Abstract
Background
Butane-2,3-diol (2,3-BD) is a fuel and platform biochemical with various industrial applications. 2,3-BD exists in three stereoisomeric forms: (2R,3R)-2,3-BD, meso-2,3-BD and (2S,3S)-2,3-BD. Microbial fermentative processes have been reported for (2R,3R)-2,3-BD and meso-2,3-BD production.
Results
The production of (2S,3S)-2,3-BD from glucose was acquired by whole cells of recombinant Escherichia coli coexpressing the α-acetolactate synthase and meso-butane-2,3-diol dehydrogenase of Enterobacter cloacae subsp. dissolvens strain SDM. An optimal biocatalyst for (2S,3S)-2,3-BD production, E. coli BL21 (pETDuet–PT7–budB–PT7–budC), was constructed and the bioconversion conditions were optimized. With the addition of 10 mM FeCl3 in the bioconversion system, (2S,3S)-2,3-BD at a concentration of 2.2 g/L was obtained with a stereoisomeric purity of 95.0 % using the metabolically engineered strain from glucose.
Conclusions
The engineered E. coli strain is the first one that can be used in the direct production of (2S,3S)-2,3-BD from glucose. The results demonstrated that the method developed here would be a promising process for efficient (2S,3S)-2,3-BD production.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-015-0324-x) contains supplementary material, which is available to authorized users.
Keywords: (2S,3S)-Butane-2,3-diol; Metabolic engineering; α-Acetolactate synthase; meso-Butane-2,3-diol dehydrogenase
Background
Butane-2,3-diol (2,3-BD) is a promising commodity biochemical that can be produced by biotechnological routes. It can be used as a starting material for the synthesis of bulk chemicals including methylethylketone, gamma-butyrolactone and 1,3-butadiene [1–3]. Additionally, the heating value of 2,3-BD (27,200 J/g) is comparable with that of other liquid fuels, e.g., methanol (22,100 J/g) and ethanol (29,100 J/g). Thus, 2,3-BD can also be used as a liquid fuel or fuel additive [1–3]. There are three isomeric forms of 2,3-BD: (2R,3R)-2,3-BD, meso-2,3-BD and (2S,3S)-2,3-BD. Optically pure 2,3-BD can act as an excellent building block in asymmetric synthesis of valuable chiral chemicals [1–3].
2,3-BD can be produced via chemical or biotechnological routes [1, 4–10]. The selective production of optically pure 2,3-BD through chemical processes is difficult to control, complicated and expensive to perform. Furthermore, the optical purity of 2,3-BD produced via chemical processes was rather low. Biotechnological routes have emerged as the preferred methods for the optically pure 2,3-BD production [1–3]. Although many microorganisms could be used to efficiently produce 2,3-BD, Paenibacillus polymyxa is the only native producer of optically pure (2R,3R)-2,3-BD [11–13]. Thus, specific strains of Escherichia coli, Saccharomyces cerevisiae, Enterobacter cloacae and Bacillus licheniformis have been engineered for optically pure 2,3-BD production in recent years [14–21]. For example, S. cerevisiae has been engineered as a heterologous host for (2R,3R)-2,3-BD production [16]. The highest concentration of (2R,3R)-2,3-BD was more than 100 g/L. 73.8 g/L of meso-2,3-BD can also be produced using systematic metabolic engineered E. coli BL21/pET-RABC from glucose [17]. However, no fermentative method for (2S,3S)-2,3-BD production has been reported.
Due to difficulties in the direct fermentative production of (2S,3S)-2,3-BD, biocatalysis has emerged as the only reported route for (2S,3S)-2,3-BD production [22–28]. Racemic acetoin, diacetyl or a mixture of three isomeric forms of 2,3-BD have been used as substrates for (2S,3S)-2,3-BD production [22–28]. The highest published concentration of (2S,3S)-2,3-BD produced from the mixture of 2,3-BD was 2.4 g/L, with a low yield of 0.12 g/g [28]. With pure diacetyl as substrate and formate for cofactor regeneration, 31.7 g/L (2S,3S)-2,3-BD was produced [23]. However, all of the above substrates were rather expensive. Thus, production of (2S,3S)-2,3-BD with high optical purity from cheap substrates, such as glucose, is rather desirable.
