Abstract
New discoveries in the last decade significantly altered our view on mitochondria. They are no longer viewed as energy-making slaves but rather individual cells-within-the-cell. In particular, it has been suggested that many important cellular mechanisms involving specific enzymes and ion channels, such as nitric oxide synthase (NOS), ATP-dependent K+ (KATP) channels, and poly-(APD-ribose) polymerase (PARP), have a distinct, mitochondrial variant. Unfortunately, exploring these parallel systems in mitochondria have technical limitations and inappropriate methods often led to inconsistent results. For example, the intriguing possibility that mitochondria are significant sources of nitric oxide (NO) via a unique mitochondrial NOS variant has attracted intense interest among research groups because of the potential for NO to affect functioning of the electron transport chain. Nonetheless, conclusive evidence concerning the existence of mitochondrial NO synthesis is yet to be presented. This review summarizes the experimental evidence gathered over the last decade in this field and highlights new areas of research that reveal surprising dimensions of NO production and metabolism by mitochondria.
Keywords: Mitochondrion, Nitric Oxide Synthase, Peroxynitrite, Reactive Nitrogen Species, Nitric Oxide, Electron Transport System, Peroxynitrite
2. INTRODUCTION
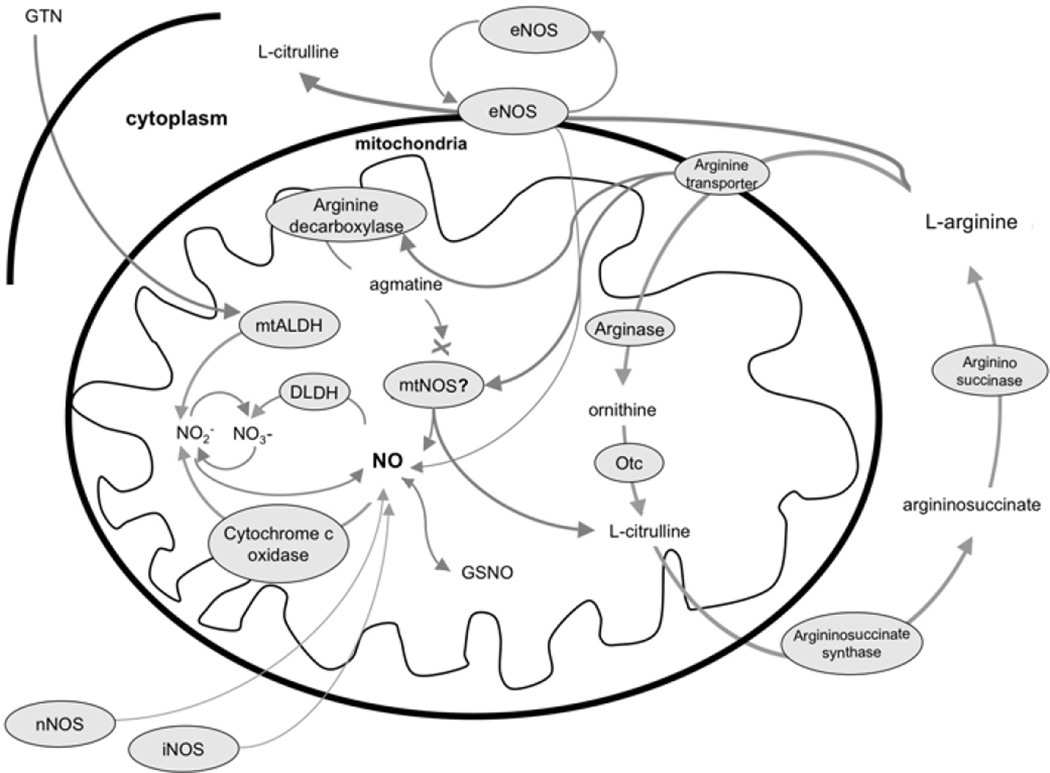
Nitric oxide, a freely diffusible gas, is produced by several metabolic pathways in the mitochondrion and in the surrounding cell and has widespread effects on mitochondrial function either as NO, or following conversion to peroxynitrite (ONOO−), or other reactive nitrogen species (RNS). Extra-mitochondrial sources of NO include the three recognized isoforms of NOS, including inducible (iNOS), endothelial NOS (eNOS), and neuronal NOS (nNOS). These enzymes convert L-arginine to NO and L-citrulline. The constitutive isoforms (eNOS and nNOS) are under tight regulatory control involving calcium availability while the inducible form (iNOS) apparently is regulated by substrate availability and transcription. Once formed, NO can diffuse into mitochondria from the surrounding cellular space or could traverse longer distances from other, more remote cells. Nitric oxide also is produced in mitochondria, although the precise metabolic pathways are controversial. In particular, the possible existence and chemical structure of a distinct NOS variant in the mitochondrion are subjects of intense debate. Additionally, it is clear that NO or RNS can be produced within mitochondria by other pathways not involving metabolism of L-arginine by NOS. These diverse mechanisms leading to availability of NO to the mitochondria as well as NO metabolism are summarized in Figure 1. In this review, we will examine the evidence for and against the existence of a distinct, functionally-active NOS isoform located in the mitochondria and also discuss other possible, non-NOS sources of mitochondrially-derived NO and RNS.
