Abstract
DNA-based taxonomic and functional profiling is widely used for the characterization of organismal communities across a rapidly increasing array of research areas that include the role of microbiomes in health and disease, biomonitoring, and estimation of both microbial and metazoan species richness. Two principal approaches are currently used to assign taxonomy to DNA sequences: DNA metabarcoding and metagenomics. When initially developed, each of these approaches mandated their own particular methods for data analysis; however, with the development of high-throughput sequencing (HTS) techniques they have begun to share many aspects in data set generation and processing. In this review we aim to define the current characteristics, goals and boundaries of each field, and describe the different software used for their analysis. We argue that an appreciation of the potential and limitations of each method can help underscore the improvements required by each field so as to better exploit the richness of current HTS-based data sets.
Keywords: DNA metabarcoding, environment, genome, metagenomics, software development
Introduction
The wide range of ‘–omics’ data sets that can now be generated, thanks to rapid developments in high-throughput sequencing (HTS) technologies, have had a major impact on a particular pair of fields that work at the ‘meta’ scale—metagenomics and DNA metabarcoding. As indicated by the Greek preposition ‘meta’, the aim of these disciplines is to move beyond the identification of single species to the identification of the total biological entities within a complex sample. In this regard, a wide range of studies has attempted to biologically characterize particular environments through extraction and sequencing of DNA taken from subsamples of the environment of interest. In brief, metagenomics could be defined as the characterization of the vast number of genomes present in an environmental sample, using both a taxonomical and a functional analytical approach. DNA metabarcoding, on the other hand, principally focuses on taxonomically describing the species present within a sample.
Given the increasing ease and reduced costs with which HTS data can be generated, these environments represent virtually any space from which a sample can be obtained, including Antarctic lakes [1, 2], hot springs [3–5], the human gut [6–10] or indeed any other part of the body [11] of any species [12–16]. Regardless of their origin, a fundamental characteristic of these samples is the complexity of the microbial communities that inhabit them and the difficulty this complexity poses for any downstream analyses [17]. Current interests focus around two kinds of analyses: (1) who is there?—the taxonomic identification of all the species present (bacteria, fungi, viruses, protozoa, mammals and plants) and (2) what are they doing?—the identification of the biological functions that those species present undertake within those particular environmental characteristics (e.g. high/low pH, extreme temperatures, salinity gradients, humidity, pressure, oxygen abundance, etc.) [18–21]. The information yielded from these two avenues can subsequently empower detailed comparisons of different microbial communities [22].
At the dawn of the fields of DNA metabarcoding and metagenomics, the respective techniques used to fulfill their goals were clearly distinguishable. DNA metabarcoding aimed to identify which species were present in a DNA extract by targeting and sequencing nucleotide barcodes, which are DNA marker sequences that have been argued as providing a unique genetic identity for each taxon of study [23–25]. Barcodes were originally detected by observing sequence alignments and locating a pair of conserved regions flanking a variable one. The most commonly used markers are 16S for bacteria, mt16S for mammals, CO1 for insects, ITS1 for fungi and rbcL, trnl and matK for plants, although non-conventional markers can also be used [24, 26, 27]. Metagenomics, on the other hand, is based around direct shotgun sequencing of DNA within an extract, and thus required the implementation of HTS technologies [28, 29] for its power to be fully exploited. A fundamental difference with DNA metabarcoding, is that the data generated using metagenomics provides additional genomic-scale information, thus enabling not only taxonomic identification, but also functional characterization of the environment [30] (Figure 1).
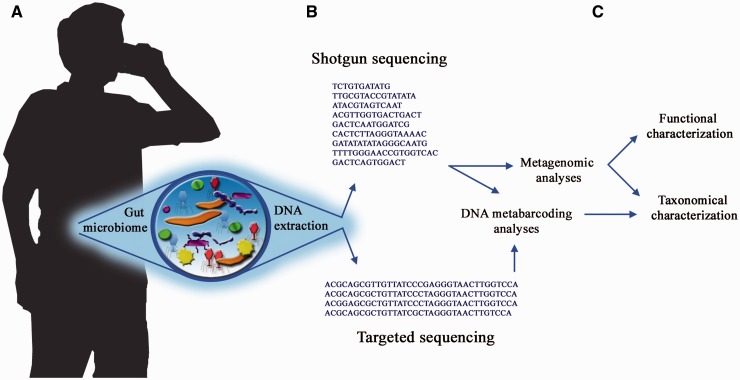
Figure 1.
Environmental sample analysis framework. (A) A sample can come from any environment that contains DNA; e.g. one of the most studied environments to date is the human gut microbiome. (B) DNA is extracted from the sample and sequenced according to the intended analyses. Shotgun sequencing produces genomic reads from the species present in the sample, while targeted sequencing produces amplicons with the aim of identifying a specific group of organisms. (C) Depending on the initial aim, whether functional and taxonomic characterization or only taxonomic characterization, the appropriate data set needs to be generated to be analyzed with the appropriate software.
Although initially distinct, recent sequencing technological developments have rapidly diminished the difference between the fields, principally because DNA metabarcoding now produces data sets with the same HTS techniques used for metagenomics [29, 31–33]. While conferring many benefits, a side effect has been a degree of confusion emerging within the research community, in particular through the labeling of some metabarcoding studies with the terms ‘metagenomics’ or ‘targeted metagenomics’ [34–39], simply due to the fact that HTS platforms are used to either decrease the cost of amplicon sequencing [40, 41] or generate barcode markers in a polymerase chain reaction (PCR)-free manner [32, 42–45]. The reason why this labeling is incorrect is due to the fact that the focus of the resulting analyses remains on barcode loci, and not on the genome as a whole.
In this regard, a distinction should be made between ‘metagenomics’ and ‘DNA metabarcoding’ as research fields, and ‘metagenomic sequencing’ as a laboratory technique. Strictly speaking, shotgun HTS of an environmental sample is the sequencing of the metagenome of the sample, regardless of the research field and the computational approach used to analyze the data set. For this reason we prefer to name metabarcoding studies that use metagenomics sequencing data as ‘PCR-free single/multiple loci metabarcoding’. Given this spreading confusion, and that we can expect an increasing number of researchers to favor PCR-free shotgun approaches [29], thanks to the continual reductions in price per base pair of HTS, we advocate that it is timely to re-examine the pros and cons of the different approaches, and re-state the specific goals of each kind of study (Table 1).
Table 1.
Methods comparison
| Type of study and aimed characterization | Metagenomics: taxonomic and functional | Metabarcoding: taxonomic | Metabarcoding: taxonomic | Metabarcoding: taxonomic | Metabarcoding: taxonomic |
|---|---|---|---|---|---|
| Laboratory method | Shotgun sequencing | Shotgun sequencing | Shotgun sequencing | PCR based | PCR based |
| Target region | Genome-wide | Multi-loci | Single locus [32, 33] | Customized barcodes | Conventional barcodes, including 16S, COI, etc. |
| DNA quantity | Care should be taken for samples coming from a body part of a macro organism so that the shotgun sequencing is not mostly host DNA | The percentage of marker genes in shotgun data sets is small [46, 47] | Only a small fraction of the reads come from a specific marker gene | Lots of customized targeted genes can be obtained | Lots of amplicons from universally targeted genes can be obtained |
| Reference database | Databases of the entire genomes can be customized | The source of the reads is largely unknown and difficult to characterize with the currently existing databases, thus many reads will not be assigned a taxonomy [48] | Single marker genes can be extracted from the data set using a reference database | There are good databases for standard barcodes, however if another region is targeted there are few and mostly not curated reported sequences. | There are several large 16S and COI databases, some of them are well curated, such as Greengenes |
| Laboratory bias | May present library build biases due to e.g. genomic nucleotide composition | May present library build biases | May present library build biases | May present primer bias if primers target wide taxonomic distributions | May present primer bias if using ‘universal’ primers for marker gene |
| Taxonomic resolution | The identification of multiple loci (marker or not) can even recover almost entire genomes of species | The phylogenies of more than one gene can provide a better consensus of the species present in the sample | It can provide good taxonomic resolution up to the species level. The taxonomic accuracy increases [33] | Sequences other than marker genes may not provide satisfactory taxonomic resolution because one sequence can be assigned to more than one species | The completeness of the well-characterized marker gene databases can provide good taxonomic resolution up to the species level |
| Cost | Deals with various challenges due to the complexity of the mixture of DNA in the sample | It may be unattainable due to the computational requirements | The ratio of used and discarded sequences that do not come from the single mined marker gene is cost inefficient | Low cost when generated on HTS platforms | Generally low cost—especially when generated on HTS platforms |
Note. Comparison of the advantages and disadvantages of various methods that are used to achieve the goals of the DNA metabarcoding and metagenomic fields.