Until now, a microorganism that could directly produce (2S,3S)-2,3-BD from glucose through bioconversion has never been reported. In this work, a recombinant E. coli was constructed through coexpressing the α-acetolactate synthase (ALS) and meso-butane-2,3-diol dehydrogenase (meso-BDH) of E. cloacae subsp. dissolvens strain SDM. Then, (2S,3S)-2,3-BD was produced from glucose using whole cells of the recombinant E. coli. The process presented in this work could be a promising alternative for the biotechnological production of optically pure (2S,3S)-2,3-BD.
Results and discussion
Design of the metabolic pathway for (2S,3S)-2,3-BD from glucose
Many bacteria such as Klebsiella pneumoniae, Klebsiella oxytoca and E. cloacae can ferment sugars to a mixture of (2R,3R)-2,3-BD, meso-2,3-BD and (2S,3S)-2,3-BD [1, 5, 29–32]. In previous studies, the mechanism of 2,3-BD stereoisomer formation in K. pneumoniae was identified [33, 34]. In the 2,3-BD-producing process, carbohydrate must first be converted to pyruvate. Then, three key enzymes, i.e., ALS, α-acetolactate decarboxylase (ALDC) and butane-2,3-diol dehydrogenase (BDH), are involved in 2,3-BD production from pyruvate. Two molecules of pyruvate are condensed to α-acetolactate by ALS. Then, ALDC catalyzes the decarboxylation of α-acetolactate to produce (3R)-acetoin. (3R)-acetoin will be reduced to meso-2,3-BD and (2R,3R)-2,3-BD by meso-BDH and (2R,3R)-BDH, respectively. α-Acetolactate is unstable and can also be catalyzed through nonenzymatic oxidative decarboxylation to produce diacetyl. (2S,3S)-2,3-BD, the target product of this study, could only be produced by the meso-BDH-catalyzed two-step reduction of diacetyl [33, 34]. Due to the low concentration of diacetyl in microbial fermentation, (2S,3S)-2,3-BD can only be produced at low concentration and stereoisomeric purity.
Based on the native (2S,3S)-2,3-BD producing process mentioned above, we designed a metabolic pathway for (2S,3S)-2,3-BD production from glucose by recombinant E. coli. Pyruvate is firstly formed from glucose via the endogenous Embden–Meyerhof–Parnas pathway (glycolysis) in E. coli. Then, α-acetolactate is produced from pyruvate by the exogenously expressed ALS. Diacetyl can be produced from α-acetolactate by nonenzymatic oxidative decarboxylation. Finally, diacetyl would be reduced to (3S)-acetoin and then to (2S,3S)-2,3-BD by the exogenously expressed meso-BDH in the recombinant E. coli (Fig. 1).
Fig. 1.
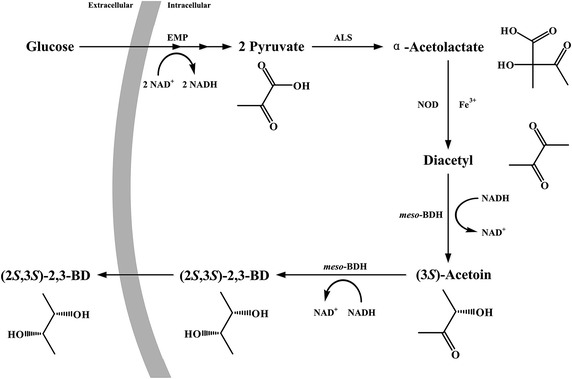
Technology road map for (2S, 3S)-2,3-BD production from glucose by recombinant E. coli. ALS α-acetolactate synthase, meso-BDH meso-butane-2,3-diol dehydrogenase, NOD non-enzymatic oxidative decarboxylation, EMP Embden–Meyerhof–Parnas pathway
Construction of recombinant plasmids for (2S,3S)-2,3-BD
In a previous report, the genes related to 2,3-BD production in different strains were cloned into E. coli BL21(DE3) and the 2,3-BD synthesis abilities of the recombinant E. coli strains were assayed [17]. The recombinant E. coli expressing the genes from E. cloacae subsp. dissolvens SDM had the best ability to produce 2,3-BD. Thus, the ALS encoded by budB in E. cloacae subsp. dissolvens SDM was selected for the production of precursors of (2S,3S)-2,3-BD (α-acetolactate and diacetyl) in recombinant E. coli. Additionally, besides the reduction of (3R)-acetoin to produce meso-2,3-BD, the meso-BDH encoded by budC in strain SDM could also catalyze the two-step reduction of diacetyl to produce (2S,3S)-2,3-BD. The recombinant E. coli strains expressing the meso-BDH have been used in the efficient production of (2S,3S)-2,3-BD from diacetyl [23]. The highly active meso-BDH was also used in the (2S,3S)-2,3-BD production process.