Figure 1.
Potential sources and metabolism of NO and related compounds in mitochondria. Arrows indicate chemical reactions, a cross indicates blocking effect. Abbreviations: DLDH: dihydrolipoamide-dehydrogenase, eNOS: endothelial nitric oxide synthase, GTN: glyceryl-trinitrate, iNOS: inducible nitric oxide synthase, GSNO: nitrosoglutathione, mtALDH: mitochondrial aldehyde dehydrogenase, mtNOS: mitochondrial nitric oxide synthase, nNOS: neuronal nitric oxide synthase, NO: nitric oxide, Oct: ornithine-citrulline transaminase.
3. DISCUSSION
3.1 Potential actions of NO in the mitochondria
Regardless of the original source, NO or derived RNS can have profound effects on mitochondrial function under physiological and pathological conditions. Nitric oxide reacts strongly with all heme moieties and thus can reversibly reduce activity of iron-containing enzymes such as cytochrome c oxidase (complex IV). In addition, nanomolar levels of NO disrupt mitochondrial respiration by inhibiting the electron transport chain by competing with O2, especially under relatively hypoxic conditions. Nitric oxide also interacts strongly with superoxide anion, which is continuously generated and released from the electron transport chain, to form peroxynitrite. Peroxynitrite can have diverse effects on mitochondrial function, such as inhibiting respiration at Complexes I and III by S-nitrosation, promoting the opening of mitochondrial KATP channels (1), and activating the mitochondrial matrix PARP cascade (2). Other RNS, such as nytroxil (HNO) can inhibit Complex II. In addition, mitochondrial enzymes can metabolize NO-containing compounds. For example, cytochrome c oxidase under appropriate conditions converts NO to NO2−, which is further oxidized to NO3− by the ketoglutarate-dehydrogenase complex (3–11). In addition, mitochondria possess a nitrate-reductase activity that converts NO2− back to NO under hypoxic conditions (12,13).
3.2. The case for a genuine mitochondrial NOS
The primary cellular source of NO is from the conversion of L-arginine to L-citrulline by the three conventional NOS isoforms. While it is generally assumed that NO is a relatively short-lived compound, NO produced in different compartments of a cell or even in adjacent cells has ready access to mitochondria. The obligatory cofactors of NOS enzymes are HEME in the active center and a flavin chain (NADPH, FAD), which serves as an electron donor. In in the case of the constitutive variants (eNOS, nNOS), but not the iNOS, a calcium-calmodulin system participates as a regulatory complex. For iNOS, substrate availability and transcription are the major regulators of NO production. Distinct nuclear genes encode the three conventional isoforms in mammals. The eNOS gene encodes a 140 kDa protein which is present primarily in endothelial cells and eNOS is the main source of NO in the vascular system. The nNOS gene encodes four isoforms: 1) the 160 kDa nNOSalpha, which is responsible for about 95% of nNOS activity in the brain (14); 2) the 155 kDa nNOSbeta which apparently represents the remaining 5% of the activity in neural tissues (15); 3) the truncated nNOSgamma with a molecular weight of approximately 125 kDa, with unknown function; and 4) a myocardial variant of nNOS, sometimes called nNOSm, which contains an extra loop in the protein chain without a clearly identified function. The iNOS gene encodes a 130 kDa protein which is typically located in a variety of inflammatory cells such as macrophages or Kupfer cells and can be induced broad pro-inflammatory stimuli (e.g. TNF, IFNg, LPS).
The more limited mitochondrial genome does not contain a conventional NOS gene, so if NOS is present in the mitochondrion, it must be relocated using traditional, well defined transport systems into the mitochondria from the cytoplasm. However, there is no evidence that the conventional NOS isoforms possess transport sequences necessary for passage through the inner mitochondrial membrane. Nonetheless, some, but not all, studies have provided evidence indicating that a mitochondrial variant, named mtNOS, may be present in the inner mitochondrial membrane or matrix. On the other hand, conventional NOS isoforms may be associated with the outer mitochondrial membrane and represent an alternative, externally regulated source of NO to the interior of the mitochondria.
The studies by Bates and collegues first reported eNOS immunoreactivity in the inner mitochondrial membrane (16,17) and these results were later reproduced by independent investigators (18,19). These researchers used immunogold electron microscopy, which only showed that there is an eNOS-like protein in the mitochondria in fixed preparations. Supporting evidence for the functional activity of mtNOS was provided via electron microscopy by the detection of NADPH diaphorase activity in mitochondria (20). Later, iNOS-like immunoreactivity was reported in mitochondrial preparations, and these studies also presented data that indicated that NO is indeed generated by isolated mitochondria preparations (21,22).