Having discussed the differences of each laboratory method to study species diversity and biological functions in an environment, it is now important to describe the characteristics of the computational methods used to analyze the data sets produced by these different approaches. Through a reminder of the technical definitions of the goals and computational methods used in metagenomics and metabarcoding, and by stating the similarities and differences of the current approaches used to analyze data sets of environmental samples, it will be possible to attain a sound understanding of these two fields. In turn, this will enable oriented software development efforts that specifically target the key questions that researchers wish to ask. Furthermore, it will facilitate the development of studies that integrate the different types of data sets, while being aware of their differences, and exploiting the full information they can provide [49–52].
While the recently produced metagenomic data sets have provided a wealth of new insights into the intricacies of microbial communities, their descriptive power remains remarkably limited by the now widely acknowledged fact that culture-based descriptions of microbial diversity underestimate the true levels of biodiversity by orders of magnitude [53, 54]. Specifically, a major problem with many current metagenomics and metabarcoding studies is that their taxonomic identification processes rely heavily on the information available for previously described species. In light of this problem, other methods have been developed to characterize the diversity in a sample without reference data sets [55]. Both approaches offer different possibilities and limitations that need to be considered before undertaking analyses (Figure 2). We discuss these in the following text, with the intention of inspiring future analytical developments. Because taxonomic profiling is the only goal shared by metabarcoding and metagenomics, we principally focus on methods regarding this aspect, although functional characterization will also be superficially explored.
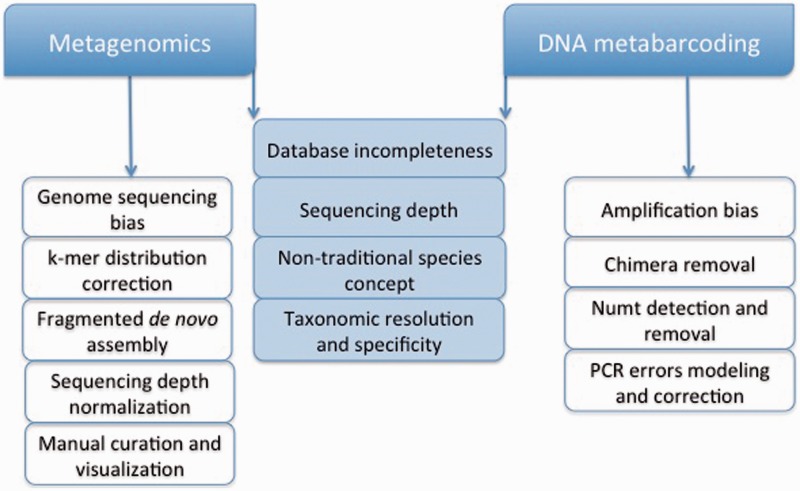
Figure 2.
Considerations and challenges for metagenomics and DNA metabarcoding. Both fields face a variety of challenges that are ideal candidates for future software development. While some of such problems are specific to one of the fields (right and left boxes), others are common to both (middle boxes).
Metabarcoding reference-based characterization
At the dawn of amplicon sequencing, reads were predominantly produced by Sanger sequencing and the data sets were small. However, data set sizes have increased by orders of magnitude, thanks to sequencing technology platforms such as the Roche GS, Ion Torrent and the Illumina series [56], each with particular commercial characteristics (e.g. cost and sequencing data yield) [57]. Thus, sequence similarity searches can today only be effectively handled through computational toolkits [58–60] that perform the necessary basic processing of the raw data. Basic processing steps in such toolkits include trimming, screening and aligning sequences against a database, clustering of sequences into operational taxonomic units (OTUs), and comparison of the sequence composition between different samples. The alignment of the reads to the reference database is probably the most important step of the analysis workflow. Different programs can be chosen for this task, such as UCLUST [61], CD-HIT [62] and BLAST [63]. After the alignment, instead of simply parsing a BLAST output, taxonomy is assigned using a predefined taxonomy map in which a reference sequence is related to the corresponding taxonomy (Figure 3A). Other methods such as obiclean from OBITools [59] and SUMATRA+SUMACLUST [64] also include steps to model and detect PCR sequencing errors to avoid incorrect taxonomic assignations by the use of clustering algorithms as UCLUST [61] and CD-HIT [62] and the sequence record counts.
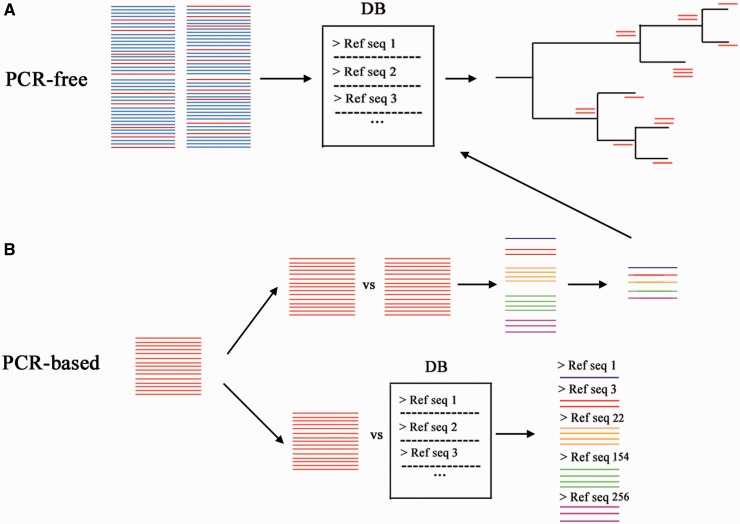
Figure 3.
Metabarcoding approaches. (A) Although PCR-free data sets are typically large, usually only a small percentage of the sequence reads map to a reference database. In such database, each entry has an assigned taxonomy so that phylogenetic placing approaches can be used for the taxonomic assignation. (B) PCR-based data sets consist of amplicon sequences that can be analyzed with the use of a reference database or without the need of it. If no database is used, the sequences are compared among themselves and are clustered by a similarity threshold; a representative sequence can be drawn from each cluster to then be compared with a reference database. On the other hand, if a database is used, the sequences are compared against the database and are assigned the taxonomy of the sequence they match under a given similarity threshold. A colour version of this figure is available online at BIB online: http://bib.oxfordjournals.org.
Based on the results of the reference database comparison, taxonomy assignation can be performed by alignment-based methods such as MEGAN [65] and MetaPhyler [66]. In this context, taxonomy is assigned against specific barcode loci databases, whether single loci such as 16S or CO1, or a set of a few phylogenetic marker loci drawn from across the genome. For example, a method called mOTU identifies what the authors call ‘metagenomics Operational Taxonomic Units’ [67], analogous to the molecular OTUs, by using 40 universal single copy phylogenetic marker genes. Some authors have referred to this approach of using multiple barcodes as metagenomics, due to the fact that the loci are drawn from the organism’s genome (nuclear plus organelle genes for eukaryotes) [66–70]. However, the total amount of sequence of the used loci is so small in comparison to the DNA content in an entire genome, that these databases cannot formally be considered genome-wide, especially for prokaryotes in which the exome is only a percentage of the genome.
Generally, alignment-based methods for shotgun data sets use an approach that can be broadly described as follows. First, the HTS reads are aligned to a backbone alignment [71, 72], subsequently each query sequence is placed into a backbone tree [73] using an extended alignment, and finally taxonomy is assigned to each read using a phylogenetic placement approach, such as the Lowest Common Ancestor (LCA) [74]. Phylogenetic placement approaches use a database and a reference tree associated to the database. LCA is one of the most commonly used algorithms in phylogenetic placement; it implements steps to address this specific issue of taxonomies coming from different database sources. In the LCA algorithm, if the read has a hit specifically to one taxon it is assigned to it, but if it has hits to different taxa it is placed higher up in the taxonomy, and reads that hit ubiquitously may even be assigned to the root node of the tree.
