Abstract
Multiple pterygium syndrome (MPS) is a phenotypically and genetically heterogeneous group of rare Mendelian conditions characterized by multiple pterygia, scoliosis, and congenital contractures of the limbs. MPS typically segregates as an autosomal-recessive disorder, but rare instances of autosomal-dominant transmission have been reported. Whereas several mutations causing recessive MPS have been identified, the genetic basis of dominant MPS remains unknown. We identified four families affected by dominantly transmitted MPS characterized by pterygia, camptodactyly of the hands, vertebral fusions, and scoliosis. Exome sequencing identified predicted protein-altering mutations in embryonic myosin heavy chain (MYH3) in three families. MYH3 mutations underlie distal arthrogryposis types 1, 2A, and 2B, but all mutations reported to date occur in the head and neck domains. In contrast, two of the mutations found to cause MPS in this study occurred in the tail domain. The phenotypic overlap among persons with MPS, coupled with physical findings distinct from other conditions caused by mutations in MYH3, suggests that the developmental mechanism underlying MPS differs from that of other conditions and/or that certain functions of embryonic myosin might be perturbed by disruption of specific residues and/or domains. Moreover, the vertebral fusions in persons with MPS, coupled with evidence of MYH3 expression in bone, suggest that embryonic myosin plays a role in skeletal development.
Main Text
Multiple pterygium syndrome (MPS) is a phenotypically and genetically heterogeneous group of rare Mendelian conditions characterized by multiple pterygia, scoliosis and congenital contractures of the limbs. Most often, MPS occurs as a simplex case, and of reported multiplex families, the majority consist of multiple affected siblings born to unaffected parents, consistent with inheritance in an autosomal-recessive pattern.1 In very rare instances, MPS has been transmitted from an affected parent to an affected child, indicative of autosomal-dominant transmission.2–5 In 1996, we revised the classification of distal arthrogryposis (DA) syndromes and categorized several additional conditions,6 including autosomal-dominant MPS, as DA syndromes because their clinical features overlap those of DA type 1 (DA1A [MIM: 108120] and DA1B [MIM: 614335]) and Freeman-Sheldon syndrome, or DA2A (MIM: 193700). Autosomal-dominant MPS, or DA8 (MIM: 178110), is one of the conditions that was added to the DA classification on the basis of the phenotypic features of four reported families in whom pterygia, camptodactyly of the hands, vertebral fusions, and scoliosis were transmitted from parent to child.2–5
Over the past decade, three previously unreported families with multiple persons who have clinical characteristics consistent with the diagnosis of DA8 and evidence of parent-to-child transmission were referred to our research program on DA syndromes (Table 1; Figures 1 and 2; Figure S1). To identify the gene(s) harboring mutations underlying DA8, we initially used Sanger sequencing to screen the proband of each family for mutations in genes known to contain mutations underlying lethal MPS (MIM: 253290) and non-lethal Escobar-variant autosomal-recessive MPS (MIM: 265000); these genes include CHRNG7,8 (MIM: 100730), CHRND9 (MIM: 100720), and CHRNA19 (MIM: 100690), as well as IRF610 (MIM: 607199), mutations in which cause popliteal pterygium syndrome (MIM: 119500). No pathogenic mutations were found in any of these candidate genes. Next, we performed exome sequencing on seven individuals in family A, three individuals in family B, and two affected individuals (III-5 and V-2) in family C (Table 1; Figures 1 and 2; Figure S1). All studies were approved by the institutional review boards of the University of Washington and Seattle Children’s Hospital, and informed consent was obtained from each participant.
Table 1.
Mutations in and Clinical Findings for Individuals with DA8