Several investigators sought to identify whether mtNOS is an independent gene entity or the product of the three conventional NOS-encoding genes. Giulivi and colleagues used anti-iNOS and anti-nNOS antibodies to detect mtNOS. This 130 kDa protein is significantly smaller than that expected from nNOS, although protein mass fingerprinting indicated that it is the alpha isoform of nNOS (23,24). Further complicating the issue was the detection of a 130 kDa mitochondrial protein band with the same antibodies, which was preserved in nNOS knockout mice (unpublished observations Lacza and colleagues). Kanai and colleagues also used knockout animals in order to find the molecular identity of mtNOS (25). They measured NOS activity in isolated heart mitochondria with an electrochemical electrode and observed that the activity was absent in nNOSalpha knockout mice. However, these observations were not supported by the direct detection of a nNOS protein in mitochondria. Using various antibodies, several groups have shown NOS-like bands in mitochondria preparations without any conclusive overall result (reviewed in (26).
We also considered the possibility that alternative isoforms of NOS, similar to those present in fungi and plants, may be represented in mammalian cells. Castello and colleagues described yeast mtNOS, and they also showed that its activity is increased under hypoxic conditions (27). The plant variant of mtNOS was first described in Arabidopsis thaliana and it was suggested that it plays a role in the protection against oxidative damage (28). On the basis of these findings an even more intriguing hypothesis emerged that this plant mtNOS (named atNOS) is also present in mammalian cells and is indeed a newly discovered NOS variant (29). However, shortly after the original publication a corrigendum appeared stating that the main finding of the paper, namely the NOS activity of the enzyme, was not reproducible, thereby invalidating the whole story (30). At present the available experimental data on plant and fungi mtNOS is too weak to support a decisive conclusion.
The common definition of mtNOS suggests that it must be located within the mitochondrial matrix or attached to the inner membrane. However, it is also possible that a cellular NOS protein is merely attached to the outer surface of the mitochondrion. Indeed, the earliest studies of mtNOS showed NADPH diaphorase activity in the vicinity of mitochondria and not in the mitochondrial matrix (20, 31, 32). Henrich and colleagues located eNOS within sensory neurons and found that the enzyme is anchored to juxta-mitochondrial smooth endoplasmic reticulum (33). Later, Gao et al. observed this phenomenon in endothelial cells and identified a pentabasic amino acid sequence in the autoinhibitory domain of eNOS, which is responsible for the mitochondrial docking of the enzyme (34). These findings indicate that mtNOS may indeed be a cellular NOS enzyme, which is loosely attached to the outer surface of mitochondria. Although there is not enough experimental evidence to prove it, one can hypothesize that mitochondrial attachment plays a role in the regulation of NOS activity, and thus docking of an active NOS on the outer membrane, with resultant NO production, regulates respiration.
3.3. The case against a genuine mitochondrial NOS
Almost ten years after the first observations raised the possibility that mitochondria possess their own internal NOS, the actual existence of mtNOS and/or an important physiological role of a putative mtNOS have not won widespread support from the research community. This reluctance to embrace the concept of mtNOS is due to: 1) the failure by other laboratories to reproduce key findings concerning the detection of mtNOS; 2) concerns that the levels of NO produced by mtNOS activity may be inadequate to have significant physiological effects; and 3) the realization that competing metabolic pathways in the mitochondria may restrict availability of L-arginine to a putative mtNOS. In addition, novel proteomic tools which can predict the cellular positioning of a protein based on N-terminal transport sequences failed to show an appropriate mitochondrial transport signal in the primary sequence of any of the known NOS isoforms, making it unlikely that a nuclear-encoded NOS is transported to the mitochondrion (35). Moreover, it is unclear whether all the usual cofactors that are needed by a functional NOS enzyme are present in correct position within the mitochondrial matrix (36) and whether traditional regulatory mechanisms controlling NOS activity are present within mitochondria.
The mitochondrial matrix has several abundant L-arginine-consuming enzymes which can effectively compete with the hypothetical mtNOS for its substrate, thereby proving a less than favorable environment for NOS. Mitochondria participate in the urea cycle and as such they contain several L-arginine-metabolizing enzymes (37). The outer mitochondrial membrane is not a barrier to diffusion of substances such as L-arginine. L-arginine enters the matrix by a specific transport process, which is catalyzed by an arginine-transporter protein in the inner membrane. Inside the matrix, L-arginine is converted to ornithine and citrulline by urea cycle enzymes, and these metabolites are then converted back to arginine in the cytosol. An alternative fate for L-arginine may involve its conversion to NO and L-citrilline by mtNOS. However, the kinetics of these two pathways favors the urea cycle even under conditions optimized for NOS activity and in the presence of arginase inhibitors. Thus, the arginase activity of liver mitochondria preparations is still 1–2 orders of magnitude higher than purported mtNOS activity (unpublished results). In addition, mitochondria contain arginine-decarboxylase, which converts arginine to agmatine (38). Agmatine is a known NOS inhibitor, making the mitochondrial matrix an even less favorable environment for a hypothetical mtNOS (39).
