Abstract
Approaches based on organismal DNA found in the environment (eDNA) have become increasingly utilized for ecological studies and biodiversity inventories as an alternative to traditional field survey methods. Such DNA-based techniques have been largely used to establish the presence of free-living organisms, but have much potential for detecting and quantifying infectious agents in the environment, which are necessary to evaluate disease risk. We developed an eDNA method to examine the distribution and abundance of the trematode Ribeiroia ondatrae, a pathogenic parasite known to cause malformations in North American amphibians. In addition to comparing this eDNA approach to classical host necropsy, we examined the detectability of R. ondatrae in water samples subject to different degradation conditions (time and temperature). Our test exhibited high specificity and sensitivity to R. ondatrae, capable of detecting as little as 14 fg of this parasite’s DNA (1/2500th of a single infectious stage) from field water samples. Compared to our results from amphibian host necropsy, quantitative PCR was ∼ 90% concordant with respect to R. ondatrae detection from 15 field sites and was also a significant predictor of host infection abundance. DNA was still detectable in lab samples after 21 days at 25 °C, indicating that our method is robust to field conditions. By comparing the advantages and disadvantages of eDNA versus traditional survey methods for determining pathogen presence and abundance in the field, we found that the lower costs and effort associated with eDNA approaches provide many advantages. The development of alternative tools is critical for disease ecology as wildlife management and conservation efforts require reliable establishment and monitoring of pathogens.
Keywords: disease, survey, environment, DNA, parasite, amphibian
Introduction
Traditional survey methods to determine the presence of particular species in the environment are often labor- and time-intensive. They are also susceptible to “false negatives”, whereby surveys fail to detect cryptic or rare species that are actually present (Dejean et al. 2011, Schmidt et al. 2013). This has pushed researchers to search for ways to quantify sampling uncertainty, such as occupancy modeling (Schmidt et al. 2013), and to develop detection tools that are less vulnerable to observer error. For the latter, methods that rely on the detection of environmental DNA (eDNA) have been advanced as a complementary approach to identify those species that are inconspicuous or difficult to find within a range of habitats (see Bohmann et al. 2014 for a review). Cellular DNA (e.g. sloughed living cells) and extracellular DNA (after cell death and destruction) are typical sources of total organismal DNA that are often very persistent in the environment (Taberlet et al, 2012). Detection of eDNA could be utilized for many types of ecological studies, including diet analysis, biodiversity inventories, and determining species distributions (Yaccoz 2012). Techniques employing eDNA are also useful for monitoring purposes, such as evaluating the status of endangered and invasive species (e.g. Goldberg et al. 2011, Piaggio et al. 2014). For example, the spread of the invasive Asian carp within North America has been effectively tracked through eDNA methods that are vital for efforts to prevent their colonization of the Great Lakes (Jerde et al. 2011).
Techniques involving eDNA also have enormous potential for monitoring disease risk, as they can facilitate our ability to establish the presence, diversity, and quantity of infectious agents. Given that infectious diseases have been recognized as a significant wildlife conservation issue (Daszak et al. 2000, Thompson et al. 2010), there is a need for effective tools that reliably evaluate the presence and abundance of pathogens in the field. Classical methods employed to detect pathogens are just as resource-intensive, if not more so, than those used for free-living organisms because they typically involve culturing (prokaryotes) or examination of host tissues via necropsy (Audemard et al. 2004, Reinitz et al. 2007). As a result, DNA-based methods have been developed to decrease the time required for in vivo detection (i.e. within hosts) while also increasing precision. For instance, the development of fluorescent probes employing real-time PCR to determine the presence of human schistosome parasites within snail hosts has led to a better understanding of local transmission dynamics and assessment of intervention efforts (Kane et al. 2013). However, this approach still requires host collection and the associated logistical issues identified above, as well as others unique to assessing infectious diseases in the field, such as extensive host sampling. Because macroparasites typically show a highly aggregated distribution in host populations, with most occurring in/on a few heavily infected hosts (Shaw et al. 1998), a large sample size is often required in order to establish their presence with confidence. Such collections become particularly challenging in remote or difficult to access locations or when rare/endangered species are involved (Bohmann et al. 2014). Host examination and the identification of parasites requires specific expertise as well (Gordon et al. 2011), collectively emphasizing the importance of detecting parasites through eDNA. To date, such methodology has been almost exclusively developed for human parasites (e.g. Worrell et al. 2011, Kao et al. 2013) but holds much promise for wildlife diseases (e.g. Audemard et al. 2004, Bridle et al. 2010), particularly to elucidate parasite ranges and determine the level of host risk.
In the present study, we aimed to develop (and apply) an eDNA method for detecting the presence of the pathogenic trematode Ribeiroia ondatrae in wetland habitats. This flatworm parasite has been documented to cause high levels of amphibian mortality and various malformations in North America (Johnson et al. 1999, 2011, Goodman and Johnson 2011a, b), and there is evidence that it has become more common over the past few decades (Johnson et al. 2003). Thus far, most reports of R. ondatrae and amphibian malformations are limited to select regions in North America; reports of malformed frogs are noticeably absent or rare in the southern United States and most of Canada (Johnson et al. 2005, Roberts and Dickinson 2012). Importantly, however, it is not clear whether this signifies a genuine absence of R. ondatrae, a presence of R. ondatrae that does not cause obvious amphibian deformities, or simply insufficient investigation in some geographic areas. Given the highly pathogenic effects of this parasite and its possible role in amphibian population declines (Johnson et al. 1999, 2011), it is crucial to determine its distribution and how its abundance is affected by anthropogenic changes, such as water temperature, nutrient concentrations, and biodiversity (Johnson et al. 2007, Paull and Johnson 2011, Koprivnikar et al. 2012).
Here our goal was to build upon a PCR-based test developed by Reinitz et al. (2007) for R. ondatrae detection within snails to create an eDNA method that would not require host collection, thus representing a relatively simple and cost-effective alternative for establishing the presence of this parasite in the field, particularly for a host group (amphibians) that is widely declining. In spite of the recent increase in ecological studies employing eDNA-related methods, standard field protocols have yet to be established and the environmental persistence of DNA is not well-understood (Barnes et al. 2014). eDNA methods may not represent a significant improvement over classic field sampling techniques if they are also prone to false negatives due to spatial or temporal issues, e.g. DNA degradation due to mistimed sampling or particular environmental conditions. We therefore compared the R. ondatrae status of 15 field sites using both amphibian host necropsy and eDNA analysis of water samples. Because trematode infectious stages have a relatively short lifespan, typically <24 hours (Combes et al. 1994), we also conducted lab experiments to determine the detectability of R. ondatrae DNA through time within water samples maintained at two different temperatures. In addition, we compared the cost and time required to establish parasite presence at field sites through our eDNA and classical methods. By considering the reliability, robustness, and resources associated with both approaches, we aimed to create a reliable field test for the presence of R. ondatrae that can be extended to other wildlife parasites.
Methods
Host and parasite collection
The life cycle of R. ondatrae is complex and involves multiple hosts. Adult worms are found within the digestive tract of avian definitive hosts and shed eggs that pass with the host’s fecal material. In water, the eggs will embryonate and hatch into free-swimming infectious stages (miracidia) that infect suitable gastropod first intermediate hosts (Planorbella/Helisoma spp. snails), undergoing multiple rounds of asexual reproduction to produce another free-swimming infectious stage (cercaria) which seeks out a second intermediate host (amphibians or fish). Within the second intermediate host, the cercaria forms a cyst and the life cycle is completed upon host consumption by an appropriate bird (see Johnson et al. 2004 for a review).