|
Family A |
Family B |
Family C |
Family D |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| III-1 | III-2 | II-2 | III-2 | II-1 | III-5a | IV-3 | V-2 | II-1 | I-1 | |
| Mutation Information | ||||||||||
| MYH3 exon | 25 | 25 | 25 | 25 | 25 | 8 | 8 | 8 | no mutation identified | no mutation identified |
| cDNA change | c.3214_3216dup | c.3214_3216dup | c.3214_3216dup | c.3224A>C | c.3224A>C | c.727_729del | c.727_729del | c.727_729del | NA | NA |
| Predicted protein alteration | p.Asn1072dup | p.Asn1072dup | p.Asn1072dup | p.Gln1075Pro | p.Gln1075Pro | p.Ser243del | p.Ser243del | p.Ser243del | NA | NA |
| GERP score | 5.64 | 5.64 | 5.64 | 5.64 | 5.64 | 4.94 | 4.94 | 4.94 | NA | NA |
| CADD score v.1.1 (phred-like) | 17.62 | 17.62 | 17.62 | 20.8 | 20.8 | 18.47 | 18.47 | 18.47 | NA | NA |
| PolyPhen-2 score (HumVar) | NA | NA | NA | 0.931 | 0.931 | NA | NA | NA | NA | NA |
| Clinical Features: Face | ||||||||||
| Downslanting palpebral fissures | + | + | + | + | + | ND | − | − | + | + |
| Ptosis | − | + | − | + | + | ND | − | − | + | + |
| Long nasal bridge | + | + | + | + | + | ND | − | − | + | + |
| Low-set, posteriorly rotated ears | − | − | − | + | + | ND | + | + | + | + |
| Clinical Features: Limbs | ||||||||||
| Hypoplastic flexion creases | + | + | + | + | + | ND | + | + | + | + |
| Camptodactyly | + | + | + | + | + | ND | + | + | + | + |
| Antecubital webbing | − | − | − | − | + | ND | − | + | − | − |
| Elbow contractures | + | + | + | + | + | ND | − | + | − | − |
| Limited forearm supination | + | + | + | ND | + | ND | ND | ND | + | + |
| Hip contractures | ND | ND | ND | ND | + | ND | − | ND | + | − |
| Knee contractures | − | + | + | + | + | + | + | + | − | − |
| Popliteal webbing | − | + | + | ND | + | ND | + | + | − | − |
| Foot contractures | − | − | − | − | metatarsus adductus | ND | limited ankle dorsiflexion | − | clubfeet | − |
| Other Clinical Features | ||||||||||
| Scoliosis | + | + | − | + | + | + | + | + | + | − |
| Short neck | + | + | + | + | + | + | + | + | + | + |
| Neck webbing | − | + | + | + | + | ND | + | + | + | + |
| Short stature | + | + | − | + (<3rd percentile) | + (<5th percentile) | + | + | ND | + | + |
| Vertebral fusion | C1–C2 | T10–L3 | − | ND | ND | C1-C2 | multiple | multiple | − | − |
| Other | webbing of fingers | NA | NA | NA | mild cervical spinal stenosis | craniosynostosis | cleft palate, craniosynostosis |
bilateral inguinal hernia, spondylolisthesis |
VSD | VSD |
This table provides a summary of clinical features of affected individuals from families in whom MYH3 mutations causing DA8 were identified and one family in whom no mutation was identified. Clinical characteristics listed in the table are primarily features that delineate DA8. cDNA positions in MYH3 (GenBank: NM_002470.3) are provided as named by the HGVS MutNomen web tool. Abbreviations are as follows: +, presence of a finding; −, absence of a finding; ND, no data; NA, not applicable; GERP, genomic evolutionary rate profiling; CADD, combined annotation-dependent depletion; VSD, ventricular septal defect.
Described on the basis of a clinician’s report.
Figure 1.
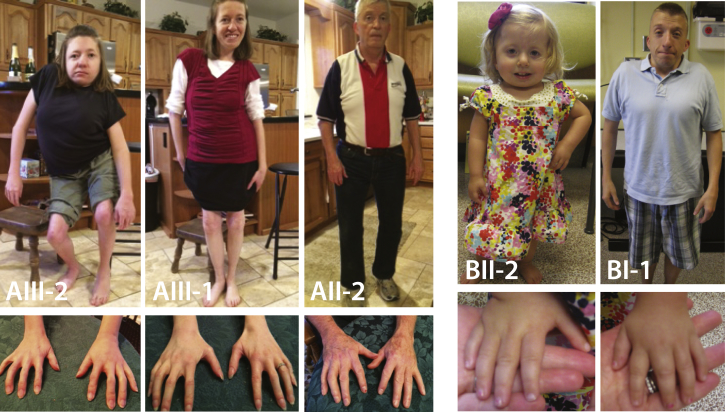
Phenotypic Characteristics of Individuals with DA8
Five individuals affected by DA8; all individuals shown have MYH3 mutations. Note the downslanting palpebral fissures, ptosis, camptodactyly of the fingers, scoliosis, short stature, and neck webbing. ID numbers for the individuals shown in this figure correspond to those in Table 1, where there is a detailed description of the phenotype of each affected individual. Figure S1 provides a pedigree of each DA8-affected family.
Figure 2.
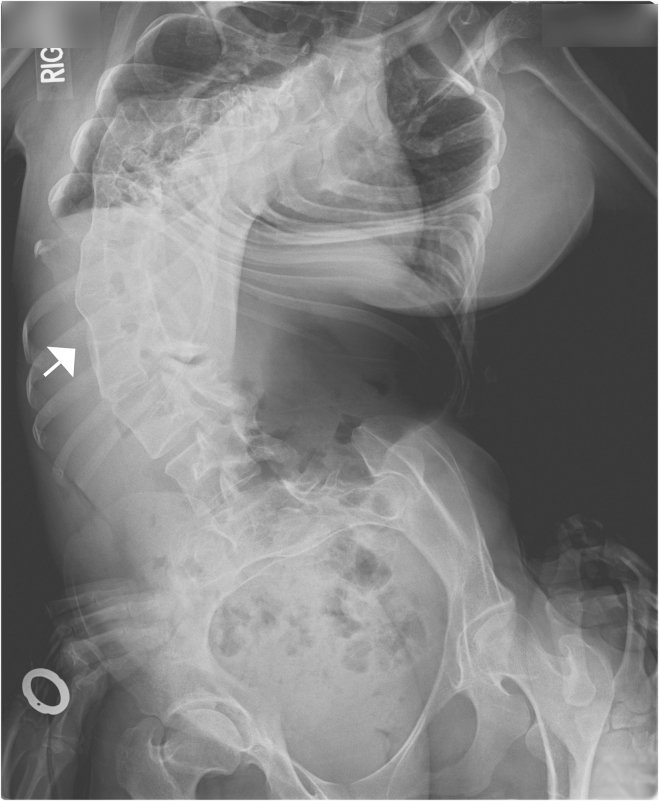
Radiograph of Severe Scoliosis and Vertebral Fusion Observed in a Person with DA8
Severe, convex rightward, rotatory scoliosis of the thoracic and lumbar spine in individual AIII-2 (Table 1 and Figure 1). Fusion of vertebral bodies from T10 to L3 is indicated by the arrow.
