Abstract
BACKGROUND
Cystic fibrosis is a life-threatening genetic disorder that has been associated with mutations in the CFTR [cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7)] gene. Hundreds of CFTR mutations have been detected to date. Current CFTR genotyping assays target a subset of these mutations, particularly a mutation panel recommended by the American College of Medical Genetics for carrier screening of the general population. Fast sequencing of the entire coding sequence in a scalable manner could expand the detection of CFTR mutations and facilitate management of costs and turnaround times in the clinical laboratory.
METHODS
We describe a proof-of-concept CFTR assay that uses PCR target enrichment and next-generation sequencing on the Ion Torrent Personal Genome Machine™ (PGM™) platform.
RESULTS
The scalability of the assay was demonstrated, with an average mean depth of coverage ranging from 500× to 3500×, depending on the number of multiplexed patient samples and the Ion Torrent chip used. In a blinded study of 79 previously genotyped patient DNA samples and cell lines, our assay detected most of the mutations, including single-nucleotide variants, small insertions and deletions, and large copy-number variants. The reproducibility was 100% for detecting mutations in independent runs. Our assay demonstrated high specificity, with only 2 false-positive calls (at 2184delA) found in 2 samples caused by a sequencing error in a homopolymer stretch of sequence. The detection rate for variants of unknown significance was very low in the targeted region.
CONCLUSIONS
With continued optimization and system refinements, PGM sequencing promises to be a powerful, rapid, and scalable means of clinical diagnostic sequencing.
Cystic fibrosis (CF)6 is a common autosomal recessive disorder caused by mutations in the CFTR [cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7)] gene, which is located at 7q31.2. More than 1900 different, heterogeneous mutations have been documented, with their distributions varying among populations (1). CF prevalence is highest among Caucasians (1 in 2500) and Ashkenazi Jews (1 in 2300), with carrier frequencies of 1 in 29 and 1 in 27, respectively (2, 3). Both the American College of Medical Genetics (ACMG) and the American College of Obstetricians and Gynecologists have recommended the use of a panethnic screening panel as a standard of care for CF carrier testing in the general population. This panel consists of 23 CFTR gene mutations with an allele frequency of at least 0.1% in the US general population (4, 5).
Screening tests covering all 23 CFTR mutations have a detection rate ranging from 94% in Ashkenazi Jews to as low as 49% in Asian Americans (5). Hence, fast and scalable screening of the entire coding sequence of the CFTR gene would increase the sensitivity for detecting “pathogenic” mutations in the general population and would allow management of costs and turnaround times in laboratory medicine. Next-generation sequencing using the Ion Torrent semiconductor technology has the potential to provide such a comprehensive, rapid, and scalable approach for detecting CFTR mutations.
We describe a proof-of-concept study to show the efficacy in the clinical laboratory of detecting different classes of mutations in the CFTR coding sequence. For this purpose, we used PCR-based target enrichment and next-generation sequencing on the Ion Torrent Personal Genome Machine™ (PGM™).
Materials and Methods
DNA SAMPLES
We tested 79 DNA samples—22 cell lines from the Coriell Institute with known CFTR mutations and 57 peripheral blood samples previously genotyped at the Dartmouth–Hitchcock Medical Center with the xTAG® Cystic Fibrosis 60 Kit v2 (Luminex). The latter assay detects 64 CFTR mutations and variants, including the 23 mutations and 4 variants (benign polymorphisms) recommended by the ACMG (4), as well as 37 additional mutations with a broad ethnicity coverage. Of the 79 samples, 24 had 2 deleterious mutations in the CFTR gene, 46 were carriers of a single pathogenic mutation, and 9 had no CFTR mutations detectable with the xTAG kit (Table 1). All human DNA samples were part of an anonymized DNA bank approved by the Institutional Committee for the Protection of Human Subjects.
Table 1.
Tested samples.