As shown in Fig. 2, the genes encoding ALDC (accession number: 392323960), ALS (accession number: 392323961) and meso-BDH (accession number: 392323962) are sequentially clustered in one operon and under control of transcriptional regulation protein AlsR in E. cloacae. Two separate plasmids, pETDuet–PT7–budB–PT7–budC and pET28a–lysR–Pabc–budB–budC, were used for expression of the budB and budC genes of E. cloacae subsp. dissolvens SDM in recombinant E. coli (Fig. 2). In a previous report, the 2,3-BD pathway genes of strain SDM were expressed under the control of different types of promoters. The recombinant E. coli strain with the native promoter (Pabc) of 2,3-BD synthesis gene cluster of strain SDM had the best ability to produce 2,3-BD [17]. Thus, in pET28a–lysR–Pabc–budB–budC, the genes lysR, budB, budC and Pabc of E. cloacae subsp. dissolvens SDM were ligated through gene splicing by overlap extension and cloned into the multiple clone site of pET28a. The expression of both budB and budC was also under the control of transcriptional regulation protein AlsR and the promoter Pabc of the 2,3-BD pathway gene cluster of strain SDM (Fig. 2b). In pETDuet–PT7–budB–PT7–budC, budB and budC were cloned into the two multiple clone sites of pETDuet-1 and under the control of the promoter PT7 (Fig. 2a).
Fig. 2.
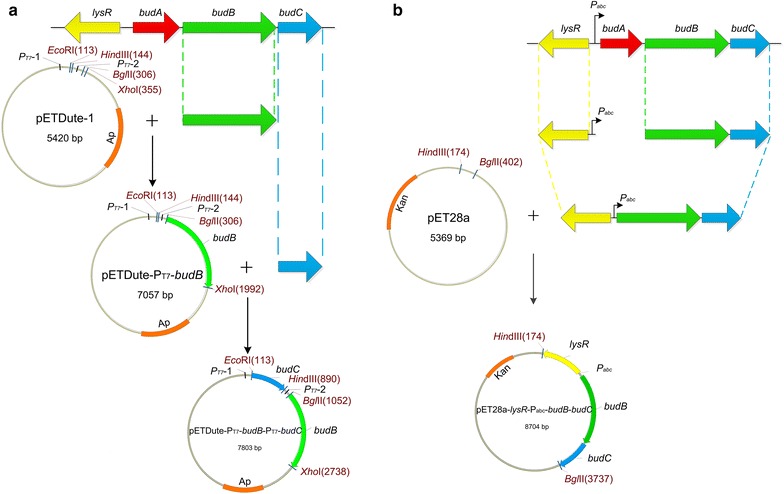
Construction of recombinant vectors for ALS and meso-BDH expression. a Construction of pETDuet–PT7–budB–PT7–budC. b Construction of pET28a–lysR–Pabc–budB–budC. lysR, the gene encoding the transcriptional regulator; budB, the gene encoding ALS; budA, the gene encoding ALDC; budC, the gene encoding meso-BDH, P abc, the predicted promoter of the 2,3-BD pathway gene cluster from E. cloacae subsp. dissolvens strain SDM
The budB and budC genes were successfully cloned from E. cloacae subsp. dissolvens SDM and then inserted into pETDuet-1 to get pETDuet–PT7–budB–PT7–budC (Additional file 1: Figure S1). The fragment budB–budC amplified from the genomic DNA of SDM was about 2500 bp; while the fragment lysR–Pabc amplified from the genomic DNA of SDM was about 1000 bp (Additioanl file 1: Figure S1). These two fragments were ligated through recombinant PCR to get fragment lysR–Pabc–budB–budC. This fragment was inserted into pET28a and resulted in plasmid pET28a–lysR–Pabc–budB–budC.