In brief, 1 μg of genomic DNA was subjected to a series of shotgun-library-construction steps, including fragmentation through acoustic sonication (Covaris), end polishing (NEBNext End Repair Module), A-tailing (NEBNext dA-Tailing Module), and PCR amplification with ligation of 8-bp barcoded sequencing adaptors (Enzymatics Ultrapure T4 Ligase) for multiplexing. 1 μg of barcoded shotgun library was hybridized to the capture probes targeting ∼36.5 Mb of coding exons (Roche Nimblegen SeqCap EZ Human Exome Library v.2.0). Library quality was determined by examination of molecular-weight distribution and sample concentration (Agilent Bioanalyzer). Pooled, barcoded libraries were sequenced via paired-end 50-bp reads and an 8-bp barcode read on Illumina HiSeq sequencers.
Demultiplexed BAM files were aligned to a human reference sequence (UCSC Genome Browser hg19) via the Burrows-Wheeler Aligner v.0.6.2. Read data from a flow-cell lane were treated independently for alignment and quality-control purposes in instances where the merging of data from multiple lanes was required. All aligned read data were subjected to (1) removal of duplicate reads (Picard MarkDuplicates v.1.70), (2) indel realignment (Genome Analysis Toolkit [GATK] IndelRealigner v.1.6-11-g3b2fab9), and (3) base-quality recalibration (GATK TableRecalibration v.1.6-11-g3b2fab9). Single-nucleotide variant (SNV) detection and genotyping were performed with the GATK UnifiedGenotyper (v.1.6-11-g3b2fab9). SNV data for each sample were formatted (variant-call format [VCF]) as “raw” calls that contained individual genotype data for one or multiple samples and flagged with the filtration walker (GATK) for marking sites that were potential false positives or of lower quality (e.g., strand bias [SB ≥ 0.1], low quality scores [QUAL ≤ 50], allelic imbalance [ABHet > 0.75], long homopolymer runs [HRun > 4], and/or low quality by depth [QD < 5]).
Because DA8 is extremely rare (only four affected families have been reported to date2–5), we excluded SNVs with an alternative allele frequency > 0.0001 in any population in the NHLBI Exome Sequencing Project Exome Variant Server dataset ESP6500, 1000 Genomes, the Exome Aggregation Consortium (ExAC v.1.0) Browser, or an internal database of ∼700 exomes. Additionally, SNVs that were flagged as low quality or potential false positives (QUAL ≤ 50, HRun > 4, QD < 5, presence within a cluster of SNPs) were also excluded from analysis. We generated copy-number variant (CNV) calls from exome data by using CoNIFER.11 Variants were annotated with the SeattleSeqAnnotation 138 server, and SNVs for which the only functional prediction label was “intergenic,” “coding-synonymous,” “utr,” “near-gene,” or “intron” were excluded. Individual genotypes with a depth < 6 or genotype quality < 20 were treated as missing in the analysis.
To exclude known causes of MPS and well-known conditions that include pterygia, we first assessed the exome data for pathogenic variants in the genes previously screened by Sanger sequencing (IRF6, CHRNG, CHRNA1, and CHRND), as well as in RAPSN9,12 (MIM: 601592) and DOK713 (MIM: 610285). No SNVs or CNVs in these genes segregated with DA8 in any of the three families tested. Next, we looked for novel and/or rare SNVs and CNVs in the same gene shared among affected probands. No genes with rare or novel variants were shared among all three families. Affected individuals in families A and B were also screened for larger CNVs via array comparative genomic hybridization on the Illumina Infinium HumanCore-24 BeadChip. No shared or overlapping CNVs were identified. Subsequently, we loosened our filtering criteria to include variants in genes that were shared by two out of three families and identified three candidate genes (SHPRH [MIM: 608048], SLC5A9, and MYH3 [MIM: 160720]) in families A and B.
The known functions of SHPRH and SLC5A9 appeared to be unrelated to the musculoskeletal phenotypes observed in all three families: SHPRH is involved in DNA repair and maintenance of genomic stability, and SLC5A9 encodes a sugar transporter primarily expressed in the small intestine and kidneys. In contrast, mutations in MYH3 frequently cause other forms of distal arthrogryposis, specifically DA2A and DA2B14 (MIM: 601680) and, more rarely, DA1.15,16 In addition to having congenital contractures, persons with DA2A and DA2B variably have short stature, scoliosis, and infrequently, pterygia of the neck.17 Accordingly, we considered MYH3 to be the most compelling candidate gene in these families.
In MYH3 (GenBank: NM_002470.3), we specifically discovered variants c.3214_3216dup (p.Asn1072dup) in family A and c.3224A>C (p.Gln1075Pro) in family B. Both variants were subsequently validated via Sanger sequencing and found to segregate with only the affected persons in each family (Figure 2). Both variants affect highly conserved amino acid residues, have identical genomic evolutionary rate profiling (GERP) scores of 5.64, and are predicted to be deleterious by multiple methods (e.g., p.Asn1072dup has a combined annotation-dependent depletion [CADD]18 score of 17.62, and p.Gln1075Pro has a CADD score of 20.8). Moreover, neither variant was found in more than 71,000 total control exomes recorded in ESP6500, 1000 Genomes phase 1 (November 2010 release), internal databases (>1,400 chromosomes), and the ExAC Browser (October 20, 2014, release).