| Mutation (+)a |
cDNA position | Variant effect | Phenotype | Identified | Variant reads, % |
Source | No. of cases |
|---|---|---|---|---|---|---|---|
| +/+ | c.1521_1523delCTT; c.1521_1523delCTT | ΔF508; ΔF508 | CF | Yes | 97 | Dartmouth | 1 |
| +/+ | c.1521_1523delCTT; c.350G>A | ΔF508; R117H | CF | Yes | 53; 50 | Dartmouth | 1 |
| +/+ | c.350G>A; c.1477C>T | R117H; Q493*b | CF | Yes | 52; 49 | Dartmouth | 1 |
| +/+ | c.1521_1523delCTT; c.1000C>T | ΔF508; R334W | CF | Yes | 49; 54 | Dartmouth | 1 |
| +/+ | c.1521_1523delCTT; c.489+1G>T | ΔF508; 621+1G>T | CF | Yes | 48; 47 | Dartmouth | 1 |
| +/+ | c.1521_1523delCTT; c.1364C>A | ΔF508; A455E | CF | Yes | 51; 46 | Dartmouth | 1 |
| +/+ | c.489+1G>T; c.2988+1G>A | 621+1G>T; 3120+1G>A | CF | Yes | 48; 49 | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.1657C>T | ΔF508; R553* | CF | Yes | 44; 49c | Coriell | 2 |
| +/+ | c.1521_1523delCTT; c.3528delC | ΔF508; 3659delC | CF | Yes | 46; 46 | Coriell | 1 |
| +/+ | c.489+1G>T; c.579+1G>T | 621+1G>T; 711+1G>T | CF | Yes | 50; 51 | Coriell | 1 |
| +/+ | c.489+1G>T; c.254G>A | 621+1G>T; G85E | CF | Yes | 50; 45 | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.1679G>C | ΔF508; R560T | CF | Yes | 44; 52 | Coriell | 1 |
| +/+ | c.489+1G>T; c.1364C>A | 621+1G>T; A455E | CF | Yes | 50; 49 | Coriell | 1 |
| +/+ | c.3909C>G; c.4046G>A | N1303K; G1349D | CF | Yes | 47; 52 | Coriell | 1 |
| +/+ | c.2657+5G>A; c.2657+5G>A | 2789+5G>A; 2789+5G>A | CF | Yes | 100 | Coriell | 1 |
| +/+ | c.1040G>C; c.1652G>A | R347P; G551D | CF | Yes | 51; 49 | Coriell | 1 |
| +/+ | c.1000C>T; c.3368–2A>T | R334W; 3500–2A>G | CF | Yes | 53; 45 | Coriell | 1 |
| +/+ | c.254G>A; c.3454G>C | G85E; D1152H | CF | Yes | 44; 47 | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.350G>A | ΔF508; R117H | CF | Yes | 49; 50 | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.54–5940_273+10250del21kb | ΔF508; CFTRdel2,3 | CF | Yes | 47; N/Ad | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.1766+1G>A | ΔF508; 1898+1G>A | CF | Yes | 47; 50 | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.2051_2052delAAinsG | ΔF508; K684Sfs | CF | Yes | 47; 50 | Coriell | 1 |
| +/+ | c.1521_1523delCTT; c.2052del | ΔF508; K684Nfs*38 | CF | Yes | 51; 55 | Coriell | 1 |
| +/− | c.1521_1523delCTT | ΔF508 | Carrier | Yes | 50c | Dartmouth | 16 |
| +/− | c.1652G>A | G551D | Carrier | Yes | 50c | Dartmouth | 5 |
| +/− | c.1519_1521delATC | ΔI507 | Carrier | Yes | 46 | Dartmouth | 1 |
| +/− | c.3454G>C | D1152H | Carrier | Yes | 50 | Dartmouth | 1 |
| +/− | c.1657C>T | R553* | Carrier | Yes | 51 | Dartmouth | 1 |
| +/− | c.178G>T | E60* | Carrier | Yes | 51 | Dartmouth | 1 |
| +/− | c.3846G>A | W1282* | Carrier | Yes | 45c | Dartmouth | 3 |
| +/− | c.1000C>T | R334W | Carrier | Yes | 51 | Dartmouth | 1 |
| +/− | c.1624G>T | G542* | Carrier | Yes | 47c | Dartmouth | 4 |
| +/− | c.3484C>T | R1162* | Carrier | Yes | 43 | Dartmouth | 1 |
| +/− | c.1766+1G>A | 1898+1G>A | Carrier | Yes | 57 | Dartmouth | 1 |
| +/− | c.3773_3774insT | 3905insT (L1258Ffs*7) | Carrier | Yes | 37 | Dartmouth | 1 |
| +/− | c.350G>A | R117H | Carrier | Yes | 50c | Dartmouth | 3 |
| +/− | c.1645A>C | S549R A>C | Carrier | No | N/A | Dartmouth | 1 |
| +/− | c.1040G>A | R347H | Carrier | Yes | 47 | Dartmouth | 1 |
| +/− | c.3909C>G | N1303K | Carrier | Yes | 46 | Dartmouth | 1 |
| +/− | c.3718–2477C>T | 3849+10kbC>T | Carrier | Yes | 51 | Coriell | 1 |
| +/− | c.2988+1G>A | 3120+1G>A | Carrier | Yes | 49 | Coriell | 1 |
| +/− | c.489+1G>T | 621+1G>T | Carrier | Yes | 50 | Coriell | 1 |
| +/− | c.1585–1G>A | 1717–1G>A | Carrier | Yes | 51 | Coriell | 1 |
| +/−e | N/Af | N/A | Normal | N/A | N/A | Dartmouth | 9 |
+/+, 2 pathogenic mutations; +/−, carrier of a single pathogenic mutation; −/−, absence of any pathogenic mutations.