Production of (2S,3S)-2,3-BD by different recombinant E. coli strains
The constructed expression vectors were transformed into E. coli BL21(DE3) and the 2,3-BD synthesis abilities of the whole cells of recombinant strains were assayed. The 20 mL reaction mixtures were incubated at 30 °C and 180 rpm in a 50-mL flask.
The concentrations of glucose and whole cells of recombinant E. coli strains were 40 g/L and 5 g dry cell weight (DCW)/L, respectively. After 24 h bioconversion, the concentrations of 2,3-BD and glucose in the reaction mixture were determined. As shown in Table 1, E. coli BL21 (pETDuet-1) and BL21 (pET28a–lysR–Pabc–budB–budC) could not produce (2S,3S)-2,3-BD within 24 h. E. coli BL21 (pETDuet–PT7–budB–PT7–budC) had a higher ability to produce 2,3-BD. 2,3-BD at a concentration of 1.14 g/L was produced by E. coli BL21 (pETDuet–PT7–budB–PT7–budC) within 24 h. The strain also showed the highest 2,3-BD yield among the strains harboring different recombinant plasmids (Table 1).
Table 1.
Glucose consumption, product and yield analyses of E. coli strains harboring different vectors in 24 h flask cultures
| Strain | Glucose consumed (g/L) | 2,3-BD (g/L) | 2,3-BD yield (g/g) |
|---|---|---|---|
| E. coli BL21 (pETDuet-1) | 12.00 ± 0.00 | ND | 0.00 |
| E. coli BL21 (pET28a–lysR–Pabc–budB–budC) | 14.00 ± 0.00 | ND | 0.00 |
| E. coli BL21 (pETDuet–PT7–budB–PT7–budC) | 13.67 ± 0.58 | 1.14 ± 0.01 | 0.08 |
Data are the mean ± standard deviations (SDs) from three parallel experiments
The activities of ALDC, ALS, and BDH in the recombinant strains were also assayed (Table 2). Consistent with the result of 2,3-BD production, E. coli BL21 (pETDuet-1) exhibited low ALS and BDH activities. E. coli BL21 (pETDuet–PT7–budB–PT7–budC) showed the highest ALS and meso-BDH activities. No ALDC activity could be detected in all of the E. coli strains. Since the concentration of 2,3-BD obtained and the ALS and meso-BDH activities of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) were higher than that of other recombinant strains, E. coli BL21 (pETDuet–PT7–budB–PT7–budC) was chosen for further investigation.
Table 2.
Enzyme activities of E. coli strains harboring different vectors in 24 h flask cultures
| Strain | ALS activity (U/mg) | ALDC activity (U/mg) | BDH activity (U/mg) | ||
|---|---|---|---|---|---|
| Reduction | Oxidation | ||||
| Diacetyl | Acetoin | meso-2,3-BD | |||
| E. coli BL21 (pETDuet-1) | 0.03 ± 0.04 | ND | 0.18 ± 0.01 | 0.20 ± 0.04 | ND |
| E. coli BL21 (pET28a–lysR–Pabc–budB–budC) | 0.16 ± 0.03 | ND | 1.79 ± 0.17 | 1.17 ± 0.09 | 0.38 ± 0.02 |
| E. coli BL21 (pETDuet–PT7–budB–PT7–budC) | 1.67 ± 0.06 | ND | 7.20 ± 1.21 | 4.20 ± 0.23 | 2.80 ± 0.20 |
Data are the mean ± standard deviations (SDs) from three parallel experiments
ALS α-acetolactate synthase, ALDC α-acetolactate decarboxylase, BDH butane-2,3-diol dehydrogenase
Effect of pH on production of (2S,3S)-2,3-BD by recombinant E. coli
The stereoisomeric composition of 2,3-BD produced by E. coli BL21 (pETDuet–PT7–budB–PT7–budC) was analyzed with GC. As shown in Fig. 3b, a mixture of 2,3-BD was obtained, which contained only 57.4 % of (2S,3S)-2,3-BD. This result is rather inconsistent with our expectation. Thus, we analyzed the bioconversion system and drew the time course of the process. As shown in Fig. 3a, during the bioconversion process, pH was decreased from 7.0 to 5.0 due to production of organic acids. Thus, we expected that the decrease in pH might be the reason of the low stereoisomeric purity of the (2S,3S)-2,3-BD.