Considerations on metabarcoding reference-based methods
Having briefly outlined current reference-based metabarcoding methods, we turn to their pros and cons. A major attraction of amplicon data sets is that they are a relatively economic way to monitor diversity, thus enabling comparison of the taxonomic composition between various environmental communities. Although marker gene databases have expanded and included genes other than 16S rDNA [26, 75, 76], the most comprehensive are for 16S rDNA [77], and to some degree for CO1. If using a relatively error-free database such as BOLD [78], SILVA [79] or Greengenes [80], the taxonomic identification can be reliable, especially if using long reads such as those from Roche GS sequencing. In contrast to metagenomic analyses, comparison of taxonomic composition can be automated for many different metabarcoded samples with the use of software such as Unifrac [22].
Despite the benefits of PCR-based methods, they face a number of challenges. Firstly, they must account for PCR and sequencing derived sequence errors, ultimately risking overestimation of biodiversity within samples [81]. Secondly, although primers used are often referred to as ‘universal’ or ‘generic’ for predetermined clades, their performance is difficult to predict on samples composed of largely unknown species, thus amplification biases may occur [82]. Other major limitations of reference-based approaches are both that of reference database incompleteness, and that different results can be obtained according to the database size. This is an aspect that has not received much attention within the majority of the taxonomic profiling studies. However, some software has been developed to deal with this issue; for example TANGO [83]. Despite these problems, considerable efforts have been made to develop phylogenetic placement methods for taxonomy profiling in metabarcoding, and various programs with different statistical bases are available [84].
PCR-free multi-locus methods represent a valuable first step through which the metabarcoding community can exploit more of the information present in shotgun data sets than is otherwise used by the mining of a single gene. However, the fact that they still largely ignore the majority of the sequence data raises the obvious challenge of better exploitation of this extra information. In this regard, it would be interesting to use composition or counts approaches to provide extra information for a more refined taxonomic assignation or to provide supporting information to the identified taxonomies.
Another major challenge for the labeling of sequences with the traditional species concept is the identification of chimeric sequences. To this end, programs such as UCHIME [85] have been developed for chimeric amplicon identification, and this has already established itself as a de facto standard step. Furthermore, the presence of nuclear mitochondrial insertion (numt) sequences is a problem that should also be taken into account. As proven by Hojun Song et al. [86], DNA metabarcoding can overestimate the number of species when nuclear mitochondrial pseudogenes are co-amplified. Several steps are suggested to deal with numts, such as BLAST search, translation of the sequences to look for indels and stop codons, comparison of the marker gene to closely related published mitochondrial genomes and examination of nucleotide usage. However, these suggestions are not straightforward to implement, and no metabarcoding toolkit has yet a program for identification of numts. On a separate matter, although it is clear that data sets of barcode amplicons do not provide functional information, it is interesting to note the development of programs such as PICRUSt [87], which predicts the functional composition of a metagenome using marker gene data and a database of reference genomes.
Metabarcoding reference-free characterization
In classical sequence characterization approaches, where a label name is assigned to sequences, and the level of attained taxonomic resolution is the most important aspect to consider [88], reference databases are the cornerstone of the analyses. However, given that the overwhelming majority of microbial diversity remains to be characterized at the genetic level [53, 54], the concept of a molecular OTU has been applied for enabling improved descriptions of the taxonomic diversity present within a sample. In this reference-free approach, reads are first clustered by a similarity threshold and a representative sequence is obtained from each cluster (Figure 3B). These clusters are not assigned a taxonomic label, but sequences within the same cluster are expected to come from the same species. Because the methodological basis of this approach is the same as that used by some reference-based programs, most of such programs offer a reference-free mode. An example of such programs is a recently developed method called UPARSE [89]. The representative sequences of the clusters resulting from the reference-free modes can be used to assess the microbial diversity of the sample or to serve as input for other reference-based methods for their taxonomy assignment.
Considerations on metabarcoding reference-free methods
Metabarcoding was born when the identification of known species was enough to characterize an environment, and it is still the best option for studies such as biodiversity monitoring [90–93] of microorganisms, as well as macro organisms like mammals and plants. In particular, the monitoring of macro organisms [94, 95] can be benefited from improvements on both reference-free and reference-based methods because the currently used generic markers for their identification are often unable to provide high taxonomic resolution [96–98]. Reference-free methods possess the clear advantage of not needing any reference database for the taxonomy assignment. However, the taxonomic assignation without the use of a database in metabarcoding also poses the challenge of the molecular OTU concept not being yet widely accepted by the community, because so far OTUs without a taxonomic classification can be only used for environment richness comparison.
A major challenge for determining the microbial species present in a sample without the need of a reference database is to use algorithms other than those also used by the methods that depend on a reference database. There is a yet underexplored alignment-free metabarcoding approach that works under a completely different methodological basis—the compression-based approach. This approach implements methods such as Universal Similarity Metric (USM) [99], an approximation of USM called Normalized Compression Distance [100] and Information-Based Distance [101] that can produce phylogenetic trees with good accuracy [99]. These kinds of analyses represent an interesting parameter and reference database free means of clustering sequences that should be further explored.
Metagenomic reference-based characterization
BLAST [63] is perhaps the most basic and widely employed method for identifying the best hit of the shotgun data set reads against databases containing taxonomically identified reference sequences. Once the BLAST output is generated, subsequent taxonomic assignation is performed using different strategies, depending on the software. BLAST is implemented in a variety of methods [65, 102, 103] that are able to undertake taxonomic and functional identification, as well as perform comparative analyses of different samples in a straightforward and interactive manner that can be clearly visualized. For example, MEGAN [65] applies the LCA algorithm on a BLAST output for taxonomic assignation. Although this approach sits on the fuzzy line that separates metabarcoding and metagenomics, when it uses reference databases consisting of complete genomic sequences and uses all the reads for comparison instead of initially fishing for marker barcodes it can be classified as a metagenomic method, otherwise it is classified as metabarcoding (Figure 4A). Shotgun Unifrac [104] as implemented in Qiime [58] is an alternate phylogenetic placement method that has proven useful for taxon identification for entities like viruses through the use of a reference database of full genomes.
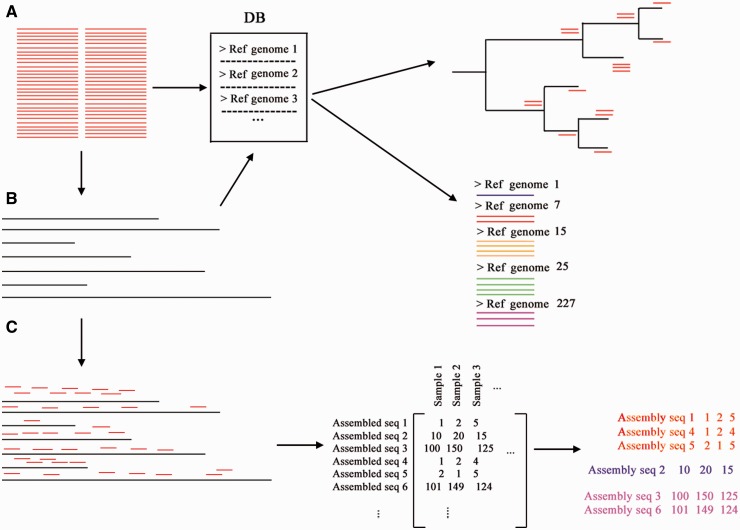
Figure 4.
Metagenomic approaches. (A) Metagenomic reference-based approaches start by mapping the reads to a genome database and then apply various algorithms to assign taxonomy, such as phylogenetic placement, or the use of unique mapping reads to the genome of a species in the database. (B) Alternatively, the reads can be de novo assembled and the scaffolds, or the open reading frames predicted on the scaffolds, can be searched against the database, thus reducing the search time. (C) Metagenomic reference-free methods usually start by de novo assembling the reads, then the number of reads mapping back to the assembled sequences (the scaffolds or the open reading frames predicted from the scaffolds) can be used to create a count matrix that can be further clustered, with each cluster representing a metagenomic species. A colour version of this figure is available online at BIB online: http://bib.oxfordjournals.org.
‘Composition-based’ approaches are an alternate strategy that exploits nucleotide usage information extracted from the reads to detect which taxonomic entities are present in the sample. Nucleotide usage is an interesting piece of information that can only be exploited if used in a metagenomic approach, because it requires information from many complete genomes. In general, composition-based methods can be considered as sitting on the interface between reference-free and reference-based methods, because they use statistical approaches such as Markov models [105], support vector machines [106], non-negative least squares [107] or mixture modeling [108]. Here however, we consider them as reference-based, simply because the database is the suite of Markov models or the required training sequence set. The methodological basis of composition-based methods can be generally explained as follows. First, models such as interpolated Markov models are generated to characterize variable-length oligonucleotides typical of a phylogenetic grouping. The models can be generated, for example, by training on chromosomes and plasmids from organisms collected from a database such as NCBI RefSeq [109]. Subsequently the model gives a score reflecting the probability of a query sequence to belong to the class of sequences on which the model was trained. There are also hybrid methods for taxonomic classification that combine the result of alignment-based and composition-based approaches in a complementary way.