, codon termination.
The mean percentage of variant reads was used for multiple samples of same genotype.
See text and Figure 1 for detection of large deletions.
Based on the 64 mutations covered by the xTAG Cystic Fibrosis Assay.
N/A, not applicable.
MULTIPLEX PCR ENRICHMENT, LIBRARY CONSTRUCTION, AND MASSIVELY PARALLEL SEQUENCING
DNA was previously extracted with the EZ1 robotic system (Qiagen). Twenty nanograms of each genomic DNA sample were used for PCR enrichment of CFTR targets by applying a custom AmpliSeq™ panel (Life Technologies). The panel consisted of 2 separate PCR primer pools; each pool required 10 ng of input DNA and produced 36 amplicons. The complete CFTR assay has 72 amplicons (mean size, 148 bp) covering all CFTR exons and 20 bp of all intron/splice sites, for a total of 10 343 targeted bases (see Table 1 in the Data Supplement that accompanies the online version of this article at http://www.clinchem.org/content/vol59/issue10). After each pool had undergone 20 PCR cycles, the PCR primers were removed, the targeted amplicon regions were end-repaired (Ion AmpliSeq Library Kit 2.0; Life Technologies), and the amplicons were ligated to sequencing adaptors containing a unique bar code and specific for the Ion Torrent sequencing platforms (Ion Xpress™ Barcode Adapters Kit; Life Technologies). After purification with AMPure XP beads (Beckman Coulter), bar-coded libraries were quantified with the Ion Library Quantification Kit (Life Technologies). Equimolar concentrations of a desired number of barcoded libraries were pooled to achieve a final concentration of 15 pmol/L.
We used the Ion OneTouch™ system to clonally amplify pooled bar-coded libraries on Ion Sphere™ particles. The subsequently enriched template-positive Ion Sphere particles were treated with sequencing primer and polymerase (Ion PGM 200 Sequencing Kit; Life Technologies), loaded onto an Ion 314™ or 318™ chip (with 10 Mb and 1 Gb of sequence data output, respectively) and sequenced on the PGM with the 200-bp single-end run configuration (see the full Ion Torrent AmpliSeq work flow at http://ioncommunity.lifetechnologies.com).
MUTATIONAL ANALYSIS
Torrent Suite™ software was used to compare base calls, read alignments, and variant calling with the reference CFTR human genomic sequence (hg19). The variants selected for further analysis met the following criteria: (a) The variant was detected in sequence reads for both strands, (b) a minimum coverage of 20× was achieved, (c) the variant frequency in the population was <1%, and (d) variant reads represented >25% of the sequence reads at a particular site (variant read threshold). Variant calls from 5 noncoding homopolymer sites within the targeted CFTR region were filtered out to avoid homopolymer-associated base-calling errors. The filtered variants were then compared with mutation databases, including the Exome Variant Server (Exome Sequencing Project, http://evs.gs.washington.edu/EVS/), dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/), the Clinical and Functional Translation of CFTR database (http://www.cftr2.org/index.php), the Cystic Fibrosis Mutation Database (http://www.genet.sickkids.on.ca/app), and GeneCards (http://www.genecards.org/cgi-bin/carddisp.pl?gene=CFTR&snp=3046#snp). All databases were last accessed in January 2013. Novel variants of unknown significance (VUS) (i.e., not found in any of the clinical databases) were analyzed with the in silico tools SIFT (6) and MutationTaster (7) to predict potential protein deleterious effects and assess evolutionary conservation.
ANALYSIS OF COPY NUMBER VARIANTS
For each sample, we derived the total coverage across each exon/target by using the Depth Of Coverage tool from the Genome Analysis Toolkit (GATK, http://www.broadinstitute.org/gatk/). These exon-level coverage values were divided by the net coverage across all exons to arrive at fractional-coverage values for each exon that could be compared across samples (i.e., exon fractional coverage = (exon coverage)/(sum of all exon coverages). We next calculated the log2 ratio of each fractional-coverage value divided by the median across the batch. Log2 ratios less than −0.7 for at least 2 consecutive targets were interpreted as deletions.