Fig. 3.
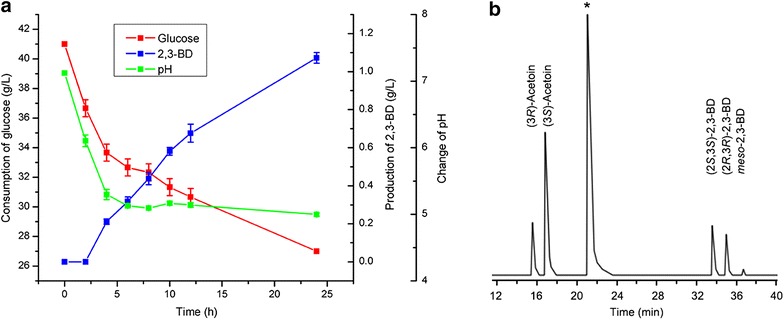
Production of 2,3-BD from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) without pH adjustment. a Time course of 2,3-BD production from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) without pH adjustment. b Chromatograph profile of 2,3-BD produced from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) without pH adjustment (asterisk isoamyl alcohol was used as the internal standard)
Then, during the bioconversion process, 10 M NaOH was added periodically to control the pH of the bioconversion system at 7.0 (Fig. 4a). As shown in Fig. 4b, pH adjustment resulted in a higher (2S,3S)-2,3-BD concentration, yield and especially a higher stereoisomeric purity of (2S,3S)-2,3-BD. Consequently, pH was maintained at 7.0 during subsequent bioconversions.
Fig. 4.
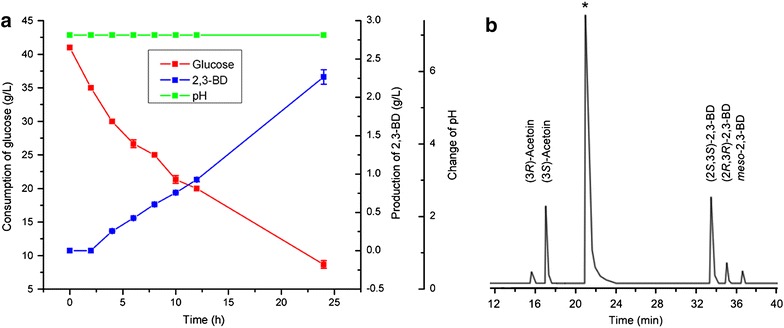
Production of 2,3-BD from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) with pH adjustment. a Time course of 2,3-BD production from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) with pH adjustment. b Chromatograph profile of 2,3-BD produced from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) with pH adjustment (asterisk isoamyl alcohol was used as the internal standard)
Effect of temperature on production of (2S,3S)-2,3-BD by recombinant E. coli
Efficiency of the bioconversion processes and non-enzymatic reaction is temperature dependent. Thus, in this study, the effects of temperature (16, 25, 30, 37, 45 and 55 °C) on (2S,3S)-2,3-BD production were also examined. As shown in Fig. 5, the highest (2S,3S)-2,3-BD concentration was obtained when the temperature was maintained at 37 °C. However, the stereoisomeric purity of (2S,3S)-2,3-BD was much lower than that of 25 and 30 °C. The stereoisomeric purity of (2S,3S)-2,3-BD is rather important for its utilization as the building block in asymmetric synthesis. Since both high product concentration and stereoisomeric purity could be obtained at 30 °C, this temperature was chosen for subsequent bioconversions.
Fig. 5.