Planorbella spp. snails infected with R. ondatrae were collected from ponds near San Jose (California, USA) and St. Catherines (Ontario, Canada). The snails were maintained in the laboratory in separate 20 L aquaria containing dechlorinated tap water, under a 16:8 light-dark cycle, and were fed a diet of boiled spinach. Trematode cercariae were collected by placing individual snails in water-filled Petri dishes overnight (for R. ondatrae) or under a lamp for 1 h during the day (for all other trematode species). The cercariae were then identified using previously published descriptions (Schell 1985, Szurocki and Richardson 2009). Trematode cercariae of each species (∼300–800 µm total body length) were pooled in groups of 25 in 1.5 ml microfuge tubes, chilled on ice to slow their movement, and we then removed most of the water using a micropipette. DNA was extracted from the pooled cercariae using the DNA Wizard Purification Kit (Promega) according the manufacturer’s protocols. The concentration was determined using a Nanodrop spectrophotometer (General Electric).
To compare the R. ondatrae detection ability of our eDNA method versus host necropsy, we also collected larval or newly-metamorphosed amphibians at field sites using sweep nets, euthanized them in a 0.2% solution of buffered MS-222 (Sigma-Aldrich), and conducted necropsies. We examined all major organ systems of the hosts, with particular focus on the skin for R. ondatrae cysts (see Fig. 1), while identifying and quantifying all macroparasites present. This allowed us to determine R. ondatrae presence at each field site (infection present in at least one host), as well as individual infection abundance (number of cysts/host – see Bush et al. 1997 for standard terminology).
Figure 1.
Equipment and methods used for eDNA and necropsy-based assessment of Ribeiroia ondatrae status of field sites: (a) sweep net collection of larval/metamorphic amphibians, (b) R. ondatrae cysts within infected host (indicated by arrows), (c) R. ondatrae cercaria - likely the largest eDNA contributor, and (d) hand-held water filtration unit for R. ondatrae eDNA collection.
PCR sensitivity and specificity to R. ondatrae DNA
A pair of PCR primers, designed by Reinitz et al. (2007), and two pairs of PCR primers designed using Primer3 (Version 0.4.0) software, were tested for their ability to PCR-amplify a 164 to 290 bp fragment of the internal transcribed spacer 2 (ITS-2; GeneBank ID: AY761142.1) sequence of R. ondatrae’s ribosomal DNA sequence (Table 1). The primers were compared to all sequences within GenBank using BLAST software to assess their specificity to the R. ondatrae sequence. End-point PCR reactions were performed using 10-fold serial dilutions of template DNA, ranging from 0.1 pg to 1 ug DNA, with primers at a concentration of 0.8 µM and EconoTaq Plus Green Master Mix (Lucigen). A gradient PCR method that systemically varied temperature and time was first used to select the optimal annealing temperature, using the following cycling conditions: 94°C for 5 minutes, 40 cycles of 94°C for 30 seconds, a variable annealing temperature ranging from 40°C to 60°C (the optimal temperature was concluded to be 46°C) for 30 seconds, 72°C for 30 seconds, and then 72°C for 5 minutes. After PCR amplification, the PCR products were resolved on 1.5% agarose gels, either stained with ethidium bromide or SYBR Gold (Invitrogen), and visualized with a BioRad Universal Hood II UV transilluminator. To avoid cross-contamination, DNA extractions and handling of samples were performed in a laminar flow cabinet and all equipment and water was UV irradiated for 15– 30 minutes between steps and prior to use. Negative control samples lacked any template DNA. A total of 5 replicate experiments were performed for each DNA dilution.
Table 1.
eDNA primer information
| Primer Pair | Sequence | Product size (bp) |
|---|---|---|
| Ro-ITS 1 | For: TCACGACGCTCAAATAGTCG Rev: GAGCATAGCTCCACCCGTAG |
240 |
| Ro-ITS 2 | For: AGTCATGGTGAGGTGCAGTGA Rev: AGACCGCTTAGATAGCAG |
290 |
|
* Ro-ITS 3 |
For: CGTGTTTGGCGATTTAGT Rev: TCAAAAATGAAGCAACAGT |
164 |
Derived using Primer3 software but identical to primers used by Reinitz et al. (2007).
The species-specificity of the primers was assessed by determining their ability to detect 0.01 to 10 individual Echinoparyphium spp. and fasciolid-type cercariae in a single PCR reaction. DNA from these two trematode species was extracted using the DNA Wizard Purification Kit (Promega) and we used the primers, thermal cycling conditions, and DNA visualization methods described above.
PCR detection of degraded R. ondatrae DNA
In addition to testing the ability of the PCR to amplify small amounts of fresh R. ondatrae DNA, the efficacy of the PCR following possible DNA degradation in water over time was evaluated. Five live R. ondatrae cercariae were placed into each of 16 different 1L glass jars containing 900 ml of dechlorinated tap water and 100ml of water from an aquarium containing only zebrafish (simulating microfauna from a fish-populated habitat). The 16 jars were divided among 4 treatment groups to examine the separate and combined effects of time (10 or 21 days) and temperature (20°C or 25°C). The water from each jar was then filtered and processed as described below for eDNA water samples to determine DNA detectability.
eDNA collection and extraction
An inexpensive (∼ $18.00 USD) hand-held eDNA collection prototype for use with water samples was developed (see Fig. 1 and Appendix for details). Ten units were constructed consisting of a filter body capable of holding 500 mL of water and a removable filter support unit to collect particulates. Water was collected from 21 different wetlands in California and southern Ontario in July 2012 and July 2013 for which R. ondatrae infection presence/absence was verified through the collection and necropsy of larval or newly-metamorphosed amphibians from each of the sites. Particulate matter (including any eDNA present) was collected by pushing 500 ml of water through the filtration support unit equipped with 3µm pore size Whatman cellulose nitrate membrane filters (Sigma-Aldrich). While smaller pore sizes capture more eDNA, clogging prevents the filtering of large water volumes (e.g. no more than 250 mL for 0.2 µm pore size) and thus we chose a larger pore size and adjusted the filtration volume to compensate (see Turner et al. 2014). A separate, unused filtration apparatus was used for each of the 10 sites in 2012 to avoid the possibility of cross-contamination and these were thoroughly washed and dried prior to re-use in 2013. Five water subsamples (5 m apart) per site near the shoreline (15–45 cm water depth) were collected by lowering the filter body just below the surface of the water and filling it to the 500 ml mark. Following attachment of the filter and its support, a bicycle pump was used to push the water through the filter, using pressures less than 40 PSI to prevent filter membrane rupture. Filters were stored in 15 ml disposable plastic tubes containing 10 ml of 70% ethanol.
The eDNA was extracted from the filters using a modified TRIZOL reagent (Invitrogen) protocol. Filters from different sites were processed separately to ensure no cross contamination. The filters from each site were air-dried in Petri dishes until all of the ethanol evaporated. The dried filters were chopped into small pieces with flame-sterilized scissors, and packed into 15 ml tubes, to which 1.5 mL of TRIZOL was added. This mixture was vortexed for 15 sec, followed by incubation for 5 minutes at room temperature to dissolve the DNA from the filters. The TRIZOL solution was then transferred to another 2 ml tube. Chloroform (300 µl) was added to the TRIZOL mixture, vortexed for 15 seconds, and incubated at room temperature for 15 minutes. The tube was then centrifuged at 15,000 xg for 15 minutes at 4°C. The aqueous phase (containing the RNA) was removed using a pipette and discarded. The DNA in the interphase and phenol phase was precipitated by adding 450 µl of 100% ethanol, followed by a gentle vortex and incubation at room temperature for 3 minutes. The DNA was pelleted by centrifuging at 15,000 xg for 5 minutes at room temperature. The DNA pellet was washed twice in 1 ml of 0.1M sodium citrate for 30 minutes on a rocking table. The DNA was then washed briefly with 1 ml of 70% ethanol and pelleted by centrifugation. The pellet was dried using a vacuum centrifuge and re-suspended in 100 µl of nuclease-free water. To enhance re-suspension, the DNA was warmed at 55°C for 10 minutes. The DNA concentrations were determined by spectrophotometry and then stored at −20°C until analyzed by PCR.
