wild-derived mouse strains by ooplasm transfer
Approximately 1-5% cytoplasm (optically estimated) from Cytochalasin B treated wild mouse oocytes was aspirated and injected into B6 zygotes, surviving zygotes were transferred with additional CD1 zygotes into pseudopregnant CD1 surrogate mothers. Details in Supplemental Experimental Procedures. Four founder females on the nuclear and mtDNA background of B6N with mtDNA heteroplasmy of the respective wild-mouse mtDNA were obtained.
mtDNA heteroplasmy levels were measured by ARMS-qPCR: LE, 5%; HB, 7%; ST, 5%; BG, 5%
Heteroplasmy quantification by Amplification Refractory Mutation System (ARMS)-qPCR
Heteroplasmy quantification was performed by ARMS-qPCR, an established method in the field. The study was conducted according to MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines (Bustin et al., 2009).
The proportion of wild-mouse derived and B6N mtDNA was determined by specific ARMS-qPCR assays based on a SNP in mt-rnr2. These assays were normalised to changes in the input mtDNA amount by a consensus assay, located in a conserved region of mt-Co2. Details in Supplemental Experimental Procedures.
Mathematical analysis
Our model describes the time evolution of heteroplasmy (defined as proportion of wild-derived mtDNA) in a given tissue in a given mouse with a sigmoidal function , where α is the (unknown) initial heteroplasmy of the mouse, β is a tissue-specific proliferation rate, and ε is Gaussian noise with variance σ2. This model is derived from considering the population fraction of two ‘species’ proliferating at different rates and is shown to be consistent with population genetic analyses and previous treatments of heteroplasmy change (see Supplemental Information). We use the data from all tissues and all mice for a given haplotype to jointly infer values of α (initial heteroplasmies for each mouse), β (proliferation rates for each tissue) and σ (noise for a haplotype) with a maximum likelihood approach, giving the most likely combination of initial mouse heteroplasmies and tissue-specific proliferation rates. We derive confidence intervals using bootstrapping with the percentile method. This model is used to infer proliferation rates, to compare models with one and two segregation rates per tissue (using the likelihood ratio test) and to analyze inter-generation segregation. For further details see Supplemental Information. Details in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
MtDNA segregation is the rule, not exception, in pairings from diverse populations
Segregation occurs prenatally, and within disease-relevant post-mitotic tissues
More diverse mtDNA pairs exhibit stronger segregation
Mathematical analysis shows modulation of segregation kinetics through development
Acknowledgements
Technical assistance of Sabine Kreidl, Silvana Klammer and Martin Hofer is gratefully acknowledged. We thank Graham Tebb for critical reading of the manuscript. This work was funded by MOEL grants no. 297 and 395 to J.P.B. (Österreichische Forschungsgemeinschaft), a start-up grant of the University of Veterinary Medicine Vienna to J.P.B., T.K. and R.S. and by a Czech Science Foundation grant to J. Piálek (206/08/0640).