Other metagenomic methods also perform taxonomic assignation based on DNA sequences from nuclear as well as mitochondrial genomes by first performing a de novo assembly and then comparing the assembled sequences to a genome-wide genes database [110–112] (Figure 4B). These methods should be considered metagenomic methods because they apply genomic algorithms such as de novo assembly, and the database can have many sequences from coding DNA sequences (CDS), markers or not, as well as non-CDS, or can consist of entire genomes, compared with those used by metabarcoding alignment-based methods using short reads as input, which are restricted to few marker genes. If the de novo assembly derives from high depth data sets, assembling them can produce almost complete genomes of high quality even from rare species [113, 114]. Other methods such as CARMA3 [115] use hidden Markov models with the Pfam database [116] to match the short reads to protein domains. The approach also uses a small percentage of the total data set, but it is to be considered metagenomic because it uses CDS from every reported gene instead of limiting itself to marker genes only. Furthermore this classification allows for functional characterization.
Functional characterization is usually performed by alignment of the sequences to already annotated proteins to find their homologous sequences [62, 117–119]. This relies on the assumption that sequence homology suggests shared function [120, 121], and it is also considered that there are different levels of functional similarity, such as pathways or protein families [122–124]. Function-oriented databases such as COG [125], Pfam [116] and TIGRFAM [126] are meant to be used for gene-level analyses, while others such as KEGG [127], MetaCyc [128] and SEED [129] are used for analyses at system or pathway level.
Considerations on metagenomics reference-based methods
Perhaps the most significant advantage of metagenomics methods that exploit reference databases is that (depending on the completeness of their reference database) they can provide reliable species identification. Furthermore, improvements to the class of methods that use a reference database for the training of the program while also allowing the discovery of new species are a significant first step into a more complete exploitation of all the information in a metagenomic data set. This aspect is of considerable importance, as it has recently been proven that the majority of microbes in the human gut (currently the best studied environment with metagenomics) are not represented by current genomic resources [9].
Although methods based on reference sequences can identify new relatives of characterized organisms, they are extremely limited when it comes to the discovery of species that remain largely uncharacterized. Furthermore, genome-based methods do not allow for genomic-scale grouping of sequences with certain characteristics most likely coming from closely related individuals, which can be used for reconstruction of species genomes. The computational time needed during the database comparison can be high, especially if using BLAST as the primary alignment tool. Another aspect to take into account with regard to the database comparison results, is that the decision of the threshold that should be used for reliably assigning a taxonomic level is somewhat arbitrary because it strongly varies for each read, and often the most reliable level is high (superkingdom or phylum). Furthermore, methods based on genome alignment require normalization by genome size in order to estimate taxonomic abundance without bias [7], something that is not possible to estimate for uncharacterized species. Lastly, metagenomic methods that start by assembling the reads into longer sequences require paired-end sequence data to perform a good-quality de novo assembly. Paired-end libraries are more expensive to generate than shotgun data sets, thus if used in this way there can be a relatively low efficiency to economic cost ratio.
Although the reliability of reference-based species identification can be better than the reference-free methods from a conventional point of view—that in which an already described species name can be assigned to a group of reads—the best taxonomic identification methods have high precision but low sensitivity. This means that they make accurate assignments but fail to classify a large portion of the input sequences, even at high taxonomic levels [130]. The development of strategies focusing on working with the unclassified reads is of paramount importance, for example, through the use of different more relaxed search parameters can be used to allow for matches to more distant relatives that would not be identified with the current stringent methods that mind the taxonomic assignation specificity.
Composition-based classifiers face more problems with regards to taxonomic assignment than alignment-based methods do, given that more reliable composition information can be obtained from longer reads [131], which is not the case of most of today’s shotgun data sets. Thus, there is room for improvement of metagenomic reference-based taxonomic assignation. Improving de novo assembly algorithms would yield long enough sequences to extract reliable composition information, but chimeric assemblies need to be identified first so as not to provide mixed up information to confound the taxonomic assignation.
So far, a number of marker genes have proven useful for prokaryotic species delineation; however, other organisms such as fungi and virus have not been genetically characterized in terms of taxonomy as deeply as prokaryotes [132, 133]. Thus, more metagenomic methods not based on genome alignment need to be developed focusing on uncharacterized species and taking into account eukaryotic [134] and virus species [135–138]. This is especially important for phages, the most abundant biological entities on the planet [133].
Regarding functional characterization of a metagenome, homology annotation is widely used in metagenomics for functional profiling, but other methods for annotating the proteome of metagenomes should be explored. This becomes an issue of importance given the incompleteness of the databases, and the huge number of proteins reported that are either uncharacterized or only assigned putative functions. To this end, context-based methods represent an area that can be further explored to refine or enhance functional annotation. These kinds of methods integrate information from genomes and pathways [87, 139–141].
A method called pseudo amino acid composition (PseAAC) [142–145] has been extensively used in computational proteomics for predicting protein structures and functions, but has been yet unexplored in metagenomics. PseAAC is a machine-learning method that uses the 20 conventional amino acids and a combination of a set of discrete sequence correlation factors obtained by using a correlation function that reflects the sequence order correlation between all the top most contiguous residues along a protein chain [142]. PseAAC has been mainly used for prediction of protein cellular attributes such as which compartment of a cell it belongs to and how it is associated to the lipid bilayer of an organelle [142], protein structural classes [146], enzyme families [147], protein–protein interactions [148], among others [149]. These attributes are closely related to the biological function of the protein.
Metagenomic reference-free characterization
Metagenomic data sets differ principally from the classical DNA metabarcoding PCR-based amplicon data sets in that they are able to exploit a key piece of information: the number of times a sequence is present [150] (Figure 4C). Although this kind of approach has yet to be widely adopted, and thus could benefit from considerable improvement, it represents a promising way for novel species and gene finding [151–154]. Currently only a handful of methods based on the notion that abundance is constant across genetic entities such as genes in a chromosome have been published [155–157]. A recent example is the method proposed by H Bjørn Nielsen et al. [55], which exploits the co-abundance profiles across metagenomic data sets from a number of samples of the same type. This method extracts groups of genes that correlate in terms of abundance to randomly picked seed genes, calling these clusters co-abundance gene groups (CAGs). Segregating a metagenome into groups of genes that have similar abundance allows the identification of biological entities like prokaryotes and phages, as well as small genetic entities representing co-inherited clonal heterogeneity. The ability of the method to discriminate between strains of the same species, even within complex metagenomics samples, indicates the power of co-abundance to segregate closely related biological entities.
Another method that uses count information is that proposed by Albertsen et al. [114]. In this method data sets from a given sample are produced using two different DNA extraction methods. The first steps are similar to those of the CAGs method, in which the reads are de novo assembled and subsequently a primary binning approach of clustering by similarity is used, thus making a non-redundant gene catalogue. Subsequently each data set’s reads are mapped to the assembled set of non-redundant scaffolds, and a normalized coverage for each scaffold is recorded. Afterwards the steps particular to this method include the binning of the scaffolds into population genomes by plotting the two coverage estimates of all scaffolds against each other. From the plot, scaffolds clustering together represent putative population genome bins.
Considerations on metagenomics reference-free methods
These methods are powerful in the sense that they allow the recovery of genomes from unknown and rare taxonomic variants, opening the possibility of finding novel enzymes and allowing a detailed functional characterization that is much broader and complete than the one that could be done on a reference-based approach. Such characterization in turn enables exploitation of a looser species definition such as that presented by the CAGs, which are of greater usefulness than the standard rigorous species definition in dealing with the widespread transfer of DNA across species boundaries. Similarly, the use of a functional assembly instead of a species taxonomic assignation to characterize an ecosystem and compare it with others represents an underexplored avenue containing considerable opportunities [9, 87]. This metagenomics species concept is useful for a complete metagenomic characterization of a sample; however, it faces the huge challenge of being understood and accepted by the scientific community that is slowly advancing to new concepts.