Qualitative end-point PCR eDNA detection
R. ondatrae eDNA (1 ul of extracted DNA) derived from filter collections was detected by PCR using the Ro-ITS 3 primer pair, as described above. Each site sub-sample was replicated 3 times for a total of 15 PCR reactions per site (5 replicate sub-samples from each site). PCR products were resolved on 1.5% agarose gels in TBE stained with ethidium bromide. A replicate was considered positive if a band of 164 bp was observed. Field sites were scored as negative for end-point PCR if <3 of 15 replicate water samples produced faintly visible PCR products after 40 cycles of PCR amplification. Sites were considered positive if 4 or more faint PCR products were observed, or one or more intensely-staining PCR products were observed in the electrophoresis gels.
Quantitative PCR eDNA detection
Quantitative PCR (q-PCR) was used to obtain estimates of the number of cercariae collected from each 500 ml sample of pond water. DNA was extracted from five cerariae as described above, and the DNA concentration was determined, to enable the amount of DNA/ceraria to be calculated. Based on three replicates, 2.5 ng of DNA could be extracted from a single cercaria. Serial dilutions of the DNA in water were then used as template for q-PCR, using BioRad’s iQ SYBR Green Supermix on a BioRad iQ5 Multicolor Real-Time Detection System as per the manufacturer’s protocols. Samples were analyzed in 10µl reactions (5µl iQ SYBR Green Supermix, 0.5µl of each Ro-ITS 3 primer, 3.5µl sterile water, and 0.5µl site DNA template) using the manufacturer’s protocol. Real time data was compiled using iQ (BioRad) software. A melt curve analysis was performed on all samples to confirm that only one amplicon was produced in each reaction. The cycle threshold (Ct) values for each DNA dilution were plotted to generate a standard curve, and linear regression was used estimate the concentrations of R. ondatrae DNA, and therefore the number of cercariae in each 500 ml pond sample. Note that relatively high Ct values correspond to a greater number of PCR cycles needed to amplify the DNA, i.e. less was initially present in a sample.
Because the q-PCR method showed greater sensitivity and accuracy compared to standard end-point PCR for our 2012 samples (detection of 5.54×10−6 versus 4.0×10−4 of a cercaria, and 90% versus 70% accuracy compared to host necropsy, respectively), only q-PCR was used to analyze the 2013 samples. Each DNA sub-sample was analyzed by q-PCR in duplicate to calculate the R. ondatrae DNA concentration for each sampling site. The q-PCR assay could consistently detect 14 fg of R. ondatrae DNA, whereas amounts below this value produced more variable Ct values. Accordingly, a sample was considered “negative” (i.e. no R. ondatrae present) below this DNA threshold, while samples with values above this threshold considered the site to be “positive.” A pond was considered positive if any one of the five sub-samples were positive.
Statistical analysis
We used a Linear Mixed Model (LMM) to determine if Ct (q-PCR Cycle Threshold) values differed between sites with R. ondatrae present or absent (categorical fixed factor based on examination of amphibians from that sampling year), with site identity and sampling year for each sub-sample as categorical random factors. To meet the assumptions of a normal distribution, the Ct values for each sub-sample were log10-transformed before analysis. We excluded sites for which we were unable to obtain any PCR reactions, or those where we could not examine at least 9 larval/newly metamorphosed amphibians from the collection year, leaving 15 sites for the LMM (Table 2). To assess whether site Ct values predicted individual infection abundance for the amphibians examined, we used a generalized linear mixed model (GLMM). As infection abundance represents count data, we used a Poisson distribution with log link function. In addition to site Ct value as a fixed effect, site identity, sampling year, and host species were included as categorical random effects. All analyses were done with SPSS 21.0.
Table 2.
Ribeiroia ondatrae presence or absence at field sites (CA = California, ON = Ontario) based on eDNA results from quantitative real-time PCR (q-PCR with Cycle threshold – Ct values) and amphibian necropsies.
| Site | # frogs examined |
Mean frog infection intensity |
Frog necropsy Ribeiroia status |
Mean Ct value |
q-PCR Ribeiroia status |
|---|---|---|---|---|---|
| 1 (CA) | 10 | 22 | Present | 32.52 | Present |
| 2 (CA) | 9 | 0 | Absent | 37.29 | Absent |
| 3 (CA) | 24 | 1.5 | Present | 25.29 | Present |
| 4 (CA) | 21 | 61.3 | Present | 32.17 | Present |
| 5 (CA) | 20 | 0.3 | Present | 33.63 | Present |
| 6 (CA) | 32 | 64.8 | Present | 32.02 | Present |
| 7 (CA) | 20 | 4.1 | Present | 30.98 | Present |
| 8 (ON) | 15 | 3 | Present | 31.69 | Present |
| 9 (ON) | 15 | 2.5 | Present | 31.23 | Present |
| 10 (ON) | 15 | 0 | Absent | 33.97 | Present |
| 11 (ON) | 19 | 0 | Absent | 36.09 | Absent |
| 12 (ON) | 24 | 0 | Absent | 36.33 | Absent |
| 13 (ON) | 20 | 1 | Present | 36.11 | Absent |
| 14 (ON) | 10 | 3 | Present | 33.75 | Present |
| 15 (ON) | 10 | 19.3 | Present | 33.13 | Present |
Cloning and sequencing of R. ondatrae ITS-2 gene
DNA of R. ondatrae cercariae was collected from snails originating from our California and southern Ontario sites, extracted from gel bands of approximately 164bp, and then purified using QIAquick Gel Extraction Kit (Qiagen) as per the manufacturer’s instructions. The PCR products were ligated into the pJET PCR cloning vector (Thermo Fisher Scientific), were transformed into Sub-cloning Efficiency™ DH5α Chemically Competent E. coli cells (Invitrogen) following the manufacturer’s protocol. Plasmid DNA was subsequently purified from three bacterial clones derived from both the Californian and Ontario R. ondatrae isolates using a Qiagen QIAprep Miniprep kit as per the manufacturer’s instructions. The PCR fragments within the plasmids were then sent to the Robarts Sequencing Facility (London, Canada) for DNA sequencing.
Comparison of eDNA and traditional methods
We determined the time and cost needed to assess the R. ondatrae status of a site (presence and abundance) using eDNA and traditional necropsy-based approaches (Table 3). For the eDNA method, we considered the time required to collect water samples, extract the DNA, and perform the q-PCR steps detailed above in order to generate a site R. ondatrae DNA concentration via the Ct value. The costs associated with both approaches were determined based on the specified materials, transport from field sites to the lab, and an estimate of labor using the time per site and a standard wage. For the traditional field-sampling approach, we estimated the time associated with collecting 18 larval/newly metamorphosed amphibians (mean sample size of our 15 retained sites), and the subsequent necropsy of each to determine R. ondatrae infection (presence and cyst abundance). As the collection and use of vertebrates in research requires both institutional and government permits, we also considered the time associated with this. We ignored time required for driving to and from field sites, as well as that to obtain access permission, as this would presumably be equivalent for both methods.
Table 3.