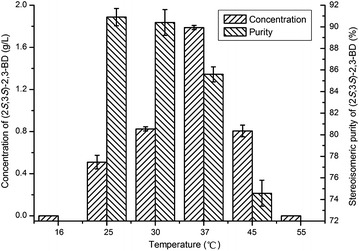
Effect of temperature on (2S,3S)-2,3-BD production by recombinant E. coli BL21 (pETDuet–PT7–budB–PT7–budC)
Effect of Fe3+ addition on (2S,3S)-2,3-BD production
To achieve higher (2S,3S)-2,3-BD concentration, non-enzymatic oxidative decarboxylation of α-acetolactate should be enhanced. It was reported that conversion of α-acetolactate into diacetyl could be enhanced by addition of Fe3+ [35]. To study the effect of the addition of Fe3+ on (2S,3S)-2,3-BD production, 10 mM FeCl3 or 10 mM ethylenediaminetetraacetic acid ferric sodium salt (EDTA-FeNa) was added at the beginning of the bioconversion process. The (2S,3S)-2,3-BD production and glucose consumption were detected after 24 h bioconversion. As shown in Table 3, addition of FeCl3 would result in a higher (2S,3S)-2,3-BD yield.
Table 3.
Effects of Fe3+ addition on glucose consumption, 2,3-BD production, yield and purity of (2S,3S)-2,3-BD
| Condition | Glucose consumed (g/L) | 2,3-BD (g/L) | 2,3-BD yield (g/g) | Purity of (2S,3S)-2,3-BD |
|---|---|---|---|---|
| Blank | 32.00 ± 1.00 | 1.93 ± 0.04 | 0.060 ± 0.001 | 0.84 ± 0.02 |
| 10 mM FeCl3 | 27.00 ± 1.00 | 1.73 ± 0.03 | 0.064 ± 0.001 | 0.86 ± 0.02 |
| 10 mM EDTA-FeNa | 24.67 ± 1.53 | 0.41 ± 0.03 | 0.017 ± 0.001 | 1.00 ± 0.00 |
Data are the mean ± standard deviations (SDs) from three parallel experiments
Batch bioconversion under optimal conditions
Combining the results mentioned above, an optimal system for the production of (2S,3S)-2,3-BD from glucose was developed. Bioconversion was conducted at 30 °C in 50-mL shake flasks containing 20 mL medium. The medium consisted of 40 g/L glucose, 10 mM FeCl3 and 5 g DCW/L whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC). The pH was maintained at 7.0 through periodical addition of 10 M NaOH.
As shown in Fig. 6, 2.2 g/L (2S,3S)-2,3-BD was obtained from 26.7 g/L glucose after 24 h of bioconversion. The yield of (2S,3S)-2,3-BD was at 16.1 % of the theoretical value (Fig. 6a). The stereoisomeric purity of the (2S,3S)-2,3-BD produced by strain E. coli BL21 (pETDuet–PT7–budB–PT7–budC) was 95 % (Fig. 6b). Just like the situation of the fermentation process, the (2S,3S)-2,3-BD was produced through a mixed acid pathway during the bioconversion process. By-products including acetate, lactate, formate, succinate, acetoin and ethanol were detected in the bioconversion system (Additional file 1: Figure S2). Further enhancement of (2S,3S)-2,3-BD production might be acquired through elimination of these by-products through gene knockout.
Fig. 6.
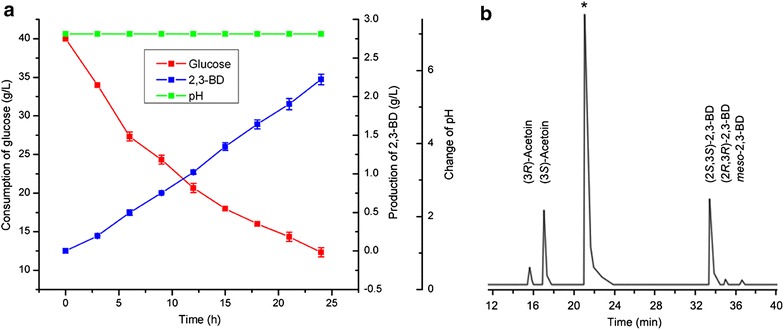
Production of 2,3-BD from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) with Fe3+ addition. a Time course of 2,3-BD production from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) with Fe3+ addition. b Chromatograph profile of 2,3-BD produced from glucose using whole cells of E. coli BL21 (pETDuet–PT7–budB–PT7–budC) with Fe3+ addition (asterisk isoamyl alcohol was used as the internal standard)
Several biotechnological routes including enzymatic or whole-cell conversion methods have been used to produce (2S,3S)-2,3-BD (Table 4). Among all of the reported biotechnological processes, Wang et al. obtained the highest (2S,3S)-2,3-BD concentration of 31.7 g/L from diacetyl using recombinant E. coli coexpressing FDH and meso-BDH [23]. Efforts have also been tried to increase (2S,3S)-2,3-BD production through biotransformation of racemic acetoin [25, 26]. However, the prices of these two substrates, acetoin and diacetyl, are too high for the actual production of (2S,3S)-2,3-BD. Using racemic 2,3-BD as the substrate, the recombinant E. coli strain that coexpressed (2R,3R)-2,3-BDH and NADH oxidase produced (2S,3S)-2,3-BD at a concentration of 2.4 g/L [28]. Due to the low content of (2S,3S)-2,3-BD in the mixture of 2,3-BD (often lower than 10 %), the yield of (2S,3S)-2,3-BD was rather low, which increased the real substrate cost of the biocatalystic process.