Footnotes
References
- Acton BM, Lai I, Shang X, Jurisicova A, Casper RF. Neutral mitochondrial heteroplasmy alters physiological function in mice. Biol Reprod. 2007;77:569–576. doi: 10.1095/biolreprod.107.060806. [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Loredo-Osti JC, Shoubridge EA. Nuclear genetic control of mitochondrial DNA segregation. Nat Genet. 2003;33:183–186. doi: 10.1038/ng1073. [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Shoubridge EA. Selection of a mtDNA sequence variant in hepatocytes of heteroplasmic mice is not due to differences in respiratory chain function or efficiency of replication. Hum Mol Genet. 2001;10:2469–2479. doi: 10.1093/hmg/10.22.2469. [DOI] [PubMed] [Google Scholar]
- Battersby BJ, Shoubridge EA. Reactive oxygen species and the segregation of mtDNA sequence variants. Nat Genet. 2007;39:571–572. doi: 10.1038/ng0507-571. author reply 572. [DOI] [PubMed] [Google Scholar]
- Blanco R, Mayordomo E, Montoya J, Ruiz-Pesini E. Rebooting the human mitochondrial phylogeny: an automated and scalable methodology with expert knowledge. BMC Bioinformatics. 2011;12:174. doi: 10.1186/1471-2105-12-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Collins ML, Eng S, Hoh R, Hellerstein MK. Measurement of mitochondrial DNA synthesis in vivo using a stable isotope-mass spectrometric technique. J Appl Physiol. 2003;94:2203–2211. doi: 10.1152/japplphysiol.00691.2002. [DOI] [PubMed] [Google Scholar]
- Craven L, Elson JL, Irving L, Tuppen HA, Lister LM, Greggains GD, Byerley S, Murdoch AP, Herbert M, Turnbull D. Mitochondrial DNA disease: new options for prevention. Hum Mol Genet. 2011;20:R168–174. doi: 10.1093/hmg/ddr373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CJ. Cell size, nuclear content, and the development of polyploidy in the Mammalian liver. Proc Natl Acad Sci U S A. 1967;57:327–334. doi: 10.1073/pnas.57.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrar GJ, Chadderton N, Kenna PF, Millington-Ward S. Mitochondrial disorders: aetiologies, models systems, and candidate therapies. Trends Genet. 2013;29:488–497. doi: 10.1016/j.tig.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Ferreira CR, Burgstaller JP, Perecin F, Garcia JM, Chiaratti MR, Meo SC, Muller M, Smith LC, Meirelles FV, Steinborn R. Pronounced segregation of donor mitochondria introduced by bovine ooplasmic transfer to the female germ-line. Biol Reprod. 2010;82:563–571. doi: 10.1095/biolreprod.109.080564. [DOI] [PubMed] [Google Scholar]
- Frederiksen AL, Andersen PH, Kyvik KO, Jeppesen TD, Vissing J, Schwartz M. Tissue specific distribution of the 3243A->G mtDNA mutation. J Med Genet. 2006;43:671–677. doi: 10.1136/jmg.2005.039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyer C, Cree LM, Mourier A, Stewart JB, Koolmeister C, Milenkovic D, Wai T, Floros VI, Hagstrom E, Chatzidaki EE, et al. Variation in germline mtDNA heteroplasmy is determined prenatally but modified during subsequent transmission. Nat Genet. 2012;44:1282–1285. doi: 10.1038/ng.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Malaspinas AS, Krause J, Briggs AW, Johnson PL, Uhler C, Meyer M, Good JM, Maricic T, Stenzel U, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EC. Regulators weigh benefits of ‘three-parent’ fertilization. Nature. 2013;502:284–285. doi: 10.1038/502284a. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA. Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet. 1996;14:146–151. doi: 10.1038/ng1096-146. [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16:93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- Jokinen R, Marttinen P, Sandell HK, Manninen T, Teerenhovi H, Wai T, Teoli D, Loredo-Osti JC, Shoubridge EA, Battersby BJ. Gimap3 regulates tissue-specific mitochondrial DNA segregation. PLoS Genet. 2010;6:e1001161. doi: 10.1371/journal.pgen.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu M, Ehler E, Perriard JC. Characterisation of postnatal growth of the murine heart. Anat Embryol (Berl) 2001;204:217–224. doi: 10.1007/s004290100206. [DOI] [PubMed] [Google Scholar]