Results
CFTR ASSAY
The objective of this study was to evaluate our AmpliSeq custom assay (PCR-based target enrichment followed by massively parallel sequencing on the PGM) for detecting inherited CFTR mutations in the clinical laboratory setting. By targeting all CFTR exons and 20 bp of all exon/intron boundaries (approximately 10.3 kb targeted bases; see Table 1 in the online Data Supplement), this design covers 159 of the 160 variants listed in the Clinical and Functional Translation of CFTR database (http://www.cftr2.org), including the panel of 23 mutations recommended for testing by the ACMG. The single intronic single-nucleotide polymorphism that was not included in this design (c.1679+1.6kbA>G) will be addressed in future iterations of this assay.
Using our custom assay, we first prepared PCR amplicon libraries for the 79 DNA samples to be sequenced in this study (Table 1). To demonstrate the scalability of our assay, we pooled different numbers of bar-coded sample libraries on either a 314 or 318 chip for sequencing on the PGM (Table 2). When 12 samples were sequenced on a 318 chip, we obtained an average mean depth of coverage of 3525×, with 99% of the targeted bases having at least 100× coverage. On the other hand, sequencing of 35 bar-coded sample libraries on a 318 chip yielded an average mean depth of coverage of 1151×, and 96% of the targeted bases had at least 100× coverage. Furthermore, an average mean read depth of approximately 500× was obtained when 12 samples were sequenced on a 314 chip, with 93% of the targeted bases at occurring with at least 100× coverage (Table 2). Hence, our scalable assay is able to multiplex a flexible number of patient samples in the clinical setting, thereby allowing better management of costs and turnaround times while maintaining the robust read depth necessary for detecting different classes of inherited CFTR mutations (see below).
Table 2.
CFTR assay scalability.a
| Chip | Pooled samples, n |
Mapped reads, n |
Reads on target |
Depth of coverage |
Coverage uniformity |
Bases at 100× |
|---|---|---|---|---|---|---|
| 314 | 5 | 123 086 | 97% | 1575× | 97% | 100% |
| 314 | 12 | 43 786 | 97% | 532× | 93% | 93% |
| 318 | 12 | 291 631 | 98% | 3525× | 96% | 99% |
| 318 | 35 | 98 670 | 93% | 1151× | 92% | 96% |
Calculated are the mean values per sample in each run.
Overall, we identified 515 variants after sequencing all 79 samples, for a mean of approximately 7 variants per sample. After applying our filters, 98 variants remained, including 90 pathogenic mutations (of the expected 91; see below), 6 VUS, and a single base-calling error (2184delA, c.2052del) in 2 samples (Table 3). Of the 6VUS,one was in a coding region (A309T), one was in the 5′ untranslated region (7:117120145G>C), and 4 were in intron regions (7:117182180A>C, 7:117199721T>C, 7:117242865C>G, 7:117305631A>G). Both the coding and intron region variants were predicted in silico to be benign. Although it remains to be confirmed, we predicted that the 5′ untranslated region VUS was disease causing. The single base-call error (2184delA; c.2052del; 7:117232266CA>C) lies in a homopolymer stretch of 7 adenines in the coding region. It is a potentially false-positive call (see below).
Table 3.
Variants detected by next-generation sequencing.
| Sample, n |
Total variants (unfiltered), n |
Filtered variants, n | |||
|---|---|---|---|---|---|
| Known pathogenic variants |
VUS | Base-calling errora |
Total | ||
| CF, 24 | 180 | 45 | 2 | 2 | 49 |
| Carrier, 46 | 298 | 45 | 3 | 0 | 48 |
| Normal, 9 | 37 | 0 | 1 | 0 | 1 |
| Total, 79 | 515 | 90 | 6 | 2 | 98 |
Due to a homopolymer-length sequencing error (7:117232266CA>C) leading to the 2184delA false-positive call.
SENSITIVITY
To assess the analytical sensitivity of the CFTR AmpliSeq assay, we blindly sequenced 70 known DNA samples: 46 carriers and 24 CF samples representing 34 unique CFTR mutations, including all of the mutations (23) recommended for carrier screening by the ACMG (Table 1). For this sample set, we obtained data from 4 sequencing runs, with a mean of 157 329 mapped reads per sample, 95% of which aligned to CFTR target regions. The mean depth of coverage was 1793×, with 96% of the targeted bases having at least 100× coverage.