Time and materials associated with eDNA (via quantitative PCR) and classical necropsy approaches to determine Ribeiroia ondatrae status (presence and quantity) for a single field site. All costs are in U.S. dollars.
| eDNA | Traditional field sampling | |
|---|---|---|
| Time (h) | ||
| Permits (vertebrate use) | n/a | 2 |
| Field sample collection | 0.8 (5 water subsamples @ 10 min each) |
1 (18 amphibians) |
| Sample processing1 | 0.5 (DNA extraction, q-PCR, and results interpretation) |
3 (18 hosts @ 10 min each necropsy) |
| TOTAL TIME | 1.3 | 6 |
| Labor subtotal | $26 (1.33 h @ $20/h) | $120 (6 h @ $20/h) |
| Transportation2 | ||
| FedEx economy (by weight) | $37 (0.5 kg from S ON to MB) $76 (0.5 kg from CA to MB) |
$37 (1 kg from S ON to MB) $87 (1 kg from CA to MB) |
| Transportation subtotal | $57 (mean) | $62 (mean) |
| Consumable materials3 | ||
| Preservatives | $4 (50 mL ethanol for 5 subsample containers) |
$8 (100 mL ethanol for 15 specimens) |
| DNA extraction & PCR4 | $12 | n/a |
| TOTAL COST | $95 | $190 |
Ten sites (5 subsamples each) with standards can be run at once as thermal cycler can hold 96 samples. Per site time calculated by dividing total processing time for 96 samples (5 h) by 10.
Based on field sites and sample processing locations in current study (S ON = southern Ontario, MB = Manitoba, CA = California)
Re-usable equipment excluded (e.g. sweep nets, hand-held filtration units, thermal cycler)
See text for specific quantities and products used
Results
PCR detection sensitivity and specificity
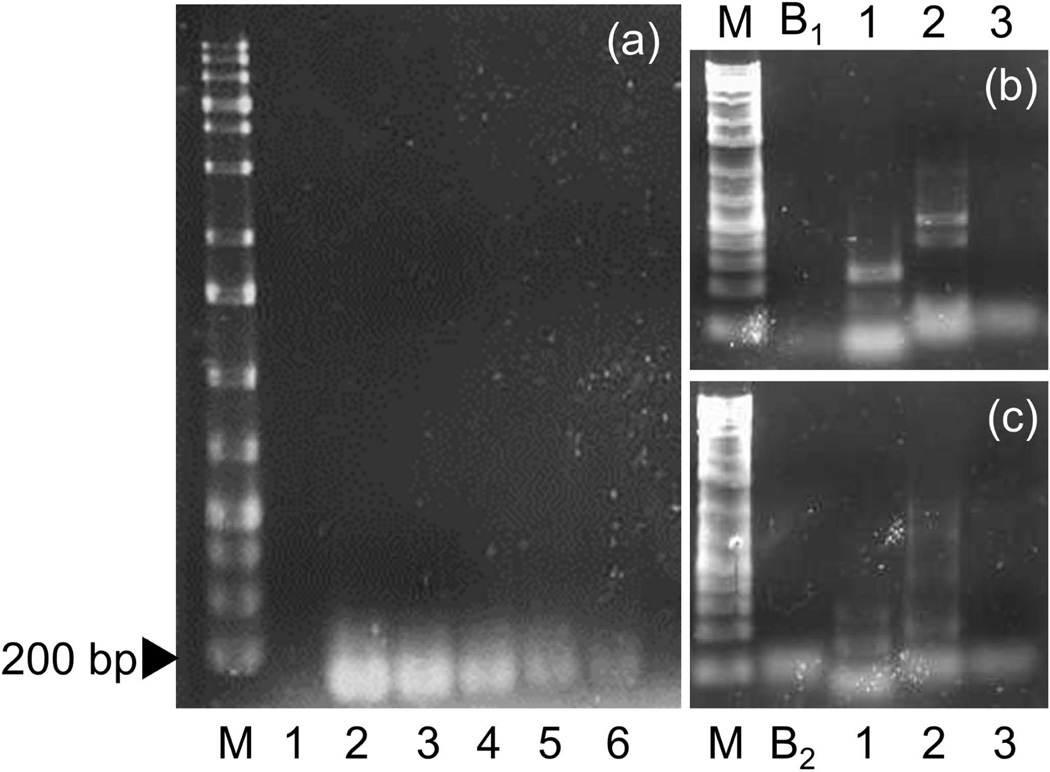
All three pairs of PCR primers amplified the expected (164–290 bp) sized PCR products from R. ondatrae, and DNA sequencing confirmed their identities. No differences in the sequences were observed between the R. ondatrae samples isolated from California and southern Ontario. Using the Ro-ITS 3 primer set, we could detect as little as 1/2500th of a single cercaria under optimal conditions using molecular grade water and directly extracting DNA without a membrane filter (Figure 2a). The Ro-ITS 3 primer set was also the most stringent as it did not amplify genes from Echinoparyphium sp. or a fasciolid-type of cercaria (Figure 2b and 2c). In contrast, primer pairs Ro-IRS 1 and Ro-ITS 2 showed reduced specificity under the PCR conditions used, both detecting 2.4 × 10−2 of a cercaria from these other trematode species. Neither time (10 or 21 days) nor temperature (20°C or 25°C) caused enough degeneration to affect the ability of the PCR to amplify the target sequence, suggesting our method was robust for detecting parasite eDNA at field sites with different conditions. Our BLAST pairwise alignment of the Ro-ITS 3 primer set amplicon (see Appendix 2) resulted in a 100% identity to the R. ondatrae ITS-2 gene in Genbank (AY761142.1) and not to any other North American species.
Figure 2.
(a) Sensitivity of the Ro-ITS 3 primer under lab conditions to dilutions of Ribeiroia ondatrae. M = ladder and 1 = primer control, with 2, 3, 4, 5, and 6 representing 4.0, 0.4, 0.04, 0.004, and 0.0004 of a cercaria, respectively. Also shown are stringency tests of primer pairs to Echinoparyphium sp. (b) and a fasciolid-type cercariae (c). M = ladder, B1 and B2 = DNA template blanks containing Ro-ITS 1 and Ro-ITS 2, respectively; 1 = Ro-ITS 1, 2 = Ro-ITS 2, and 3 = Ro-ITS 3.
eDNA presence and abundance
The end-point PCR method was determined to be 70% accurate for our 2012 field samples when compared to R. ondatrae infections detected via amphibian necropsy. Thus, three of the 2012 sites appeared as “false negatives” (i.e. R. ondatrae infection was found in amphibians but not via PCR); however, the end-point PCR method did not show any false positives.
The calibration curve generated for our q-PCR tests indicated the effective range of this method to be from 10 to 5.54 × 10−6 cercariae; below this range, the Ct values were too high and quite variable, making the test inaccurate. To reduce the chances of a false positive, we consequently considered sites as negative if the mean Ct value was 35 or higher (5.54 × 10−6 cercariae or less). We considered Ct scores of <35 to indicate R. ondatrae presence and our line of best fit had a R2 value of 0.81, indicating that Ct values lower than 35 provided a reasonably strong estimation of cercariae abundance in field samples. The q-PCR technique showed 86.6% accuracy (correct for 13/15 field sites in 2012 and 2013) when compared to the host necropsy data (Table 2).