Independently of the effort at the University of Washington to identify a gene associated with DA8, investigators at Baylor College of Medicine identified a five-generation family in whom ten persons were reported to be variably affected by a dominantly inherited condition characterized by short stature, scoliosis, multiple vertebral fusions, camptodactyly of the fingers, and multiple pterygia (Table 1; Figure S1), consistent with the diagnosis of DA8. Four individuals from this family were enrolled in a research protocol approved by the institutional review board at Baylor College of Medicine. Informed consent was obtained from each of these individuals prior to enrollment and prior to initiation of research studies. VCRome v.2.1 target-capture reagents (Roche Nimblegen) were used for performing whole-exome sequencing of DNA obtained from peripheral-blood monocytes from the proband (individual IV-3, Figure S1) and her father (individual III-5, Figure S1). 19 Sequencing was conducted on the Illumina HiSeq 2000 as previously described.19 Alignment, variant calling, and annotation were completed with the Mercury pipeline.20 Comparison of exome sequence data from the proband and her affected father (Table 1; Figure S1) identified 46 shared rare variants (i.e., frequency < 0.0001 in ESP6500, 1000 Genomes phase 1 [November 2010 release], and the ExAC Browser [October 20, 2014, release]). No variants were identified in SHPRH or SLC5A9; however, one MYH3 variant not found in any other databases, c.727_729del (p.Ser243del), was found to affect a highly conserved amino acid residue (GERP score of 4.94) and was predicted to be highly deleterious (CADD v.1.1 score of 18.47). This variant was validated by Sanger sequencing and segregated in all of the affected individuals for whom DNA was available for testing. This variant was not observed in ESP6500, the ExAC Browser (v.0.3, January 13, 2015, release), or 1000 Genomes (October 2014 release).
In 2006, we reported that mutations in MYH3 cause DA2A and DA2B.14 Nevertheless, MYH3 was never considered a high-priority candidate gene in individuals with DA8 because affected individuals had multiple pterygia of the limbs, severe scoliosis, and vertebral fusions and did not have contractures of the facial muscles. More recently, we and others identified mutations in MYH3 in DA1-affected persons who had no facial contractures15,16 and DA2B-affected persons who had vertebral fusions and severe congenital scoliosis,21 indicating that the phenotypic spectrum associated with MYH3 variants is broader than originally considered. This inference is now supported by our observation that variants in MYH3 cause DA8 and a wide range of phenotypic connective-tissue abnormalities, including congenital contractures, multiple pterygia, and bony fusions.
MYH3 encodes muscle embryonic myosin heavy chain (MYH3), a skeletal-muscle myosin composed of a globular motor domain (amino acid residues ∼1–779) attached by short neck (∼779–840) and hinge (∼840) regions to a long coiled-coil rod domain (∼840–1,940). The majority of the rod region comprises the myosin tail domain (amino acid residues ∼1,070–1,940). Embryonic myosin exists as a dimer in which the tail domains are intertwined (Figure 3). Hundreds of myosin dimers assemble with one another and other proteins to form the thick filaments of the sarcomere, which is the subcellular contractile apparatus of cardiac-muscle and skeletal-muscle cells.
Figure 3.
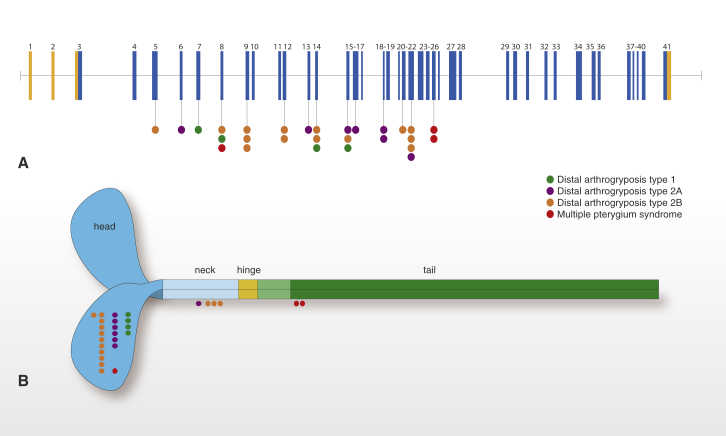
Genomic Model of MYH3 and Stylized Structure of Embryonic Myosin Illustrate the Spectrum of Mutations that Cause DA Syndromes
MYH3 is composed of 41 exons, 39 of which are protein coding (blue) and two of which are non-coding (orange).
(A) Lines with attached dots indicate the approximate locations of MYH3 mutations that cause DA1 (green), DA2A (purple), DA2B (orange), and DA8 (red). The color of each dot reflects the diagnosis.
(B) Schematic of an embryonic myosin dimer. The approximate locations of MYH3 mutations relative to each affected amino acid residue in the myosin domain are indicated by colored circles. All mutations causing DA1, DA2A, and DA2B affect amino acid residues located in the motor and neck of embryonic myosin, whereas the two mutations causing DA8 are the only mutations that alter amino acid residues located in the tail. (Both variants in the tail of MYH3 [c.4934A>C (p.Asp1622Ala) and c.2979C>T (p.Ala1637Val)] and reported by Toydemir et al.14 are now thought to be polymorphisms on the basis of additional family information and frequencies in large databases of control populations.)