Both reference-based and reference-free metagenomic methods face the problem that comparing many different samples requires manual inspection. Interesting methods such as the differential depth binning cannot be implemented automatically for many samples because it requires manual examination of the plot [158], and others like MEGAN [65] and MG-RAST [103] are based on visual and interactive analyses. However, the canopy clustering-based method [55] represents a first step into comparison of multiple samples in an automated way.
On a separate subject, there is an approach similar to the PseAAC but applied to DNA/RNA sequences called pseudo K-tuple nucleotide composition (PseKNC) [159, 160]. PseKNC has been used for example to infer recombination spots [161, 162], promoters [163], nucleosome positioning [164], and also CDNA-related features as splicing sites [165], and translation initiation sites [166]. These kinds of methods could be used to annotate genomic features on genomes drawn from reference-free metagenomics methods.
Finally, the complexity of the data set has a big impact on the accuracy of the results and the effect of the sequencing depth on the results has a significant impact on the results [113, 167]. Further exploration on the impact of sequencing depth and proper integration of data sets coming from different sequencing technologies, each one with different nucleotide miscalling problems, would provide more information on how to optimally exploit the information, and on where to delimit how much information one can draw from them.
Final remarks
Although it is difficult to accurately predict how many of the methodological challenges will be addressed in the future and which new tools will be developed, it is expected that reference-based methods will rapidly benefit from the accumulation of information in the data sets, while reference-free methods would be helped by an extended acceptance of a broader concept of species definition. Computational technological advances are also expected to play an impact on the kind of programs developed. For example, a wider usage of cloud computing [168] or the higher feasibility of acquiring computational resources with much more power would enable the processing of even larger data sets with more computationally demanding algorithms.
Another important issue that remains to be discussed is the development of computational tools in concert with laboratory method development. For example, the laboratory method called Hi-C which was developed for generating chromatin-level contact probability maps [169] has been successfully applied to reconstruct individual genomes of microbial species present within a synthetic metagenome sample [170]. Although this method still needs to be further modified to be more widely applied to complex real metagenomics samples, computational method developers can influence in their refinement. The metabarcoding field has also been benefited from the development of refined laboratory methods that include the use of double tagged amplicons coupled to multiple PCR replicates, which are HTS sequenced in a multiplexed manner [92]. Although this protocol provides more information for distinguishing PCR/sequencing errors and chimeras, there is currently only one method developed to analyze specifically that kind of data sets (in the form Zepeda Mendoza ML, Carmona Baez A, Bohmann K, Gilbert MTP, submitted for publication). Other techniques such as HITChip [171] have benefited the large-scale taxonomic profiling in reference-based metabarcoding studies. However, development of programs to customize the design of chip probes based on non-standard resources, such as in-house databases, is still needed.
In summary, the primary message that we hope to have conveyed with the description and definition of the methods boundaries presented here, is the need for future software development for metagenomics and DNA metabarcoding data analyses. Secondly, we seek to clarify the confusion regarding the mislabeling of some metabarcoding studies with the terms ‘metagenomics’ or ‘targeted metagenomics’. Thirdly, we intend to draw researchers’ attention to the challenges that the current methods face, and suggest avenues of method development for further exploration. For example, we believe that metagenomics would greatly benefit from a closer collaboration with the algorithms used in computational proteomics, such as PseAAC and PseKNC. Finally, we believe that consideration of the pros and cons of the different approaches, and the specific goals of the two ‘meta-scale’ research fields, will help researchers choose the appropriate methods to use to address the specific questions of their studies (Figure 5).
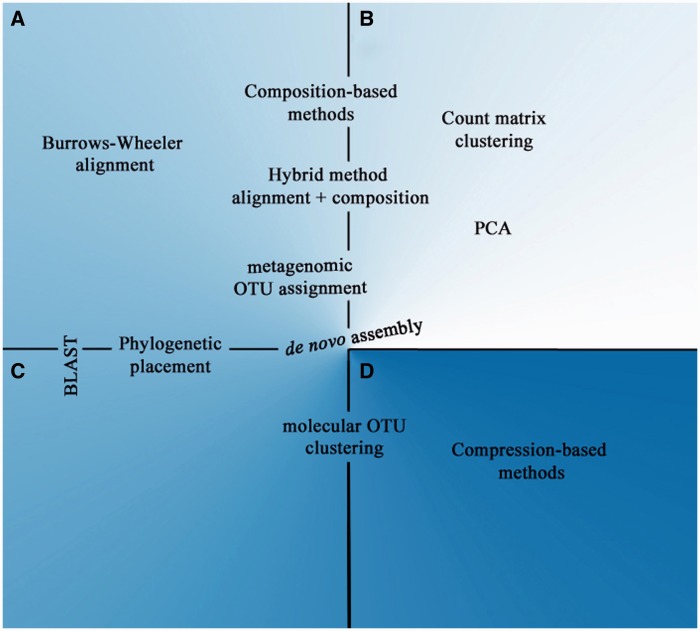
Figure 5.
Method classification placement map. As observed in the placement of the methods, there is lack of software in some areas while there is wealth in others, especially at the borderlines where at first they might seem difficult to classify. (A) Metagenomic reference based. (B) Metagenomic reference free. (C) DNA metabarcoding reference based. (D) DNA metabarcoding reference free.
Key Points.
Metabarcoding and metagenomics share many aspects of their software and this has led to a misunderstanding of their meaning and goals.
To distinguish ‘metagenomics’ and ‘DNA metabarcoding’ as research fields, and ‘metagenomic sequencing’ as a laboratory technique, DNA metabarcoding can be subdivided by how many barcodes are used (single and multiple loci) and which sequencing technique is used (PCR-based or PCR-free).
In general, metagenomics and DNA metabarcoding software can be divided based on whether they use a reference database or not, both types posing different challenges.
Re-examination of the pros and cons of the different approaches in metagenomics and metabarcoding is important to decide on the method to use for the study.
Method development in metagenomics and metabarcoding would benefit from considering recently emerging techniques in other disciplines.
Funding
This work was supported by the Lundbeck Foundation grant number R52-A5062.
Biographies
Marie Lisandra Zepeda Mendoza is a PhD student at the Centre for GeoGenetics, Natural History Museum of Denmark, University of Copenhagen, Denmark.
Thomas Sicheritz-Pontén is the Head of Metagenomics group and professor at the Center for Biological Sequence Analysis, Department of Systems Biology, Technical University of Denmark, Denmark.