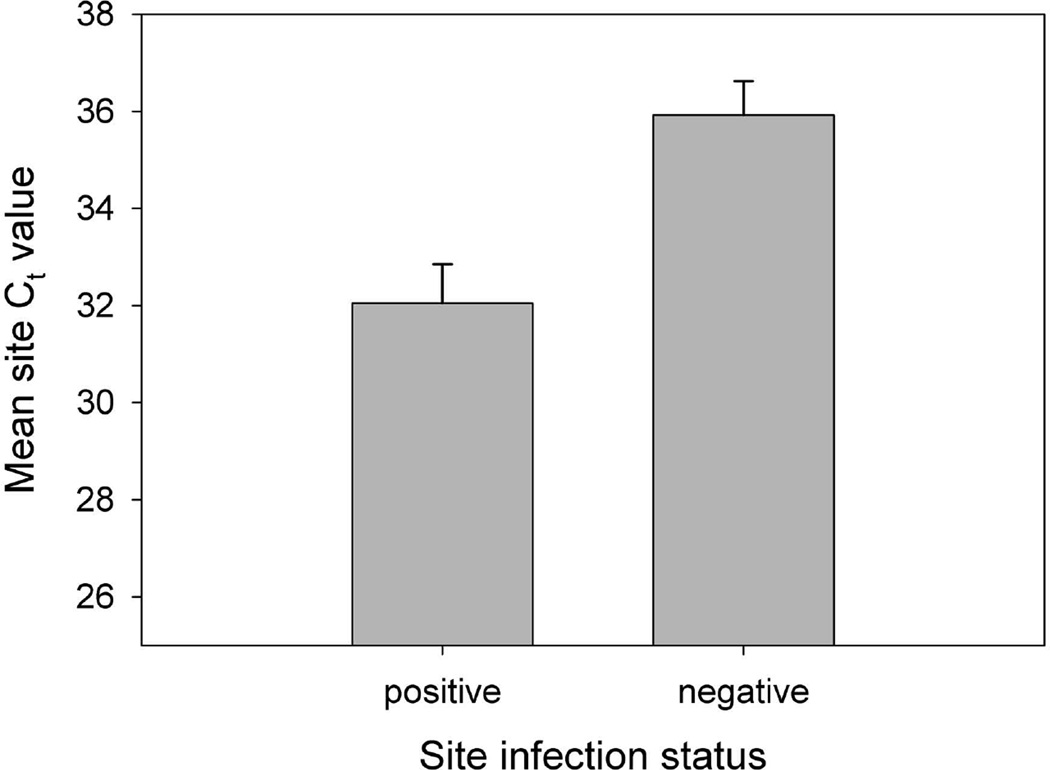
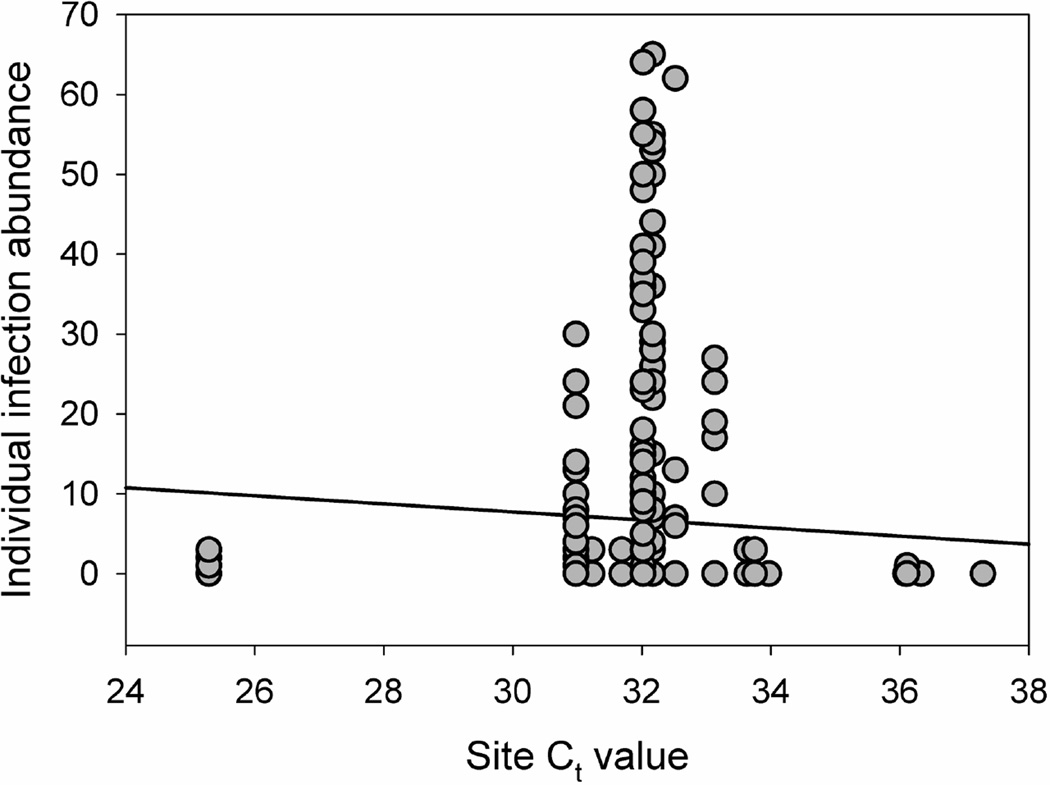
The results of the LMM indicated a significant difference in the mean q-PCR Ct value between field sites characterized as R. ondatrae positive or negative based on amphibian necropsy (F1,83 = 7.101, P = 0.009). Sites with R. ondatrae-infected amphibians had a mean Ct value of 32.51 (± 0.34 S.E.) compared to a mean of 35.04 (± 0.44 S.E.) for those where infected frogs were not found (Figure 3). In addition, the GLMM results showed that mean site Ct value was a significant predictor of individual R. ondatrae infection abundance within examined amphibians (F1,266 = 4.721, P = 0.031, coefficient = −0.518; Figure 4). Our calculation of the average cost and time needed to assess the R. ondatrae status of each field site in the present study indicates that the necropsy-based approach took almost 5 times as long and cost double compared to using eDNA (Table 3).
Figure 3.
Mean Cycle Threshold (Ct) value (± S.E.) resulting from quantitative PCR of field-collected Ribeiroia ondatrae eDNA in water samples. Note that higher Ct values correspond to a greater number of PCR cycles required to amplify the DNA, i.e. less was initially present in a sample. Site status (negative or positive) reflects the absence or presence of R. ondatrae-infected amphibians as determined by necropsy.
Figure 4.
Relationship between R ondatrae infection abundance within individual amphibians and mean site concentration of R. ondatrae cercariae as determined through quantitative PCR (Ct = Cycle Threshold value). Note that line of best fit is for illustrative purposes only.
Discussion
Our eDNA method for field-detection of the pathogenic amphibian parasite R. ondatrae proved to be both sensitive and specific, closely matching results derived from traditional host necropsy, and illustrating the broad potential of this approach for large-scale spatial investigations related to disease ecology. We were able to detect as little as 14 fg of R. ondatrae DNA from environmental water samples compared to the detection limit of 100 fg for PCR of snail host tissues described by Reinitz et al. (2007). This eDNA approach also allowed us to confirm the presence of R ondatrae in southern Ontario, considerably expanding the known range of this parasite and illustrating how such methodology can be used to investigate pathogen distribution. These results further demonstrate the complexity of evaluating pathogen presence based on restricted criteria. The presence of malformed frogs has often prompted a subsequent examination to determine whether R. ondatrae is present; however, amphibian species vary in their propensity to develop malformations following exposure (Johnson et al. 2012). In southern Ontario, only grey tree frogs (Hyla versicolor), green frogs (Lithobates clamitans), and American bullfrogs (Lithobates catesbeianus) have been detected at our field sites, all of which are highly resistant to R. ondatrae -induced malformations (Johnson et al. 2012, LaFonte and Johnson 2013). While amphibian deformities have been reported in eastern Canada (Ouellet et al. 1997), the role of R. ondatrae has not been specifically examined in relation to these observations, and has only been confirmed as a causative agent in a host species (Pseudacris regilla) restricted to western Canada (Roberts and Dickinson 2012).