In an earlier paper, we reported a small but significant mtNOS activity and detected eNOS-like immunoreactivity by immunogold electron microscopy in isolated liver mitochondria (18). Unfortunately, follow-up experiments in mitochondria from other tissues failed to support the conclusions drawn from these early experiments. Although NOS activity increased after mild hypoxia, this was not explained by either the increased amount of eNOS protein or a change in eNOS phosphorylation (unpublished observations Lacza and colleagues). In a subsequent study, we found that L-arginine-to-L-citrulline conversion in liver mitochondria is not dependent on NOS activity34. An independent group reported similar results using different methodologies, confirming that mitochondria do not produce significant amounts of NOS-derived NO (40). Additionally, we confirmed these results in brain and heart tissues of several species (41, 42). Most importantly, we were not able to detect significant NOS-dependent NO production by human heart mitochondria, thereby questioning the importance of mtNOS in general (42).
It is commonly accepted that mitochondria preparations typically contain about 1–4% of cellular elements, and basically all cells contain some variant of the conventional NOS (43). Therefore, full tissue NOS activity must be measured in parallel with the isolated mitochondria and if the mitochondrial NOS signal amounts to a few percent of the full tissue value it must be discarded as possible contamination. The same method can be used in Western blots and other methods, which aim to detect NOS protein levels in mitochondrial membranes. Unfortunately, the papers, which support the existence of mtNOS, do not provide complete data on full tissue values, making the interpretation of their results difficult.
With the help of extensive positive and negative controls we were able to exclude all known conventional mammalian NOS proteins as candidates for the proposed mtNOS (35). While one could argue that negative findings may be the result of inappropriate techniques, the combined findings from at least three independent laboratories and published in five full-length papers provide strong evidence that NOS activity in mitochondria is either nonexistent or falls below the threshold needed to induce physiological responses (35, 40–42, 44). After thoroughly reviewing the literature, Brookes also came to this conclusion (36). The methodological issues regarding the measurement of NOS in mitochondria were reviewed in our previous paper and no technological advances have emerged since then which may provide definitive evidence for or against mtNOS (45).
3.4. Alternative sources of NO in the mitochondrion
A bacterial NOS-variant has been described in Bacillus subtilis (46) and its characteristics may provide insight to the possible nature of a non-conventional mammalian NOS adapted to the physically more restricted environment of the mitochondrial matrix. This bacterial enzyme shows homology only with the oxygenase domain of mammalian NOS and therefore is a significantly smaller protein. In the presence of a recombinant NOS reductase domain, this enzyme binds HEME and converts L-arginine to L-citrulline with the production of NO. Although intriguing, there is no evidence that a similar, nontraditional arrangement is present in the mitochondria of higher organisms.
The first reports that initiated the hypothesis that the mitochondrial respiratory chain is a source of NO were performed by Kozlov and colleagues (12, 13). In these studies isolated liver mitochondria produced detectable amounts of NO through the reduction of nitrate under anoxic conditions. Using selective inhibitors of the respiratory chain complexes, the location of nitrate reductase activity was identified as the unbiquinone cycle. Unfortunately, this phenomenon was only present in the absence of oxygen, so its physiological importance is unclear. Later, a similar mechanism was described in green algae, indicating that the nitrate reductase activity of the mitochondrial respiratory chain is a general phenomenon (47). In a recent study, we detected a strong NO-related signal in mitochondria using fluorescent probes, which was also dependent on the ubiquinone cycle under physiological conditions (48). In our study, we showed that the signal is independent of L-arginine, Ca++, or NOS enzymes, but it is completely dependent on the electron flow of the first half of the respiratory chain. Due to technical reasons it was impossible to test whether the substrate of this reaction is the nitrate anion or other organic nitrates. It should be noted, that diaminofluoresceins (DAF-2 or DAF-FM), which are widely used as NO-probes, react poorly with NO but have a stronger affinity to RNS, especially N2O3 (49). This apolar compound can be formed by the nonenzymatic reaction of NO with molecular oxygen, but as we showed, it can also be the by-product of mitochondrial respiration. Since the DAF signal is typically concentrated in mitochondria, it can be argued that mitochondria are very prominent producers of RNS but are insignificant sources of NO (35, 41,48, 50–55). A recent report from Boveris and colleagues lends support to this hypothesis; they show that mitochondrial NO production is a voltage-dependent event (56).
Mitochondrial proteins and the tripeptide glutathione contain thiol residues, which reversibly bind NO. This pool of nitrated thiols acts as an endogenous NO storage system that can absorb NO from any source and release it into the mitochondrial matrix. Both nitrated tyrosine residues and S-NO-glutathione (GSNO) were shown to be present in mitochondria (57, 58). Under experimental conditions, the nitrosothiol bond can be released by heat or light, which makes the investigation of these NO sources relatively simple, however, it does not yield any information about the physiological mechanisms which control nitrosothiol formation and metabolism.