Table 4.
Researches on the production of (2S,3S)-2,3-BD using varying substrates
| Biocatalyst | Substrate | (2S,3S)-2,3-BD (g/L) | Yield (g/g) | Co-substrate | Cost ($/kg) | References |
|---|---|---|---|---|---|---|
| E. coli coexpressing formate dehydrogenase from Candida boidinii NCYC 1513 and meso-BDH from E. cloacae subsp. dissolvens SDM | Diacetyl | 31.7 | 0.90 | Formate | 24.4 | [23] |
| E. coli expressing meso-BDH from E. cloacae subsp. dissolvens SDM | Diacetyl | 26.8 | 0.67 | Glucose | 32.8 | [24] |
| E. coli JM109 coexpressing meso-BDH from K. pneumoniae and (2S,3S)-2,3-BDH from Brevibacterium saccharolyticum C-1012 | Diacetyl | 2.2 | 0.93 | Glucose | 23.7 | [25] |
| E. coli JM109 expressing (2S,3S)-2,3-BDH from Br. saccharolyticum | Racemic acetoin | 3.7 | 0.37 | Glucose | 85.7 | [26] |
| E. coli expressing acetoin reductase from Rhodococcus erythropolis WZ010 | Diacetyl | 5.3 | 1.03 | NADH | 21.4 | [27] |
| E. coli BL21 coexpressing (2R,3R)-2,3-BDH from Bacillus subtilis 168 and NADH oxidase from Lactobacillus brevis CICC 6004 | 2,3-BD | 2.4 | 0.12 | 13.3 | [28] | |
| E. coli BL21 (DE3) coexpressing ALS and meso-BDH from E. cloacae subsp. dissolvens SDM | Glucose | 2.2 | 0.08 | 9.4 | This work |
Cost analyses of substrates were performed as described in Additional file 1
It has been reported that the cost of substrates accounted for more than about 30 % of total cost of 2,3-BD production. In this work, we constructed a recombinant E. coli coexpressing ALS and meso-BDH for the production of (2S,3S)-2,3-BD from a cheap substrate, glucose. The product concentration of our system was lower than that of processes using other substrates (Table 4). However, the cost analyses indicated that our process could produce (2S,3S)-2,3-BD at a rather low substrate cost (Additional file 1: Table S1). Thus, the method presented in this work would be a promising process for (2S,3S)-2,3-BD production.
Conclusions
An efficient process for (2S,3S)-2,3-BD production from glucose was developed by using recombinant E. coli coexpressing ALS and meso-BDH. Under optimal conditions, the bioconversion process could produce 2.2 g/L (2S,3S)-2,3-BD with a yield of 0.08 g/g glucose. The stereoisomeric purity of (2S,3S)-2,3-BD produced from glucose was 95 %. The cost analysis suggests that the novel bioconversion system can serve as a promising choice for the industrial production of (2S,3S)-2,3-BD.
Methods
Materials
(2S,3S)-2,3-BD (99.0 %), (2R,3R)-2,3-BD (98.0 %) and meso-2,3-BD (98.0 %) were purchased from ACROS (The Kingdom of Belgium). Racemic acetoin (99.0 %) was purchased from Apple Flavor & Fragrance Group Co. (Shanghai, China). Diacetyl, ampicillin, isopropyl-β-d-thiogalactoside (IPTG), reduced nicotinamide adenine dinucleotide (NADH) and nicotinamide adenine dinucleotide (NAD) were purchased from Sigma. PCR primers were prepared by Sangon (Shanghai, China). Fast Pfu DNA polymerase was purchased from TransGen Biotech (Beijing, China). T4 DNA ligase and restriction endonucleases were obtained from Fermentas (Lithuania). All other chemicals were of analytical grade and commercially available.