We had one “false negative” whereby R. ondatrae infection was found in hosts from a site but not in the collected from among the 5 site subsamples. This probably reflects the highly heterogeneous distribution of trematode-infected snails (and therefore of infectious stages) within water bodies (Fernandez and Esch 1991) but can also result from low sample size. The results of a recent eDNA study to detect an amphibian fungal pathogen (Batrachochytrium dendrobatidis) indicated that 6 subsamples were sufficient to ensure high accuracy (Schmidt et al. 2013), but the optimal number is likely pathogen-dependent and highly influenced by target organism density (Moyer et al. 2014). Consequently, large volumes of water are required to achieve a high probability of detecting the eDNA of species with low abundance, thereby minimizing false negatives (Moyer et al. 2014). DNA degradation is a less likely explanation for host versus water sample differences given our ability to detect R. ondatrae after 21 days in lab water samples kept at 25°C, conditions in which breakdown should be rapid (Dejean et al. 2011, Barnes et al. 2014). Our choice of target DNA probably facilitated detection under such circumstances given its relative abundance compared to other sequences within the genomic DNA. Notably, the ITS-2 sequence is tandemly repeated within the ribosomal DNA (Prokopowich et al. 2003), often in thousands of copies, resulting in a high probability that some of this relatively short target sequence will be intact after prolonged exposure in an aquatic environment (Deagle et al. 2006). Because our eDNA field collections took place in mid-summer when we have previously collected snails with active R. ondatrae infections from some sites included in the present study, it is doubtful that no cercariae had emerged for over 21 days, or site water temperatures were so high that DNA breakdown occurred much more rapidly than what we observed at 25°C. Environmental inhibitors of the PCR reaction, such as humic acids and heavy metals, may instead have played a role (Wilson 1997, Matheson et al. 2010). While commercially available kits can deal with such inhibitors to a certain extent, this increases the cost of sample processing and our aim was to develop a test that was as economical as possible in order to allow future large-scale field testing.
We also had a possible “false positive” for one southern Ontario site in which R. ondatrae DNA was detected but infected amphibians were not found. If the prevalence of infection was low at that site, the number of frogs examined may have been inadequate to establish parasite presence. There is wide variation in the proportion of amphibians infected with R. ondatrae across field sites in North America, ranging from 0–100% (Johnson and McKenzie 2009). Given that our chosen primer set had a high specificity, failing to amplify the DNA of two other trematode species commonly found in our field sites (Koprivnikar and Redfern 2012, Johnson et al. 2013), our results likely indicate true R. ondatrae presence rather than that of other parasites; however, further testing with other parasites will allow us to assess this possibility. Rather, host species identity is likely the primary explanation. As discussed above, grey tree frogs were primarily sampled in the southern Ontario wetlands, and are highly resistant to R. ondatrae infection (Johnson et al. 2012). Because tadpoles of this species are capable of actively clearing cysts within 72 hours (LaFonte and Johnson 2013), collected hosts would have to represent recent infection events in order to detect R. ondatrae through necropsy. The eDNA results may therefore provide a more accurate assessment of parasite presence in this case.
However, the detection of R. ondatrae DNA from a site is not necessarily indicative of amphibian infection given the complex life cycle of most trematodes, particularly the occurrence of multiple life history stages. Avian definitive hosts may deposit trematode eggs at sites lacking suitable gastropod first intermediate hosts. Consequently, the DNA of eggs or hatched miracidia could be collected in water samples and amplified through the same PCR protocol as for the stage infectious to amphibians (cercariae). Environmental RNA (eRNA) represents another approach to determine which stages of the trematode life cycle are present. Due to the relatively rapid degradation of RNA compared to DNA, there is a smaller temporal window of collection, possibly permitting the detection of recently-emerged cercariae rather than eggs originating from transient birds. In addition, the identification of genes up-regulated during specific points of the life cycle may allow detection of relevant parasite stages (Juthikumar et al. 2010).
More importantly, eDNA tests for pathogens requiring multiple hosts can be used not only to assess whether a location is currently functionally colonized (i.e. the entire life cycle is maintained on site), but also the potential risk of host disease under the right circumstances. For instance, a site may be considered low-risk for R. ondatrae-induced malformations if infected amphibians are not found, or appropriate intermediate host species not present. However, avian definitive hosts may continually introduce parasite eggs that are detectable via eDNA, indicating future risk if the site is colonized by competent first or second intermediate hosts. In this case, the development of a test with combined primers to simultaneously detect the presence of both R. ondatrae and the required first intermediate host (Planorbella spp. snails) would be ideal to best evaluate amphibian infection risk. Such multi-faceted tests will be important in order to extend from the current focus on single-species detection (free-living or symbiotic) and allow assessment of community composition (Goldberg et al. 2011, Lodge et al. 2012, Thomsen et al. 2012). Collected eDNA can also be useful for other purposes, such as assessing pathogen diversity through metagenomics approaches (Smith et al. 2012).
Our eDNA-based detection of R. ondatrae constitutes an effective replacement for traditional host collection and examination for macroparasite infection. In terms of accuracy, it would appear that the two methods are relatively comparable. We had an almost 90% match between these approaches but classical sampling methods are also prone to methodological issues, especially for rare or cryptic species, and are thus not completely reliable either (Schmidt et al. 2013). If our “false positive” indicates true R. ondatrae presence, then this balances the false negative, making the two methods equal. This is comparable to previous studies contrasting eDNA and classic sampling approaches for free-living organisms (e.g., Jerde et al. 2011, Pilliod et al. 2011). Consequently, a site-occupancy approach with repeated sampling should generally be considered for eDNA studies (Schmidt et al. 2013), although it may not be appropriate for detecting the presence of certain wildlife pathogens such as R. ondatrae given the brief window for collecting infected hosts (larval and newly-metamorphosed amphibians). We note that while we found our q-PCR approach more sensitive than end-point PCR, these platforms often provide similar non-quantitative results and the latter approach is less expensive and typically more accessible (Nathan et al. 2014).
In view of the comparable accuracy of our eDNA and necropsy approaches, the advantages of the former primarily relate to the ease of obtaining permits and conducting field collection, particularly time and sample transport, and expense. Eliminating the need to collect and euthanize also obviously spares wild hosts, which can be critical for populations of at-risk and endangered species. Unlike the detection of most free-living organisms, assessing the presence and intensity of pathogens typically requires hosts to be collected in the field and then later examined in a lab setting. Given the high among-host aggregation observed for many parasites (Shaw et al. 1998), an adequate number of individuals must also be gathered. This number will vary for different parasites but will likely exceed the number of environmental subsamples for each site. We gathered 5 water subsamples at each of our field sites and this generally took less than 30 person-minutes with our pump-action collection apparatus. However, the time needed to collect 9–32 amphibians/site was often considerably longer (sometimes double or more). Once collected, there is also the issue of properly preserving/storing host tissues for later necropsy, necessitating the use of chemicals or access to freezers, as well as shipping costs, adequate storage space, and expertise for identification. For remote sites, or when dealing with host species that are endangered, this could prove to be extremely challenging, if not impossible. In comparison, our cellulose nitrate membrane filters were preserved in small amounts of ethanol and appeared to remain stable until processing.
With respect to cost, eDNA-based tests are generally considered more affordable than traditional sampling methods; our materials costs were approximately $12 (USD) per site, which is comparable to costs reported by other studies (Goldberg et al. 2011, Worrell et al. 2011). By considering the total cost and effort to assess the R. ondatrae status of a site, we still found an overall advantage to using an eDNA approach. This is primarily driven by the greater labor associated with obtaining permits, sample collection, and processing time for the classical necropsy-based approach (almost five times more), particularly since the q-PCR method allows multiple sites to be run at the same time. As a result, the cost per site using the traditional method was double. While there are obvious advantages to eDNA methods for detecting wildlife pathogens, we caution that necropsies should not be replaced. For instance, our site Ct values were strongly related to host infection abundance but not a perfect predictor (coefficient of −0.518), illustrating the value of retaining classical methods in order to collect certain data. Because macroparasite pathology is often intensity-dependent, as is the case for R. ondatrae (Johnson et al. 2012), such information can be critical to understanding the effects of parasites on individual hosts and populations.