4. PERSPECTIVES
It is our view that no conclusive evidence has been provided which can prove the existence of an authentic mtNOS to the satisfaction of all interested parties. However, as is often the case, the efforts in this area have led to new discoveries regarding the role of NO in mitochondrial metabolism. It has been shown that eNOS can dock to the outer mitochondrial membrane and provide a source of NO in close proximity to mitochondrial respiratory enzymes. Although it is not a mtNOS in the classic case, juxta-mitochondrial eNOS nonetheless has the same functional profile, which is even more interesting since enzyme activity can be regulated from the cellular side. In addition, new enzymatic mechanisms were revealed which can generate NO or RNS with the involvement of the ubiquinone cycle. Further experimental work is required to define the precise biochemistry of such reactions, and especially to find the immediate endogenous substrates. The discovery that nitroglycerin is converted to active nitrates by a mitochondrial enzyme adds further clinical importance to this line of research. Although just as complicated as the issue of mtNOS, other newly discovered mechanisms can be related to the “mitochondrial-derived NO story.” For example, it has been shown that a mitochondrial variant of PARP can be activated by nitrosative stress and lead to cell death (2). In contrast, NO or peroxynitrite has been shown to open mitochondrial KATP channels and thus participate in cellular preconditioning against lethal stresses (53, 59). We conclude that even if the existence of a genuine mtNOS seems to be elusive, mitochondrial derived NO and related RNS can play a crucial role in several important physiological and pathological pathways.
ACKNOWLEDGEMENTS
This study was supported by grants from the American Heart Association Bugher Foundation Award (0270114N), the National Institutes of Health (HL30260, HL077731, HL06380), the Hungarian OTKA (D-45933, T-049621) and the Öveges and the Bolyai fellowships.
Abbreviations
- atNOS
Arabidopsis Thaliana NOS
- DAF
diaminofluorescein
- DLDH
dihydrolipoamide-dehydrogenase
- eNOS
endothelial nitric oxide synthase
- GTN
glyceryl-trinitrate
- iNOS
inducible nitric oxide synthase
- GSNO
nitrosoglutathione
- KATP
ATP-dependent K+ channels
- KGDH
ketoglutarate-dehydrogenase dehydrogenase
- mtNOS
mitochondrial nitric oxide synthase
- nNOS
neuronal nitric oxide synthase
- NO
nitric oxide
- Oct
ornithine-citrulline transaminase
- PARP
poly-(APD-ribose) polymerase
- RNS
reactive nitrogen species
REFERENCES
- 1.Lacza Z, Snipes JA, Kis B, Szabo C, Grover G, Busija DW. Investigation of the subunit composition and the pharmacology of the mitochondrial ATP-dependent K+ channel in the brain. Brain Res. 2003;994(1):27–36. doi: 10.1016/j.brainres.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 2.Du L, Zhang X, Han YY, Burke NA, Kochanek PM, Watkins SC, Graham SH, Carcillo JA, Szabo C, Clark RS. Intra-mitochondrial poly(ADP-ribosylation) contributes to NAD+ depletion and cell death induced by oxidative stress. J Biol Chem. 2003;278:18426–18433. doi: 10.1074/jbc.M301295200. [DOI] [PubMed] [Google Scholar]
- 3.Lizasoain I, Moro MA, Knowles RG, Darley-Usmar V, Moncada S. Nitric oxide and peroxynitrite exert distinct effects on mitochondrial respiration which are differentially blocked by glutathione or glucose. Biochem J. 1996;314:877–880. doi: 10.1042/bj3140877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown GC. Nitric oxide inhibition of cytochrome oxidase and mitochondrial respiration: implications for inflammatory, neurodegenerative and ischaemic pathologies. Mol Cell Biochem. 1997;174:189–192. [PubMed] [Google Scholar]
- 5.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proc Natl Acad Sci U S A. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boveris A, Costa LE, Cadenas E, Poderoso JJ. Regulation of mitochondrial respiration by adenosine diphosphate, oxygen, and nitric oxide. Methods Enzymol. 1999;301:188–198. doi: 10.1016/s0076-6879(99)01082-4. [DOI] [PubMed] [Google Scholar]
- 7.Sarti P, Lendaro E, Ippoliti R, Bellelli A, Benedetti PA, Brunori M. Modulation of mitochondrial respiration by nitric oxide: investigation by single cell fluorescence microscopy. FASEB J. 1999;13:191–197. doi: 10.1096/fasebj.13.1.191. [DOI] [PubMed] [Google Scholar]