All of the mutations known to cause DA1, DA2A, and DA2B16 affect amino acid residues in the head and neck domains of embryonic myosin, whereas two of the three mutations that cause DA8 affect the tail domain. The clinical characteristics of the DA8-affected individuals with a mutation affecting the tail domain were more alike than the features of DA8-affected persons with a mutation affecting the head domain. This observation is limited, of course, by the fact that our dataset contained a small number of both families and individuals with DA8 caused by MYH3 mutations. Nevertheless, speculation on domain-specific, or at least domain-predominant, phenotypes has been reported previously for mutations in both muscle and non-muscle myosins. For example, mutations affecting the head domain of MYH9 (encoded by MYH9 [MIM: 160775]) cause severe thrombocytopenia and are associated with nephritis and deafness, whereas mutations affecting the tail domain cause less severe thrombocytopenia, such that median platelet counts are more than two times higher than those associated with head-domain mutations.22 Similarly, mutations throughout MYH7 (MIM: 160760) can cause hypertrophic (MIM: 192600) and/or dilated cardiomyopathy (MIM: 613426), whereas skeletal myopathies (e.g., myosin-storage myopathy [MIM: 608358] and Laing distal myopathy [MIM: 160500]) are caused almost exclusively by mutations affecting the tail domain of MYH7.23 Moreover, mutations affecting the tail of MYH7 disrupt stability or self-assembly of sarcomere filaments or protein-protein interactions mediated through the tail, whereas mutations affecting the motor appear to alter the ATPase and actin-binding properties of myosin.24 This precedent of domain-specific phenotypic differences associated with mutations in other myosin-encoding genes suggests that similar genotype-phenotype relationships might exist in MYH3.
Skeletal abnormalities have not commonly been observed, or at least reported, in persons with DA1, DA2A, or DA2B and with mutations in MYH3. Yet, skeletal defects were found in each of the three DA8-affected families carrying an MYH3 mutation in our study. The most common defects observed were vertebral abnormalities, including hemivertebrae and vertebral fusions of C1 and C2 or vertebrae of the thoracolumbar spine (Figure 2). A similar range of vertebral abnormalities, as well as both carpal and tarsal fusions, has been reported previously2–5 in families affected by dominantly inherited MPS. In addition, two persons in family C had craniosynostosis. These observations suggest that embryonic myosin plays a role in skeletal development, which is perturbed by the MYH3 mutations that cause DA8. Such a role for embryonic myosin would not be unprecedented given that other proteins of the contractile apparatus of skeletal muscle, such as that encoded by troponin I type 2 (skeletal, fast) (TNNI2 [MIM: 191043]), are present in osteoblasts and chondrocytes in long-bone growth plates and play a role in skeletal development.25 Moreover, multiple myosins have been reported to interact with Runx2 in rat osteoblasts, and osteoblast differentiation in vitro has been associated with the expression of myosin.26
To investigate this possibility further, we used MYH3-cDNA-specific primers that we had previously developed27 to test for MYH3 expression in a variety of human fetal tissues. As expected, MYH3 was expressed in the thymus, placenta, heart, and liver (Figure 4A). However, whereas MYH3 expression was not detectable in cartilage, it was expressed in bone (Figure 4A). Next, to determine the relative expression levels of MYH3, we conducted a qPCR assay and confirmed MYH3 expression in bone at levels comparable to those in several other tissues, including the brain, with known MYH3 expression28 (Figure 4B). These preliminary findings suggest that the bony fusions in individuals with DA8 result from disruption of an unappreciated role of embryonic myosin in vertebral development, and perhaps in skeletal development in general.
Figure 4.
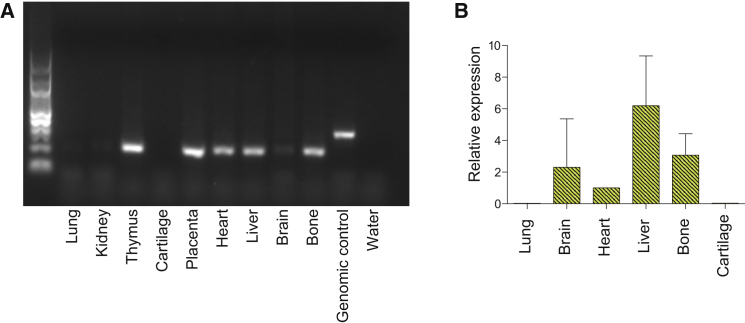
Expression of MYH3 in Bone and Cartilage
(A) RNA was isolated from a 17-week-old fetus, and cDNA was generated according to standard protocols.27MYH3 primers were predicted to yield 313-bp amplicons from genomic DNA (control) and 206-bp amplicons from cDNA. MYH3 cDNA expression was detected in the bone, thymus, placenta, heart, brain, and liver. Expression was not detected in cartilage, lungs, or kidneys.
(B) qPCR was used for assessing the relative expression levels of MYH3 cDNA in bone, cartilage, brain, heart, liver, and lungs. MYH3 cDNA expression was detected in bone at a level similar to that observed in the brain and heart and was barely detectable in the lungs and cartilage.