Here we present a reliable and resource-minimizing approach to detect the trematode R. ondatrae via eDNA that will enable us to better understand the distribution of this highly pathogenic macroparasite and the risk it poses to amphibians, as well as aiding in the investigation of other wildlife diseases. This will facilitate a reduction in the number of hosts collected, an increase in the number of inspected sites, an expansion into areas that are difficult to access, and the inclusion of multiple temporal scales. Given the documented and projected effects of environmental perturbations on wildlife diseases (Daszak et al. 2001, Altizer et al. 2013), it is essential that we develop tools to determine current pathogen distributions and monitor these for changes as such considerations are critical to conservation and management efforts.
Supplementary Material
Acknowledgments
We thank Travis McDevitt-Galles for assistance with field collections, Bryan LaFonte, Esra Kellermanns, Julia Redfern, and Hannah Mazier for their necropsy work, and two anonymous reviewers for helpful comments on earlier versions of the manuscript. This research was supported by grants from NSERC (JK and SW), the NSF (DEB-1149308) and NIH (R01GM109499), and a fellowship from the David and Lucile Packard Foundation (PTJJ).
Footnotes
PREPRINT
This preprint is a PDF of a manuscript that has been accepted for publication in an ESA journal. It is the final version that was uploaded and approved by the author(s). While the paper has been through the usual rigorous peer review process of ESA journals, it has not been copy-edited, nor have the graphics and tables been modified for final publication. Also note that the paper may refer to online Appendices and/or Supplements that are not yet available. We have posted this preliminary version of the manuscript online in the interest of making the scientific findings available for distribution and citation as quickly as possible following acceptance. However, readers should be aware that the final, published version will look different from this version and may also have some differences in content.
Ecological Archives material:
Appendix A (hand-held filter unit details)
Literature cited
- Altizer S, Ostfeld RS, Harvell CD, Johnson PTJ, Kutz S. Climate change and infectious diseases: from evidence to a predictive framework. Science. 2013;341:514–519. doi: 10.1126/science.1239401. [DOI] [PubMed] [Google Scholar]
- Audemard C, Reece KS, Burreson EM. Real-Time PCR for detection and quantification of the protistan parasite Perkinsus marinus in environmental waters. Applied and Environmental Microbiology. 2004;70:6611–6618. doi: 10.1128/AEM.70.11.6611-6618.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes MA, Turner CR, Jerde CL, Renshaw MA, Chadderton WL, Lodge DM. Environmental conditions influence eDNA persistence in aquatic systems. Environmental Science and Technology. 2014;48:1819–1827. doi: 10.1021/es404734p. [DOI] [PubMed] [Google Scholar]
- Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Yu DW, de Bruyn M. Environmental DNA for wildlife biology and biodiversity monitoring. Trends in Ecology and Evolution. 2014;29:358–367. doi: 10.1016/j.tree.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Bridle AR, Crosbie PBB, Cadoret K, Nowak BF. Rapid detection and quantification of Neoparamoeba perurans in the marine environment. Aquaculture. 2010;309:56–61. [Google Scholar]
- Bush AO, Lafferty KD, Lotz JM, Shostak AW. Parasitology meets ecology on its own terms: Margolis et al. revisited. Journal of Parasitology. 1997;83:575–583. [PubMed] [Google Scholar]
- Combes C, Fournier A, Mone H, Theron A. Behaviors in trematode cercariae that enhance parasite transmission: patterns and processes. Parasitology. 1994;109:S3–S13. doi: 10.1017/s0031182000085048. [DOI] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Emerging infectious diseases of wildlife-threats to biodiversity and human health. Science. 2000;287:443–449. doi: 10.1126/science.287.5452.443. [DOI] [PubMed] [Google Scholar]
- Daszak P, Cunningham AA, Hyatt AD. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Tropica. 2001;78:103–116. doi: 10.1016/s0001-706x(00)00179-0. [DOI] [PubMed] [Google Scholar]
- Deagle B, Eveson JP, Jarman S. Quantification of damage in DNA recovered from highly degraded samples - a case study on DNA in faeces. Frontiers in Zoology. 2006;3:11. doi: 10.1186/1742-9994-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejean T, Valentini A, Duparc A, Pellier-Cuit S, Pompanon F, Taberlet P, Miaud C. Persistence of environmental DNA in freshwater ecosystems. PLoS ONE. 2011;6:e23398. doi: 10.1371/journal.pone.0023398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Esch GW. Guild structure of larval trematodes in the snail, Helisoma anceps: patterns and processes at the individual host level. Journal of Parasitology. 1991;77:528–539. [PubMed] [Google Scholar]
- Goldberg CS, Pilliod DS, Arkle RS, Waits LP. Molecular detection of vertebrates in stream water: a demonstration using rocky mountain tailed frogs and Idaho giant salamanders. PLoS ONE. 2011;6:e22746. doi: 10.1371/journal.pone.0022746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman BA, Johnson PTJ. Disease and the extended phenotype: parasites control host performance and survival through induced changes in body plan. PLoS ONE. 2011a;6:e20193. doi: 10.1371/journal.pone.0020193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman BA, Johnson PTJ. Ecomorphology and disease: understanding the cryptic effects of parasitism on host habitat use, thermoregulation, and predator avoidance. Ecology. 2011b;92:542–548. doi: 10.1890/10-0516.1. [DOI] [PubMed] [Google Scholar]
- Gordon CA, Gray DJ, Gobert GN, McManus DP. DNA amplification approaches for the diagnosis of key parasitic helminth infections of humans. Molecular and Cellular Probes. 2011;25:143–152. doi: 10.1016/j.mcp.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Jerde CL, Mahon AR, Chadderton WL, Lodge DM. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conservation Letters. 2011;4:150–157. [Google Scholar]
- Johnson PTJ, McKenzie VJ. In: Effects of environmental change on helminth infections in amphibians: exploring the emergence of Ribeiroia and Echinostoma infections in North America. Fried B, Toledo R, editors. New York: The Biology of Echinostomes Springer; 2009. pp. 249–280. [Google Scholar]
- Johnson PTJ, Lunde KB, Ritchie EG, Launer AE. The effect of trematode infection on amphibian limb development and survivorship. Science. 1999;284:802–804. doi: 10.1126/science.284.5415.802. [DOI] [PubMed] [Google Scholar]
- Johnson PTJ, Lunde KB, Zelmer DA, Werner JK. Limb deformities as an emerging parasitic disease in amphibians: evidence from museum specimens and resurvey data. Conservation Biology. 2003;17:1724–1737. [Google Scholar]
- Johnson PTJ, Sutherland DR, Kinsella JM, Lunde KB. Review of the trematode genus Ribeiroia (Psilostomidae): ecology, life history and pathogenesis with special emphasis on the amphibian malformation problem. Advances in Parasitology. 2004;57:192–253. doi: 10.1016/S0065-308X(04)57003-3. [DOI] [PubMed] [Google Scholar]
- Johnson PTJ, Chase JM, Dosch KL, Gross J, Hartson RB, Larson D, Sutherland DR, Carpenter SR. Aquatic eutrophication promotes pathogenic infection in amphibians. Proceedings of the National Academy of Sciences USA. 2007;104:15781–15786. doi: 10.1073/pnas.0707763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson PTJ, Kellermanns E, Bowerman J. Critical windows of disease risk: amphibian pathology driven by developmental changes in host resistance and tolerance. Functional Ecology. 2011;25:726–734. [Google Scholar]