- 8.Brookes PS, Bolanos JP, Heales SJ. The assumption that nitric oxide inhibits mitochondrial ATP synthesis is correct. FEBS Lett. 1999;446:261–263. doi: 10.1016/s0014-5793(99)00217-3. [DOI] [PubMed] [Google Scholar]
- 9.Brown GC. Nitric oxide and mitochondrial respiration. Biochim Biophys Acta. 1999;1411:351–369. doi: 10.1016/s0005-2728(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 10.Boveris A, Costa LE, Poderoso JJ, Carreras MC, Cadenas E. Regulation of mitochondrial respiration by oxygen and nitric oxide. Ann N Y Acad Sci. 2000;899:121–135. doi: 10.1111/j.1749-6632.2000.tb06181.x. [DOI] [PubMed] [Google Scholar]
- 11.Brown GC. Nitric oxide as a competitive inhibitor of oxygen consumption in the mitochondrial respiratory chain. Acta Physiol Scand. 2000;168:667–674. doi: 10.1046/j.1365-201x.2000.00718.x. [DOI] [PubMed] [Google Scholar]
- 12.Kozlov AV, Staniek K, Nohl H. Nitrite reductase activity is a novel function of mammalian mitochondria. FEBS Lett. 1999;454:127–130. doi: 10.1016/s0014-5793(99)00788-7. [DOI] [PubMed] [Google Scholar]
- 13.Nohl H, Staniek K, Sobhian B, Bahrami S, Redl H, Kozlov AV. Mitochondria recycle nitrite back to the bioregulator nitric monoxide. Acta Biochim Pol. 2000;47:913–921. [PubMed] [Google Scholar]
- 14.Huang PL, Lo EH. Genetic analysis of NOS isoforms using nNOS and eNOS knockout animals. Prog Brain Res. 1998;118:13–25. doi: 10.1016/s0079-6123(08)63197-0. [DOI] [PubMed] [Google Scholar]
- 15.Huang PL, Dawson TM, Bredt DS, Snyder SH, Fishman MC. Targeted disruption of the neuronal nitric oxide synthase gene. Cell. 1993;75:1273–1286. doi: 10.1016/0092-8674(93)90615-w. [DOI] [PubMed] [Google Scholar]
- 16.Bates TE, Loesch A, Burnstock G, Clark JB. Mitochondrial nitric oxide synthase: a ubiquitous regulator of oxidative phosphorylation? Biochem Biophys Res Commun. 1996;218:40–44. doi: 10.1006/bbrc.1996.0008. [DOI] [PubMed] [Google Scholar]
- 17.Bates TE, Loesch A, Burnstock G, Clark JB. Immunocytochemical evidence for a mitochondrially located nitric oxide synthase in brain and liver. Biochem Biophys Res Commun. 1995;213:896–900. doi: 10.1006/bbrc.1995.2213. [DOI] [PubMed] [Google Scholar]
- 18.Lacza Z, Puskar M, Figueroa JP, Zhang J, Rajapakse N, Busija DW. Mitochondrial nitric oxide synthase is constitutively active and is functionally upregulated in hypoxia. Free Radic Biol Med. 2001;31:1609–1615. doi: 10.1016/s0891-5849(01)00754-7. [DOI] [PubMed] [Google Scholar]
- 19.Hotta Y, Otsuka-Murakami H, Fujita M, Nakagawa J, Yajima M, Liu W, Ishikawa N, Kawai N, Masumizu T, Kohno M. Protective role of nitric oxide synthase against ischemia-reperfusion injury in guinea pig myocardial mitochondria. Eur J Pharmacol. 1999;380:37–48. doi: 10.1016/s0014-2999(99)00531-2. [DOI] [PubMed] [Google Scholar]
- 20.Loesch A, Belai A, Burnstock G. An ultrastructural study of NADPH-diaphorase and nitric oxide synthase in the perivascular nerves and vascular endothelium of the rat basilar artery. J Neurocytol. 1994;23:49–59. doi: 10.1007/BF01189816. [DOI] [PubMed] [Google Scholar]
- 21.Tatoyan A, Giulivi C. Purification and characterization of a nitric-oxide synthase from rat liver mitochondria. J Biol Chem. 1998;273:11044–11048. doi: 10.1074/jbc.273.18.11044. [DOI] [PubMed] [Google Scholar]
- 22.Ghafourifar P, Richter C. Nitric oxide synthase activity in mitochondria. FEBS Lett. 1997;418:291–296. doi: 10.1016/s0014-5793(97)01397-5. [DOI] [PubMed] [Google Scholar]
- 23.Elfering SL, Sarkela TM, Giulivi C. Biochemistry of mitochondrial nitric-oxide synthase. J Biol Chem. 2002;277:38079–38086. doi: 10.1074/jbc.M205256200. [DOI] [PubMed] [Google Scholar]
- 24.Giulivi C. Characterization and function of mitochondrial nitric-oxide synthase. Free Radic Biol Med. 2003;34:397–408. doi: 10.1016/s0891-5849(02)01298-4. [DOI] [PubMed] [Google Scholar]