M. Thomas P. Gilbert is Professor of Palaeogenomics at the Natural History Museum of Denmark, University of Copenhagen.
References
- 1.Shtarkman YM, Koçer ZA, Edgar R, et al. Subglacial Lake Vostok (Antarctica) accretion ice contains a diverse set of sequences from aquatic, marine and sediment-inhabiting bacteria and eukarya. PLoS One 2013;8(7):e67221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yau S, Lauro FM, Williams TJ, et al. Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J 2013;7(10):1944–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolduc B, Shaughnessy DP, Wolf YI, et al. Identification of novel positive-strand RNA viruses by metagenomic analysis of archaea-dominated Yellowstone hot springs. J Virol 2012;86(10):5562–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schoenfeld T, Patterson M, Richardson PM, et al. Assembly of viral metagenomes from Yellowstone hot springs. Appl Environ Microbiol 2008;74(13):4164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eme L, Reigstad LJ, Spang A, et al. Metagenomics of Kamchatkan hot spring filaments reveal two new major (hyper)thermophilic lineages related to Thaumarchaeota. Res Microbiol 2013;164(5):425–38. [DOI] [PubMed] [Google Scholar]
- 6.Nelson KE, Weinstock GM, Highlander SK, et al. A catalog of reference genomes from the human microbiome. Science 2010;328(5981):994–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature 2011;473(7346):174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500(7464):541–6. [DOI] [PubMed] [Google Scholar]
- 9.Shafquat A, Joice R, Simmons SL, et al. Functional and phylogenetic assembly of microbial communities in the human microbiome. Trends Microbiol 2014;22(5):261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464(7285):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuy AK, David MS, Li L, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 2014;9(3):e90899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips CD, Phelan G, Dowd SE, et al. Microbiome analysis among bats describes influences of host phylogeny, life history, physiology and geography. Mol Ecol 2012;21(11):2617–27. [DOI] [PubMed] [Google Scholar]
- 13.Hu Y, Lukasik P, Moreau CS, et al. Correlates of gut community composition across an ant species (Cephalotes varians) elucidate causes and consequences of symbiotic variability. Mol Ecol 2014;23(6):1284–300. [DOI] [PubMed] [Google Scholar]
- 14.Warnecke F, Luginbühl P, Ivanova N, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 2007;450(7169):560–5. [DOI] [PubMed] [Google Scholar]
- 15.Delsuc F, Metcalf JL, Wegener Parfrey L, et al. Convergence of gut microbiomes in myrmecophagous mammals. Mol Ecol 2014;23(6):1301–17. [DOI] [PubMed] [Google Scholar]
- 16.Singh KM, Shah T, Deshpande S, et al. High through put 16S rRNA gene-based pyrosequencing analysis of the fecal microbiota of high FCR and low FCR broiler growers. Mol Biol Rep 2012;39(12):10595–602. [DOI] [PubMed] [Google Scholar]
- 17.Rondon MR, August PR, Bettermann AD, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 2000;66(6):2541–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Filippo C, Ramazzotti M, Fontana P, et al. Bioinformatic approaches for functional annotation and pathway inference in metagenomics data. Brief Bioinform 2012;13(6):696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster JA, Bunge J, Gilbert JA, et al. Measuring the microbiome: perspectives on advances in DNA-based techniques for exploring microbial life. Brief Bioinform 2012;13(4):420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinstock GM. Genomic approaches to studying the human microbiota. Nature 2012;489(7415):250–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida M, Takaki Y, Eitoku M, et al. Metagenomic analysis of viral communities in (hado)pelagic sediments. PLoS One 2013;8(2):e57271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lozupone C, Knight R. UniFrac: a New Phylogenetic Method for Comparing Microbial Communities. Appl Environ Microbiol 2005;71(12):8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Willerslev E, Davison J, Moora M, et al. Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 2014;506(7486):47–51. [DOI] [PubMed] [Google Scholar]
- 24.Blaalid R, Kumar S, Nilsson RH, et al. ITS1 versus ITS2 as DNA metabarcodes for fungi. Mol Ecol Resour 2013;13(2):218–24. [DOI] [PubMed] [Google Scholar]
- 25.Aylagas E, Borja A, Rodríguez-Ezpeleta N. Environmental status assessment using DNA metabarcoding: towards a genetics based marine biotic index (gAMBI). PLoS One 2014;9(3):e90529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riaz T, Shehzad W, Viari A, et al. ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res 2011;39(21):e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Epp LS, Boessenkool S, Bellemain EP, et al. New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Mol Ecol 2012;21(8):1821–33. [DOI] [PubMed] [Google Scholar]
- 28.Giampaoli S, Berti A, Di Maggio RM, et al. The environmental biological signature: NGS profiling for forensic comparison of soils. Forensic Sci Int 2014;240:41–7. [DOI] [PubMed] [Google Scholar]
- 29.Taberlet P, Coissac E, Pompanon F, et al. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 2012;21(8):2045–50. [DOI] [PubMed] [Google Scholar]
- 30.Su X, Pan W, Song B, et al. Parallel-META 2.0: enhanced metagenomic data analysis with functional annotation, high performance computing and advanced visualization. Zhang Z, ed. PLoS One 2014;9(3):e89323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binladen J, Gilbert MTP, Bollback JP, et al. The use of coded PCR primers enables high-throughput sequencing of multiple homolog amplification products by 454 parallel sequencing. Hahn M, ed. PLoS One 2007;2(2):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Li Y, Liu S, et al. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. Gigascience 2013;2(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ong SH, Kukkillaya VU, Wilm A, et al. Species identification and profiling of complex microbial communities using shotgun Illumina sequencing of 16S rRNA amplicon sequences. Parkinson J, ed. PLoS One 2013;8(4):e60811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choudhari S, Lohia R, Grigoriev A. Comparative metagenome analysis of an Alaskan glacier. J Bioinform Comput Biol 2014;12(2):1441003. [DOI] [PubMed] [Google Scholar]
- 35.Sturgeon A, Stull JW, Costa MC, et al. Metagenomic analysis of the canine oral cavity as revealed by high-throughput pyrosequencing of the 16S rRNA gene. Vet Microbiol 2013;162(2-4):891–8. [DOI] [PubMed] [Google Scholar]
- 36.Tan L, Wang H, Li C, et al. 16S rDNA-based metagenomic analysis of dental plaque and lung bacteria in patients with severe acute exacerbations of chronic obstructive pulmonary disease. J Periodontal Res 2014;49(6):769-9. [DOI] [PubMed] [Google Scholar]
- 37.Keshri J, Mishra A, Jha B. Microbial population index and community structure in saline-alkaline soil using gene targeted metagenomics. Microbiol Res 2013;168(3):165–73. [DOI] [PubMed] [Google Scholar]
- 38.Yousuf B, Keshri J, Mishra A, et al. Application of targeted metagenomics to explore abundance and diversity of CO-fixing bacterial community using cbbL gene from the rhizosphere of Arachis hypogaea. Gene 2012;506(1):18–24. [DOI] [PubMed] [Google Scholar]
- 39.Machado VS, Oikonomou G, Bicalho MLS, et al. Investigation of postpartum dairy cows’ uterine microbial diversity using metagenomic pyrosequencing of the 16S rRNA gene. Vet Microbiol 2012;159(3-4):460–9. [DOI] [PubMed] [Google Scholar]
- 40.Marlow MA, Boité MC, Ferreira GEM, et al. Multilocus sequence analysis for Leishmania braziliensis outbreak investigation. PLoS Negl Trop Dis 2014;8(2):e2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrow LN, Ralicki HF, Emme SA, et al. Species tree estimation of North American chorus frogs (Hylidae: Pseudacris) with parallel tagged amplicon sequencing. Mol Phylogenet Evol 2014;75:78–90. [DOI] [PubMed] [Google Scholar]
- 42.Campanaro S, Treu L, Vendramin V, et al. Metagenomic analysis of the microbial community in fermented grape marc reveals that Lactobacillus fabifermentans is one of the dominant species: insights into its genome structure. Appl Microbiol Biotechnol 2014;98(13):6015–37. [DOI] [PubMed] [Google Scholar]
- 43.McCann JC, Wickersham TA, Loor JJ. High-throughput Methods Redefine the Rumen Microbiome and Its Relationship with Nutrition and Metabolism. Bioinform Biol Insights 2014;8:109–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rascovan N, Carbonetto B, Revale S, et al. The PAMPA datasets: a metagenomic survey of microbial communities in Argentinean pampean soils. Microbiome 2013;1(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Puritz JB, Toonen RJ. Next-generation sequencing for high-throughput molecular ecology: a step-by-step protocol for targeted multilocus genotyping by pyrosequencing. Methods Mol Biol 2013;1006:89–99. [DOI] [PubMed] [Google Scholar]
- 46.García Martín H, Ivanova N, Kunin V, et al. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat Biotechnol 2006;24(10):1263–9. [DOI] [PubMed] [Google Scholar]
- 47.Venter JC, Remington K, Heidelberg JF, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004;304(5667):66–74. [DOI] [PubMed] [Google Scholar]
- 48.Krause L, Diaz NN, Goesmann A, et al. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res 2008;36(7):2230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagkouvardos I, Weinmaier T, Lauro FM, et al. Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J 2014;8(1):115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med 2014;6(237):237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poretsky R, Rodriguez-R LM, Luo C, et al. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One 2014;9(4):e93827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitra S, Förster-Fromme K, Damms-Machado A, et al. Analysis of the intestinal microbiota using SOLiD 16S rRNA gene sequencing and SOLiD shotgun sequencing. BMC Genomics 2013;14(Suppl 5):S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Torsvik V, Goksoyr J, Daae FL. High diversity in DNA of soil bacteria. Appl Envir Microbiol 1990;56(3):782–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 2004;68(4):669–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nielsen HB, Almeida M, Juncker AS, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol 2014;32(8):822–8. [DOI] [PubMed] [Google Scholar]