- Johnson PTJ, Rohr JR, Hoverman JT, Kellermanns E, Bowerman J, Lunde KB. Living fast and dying of infection: host life history drives interspecific variation in infection and disease risk. Ecology Letters. 2012;15:235–242. doi: 10.1111/j.1461-0248.2011.01730.x. [DOI] [PubMed] [Google Scholar]
- Johnson PTJ, Preston DL, Hoverman JT, Richgels KLD. Biodiversity reduces disease through predictable changes in host community competence. Nature. 2013;494:230–234. doi: 10.1038/nature11883. [DOI] [PubMed] [Google Scholar]
- Juthikumar N, Sobsey MD, Cromeans TL. Development of an RNA extraction protocol for detection of waterborne viruses by reverse transcriptase quantitative PCR (RT-qPCR) Journal of Virological Methods. 2010;169:8–12. doi: 10.1016/j.jviromet.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Kane RA, Stothard JR, Rollinson D, Leclipteux T, Evraerts J, Standley CJ, Allan F, Betson M, Kaba R, Mertens P, Laurent T. Detection and quantification of schistosome DNA in freshwater snails using either fluorescent probes in real-time PCR or oligochromatographic dipstick assays targeting the ribosomal intergenic spacer. Acta Tropica. 2013;128:241–249. doi: 10.1016/j.actatropica.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Kao P-M, Tung M-C, Hsu B-M, Tsai H-L, She C-Y, Shen S-M, Huang W-C. Real-time PCR method for the detection and quantification of Acanthamoeba species in various types of water samples. Parasitology Research. 2013;112:1131–1136. doi: 10.1007/s00436-012-3242-x. [DOI] [PubMed] [Google Scholar]
- Koprivnikar J, Redfern JC. Agricultural effects on amphibian parasitism: importance of general habitat perturbations and parasite life cycles. Journal of Wildlife Diseases. 2012;48:925–36. doi: 10.7589/2011-09-258. [DOI] [PubMed] [Google Scholar]
- Koprivnikar J, Marcogliese DJ, Rohr JR, Orlofske SA, Raffel TR, Johnson PTJ. Macroparasite infections of amphibians: what can they tell us? EcoHealth. 2012;9:342–360. doi: 10.1007/s10393-012-0785-3. [DOI] [PubMed] [Google Scholar]
- LaFonte BE, Johnson PTJ. Experimental infection dynamics: using immunosuppression and in vivo parasite tracking to understand host resistance in an amphibian-trematode system. Journal of Experimental Biology. 2013;216:3700–3708. doi: 10.1242/jeb.088104. [DOI] [PubMed] [Google Scholar]
- Lodge DM, Turner CR, Jerde CL, Barnes MA, Chadderton L, Egan SP, Feder JL, Mahon AR, Pfrender ME. Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Molecular Ecology. 2012;21:2555–2558. doi: 10.1111/j.1365-294X.2012.05600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson CD, Gurney C, Esau N, Lehto R. Assessing PCR inhibition from humic substances. The Open Enzyme Inhibition Journal. 2010;3:38–45. [Google Scholar]
- Moyer GR, Díaz-Ferguson E, Hill JE, Shea C. Assessing environmental DNA detection in controlled lentic systems. PLoS ONE. 2014;9:e103767. doi: 10.1371/journal.pone.0103767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan LM, Simmons M, Wegleitner BJ, Jerde CL, Mahon AR. Quantifying environmental DNA signals for aquatic invasive species across multiple detection platforms. Environmental Science and Technology. 2014 doi: 10.1021/es5034052. [DOI] [PubMed] [Google Scholar]
- Ouellet M, Bonin J, Rodrigue J, DesGranges JL, Lair S. Hindlimb deformities (ectromelia, ectrodactyly) in free-living anurans from agricultural habitats. Journal of Wildlife Diseases. 1997;33:95–104. doi: 10.7589/0090-3558-33.1.95. [DOI] [PubMed] [Google Scholar]
- Paull SH, Johnson PTJ. How will climate change affect host-parasite interactions? Understanding differential responses of hosts and parasites. Freshwater Biology. 2011;56:767–778. [Google Scholar]
- Piaggio AJ, Engeman RM, Hopken MW, Humphrey JS, Keacher KL, Bruce WE, Avery ML. Detecting an elusive invasive species: a diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Molecular Ecology Resources. 2014;14:374–380. doi: 10.1111/1755-0998.12180. [DOI] [PubMed] [Google Scholar]
- Pilliod DS, Goldberg CS, Arkle RS, Waits LP. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Canadian Journal of Fisheries and Aquatic Sciences. 2011;70:1123–1130. [Google Scholar]
- Prokopowich CD, Gregory TR, Crease TJ. Theo corrlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46:48–50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- Reinitz DM, Yoshino TP, Cole RA. A Ribeiroia spp. (Class: Trematoda) -specific PCR-based diagnostic. Journal of Parasitology. 2007;93:1234–1238. doi: 10.1645/GE-3584RN.1. [DOI] [PubMed] [Google Scholar]
- Roberts CD, Dickinson TE. Ribeiroia ondatrae causes limb abnormalities in a Canadian amphibian community. Canadian Journal of Zoology. 2012;90:808–814. [Google Scholar]
- Schell SC. Handbook of trematodes of North America North of Mexico. Moscow, Idaho, USA: University Press of Idaho; 1985. [Google Scholar]
- Schmidt BR, Kéry M, Ursenbacher S, Hyman OJ, Collins JP. Site occupancy models in the analysis of environmental DNA presence/absence surveys: a case study of an emerging amphibian pathogen. Methods in Ecology and Evolution. 2013;4:646–653. [Google Scholar]
- Shaw DJ, Grenfell BT, Dobson AP. Patterns of macroparasite aggregation in wildlife host populations. Parasitology. 1998;117:597–610. doi: 10.1017/s0031182098003448. [DOI] [PubMed] [Google Scholar]
- Smith KF, Schmidt V, Rosen GE, Amaral-Zettler L. Microbial diversity and potential pathogens in ornamental fish aquarium water. PLoS ONE. 2012;7:e39971. doi: 10.1371/journal.pone.0039971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szuroczki D, Richardson JML. The role of trematode parasites in larval anuran communities: an aquatic ecologist’s guide to the major players. Oecologia. 2009;161:371–385. doi: 10.1007/s00442-009-1388-8. [DOI] [PubMed] [Google Scholar]
- Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH. Environmental DNA. Molecular Ecology. 2012;21:1789–1793. doi: 10.1111/j.1365-294X.2012.05542.x. [DOI] [PubMed] [Google Scholar]
- Thompson RCA, Lymbery AJ, Smith A. Parasites, emerging disease and wildlife conservation. International Journal for Parasitology. 2010;40:1163–1170. doi: 10.1016/j.ijpara.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Thomsen P, Kielgast JOS, Iversen LL, Wiuf C, Rasmussen M, Gilbert MTP, Orlando L, Willerslev E. Monitoring endangered freshwater biodiversity using environmental DNA. Molecular Ecology. 2012;21:2565–2573. doi: 10.1111/j.1365-294X.2011.05418.x. [DOI] [PubMed] [Google Scholar]
- Turner CR, Barnes MA, Xu CC, Jones SE, Jerde CL, Lodge DM. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods in Ecology and Evolution. 2014;5:676–684. [Google Scholar]
- Wilson IG. Minireview: Inhibition and facilitation of nucleic acid amplification. Applied and Environmental Microbiology. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell C, Xiao N, Vidal JE, Chen L, Zhong B, Remais J. Field detection of Schistosoma japonicum cercariae in environmental water samples by quantitative PCR. Applied and Environmental Microbiology. 2011;77:2192–2195. doi: 10.1128/AEM.01561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaccoz NG. The future of environmental DNA in ecology. Molecular Ecology. 2012;21:2031–2038. doi: 10.1111/j.1365-294X.2012.05505.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.