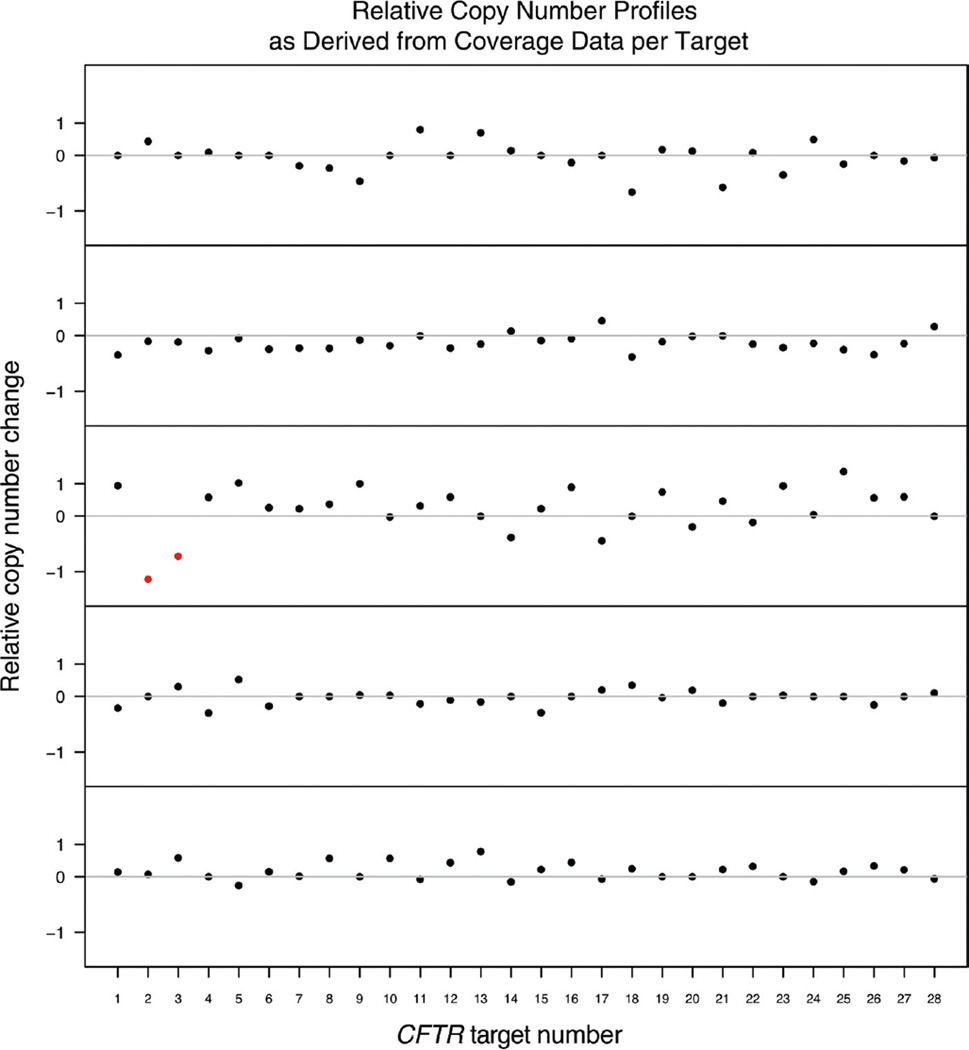
Our assay correctly identified all CF cases known to be either homozygous or compound heterozygous (24 of 24, 100% sensitivity; see Table 1). We detected apparent deletion of exons 2 and 3 in 1 sample (Fig. 1), a result consistent with a previously described 21-kb deletion of these exons [CFTRdele2,3(21kb)] (8). We inferred the presence of this event from the decreased coverage of consecutive exons (log2 ratios of less than −0.7) compared with the median observed for samples sequenced in the same batch. This observation demonstrates the semiquantitative nature of our assay, although additional assay and algorithm development is needed to reduce the variation in coverage across amplicons and to facilitate the use of larger reference sets for normalization beyond those used for normalization with a batch. Notably, the calculation of fractional-coverage values from a relatively few targets (28 in our assay) can be skewed by copy number variation that affects a few probes. In our deletion case, the loss of 2 of the 28 exons/targets increased fractional-coverage values, because intact targets received an increased fraction of the overall coverage. This effect, which was manifested as artificially inflated log2 ratios for this sample, has the potential for identifying false-positive copy number gains. This limitation reflects the small number of targets queried by our assay, and this problem may be reduced by including additional targets or by developing a more robust normalization method. Although the exact breakpoints could not be determined from our exon-centric assay, we suspect that the breakpoints are likely in repetitive DNA elements not targeted by our design. Our results, however, do demonstrate a proof of principle for detecting exon-level deletions in CFTR.