- 56.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol 2008;26(10):1135–45. [DOI] [PubMed] [Google Scholar]
- 57.Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour 2011;11(5):759–69. [DOI] [PubMed] [Google Scholar]
- 58.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7(5):335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyer F, Mercier C, Bonin A, et al. OBITools: a Unix-inspired software package for DNA metabarcoding. Molecular Ecology Resources 2014, submitted. http://metabarcoding.org/obitools/doc/welcome.html. [DOI] [PubMed] [Google Scholar]
- 60.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26(19):2460–1. [DOI] [PubMed] [Google Scholar]
- 62.Huang Y, Niu B, Gao Y, et al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 2010;26(5):680–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol 1990;215(3):403–10. [DOI] [PubMed] [Google Scholar]
- 64.Mercier C, Boyer F, Bonin A, et al. SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences. In: Programs and Abstracts of the SeqBio 2013 workshop. Abstract, pp. 27–29. GdRBIM and gdrIM. Montpellier, France [Google Scholar]
- 65.Huson DH, Weber N. Microbial community analysis using MEGAN. Methods Enzymol 2013;531:465–85. [DOI] [PubMed] [Google Scholar]
- 66.Liu B, Gibbons T, Ghodsi M, et al. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics 2011;12(Suppl 2):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sunagawa S, Mende DR, Zeller G, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods 2013;10(12):1196–9. [DOI] [PubMed] [Google Scholar]
- 68.Nguyen N-P, Mirarab S, Liu B, et al. TIPP: taxonomic identification and phylogenetic profiling. Bioinformatics 2014;30(24):3548–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 2012;9(8):811–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berendzen J, Bruno WJ, Cohn JD, et al. Rapid phylogenetic and functional classification of short genomic fragments with signature peptides. BMC Res Notes 2012;5(1):460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu K, Warnow T. Large-scale multiple sequence alignment and tree estimation using SATé. Methods Mol Biol 2014;1079:219–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu K, Warnow TJ, Holder MT, et al. SATe-II: very fast and accurate simultaneous estimation of multiple sequence alignments and phylogenetic trees. Syst Biol 2012;61(1):90–106. [DOI] [PubMed] [Google Scholar]
- 73.Mirarab S, Nguyen N, Warnow T. SEPP: SATé-enabled phylogenetic placement. Pac Symp Biocomput 2012:247–58. [DOI] [PubMed] [Google Scholar]
- 74.Bender MA, Farach-Colton M, Pemmasani G, et al. Lowest common ancestors in trees and directed acyclic graphs. J Algorithms 2005;57(2):75–94. [Google Scholar]
- 75.Graham DE, Overbeek R, Olsen GJ, et al. An archaeal genomic signature. Proc Natl Acad Sci USA 2000;97(7):3304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Feau N, Decourcelle T, Husson C, et al. Finding single copy genes out of sequenced genomes for multilocus phylogenetics in non-model fungi. Butler G, ed. PLoS One 2011;6(4):e18803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cole JR, Chai B, Farris RJ, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res 2005;33(Database issue):D294–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ratnasingham S, Hebert PDN. BOLD: The barcode of life data system ( http://www.barcodinglife.org). Mol Ecol Notes 2007;7(3):355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006;72(7):5069–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Douglas W, Yu YJ. Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol Evol 2012;3:613–23. [Google Scholar]
- 82.Bellemain E, Carlsen T, Brochmann C, et al. ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 2010;10:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alonso-Alemany D, Barré A, Beretta S, et al. Further steps in TANGO: improved taxonomic assignment in metagenomics. Bioinformatics 2014;30(1):17–23. [DOI] [PubMed] [Google Scholar]
- 84.Zhang J, Kapli P, Pavlidis P, et al. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013;29(22):2869–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27(16):2194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Song H, Buhay JE, Whiting MF, et al. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA 2008;105(36):13486–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langille MGI, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31(9):814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hibert F, Taberlet P, Chave J, et al. Unveiling the diet of elusive rainforest herbivores in next generation sequencing era? The tapir as a case study. PLoS One 2013;8(4):e60799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10(10):996–8. [DOI] [PubMed] [Google Scholar]
- 90.Bohmann K, Monadjem A, Lehmkuhl Noer C, et al. Molecular diet analysis of two african free-tailed bats (molossidae) using high throughput sequencing. PLoS One 2011;6(6):e21441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burgar JM, Murray DC, Craig MD, et al. Who’s for dinner? High-throughput sequencing reveals bat dietary differentiation in a biodiversity hotspot where prey taxonomy is largely undescribed. Mol Ecol 2013;4(1):27. [DOI] [PubMed] [Google Scholar]
- 92.Hope PR, Bohmann K, Gilbert MTP, et al. Second generation sequencing and morphological faecal analysis reveal unexpected foraging behaviour by Myotis nattereri (Chiroptera, Vespertilionidae) in winter. Front Zool 2014;11(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bohmann K, Evans A, Gilbert MTP, et al. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol Evol 2014;29(6):358–67. [DOI] [PubMed] [Google Scholar]
- 94.Schnell IB, Thomsen PF, Wilkinson N, et al. Screening mammal biodiversity using DNA from leeches. Curr Biol 2012;22(8):R262–3. [DOI] [PubMed] [Google Scholar]
- 95.Coissac E, Riaz T, Puillandre N. Bioinformatic challenges for DNA metabarcoding of plants and animals. Mol Ecol 2012;21(8):1834–47. [DOI] [PubMed] [Google Scholar]
- 96.Janssen T, Schneider H. Exploring the evolution of humus collecting leaves in drynarioid ferns (Polypodiaceae, Polypodiidae) based on phylogenetic evidence. Plant Syst Evol 2005;252(3–4):175–97. [Google Scholar]
- 97.Schneider H, Ranker TA, Russell SJ, et al. Origin of the endemic fern genus Diellia coincides with the renewal of Hawaiian terrestrial life in the Miocene. Proc Biol Sci 2005;272(1561):455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhan A, Bailey SA, Heath DD, et al. Performance comparison of genetic markers for high-throughput sequencing-based biodiversity assessment in complex communities. Mol Ecol Resour 2014;14(5):1049–59. [DOI] [PubMed] [Google Scholar]
- 99.La Rosa M, Fiannaca A, Rizzo R, et al. Alignment-free analysis of barcode sequences by means of compression-based methods. BMC Bioinformatics 2013;14(Suppl 7):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cilibrasi R, Vitanyi PMB. Clustering by compression. IEEE Trans Inf Theory 2005;51(4):1523–45. [Google Scholar]
- 101.Li M, Badger JH, Chen X, et al. An information-based sequence distance and its application to whole mitochondrial genome phylogeny. Bioinformatics 2001;17(2):149–54. [DOI] [PubMed] [Google Scholar]
- 102.Monzoorul Haque M, Ghosh TS, Komanduri D, et al. SOrt-ITEMS: sequence orthology based approach for improved taxonomic estimation of metagenomic sequences. Bioinformatics 2009;25(14):1722–30. [DOI] [PubMed] [Google Scholar]
- 103.Glass EM, Wilkening J, Wilke A, et al. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc 2010;2010(1):pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 104.Caporaso JG, Knight R, Kelley ST. Host-associated and free-living phage communities differ profoundly in phylogenetic composition. Gilbert J, ed. PLoS One 2011;6(2):e16900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods 2009;6(9):673–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McHardy AC, Martín HG, Tsirigos A, et al. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods 2007;4(1):63–72. [DOI] [PubMed] [Google Scholar]
- 107.Silva GGZ, Cuevas DA, Dutilh BE, et al. FOCUS: an alignment-free model to identify organisms in metagenomes using non-negative least squares. PeerJ 2014;2:e425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Meinicke P, Asshauer KP, Lingner T. Mixture models for analysis of the taxonomic composition of metagenomes. Bioinformatics 2011;27(12):1618–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res 2007;35(Database issue):D61–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kultima JR, Sunagawa S, Li J, et al. MOCAT: A Metagenomics Assembly and Gene Prediction Toolkit. PLoS ONE 2012;7(10):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Peng Y, Leung HCM, Yiu SM, et al. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012;28(11):1420–8. [DOI] [PubMed] [Google Scholar]
- 112.Boisvert S, Raymond F, Godzaridis E, et al. Ray Meta: scalable de novo metagenome assembly and profiling. Genome Biol 2012;13(12):R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Luo C, Tsementzi D, Kyrpides NC, et al. Individual genome assembly from complex community short-read metagenomic datasets. ISME J 2012;6(4):898–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Albertsen M, Hugenholtz P, Skarshewski A, et al. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 2013;31(6):533–8. [DOI] [PubMed] [Google Scholar]