- Lippold S, Xu H, Ko A, Li M, Renaud G, Butthof A, Schroeder R, Stoneking M. Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences. bioRxiv. 2014 doi: 10.1186/2041-2223-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama M, Iida R, Takatsuka H, Yasuda T, Matsuki T. Quantitative change in mitochondrial DNA content in various mouse tissues during aging. Biochim Biophys Acta. 2005;1723:302–308. doi: 10.1016/j.bbagen.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Meirelles FV, Smith LC. Mitochondrial genotype segregation in a mouse heteroplasmic lineage produced by embryonic karyoplast transplantation. Genetics. 1997;145:445–451. doi: 10.1093/genetics/145.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Loshuertos R, Acin-Perez R, Fernandez-Silva P, Movilla N, Perez-Martos A, Rodriguez de Cordoba S, Gallardo ME, Enriquez JA. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- Mueller EE, Brunner SM, Mayr JA, Stanger O, Sperl W, Kofler B. Functional differences between mitochondrial haplogroup T and haplogroup H in HEK293 cybrid cells. PLoS One. 2012;7:e52367. doi: 10.1371/journal.pone.0052367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Boyer SN, Aquadro CF. Nonneutral evolution at the mitochondrial NADH dehydrogenase subunit 3 gene in mice. Proc Natl Acad Sci U S A. 1994;91:6364–6368. doi: 10.1073/pnas.91.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne BA, Wilson IJ, Yu-Wai-Man P, Coxhead J, Deehan D, Horvath R, Taylor RW, Samuels DC, Santibanez-Koref M, Chinnery PF. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22:384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poovathingal SK, Gruber J, Halliwell B, Gunawan R. Stochastic drift in mitochondrial DNA point mutations: a novel perspective ex silico. PLoS Comput Biol. 2009;5:e1000572. doi: 10.1371/journal.pcbi.1000572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J, Chiaratti MR, Meirelles FV, Kennedy S, Wells D, Holt IJ. Transmission of mitochondrial DNA diseases and ways to prevent them. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Poulton J, Marchington D, Suomalainen A. Decrease of 3243 A-->G mtDNA mutation from blood in MELAS syndrome: a longitudinal study. Am J Hum Genet. 2001;68:238–240. doi: 10.1086/316930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasimha HK, Chinnery PF, Samuels DC. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A-->G mutation in blood. Am J Hum Genet. 2008;82:333–343. doi: 10.1016/j.ajhg.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt K, Dowling DK, Morrow EH. Medicine. Mitochondrial replacement, evolution, and the clinic. Science. 2013;341:1345–1346. doi: 10.1126/science.1237146. [DOI] [PubMed] [Google Scholar]
- Sakata H, Takayama H, Sharp R, Rubin JS, Merlino G, LaRochelle WJ. Hepatocyte growth factor/scatter factor overexpression induces growth, abnormal development, and tumor formation in transgenic mouse livers. Cell Growth Differ. 1996;7:1513–1523. [PubMed] [Google Scholar]
- Samuels DC, Li C, Li B, Song Z, Torstenson E, Boyd Clay H, Rokas A, Thornton-Wells TA, Moore JH, Hughes TM, et al. Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 2013;9:e1003929. doi: 10.1371/journal.pgen.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Nakada K, Shitara H, Kasahara A, Yonekawa H, Hayashi J. Deletion-mutant mtDNA increases in somatic tissues but decreases in female germ cells with age. Genetics. 2007;177:2031–2037. doi: 10.1534/genetics.107.081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, Crimi M, Waymire K, Lin CS, Masubuchi S, Friend N, Koike M, et al. Heteroplasmy of Mouse mtDNA Is Genetically Unstable and Results in Altered Behavior and Cognition. Cell. 2012;151:333–343. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JC. Ooplasm donation in humans: the need to investigate the transmission of mitochondrial DNA following cytoplasmic transfer. Hum Reprod. 2002;17:1954–1958. doi: 10.1093/humrep/17.8.1954. [DOI] [PubMed] [Google Scholar]
- St John JC, Campbell KH. The battle to prevent the transmission of mitochondrial DNA disease: is karyoplast transfer the answer? Gene Ther. 2010;17:147–149. doi: 10.1038/gt.2009.164. [DOI] [PubMed] [Google Scholar]
- St John JC, Facucho-Oliveira J, Jiang Y, Kelly R, Salah R. Mitochondrial DNA transmission, replication and inheritance: a journey from the gamete through the embryo and into offspring and embryonic stem cells. Hum Reprod Update. 2010;16:488–509. doi: 10.1093/humupd/dmq002. [DOI] [PubMed] [Google Scholar]
- Takeda K, Takahashi S, Onishi A, Hanada H, Imai H. Replicative advantage and tissue-specific segregation of RR mitochondrial DNA between C57BL/6 and RR heteroplasmic mice. Genetics. 2000;155:777–783. doi: 10.1093/genetics/155.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Med. 2013;3:a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa T, Hasegawa M. Extreme nearly neutral evolution in mitochondrial genomes of laboratory mouse strains. Gene. 2014;534:444–448. doi: 10.1016/j.gene.2013.08.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.