Five simplex MPS cases referred to our center included clinical characteristics that overlapped with DA8 and in several instances were indistinguishable from DA8. Exome sequencing did not reveal MYH3 mutations in any of the affected individuals, but three of their cases were explained by mutations in CHRNG. Specifically, two persons were compound heterozygous (c.[753_754delCT];[805+1G>A], p.[Val253Alafs∗44];[?] and c.[459dupA];[1082dupC], p.[Val265Serfs∗24];[Leu362Alafs∗36]) and one was homozygous (c.459dupA [p.Val154Serfs∗24]) for CHRNG mutations (GenBank: NM_005199.4). Two of these mutations, c.753_754delCT and c.459dupA, have been reported in multiple individuals with recessively inherited MPS1,8 and have been described as “recurrent.” Yet, both of these mutations are in ESP6500 and the ExAC Browser at frequencies as high as 0.002 (e.g., c.753_754delCT in European Americans in ESP6500), suggesting that standing rather than de novo variation contributes to the relatively high incidence of MPS caused by mutations in CHRNG. Moreover, c.459dupA occurs on an identical ∼700-kb haplotype in each of the apparently unrelated MPS-affected persons whom we tested. Therefore, the c.459dupA mutation most likely arose many generations ago, and previous attempts to identify the shared haplotype failed because of its small size.1
Our findings collectively suggest that features such as scoliosis and pterygia observed in individuals with DA8 most likely have a different underlying molecular mechanism(s) than that in recessive forms of MPS caused by mutations in the genes CHRNG, CHRND, and CHRNA1,9 encoding subunits of the skeletal-muscle nicotinic acetylcholine receptor (AChR). AChR is required for normal muscle development, and muscle biopsies from individuals with recessive MPS have demonstrated abnormal distribution of AChR and acetylcholinesterase.29 Because AChR is necessary for proper organization and establishment of the neuromuscular junction,30 as well as muscle and synaptic maturation,31 it has been suggested that many features of recessive MPS, such as joint contractures and pterygia, are ultimately caused by fetal akinesia7 as a result of abnormal muscle and synaptic development. In contrast, MYH3 mutations that cause DA2A and/or DA2B are hypothesized to alter contractile function of skeletal myofibers during development, resulting in congenital contractures and altered movement in utero. Consistent with this hypothesis, we have recently demonstrated that the MYH3 variant c.2014C>T (p.Arg672Cys), the most common cause of DA2A, causes reduced contractile force and prolonged relaxation of human skeletal myofibers.27 Together, these findings suggest that pterygia in persons with DA8 develop differently than in persons with autosomal-recessive MPS.
We failed to identify the cause of dominantly inherited MPS in one family originally diagnosed with DA8 (family D in Table 1 and Figure S1). In retrospect, the phenotype in this family differs from that found in the other DA8-affected families. Specifically, neither of the affected persons in this family have popliteal or antecubital ptergyia, and both persons lack vertebral fusions. Additionally, each individual has a ventriculoseptal defect. Accordingly, although the lack of an MYH3 mutation in this family could be interpreted as evidence that DA8 is genetically heterogeneous, the alternative is that the phenotype in this family is distinctive enough to prompt delineation of a separate Mendelian condition.
In summary, we used exome sequencing to discover that mutations in MYH3 cause DA8. The phenotypic overlap among persons with DA8, coupled with physical findings distinct from other DA conditions caused by mutations in MYH3, suggests that the developmental mechanism underlying DA8 differs from that underlying other DA conditions and/or that certain functions of embryonic myosin might be perturbed by disruption of specific residues and/or domains.
Consortia
The members of the University of Washington Center for Mendelian Genomics are Michael J. Bamshad, Jay Shendure, Deborah A. Nickerson, Gonçalo R. Abecasis, Peter Anderson, Elizabeth Marchani Blue, Marcus Annable, Brian L. Browning, Kati J. Buckingham, Christina Chen, Jennifer Chin, Jessica X. Chong, Gregory M. Cooper, Colleen P. Davis, Christopher Frazar, Tanya M. Harrell, Zongxiao He, Preti Jain, Gail P. Jarvik, Guillaume Jimenez, Eric Johanson, Goo Jun, Martin Kircher, Tom Kolar, Stephanie A. Krauter, Niklas Krumm, Suzanne M. Leal, Daniel Luksic, Colby T. Marvin, Margaret J. McMillin, Sean McGee, Patrick O’Reilly, Bryan Paeper, Karynne Patterson, Marcos Perez, Sam W. Phillips, Jessica Pijoan, Christa Poel, Frederic Reinier, Peggy D. Robertson, Regie Santos-Cortez, Tristan Shaffer, Cindy Shephard, Kathryn M. Shively, Deborah L. Siegel, Joshua D. Smith, Jeffrey C. Staples, Holly K. Tabor, Monica Tackett, Jason G. Underwood, Marc Wegener, Gao Wang, Marsha M. Wheeler, and Qian Yi.
Acknowledgments
We thank the families for their participation and support; Christa Poel, Karynne Patterson, Jennifer Chin, Yuqing Chen, Alyssa Tran, Philippe Campeau, and James Lu for technical assistance; and Alice Ward Racca for helpful comments. Our work was supported in part by grants from the NIH National Human Genome Research Institute and NHLBI (1U54HG006493 to M.B., D.N., and J.S.; 1RC2HG005608 to M.B., D.N., and J.S.; 5R000HG004316 to H.K.T.), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD; 1R01HD048895 to M.J.B.), the Life Sciences Discovery Fund (2065508 and 0905001), and the Washington Research Foundation. This work was also supported by the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (HD024064), funded by the Eunice Kennedy Shriver NICHD (U54HG003273 and U54 U54HG006542 to R.A.G.; T32GM07526 to M.J. and L.C.B.; P01HD070394 to B.L.). L.C.B. was also supported by the Medical Genetics Training Award in Clinical Biochemical Genetics from Genzyme and the Annual Clinical Genetics Meeting Foundation for Genetic and Genomic Medicine, the National Urea Cycle Disorders Foundation Fellowship, and a fellowship from the Urea Cycle Disorders Consortium (U54HD061221), which is a part of the NIH Rare Disease Clinical Research Network, supported through collaboration among the Office of Rare Diseases Research, the National Center for Advancing Translational Science, and the Eunice Kennedy Shriver NICHD. D.K. was supported by the NIH National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR066124).