Fig. 1. Changes in relative copy number calls at 28 CFTR targets as inferred from log2 ratios of sample/batch median coverage.
Each panel corresponds to one of 5 samples sequenced in a single assay run and normalized as a batch. An apparent deletion is marked in red on the third panel.
Our assay also detected all but 1 case with a single mutation (45 of 46, 98% sensitivity). The missed mutation (S549R; A>C, c.1645A>C; Table 1) was expected, because it was masked by one of the primers in our design, a limitation that will be addressed in the next iteration of this assay. Our assays correctly identified all ACMG mutations (23 of 23) recommended for CFTR carrier screening (Table 4). Overall, our assay showed a sensitivity of 98.6% (69 of 70; Table 5) and demonstrated an ability to correctly identify different classes of mutations, including nonsense, missense, frameshift, in-frame deletion, splice site, and large-deletion mutations (Table 1).
Table 4.
ACMG mutation panel.
| Mutation | cDNA position | Mutation class | Sensitivity (TPa) | Specificity (TN) | Accuracyb |
|---|---|---|---|---|---|
| G85E | c.254G>A | Missense | Detected (2/2) | 100% (77/77) | 100% |
| R117H | c.350G>A | Missense | Detected (6/6) | 100% (73/73) | 100% |
| 621+1G>T | c.489+1G>T | Splice site | Detected (6/6) | 100% (73/73) | 100% |
| 711+1G>T | c.579+1G>T | Splice site | Detected (1/1) | 100% (78/78) | 100% |
| R334W | c.1000C>T | Missense | Detected (3/3) | 100% (76/76) | 100% |
| R347P | c.1040G>C | Missense | Detected (1/1) | 100% (78/78) | 100% |
| A455E | c.1364C>A | Missense | Detected (2/2) | 100% (77/77) | 100% |
| ΔI507 | c.1519_1521delATC | In-frame deletion | Detected (1/1) | 100% (78/78) | 100% |
| ΔF508 | c.1521_1523delCTT | In-frame deletion | Detected (30/30) | 100% (49/49) | 100% |
| G542* | c.1624G>T | Nonsense | Detected (4/4) | 100% (75/75) | 100% |
| G551D | c.1652G>A | Missense | Detected (6/6) | 100% (73/73) | 100% |
| R553* | c.1657C>T | Nonsense | Detected (3/3) | 100% (76/76) | 100% |
| R560T | c.1679G>C | Missense | Detected (1/1) | 100% (78/78) | 100% |
| 1898+1G>A | c.1766+1G>A | Splice site | Detected (2/2) | 100% (77/77) | 100% |
| 2789+5G>A | c.2657+5G>A | Splice site | Detected (1/1) | 100% (78/78) | 100% |
| 3120+1G>A | c.2988+1G>A | Splice site | Detected (2/2) | 100% (77/77) | 100% |
| R1162* | c.3484C>T | Nonsense | Detected (1/1) | 100% (78/78) | 100% |
| 3659delC | c.3528del | Frameshift deletion | Detected (1/1) | 100% (78/78) | 100% |
| 3849+10kbC>T | c.3718–2477C>T | Splice site | Detected (1/1) | 100% (78/78) | 100% |
| W1282* | c.3846G>A | Nonsense | Detected (3/3) | 100% (75/75) | 100% |
| N1303K | c.3909C>G | Missense | Detected (1/1) | 100% (78/78) | 100% |
| 2184delA | c.2052del | Frameshift deletion | Detected (1/1) | 97% (76/78) | 97% |
| 1717-1G>A | c.1585-1G>A | Splice site | Detected (1/1) | 100% (78/78) | 100% |
TP, true-positive rate; TN, true-negative rate.
Calculated from the rates of true positives and true negatives.
Table 5.
Analytical performance of the CFTR assay.
24 of 24 CF samples and 45 of 46 carrier samples.
9 of 9 normal samples.
Forty-seven samples were sequenced in 3 independent runs.