- 25.Kanai AJ, Pearce LL, Clemens PR, Birder LA, VanBibber MM, Choi SY, de GWC, Peterson J. Identification of a neuronal nitric oxide synthase in isolated cardiac mitochondria using electrochemical detection. Proc Natl Acad Sci U S A. 2001;98:14126–14131. doi: 10.1073/pnas.241380298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lacza Z, Pankotai E, Csordas A, Gero D, Kiss L, Horvath EM, Kollai M, Busija DW, Szabo C. Mitochondrial NO and reactive nitrogen species production: does mtNOS exist? Nitric Oxide. 2006;14:162–168. doi: 10.1016/j.niox.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Castello PR, David PS, McClure T, Crook Z, Poyton RO. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab. 2006;3:277–287. doi: 10.1016/j.cmet.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 28.Guo FQ, Crawford NM. Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell. 2005;17:3436–3450. doi: 10.1105/tpc.105.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemojtel T, Kolanczyk M, Kossler N, Stricker S, Lurz R, Mikula I, Duchniewicz M, Schuelke M, Ghafourifar P, Martasek P, Vingron M, Mundlos S. Mammalian mitochondrial nitric oxide synthase: characterization of a novel candidate. FEBS Lett. 2006;580:455–462. doi: 10.1016/j.febslet.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 30.Zemojtel T, Kolanczyk M, Kossler N, Stricker S, Lurz R, Mikula I, Duchniewicz M, Schuelke M, Martaseke P, Ghafourifar P, Vingron M, Mundlos S. Corrigendum to “Mammalian mitochondrial nitric oxide synthase: Characterization of a novel candidate” [FEBS Lett. 580 (2006) 455–462] FEBS Lett. 2007;581:2072–2073. doi: 10.1016/j.febslet.2005.12.038. [DOI] [PubMed] [Google Scholar]
- 31.Frandsen U, Lopez-Figueroa M, Hellsten Y. Localization of nitric oxide synthase in human skeletal muscle. Biochem Biophys Res Commun. 1996;227:88–93. doi: 10.1006/bbrc.1996.1472. [DOI] [PubMed] [Google Scholar]
- 32.Rothe F, Huang PL, Wolf G. Ultrastructural localization of neuronal nitric oxide synthase in the laterodorsal tegmental nucleus of wild-type and knockout mice. Neuroscience. 1999;94:193–201. doi: 10.1016/s0306-4522(99)00263-8. [DOI] [PubMed] [Google Scholar]
- 33.Henrich M, Hoffmann K, Konig P, Gruss M, Fischbach T, Godecke A, Hempelmann G, Kummer W. Sensory neurons respond to hypoxia with NO production associated with mitochondria. Mol Cell Neurosci. 2002;20:307–322. doi: 10.1006/mcne.2002.1111. [DOI] [PubMed] [Google Scholar]
- 34.Gao S, Chen J, Brodsky SV, Huang H, Adler S, Lee JH, Dhadwal N, Cohen-Gould L, Gross SS, Goligorsky MS. Docking of endothelial nitric oxide synthase (eNOS) to the mitochondrial outer membrane: a pentabasic amino acid sequence in the autoinhibitory domain of eNOS targets a proteinase K-cleavable peptide on the cytoplasmic face of mitochondria. J Biol Chem. 2004;279:15968–15974. doi: 10.1074/jbc.M308504200. [DOI] [PubMed] [Google Scholar]
- 35.Lacza Z, Snipes JA, Zhang J, Horvath EM, Figueroa JP, Szabo C, Busija DW. Mitochondrial nitric oxide synthase is not eNOS, nNOS or iNOS. Free Radic Biol Med. 2003;35:1217–1228. doi: 10.1016/s0891-5849(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 36.Brookes PS. Mitochondrial nitric oxide synthase. Mitochondrion. 2004;3:187–204. doi: 10.1016/j.mito.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Wu G, Morris SMJ. Arginine metabolism: nitric oxide and beyond. Biochem J. 1998;336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Regunathan S, Reis DJ. Agmatine is synthesized by a mitochondrial arginine decarboxylase in rat brain. Ann N Y Acad Sci. 1995;763:325–329. doi: 10.1111/j.1749-6632.1995.tb32418.x. [DOI] [PubMed] [Google Scholar]
- 39.Galea E, Regunathan S, Eliopoulos V, Feinstein DL, Reis DJ. Inhibition of mammalian nitric oxide synthases by agmatine, an endogenous polyamine formed by decarboxylation of arginine. Biochem J. 1996;316:247–249. doi: 10.1042/bj3160247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.L KS, Tay A1 Yvonne Mei Sian, Sheu F-S, Jenner A, Whiteman M, Wong KP, Halliwell B. Do Mitochondria make Nitric Oxide? No. Free Radical Res. 2004;38:591–599. doi: 10.1080/10715760410001694008. [DOI] [PubMed] [Google Scholar]
- 41.Lacza Z, Horn TF, Snipes JA, Zhang J, Roychowdhury S, Horvath EM, Figueroa JP, Kollai M, Szabo C, Busija DW. Lack of mitochondrial nitric oxide production in the mouse brain. J Neurochem. 2004;90:942–951. doi: 10.1111/j.1471-4159.2004.02553.x. [DOI] [PubMed] [Google Scholar]