- 115.Gerlach W, Stoye J. Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Res 2011;39(14):e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bateman A. The Pfam protein families database. Nucleic Acids Res 2002;30(1):276–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yooseph S, Li W, Sutton G. Gene identification and protein classification in microbial metagenomic sequence data via incremental clustering. BMC Bioinformatics 2008;9:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25(17):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Rychlewski L, Jaroszewski L, Li W, et al. Comparison of sequence profiles. strategies for structural predictions using sequence information. Protein Sci 2000;9(2):232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Murzin AG, Brenner SE, Hubbard T, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 1995;247(4):536–40. [DOI] [PubMed] [Google Scholar]
- 121.Orengo CA, Michie AD, Jones S, et al. CATH–a hierarchic classification of protein domain structures. Structure 1997;5(8):1093–108. [DOI] [PubMed] [Google Scholar]
- 122.Sharon I, Bercovici S, Pinter RY, et al. Pathway-based functional analysis of metagenomes. J Comput Biol 2011;18(3):495–505. [DOI] [PubMed] [Google Scholar]
- 123.Overbeek R, Olson R, Pusch GD, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 2014;42(Database issue):D206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.An L, Pookhao N, Jiang H, et al. Statistical approach of functional profiling for a microbial community. PLoS One 2014;9(9):e106588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tatusov RL. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res 2001;29(1):22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res 2003;31(1):371–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28(1):27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Caspi R, Altman T, Dale JM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 2010;38(Database issue):D473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Overbeek R, Begley T, Butler RM, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 2005;33(17):5691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mande SS, Mohammed MH, Ghosh TS. Classification of metagenomic sequences: methods and challenges. Brief Bioinform 2012;13(6):669–81. [DOI] [PubMed] [Google Scholar]
- 131.Mitra S, Schubach M, Huson DH. Short clones or long clones? A simulation study on the use of paired reads in metagenomics. BMC Bioinformatics 2010;11(Suppl 1):S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Guarro J, Gene J, Stchigel AM. Developments in fungal taxonomy. Clin Microbiol Rev 1999;12(3):454–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Dutilh BE, Cassman N, McNair K, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 2014;5:4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kosakovsky Pond S, Wadhawan S, Chiaromonte F, et al. Windshield splatter analysis with the Galaxy metagenomic pipeline. Genome Res. 2009;19(11):2144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hurwitz BL, Deng L, Poulos BT, et al. Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ Microbiol 2013;15(5):1428–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Roossinck MJ. Plant virus metagenomics: biodiversity and ecology. Annu Rev Genet 2012;46:359–69. [DOI] [PubMed] [Google Scholar]
- 137.Alavandi SV, Poornima M. Viral metagenomics: a tool for virus discovery and diversity in aquaculture. Indian J Virol 2012;23(2):88–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol 2012;2(1):63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ye Y, Osterman A, Overbeek R, et al. Automatic detection of subsystem/pathway variants in genome analysis. Bioinformatics 2005;21(Suppl 1):i478–86. [DOI] [PubMed] [Google Scholar]
- 140.Kelley BP, Sharan R, Karp RM, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci USA 2003;100(20):11394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Green ML, Karp PD. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics 2004;5:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chou KC. Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins 2001;43(3):246–55. [DOI] [PubMed] [Google Scholar]
- 143.Du P, Gu S, Jiao Y. PseAAC-General: fast building various modes of general form of Chou’s pseudo-amino acid composition for large-scale protein datasets. Int J Mol Sci 2014;15(3):3495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Du P, Wang X, Xu C, et al. PseAAC-Builder: a cross-platform stand-alone program for generating various special Chou’s pseudo-amino acid compositions. Anal Biochem 2012;425(2):117–19. [DOI] [PubMed] [Google Scholar]
- 145.Cao D-S, Xu Q-S, Liang Y-Z. Propy: a tool to generate various modes of Chou’s PseAAC. Bioinformatics 2013;29(7):960–2. [DOI] [PubMed] [Google Scholar]
- 146.Li Z-C, Zhou X-B, Dai Z, et al. Prediction of protein structural classes by Chou’s pseudo amino acid composition: approached using continuous wavelet transform and principal component analysis. Amino Acids 2009;37(2):415–25. [DOI] [PubMed] [Google Scholar]
- 147.Chou K-C. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics 2005;21(1):10–19. [DOI] [PubMed] [Google Scholar]
- 148.Bock JR, Gough DA. Predicting protein-protein interactions from primary structure. Bioinformatics 2001;17(5):455–60. [DOI] [PubMed] [Google Scholar]
- 149.Chou K-C. Pseudo amino acid composition and its applications in bioinformatics, proteomics and system biology. Curr Proteomics 2009;6(4):262–274. [Google Scholar]
- 150.Allen HK, Bunge J, Foster JA, et al. Estimation of viral richness from shotgun metagenomes using a frequency count approach. Microbiome 2013;1(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bayer S, Kunert A, Ballschmiter M, et al. Indication for a new lipolytic enzyme family: isolation and characterization of two esterases from a metagenomic library. J Mol Microbiol Biotechnol 2010;18(3):181–7. [DOI] [PubMed] [Google Scholar]
- 152.Kazimierczak KA, Rincon MT, Patterson AJ, et al. A new tetracycline efflux gene, tet(40), is located in tandem with tet(O/32/O) in a human gut firmicute bacterium and in metagenomic library clones. Antimicrob Agents Chemother 2008;52(11):4001–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jiang X, Langille MGI, Neches RY, et al. Functional biogeography of ocean microbes revealed through non-negative matrix factorization. PLoS One 2012;7(9):e43866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Holdeman LV, Moore WEC. New genus, coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. Int J Syst Bacteriol 1974;24(2):260–77. [Google Scholar]
- 155.Carr R, Shen-Orr SS, Borenstein E. Reconstructing the genomic content of microbiome taxa through shotgun metagenomic deconvolution. Ouzounis CA, ed. PLoS Comput Biol 2013;9(10):e1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Devarajan K. Nonnegative matrix factorization: an analytical and interpretive tool in computational biology. PLoS Comput Biol 2008;4(7):e1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012;490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- 158.Gonzalez A, Knight R. Advancing analytical algorithms and pipelines for billions of microbial sequences. Curr Opin Biotechnol 2012;23(1):64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen W, Lei T-Y, Jin D-C, et al. PseKNC: a flexible web server for generating pseudo K-tuple nucleotide composition. Anal Biochem 2014;456:53–60. [DOI] [PubMed] [Google Scholar]
- 160.Chen W, Zhang X, Brooker J, et al. PseKNC-General: a cross-platform package for generating various modes of pseudo nucleotide compositions. Bioinformatics 2015;31(1):119–20. [DOI] [PubMed] [Google Scholar]
- 161.Qiu W-R, Xiao X, Chou K-C. iRSpot-TNCPseAAC: identify recombination spots with trinucleotide composition and pseudo amino acid components. Int J Mol Sci 2014;15(2):1746–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chen W, Feng P-M, Lin H, et al. iRSpot-PseDNC: identify recombination spots with pseudo dinucleotide composition. Nucleic Acids Res 2013;41(6):e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lin H, Deng E-Z, Ding H, et al. iPro54-PseKNC: a sequence-based predictor for identifying sigma-54 promoters in prokaryote with pseudo k-tuple nucleotide composition. Nucleic Acids Res 2014;42(21):12961–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Guo S-H, Deng E-Z, Xu L-Q, et al. iNuc-PseKNC: a sequence-based predictor for predicting nucleosome positioning in genomes with pseudo k-tuple nucleotide composition. Bioinformatics 2014;30(11):1522–9. [DOI] [PubMed] [Google Scholar]
- 165.Chen W, Feng P-M, Lin H, et al. iSS-PseDNC: identifying splicing sites using pseudo dinucleotide composition. Biomed Res Int 2014;2014:623149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen W, Feng P-M, Deng E-Z, et al. iTIS-PseTNC: a sequence-based predictor for identifying translation initiation site in human genes using pseudo trinucleotide composition. Anal Biochem 2014;462:76–83. [DOI] [PubMed] [Google Scholar]
- 167.Knight R, Jansson J, Field D, et al. Unlocking the potential of metagenomics through replicated experimental design. Nat Biotechnol 2012;30(6):513–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Schatz MC, Langmead B, Salzberg SL. Cloud computing and the DNA data race. Nat Biotechnol. 2010;28(7):691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009;326(5950):289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Burton JN, Liachko I, Dunham MJ, et al. Species-level deconvolution of metagenome assemblies with Hi-C-based contact probability maps. G3 (Bethesda). 2014;4(7):1339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rajilić-Stojanović M, Heilig HGHJ, Molenaar D, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 2009;11(7):1736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]