SPECIFICITY
We determined the analytical specificity of our assay by sequencing 9 samples that had been classified as normal noncarriers according the xTAG Cystic Fibrosis 60 Kit v2. The mean number of reads mapping to CFTR targeted regions in this data set was 129 749 per sample. The mean depth of coverage was 1729×, with 100% of the targeted bases having at least 100× coverage. None of the 64 mutations covered by the xTAG assay were detected in our assay, for a specificity of 100% at all 64 positions for this sample set (Table 5). Furthermore, no other known pathogenic mutations were detected in these 9 samples. Interestingly, only 1 intronic VUS (7: 117199721T>C) was identified in 1 sample and was predicted in silico to be benign.
Nonetheless, of the 79 sequenced DNA samples, only a single false positive (2184delA; c.2052del) was called twice in 2 CF samples. This false positive was due to a homopolymer-sequencing error (Table 3). This error was found in approximately 30% of the reads spanning this homopolymer stretch. Given that it was erroneously called in only 2 of the 79 sequenced samples, the specificity of this assay at this position is 97% (Table 4). Nevertheless, it is one of the mutations in the ACMG’s recommended panel; therefore, a higher variant threshold may be necessary at this position to avoid such false-positive calls. The remaining 22 mutations were not detected (100% specificity) in samples that were otherwise negative for the respective mutation (Table 4).
PRECISION
To determine the reproducibility of our assay, we sequenced 47 samples in 3 independent runs at 3 different sites. The same 53 pathogenic mutations and 3 VUS were reproducibly detected in all 3 separate runs, for a between-run reproducibility of 100% (Table 5).
Discussion
We have presented data on the first version of our target-enrichment assay designed to generate a CFTR PCR amplicon library for sequencing on the PGM platform. Our design covers 159 of 160 mutations listed on the Clinical and Functional Translation of CFTR website, including the 23-mutation panel recommended by the ACMG.
Our assay has several advantages for diagnostic CFTR sequencing, including speed and scalability. Because approximately 10 kb of the CFTR coding region were targeted with our design, we were able to pool different numbers of bar-coded amplicon libraries on Ion chips with different sequencing-output capacities (314 and 318 chips) while maintaining the high depth of coverage necessary for reliable variant calling. In the clinical setting, such flexibility offers the capability to manage costs and turnaround times. Our cost analysis suggests that the costs with a 318 chip are $154 and $103 per sample when 12 and 35 samples, respectively, are pooled in a run. For the 314 chip, the cost per sample is $166 and $121 when multiplexing 5 and 12 samples, respectively. We expect additional reductions in costs when more samples are pooled on a 314, 316, or 318 chip, given the high depth of coverage that can still be obtained.
Furthermore, our assay demonstrated high sensitivity in detecting different classes of mutations, including nonsense, missense, frameshift, in-frame deletion, splice site, and large-deletion mutations. All 23 ACMG-recommended mutations were identified in our assay with 100% accuracy (except for 2184delA, which had a 97% accuracy; Table 4). Although we have demonstrated that analysis of relative sequence coverage can be used to infer relative copy-number states, additional work is necessary to develop a robust, clinical-grade algorithm for systematic detection of copy number variants. It is worth noting that compared with algorithms for single-nucleotide variants, the existing algorithms for detecting copy number variants from sequencing data are still less mature and not yet user-friendly (9). This area is still under development, however, and we expect more copy number variant algorithms tuned for optimal performance to be available very soon.
Another major advantage is the low detection rate for VUS, which can be a source of stress for patients, providers, and counselors. In the analysis of normal noncarrier cases, we detected only 1 VUS in 9 samples and a total of 6 VUS in all 79 sequenced DNA samples. This low VUS frequency is also likely due to the availability of well-annotated CFTR mutation databases. In general, VUS detection is one of the challenges that might impede the introduction of next-generation sequencing– based testing into clinical laboratories; however, a better understanding of such variants requires interlaboratory sharing of genomic and phenotypic data, which has recently begun with several initiatives, including the International Collaboration for Clinical Genomics (10). With such endeavors, our knowledge about variants and their impact on human health and disease will likely increase and ultimately lead to a lower frequency of VUS detection.
A major limitation of this assay using the Ion Torrent technology is the sequencing errors associated with homopolymer stretches. This limitation for detecting an important class of variants, including the poly(T) tract polymorphism, would require a separate means of testing. Another example is the 2184delA mutation, which a previous study found to have a high rate of false-positive mutant reads at this position (11). In our study, we have set our variant read threshold at 25%, and this filter allowed the only 2 false positives out of the 79 mutant-negative samples at this position, for a specificity of 97%; however, increasing the variant read threshold in this assay did not affect the reliability of variant calling at any of the other positions tested in our study.