- 42.Csordas A, Pankotai E, Snipes JA, Cselenyak A, Sarszegi Z, Cziraki A, Gaszner B, Papp L, Benko R, Kiss L, Kovacs E, Kollai M, Szabo C, Busija DW, Lacza Z. Human heart mitochondria do not produce physiologically relevant quantities of nitric oxide. Life Sci. 2007;80:633–637. doi: 10.1016/j.lfs.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Sims NR. Rapid isolation of metabolically active mitochondria from rat brain and subregions using Percoll density gradient centrifugation. J Neurochem. 1990;55:698–707. doi: 10.1111/j.1471-4159.1990.tb04189.x. [DOI] [PubMed] [Google Scholar]
- 44.French S, Giulivi C, Balaban RS. Nitric oxide synthase in porcine heart mitochondria: evidence for low physiological activity. Am J Physiol Heart Circ Physiol. 2001;280:H2863–7. doi: 10.1152/ajpheart.2001.280.6.H2863. [DOI] [PubMed] [Google Scholar]
- 45.Lacza Z, Pankotai E, Csordas A, Gero D, Kiss L, Horvath EM, Kollai M, Busija DW, Szabo C. Mitochondrial NO and reactive nitrogen species production: Does mtNOS exist? Nitric Oxide. 2006;14(2):162–168. doi: 10.1016/j.niox.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 46.Adak S, Aulak KS, Stuehr DJ. Direct evidence for nitric oxide production by a nitric-oxide synthase-like protein from Bacillus subtilis. J Biol Chem. 2002;277:16167–16171. doi: 10.1074/jbc.M201136200. [DOI] [PubMed] [Google Scholar]
- 47.Tischner R, Planchet E, Kaiser WM. Mitochondrial electron transport as a source for nitric oxide in the unicellular green alga Chlorella sorokiniana. FEBS Lett. 2004;576:151–155. doi: 10.1016/j.febslet.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 48.Lacza Z, Kozlov AV, Pankotai E, Csordas A, Wolf G, Redl H, Kollai M, Szabo C, Busija DW, Horn TF. Mitochondria produce reactive nitrogen species via an arginine-independent pathway. Free Radic Res. 2006;40:369–378. doi: 10.1080/10715760500539139. [DOI] [PubMed] [Google Scholar]
- 49.Lacza Z, Horvath EM, Pankotai E, Csordas A, Kollai M, Szabo C, Busija DW. The novel red-fluorescent probe DAR-4M measures reactive nitrogen species rather than NO. J Pharmacol Toxicol Methods. 2005;52:335–340. doi: 10.1016/j.vascn.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 50.Lopez-Figueroa MO, Caamano C, Morano MI, Ronn LC, Akil H, Watson SJ. Direct evidence of nitric oxide presence within mitochondria. Biochem Biophys Res Commun. 2000;272:129–133. doi: 10.1006/bbrc.2000.2748. [DOI] [PubMed] [Google Scholar]
- 51.Zanella B, Calonghi N, Pagnotta E, Masotti L, Guarnieri C. Mitochondrial nitric oxide localization in H9c2 cells revealed by confocal microscopy. Biochem Biophys Res Commun. 2002;290:1010–1014. doi: 10.1006/bbrc.2001.6284. [DOI] [PubMed] [Google Scholar]
- 52.Dennis J, Bennett JPJ. Interactions among nitric oxide and Bcl-family proteins after MPP+ exposure of SH-SY5Y neural cells I: MPP+ increases mitochondrial NO and Bax protein. J Neurosci Res. 2003;72:76–88. doi: 10.1002/jnr.10539. [DOI] [PubMed] [Google Scholar]
- 53.Lebuffe G, Schumacker PT, Shao ZH, Anderson T, Iwase H, Vanden HTL. ROS and NO trigger early preconditioning: relationship to mitochondrial KATP channel. Am J Physiol Heart Circ Physiol. 2003;284:H299–H308. doi: 10.1152/ajpheart.00706.2002. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto Y, Henrich M, Snipes RL, Kummer W. Altered production of nitric oxide and reactive oxygen species in rat nodose ganglion neurons during acute hypoxia. Brain Res. 2003;961:1–9. doi: 10.1016/s0006-8993(02)03826-x. [DOI] [PubMed] [Google Scholar]
- 55.Dedkova EN, Ji X, Lipsius SL, Blatter LA. Mitochondrial calcium uptake stimulates nitric oxide production in mitochondria of bovine vascular endothelial cells. Am J Physiol Cell Physiol. 2004;286:C406–C415. doi: 10.1152/ajpcell.00155.2003. [DOI] [PubMed] [Google Scholar]
- 56.Valdez LB, Boveris A. Mitochondrial nitric oxide synthase, a voltage-dependent enzyme, is responsible for nitric oxide diffusion to cytosol. Frontiers in Bioscience. 2007;12:1210–1219. doi: 10.2741/2139. [DOI] [PubMed] [Google Scholar]
- 57.Steffen M, Sarkela TM, Gybina AA, Steele TW, Trasseth NJ, Kuehl D, Giulivi C. Metabolism of S-nitrosoglutathione in intact mitochondria. Biochem J. 2001;356:395–402. doi: 10.1042/0264-6021:3560395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 59.Dahlem YA, Horn TF, Buntinas L, Gonoi T, Wolf G, Siemen D. The human mitochondrial KATP channel is modulated by calcium and nitric oxide: a patch-clamp approach. Biochim Biophys Acta. 2004;1656:46–56. doi: 10.1016/j.bbabio.2004.01.003. [DOI] [PubMed] [Google Scholar]