Published: May 7, 2015
Footnotes
Supplemental Data include one figure and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.04.004.
Contributor Information
Michael J. Bamshad, Email: mbamshad@uw.edu.
University of Washington Center for Mendelian Genomics:
Michael J. Bamshad, Jay Shendure, Deborah A. Nickerson, Gonçalo R. Abecasis, Peter Anderson, Elizabeth Marchani Blue, Marcus Annable, Brian L. Browning, Kati J. Buckingham, Christina Chen, Jennifer Chin, Jessica X. Chong, Gregory M. Cooper, Colleen P. Davis, Christopher Frazar, Tanya M. Harrell, Zongxiao He, Preti Jain, Gail P. Jarvik, Guillaume Jimenez, Eric Johanson, Goo Jun, Martin Kircher, Tom Kolar, Stephanie A. Krauter, Niklas Krumm, Suzanne M. Leal, Daniel Luksic, Colby T. Marvin, Margaret J. McMillin, Sean McGee, Patrick O’Reilly, Bryan Paeper, Karynne Patterson, Marcos Perez, Sam W. Phillips, Jessica Pijoan, Christa Poel, Frederic Reinier, Peggy D. Robertson, Regie Santos-Cortez, Tristan Shaffer, Cindy Shephard, Kathryn M. Shively, Deborah L. Siegel, Joshua D. Smith, Jeffrey C. Staples, Holly K. Tabor, Monica Tackett, Jason G. Underwood, Marc Wegener, Gao Wang, Marsha M. Wheeler, and Qian Yi
Web Resources
The URLs for data presented herein are as follows:
ExAC Browser, http://exac.broadinstitute.org/
Human Genome Variation Society, http://www.hgvs.org/mutnomen/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
SeattleSeq Annotation 138, http://snp.gs.washington.edu/SeattleSeqAnnotation138/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Vogt J., Morgan N.V., Rehal P., Faivre L., Brueton L.A., Becker K., Fryns J.P., Holder S., Islam L., Kivuva E. CHRNG genotype-phenotype correlations in the multiple pterygium syndromes. J. Med. Genet. 2012;49:21–26. doi: 10.1136/jmedgenet-2011-100378. [DOI] [PubMed] [Google Scholar]
- 2.Frias, J.L., Holahan, J.R., Rosenbloom, A.L., and Felman, A.H. (1973). An autosomal dominant syndrome of multiple pterygium, ptosis, and skeletal abnormalities. Proceedings of the Fourth International Conference on Birth Defects. Excerpta Medica 19.
- 3.Kawira E.L., Bender H.A. An unusual distal arthrogryposis. Am. J. Med. Genet. 1985;20:425–429. doi: 10.1002/ajmg.1320200302. [DOI] [PubMed] [Google Scholar]
- 4.McKeown C.M., Harris R. An autosomal dominant multiple pterygium syndrome. J. Med. Genet. 1988;25:96–103. doi: 10.1136/jmg.25.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prontera P., Sensi A., Merlo L., Garani G., Cocchi G., Calzolari E. Familial occurrence of multiple pterygium syndrome: expression in a heterozygote of the recessive form or variability of the dominant form? Am. J. Med. Genet. A. 2006;140:2227–2230. doi: 10.1002/ajmg.a.31428. [DOI] [PubMed] [Google Scholar]
- 6.Bamshad M., Jorde L.B., Carey J.C. A revised and extended classification of the distal arthrogryposes. Am. J. Med. Genet. 1996;65:277–281. doi: 10.1002/(SICI)1096-8628(19961111)65:4<277::AID-AJMG6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 7.Hoffmann K., Müller J.S., Stricker S., Mégarbané A., Rajab A., Lindner T.H., Cohen M., Chouery E., Adaimy L., Ghanem I. Escobar syndrome is a prenatal myasthenia caused by disruption of the acetylcholine receptor fetal γ subunit. Am. J. Hum. Genet. 2006;79:303–312. doi: 10.1086/506257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan N.V., Brueton L.A., Cox P., Greally M.T., Tolmie J., Pasha S., Aligianis I.A., van Bokhoven H., Marton T., Al-Gazali L. Mutations in the embryonal subunit of the acetylcholine receptor (CHRNG) cause lethal and Escobar variants of multiple pterygium syndrome. Am. J. Hum. Genet. 2006;79:390–395. doi: 10.1086/506256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michalk A., Stricker S., Becker J., Rupps R., Pantzar T., Miertus J., Botta G., Naretto V.G., Janetzki C., Yaqoob N. Acetylcholine receptor pathway mutations explain various fetal akinesia deformation sequence disorders. Am. J. Hum. Genet. 2008;82:464–476. doi: 10.1016/j.ajhg.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo S., Schutte B.C., Richardson R.J., Bjork B.C., Knight A.S., Watanabe Y., Howard E., de Lima R.L.L.F., Daack-Hirsch S., Sander A. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat. Genet. 2002;32:285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E., NHLBI Exome Sequencing Project Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogt J., Harrison B.J., Spearman H., Cossins J., Vermeer S., ten Cate L.N., Morgan N.V., Beeson D., Maher E.R. Mutation analysis of CHRNA1, CHRNB1, CHRND, and RAPSN genes in multiple pterygium syndrome/fetal akinesia patients. Am. J. Hum. Genet. 2008;82:222–227. doi: 10.1016/j.ajhg.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogt J., Morgan N.V., Marton T., Maxwell S., Harrison B.J., Beeson D., Maher E.R. Germline mutation in DOK7 associated with fetal akinesia deformation sequence. J. Med. Genet. 2009;46:338–340. doi: 10.1136/jmg.2008.065425. [DOI] [PubMed] [Google Scholar]