Nonetheless, substantial improvements in homopolymer sequencing are required to avoid erroneous calls. Although filtering out variants from certain homopolymer sites would reduce false-positive calls, it would also decrease the sensitivity of mutation detection if mutations were present at those sites. Furthermore, although increasing the variant read threshold to 25% might be tolerated for detecting inherited mutations, that might not be the case for detecting somatic mutations, for which a 5%threshold is set in many cases.
In conclusion, our assay has the ability to detect different classes of inherited CFTR mutations with not only high sensitivity and precision but also limited numbers of VUS. In addition, the scalability of the assay and the rapid data generation on the PGM could make it the platform of choice in the clinical laboratory, assuming that homopolymer sequencing will be improved in the near future.
Supplementary Material
Acknowledgments
The authors thank the staff of the Dartmouth–Hitchcock Medical Center Molecular Pathology Laboratory and the Translational Research Program. The data presented in this report was generated in part through the Department of Pathology Translational Research Shared Resource Laboratory of the Geisel School of Medicine at Dartmouth, the Dartmouth–Hitchcock Medical Center, and the Norris Cotton Cancer Center.
Footnotes
Nonstandard abbreviations: CF, cystic fibrosis; ACMG, American College of Medical Genetics; PGM, Ion Torrent Personal Genome Machine; VUS, variant(s) of unknown significance
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: T. Ross, Life Technologies; M. Shah, Life Technologies; C.C. Lee, Life Technologies; T.T. Harkins, Life Technologies; L.J. Tafe, Dartmouth–Hitchcock Medical Center.
Consultant or Advisory Role: None declared.
Stock Ownership: C.C. Lee, Life Technologies; T.T. Harkins, Life Technologies.
Honoraria: None declared.
Research Funding: None declared.
Expert Testimony: None declared.
Patents: None declared.
Role of Sponsor: No sponsor was declared.
References
- 1.Cystic Fibrosis Centre. Hospital for Sick Children, Toronto, Ontario, Canada. [Accessed January 2013];Cystic Fibrosis Mutation Database. http://www.genet.sickkids.on.ca/cftr/ [Google Scholar]
- 2.Palomaki GE, FitzSimmons SC, Haddow JE. Clinical sensitivity of prenatal screening of cystic fibrosis via CFTR carrier testing in a United States panethnic population. Genet Med. 2004;6:405–415. doi: 10.1097/01.gim.0000139505.06194.39. [DOI] [PubMed] [Google Scholar]
- 3.Strom CM, Crossley B, Buller-Buerkle A, Jarvis M, Quan F, Peng M, et al. Cystic fibrosis testing 8 years on: lessons learned from carrier screening and sequencing analysis. Genet Med. 2011;13:166–172. doi: 10.1097/GIM.0b013e3181fa24c4. [DOI] [PubMed] [Google Scholar]
- 4.Watson MS, Cutting GR, Desnick RJ, Driscoll DA, Klinger K, Mennuti M, et al. Cystic fibrosis population carrier screening, 2004 revision of American College of Medical Genetics mutation panel. Genet Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.American College of Obstetricians and Gynecologists (ACOG), ACMG. Preconception and prenatal carrier screening for cystic fibrosis: clinical and laboratory guidelines. Washington (DC): ACOG; 2001. [Google Scholar]
- 6.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding nonsynonymous variants on protein function using the SIFT algorithm. Nature Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 7.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 8.Dörk T, Macek M, Jr, Mekus F, Tümmler B, Tzountzouris J, Casals T, et al. Characterization of a novel 21-kb deletion, CFTRdele2,3(21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–268. doi: 10.1007/s004390000246. [DOI] [PubMed] [Google Scholar]
- 9.Duan J, Zhang J, Deng H, Wang Y. Comparative studies of copy number variation detection methods for next-generation sequencing technologies. PLoS ONE. 2013;8:e59128. doi: 10.1371/journal.pone.0059128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riggs ER, Wain KE, Riethmaier D, Savage M, Smith-Packard B, Kaminsky E, et al. Towards a universal clinical genomics database: The 2012 International Standards for Cytogenomic Arrays Consortium Meeting. Hum Mutat. 2013;34:915–919. doi: 10.1002/humu.22306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott AM, Radecki J, Moghis B, Li X, Kammesheidt A. Rapid detection of the ACMG/ACOG-recommended 23 CFTR disease-causing mutations using Ion Torrent semiconductor sequencing. J Biomol Tech. 2012;23:24–30. doi: 10.7171/jbt.12-2301-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.