- 14.Toydemir R.M., Rutherford A., Whitby F.G., Jorde L.B., Carey J.C., Bamshad M.J. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nat. Genet. 2006;38:561–565. doi: 10.1038/ng1775. [DOI] [PubMed] [Google Scholar]
- 15.Alvarado D.M., Buchan J.G., Gurnett C.A., Dobbs M.B. Exome sequencing identifies an MYH3 mutation in a family with distal arthrogryposis type 1. J. Bone Joint Surg. Am. 2011;93:1045–1050. doi: 10.2106/JBJS.J.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck A.E., McMillin M.J., Gildersleeve H.I., Kezele P.R., Shively K.M., Carey J.C., Regnier M., Bamshad M.J. Spectrum of mutations that cause distal arthrogryposis types 1 and 2B. Am. J. Med. Genet. A. 2013;161A:550–555. doi: 10.1002/ajmg.a.35809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevenson D.A., Carey J.C., Palumbos J., Rutherford A., Dolcourt J., Bamshad M.J. Clinical characteristics and natural history of Freeman-Sheldon syndrome. Pediatrics. 2006;117:754–762. doi: 10.1542/peds.2005-1219. [DOI] [PubMed] [Google Scholar]
- 18.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupski J.R., Gonzaga-Jauregui C., Yang Y., Bainbridge M.N., Jhangiani S., Buhay C.J., Kovar C.L., Wang M., Hawes A.C., Reid J.G. Exome sequencing resolves apparent incidental findings and reveals further complexity of SH3TC2 variant alleles causing Charcot-Marie-Tooth neuropathy. Genome Med. 2013;5:57. doi: 10.1186/gm461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beck A.E., McMillin M.J., Gildersleeve H.I., Shively K.M., Tang A., Bamshad M.J. Genotype-phenotype relationships in Freeman-Sheldon syndrome. Am. J. Med. Genet. A. 2014;164A:2808–2813. doi: 10.1002/ajmg.a.36762. [DOI] [PubMed] [Google Scholar]
- 22.Pecci A., Panza E., Pujol-Moix N., Klersy C., Di Bari F., Bozzi V., Gresele P., Lethagen S., Fabris F., Dufour C. Position of nonmuscle myosin heavy chain IIA (NMMHC-IIA) mutations predicts the natural history of MYH9-related disease. Hum. Mutat. 2008;29:409–417. doi: 10.1002/humu.20661. [DOI] [PubMed] [Google Scholar]
- 23.Tajsharghi H., Oldfors A. Myosinopathies: pathology and mechanisms. Acta Neuropathol. 2013;125:3–18. doi: 10.1007/s00401-012-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armel T.Z., Leinwand L.A. Mutations in the beta-myosin rod cause myosin storage myopathy via multiple mechanisms. Proc. Natl. Acad. Sci. USA. 2009;106:6291–6296. doi: 10.1073/pnas.0900107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X., Wang F., Zhao Y., Yang P., Chen J., Sun H., Liu L., Li W., Pan L., Guo Y. A gain-of-function mutation in Tnni2 impeded bone development through increasing Hif3a expression in DA2B mice. PLoS Genet. 2014;10:e1004589. doi: 10.1371/journal.pgen.1004589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J., Lazarenko O.P., Blackburn M.L., Shankar K., Badger T.M., Ronis M.J.J., Chen J.-R. Feeding blueberry diets in early life prevent senescence of osteoblasts and bone loss in ovariectomized adult female rats. PLoS ONE. 2011;6:e24486. doi: 10.1371/journal.pone.0024486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Racca A.W., Beck A.E., McMillin M.J., Korte F.S., Bamshad M.J., Regnier M. The embryonic myosin R672C mutation that underlies Freeman-Sheldon syndrome impairs cross-bridge detachment and cycling in adult skeletal muscle. Hum. Mol. Genet. 2015 doi: 10.1093/hmg/ddv084. Published online March 3, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong J.X., McMillin M.J., Shively K.M., Beck A.E., Marvin C.T., Armenteros J.R., Buckingham K.J., Nkinsi N.T., Boyle E.A., Berry M.N., University of Washington Center for Mendelian Genomics De Novo Mutations in NALCN Cause a Syndrome Characterized by Congenital Contractures of the Limbs and Face, Hypotonia, and Developmental Delay. Am. J. Hum. Genet. 2015;96:462–473. doi: 10.1016/j.ajhg.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson K.G., Viereck M.J., Margiotta M.V., Gripp K.W., Abdul-Rahman O.A., Akins R.E. Neuromotor synapses in Escobar syndrome. Am. J. Med. Genet. A. 2013;161A:3042–3048. doi: 10.1002/ajmg.a.36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koenen M., Peter C., Villarroel A., Witzemann V., Sakmann B. Acetylcholine receptor channel subtype directs the innervation pattern of skeletal muscle. EMBO Rep. 2005;6:570–576. doi: 10.1038/sj.embor.7400429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi M., Kubo T., Mizoguchi A., Carlson C.G., Endo K., Ohnishi K. Spontaneous muscle action potentials fail to develop without fetal-type acetylcholine receptors. EMBO Rep. 2002;3:674–681. doi: 10.1093/embo-reports/kvf128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.