Bacterial strains and plasmids
The bacterial strains and plasmids used in this study are listed in Table 5. E. coli DH5α and BL21 (DE3) were used as cloning and expression host, respectively. The pEASY-Blunt cloning vector (TransGen Biotech, China) was used for gene cloning, pETDuet-1 and pET28a were used for gene expression. E. cloacae strain SDM was cultured in a medium containing the following (g/L) at pH 7.0: glucose, 15; peptone, 10; yeast extract, 5; KCl, 5. Luria–Bertani (LB) medium was used for E. coli cultivations. Ampicillin was used at a concentration of 100 μg/mL and kanamycin was used at a concentration of 50 μg/mL.
Table 5.
Bacterial strains, plasmids and primers used in this study
| Name | Characteristic | References |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80 lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rk−, mk+) supE44λ-thi-1 | Novagen |
| E. coli BL21(DE3) | F− ompT gal dcm lon hsdSB(r−Bm−B) λ(DE3) | Novagen |
| E. cloacae subsp. dissolvens SDM | Wild type | [5] |
| E. coli BL21 (pET28a–lysR–Pabc–budB–budC) | E. coli BL21(DE3) harboring pET28a–lysR–Pabc–budB–budC | This study |
| E. coli BL21 (pETDuet–PT7–budB–PT7–budC) | E. coli BL21(DE3) harboring pETDuet–PT7–budB–PT7–budC | This study |
| Plasmids | ||
| pET28a | Expression vector, Kanr | Novagen |
| pETDute-1 | Expression vector, Ampr | Novagen |
| pEasy-Blunt | Kanr Ampr oripUC | Transgen |
| pET28a–lysR–Pabc–budB–budC | lysR, Pabc, budB and budC of E. cloacae subsp. dissolvens SDM were ligated and cloned into the multiple clone site of pET28a | This study |
| pETDuet–PT7–budB–PT7–budC | budB and budC from Enterobacter cloacae subsp. dissolvens SDM were cloned into the two multiple clone sites of pETDuet-1 | This study |
| Primers | ||
| budB-F (BglII) | 5′-AGATCTAGTGAACAGTGATAAACAG-3′ | This study |
| budB-R (XhoI) | 5′-CTCGAGTCACAAAATCTGGCTGAGA-3′ | This study |
| budC-F (EcoRI) | 5′-GAATTCAATGCAAAAAGTTGCTCTCG-3′ | This study |
| budC-R (HindIII) | 5′-AAGCTTTTAATTGAATACCATCCCACCGT-3′ | This study |
| lysR–P abc-F (BglII) | 5′-CGGTAGATCTCTACTCCTCGCTTATCATCG-3′ | This study |
| lysR–P abc-R | 5′-CTCACTGTTCATGCTCGTCCTCTTC-3′ | This study |
| budB–budC-F | 5′-GAAGAGGACGAGCATGAACAGTGAG-3′ | This study |
| budB–budC-R (HindIII) | 5′-GCCTAAGCTTTTAGTTGAACACCATCCCA-3′ | This study |
Construction of plasmid pETDuet–PT7–budB–PT7–budC
E. cloacae strain SDM genomic DNAs were extracted with the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). The budB gene was amplified by PCR using forward primer budB-F with a BglII restriction site insertion and reverse primer budB-R with a XhoI restriction site insertion (Table 5). The PCR product was firstly ligated to the pEASY-Blunt vector, and the resulting plasmid was designated pEASY-Blunt–budB. Next, pEASY-Blunt–budB was digested with BglII and XhoI, and the gel-purified budB fragment was ligated to the pETDuet-1 vector digested with the same restriction enzymes. The resulting plasmid was designated pETDuet–PT7–budB. Using the same process as described above, the budC gene fragment was obtained from the genome of E. cloacae strain SDM using primers budC-F (with the EcoRI restriction site) and budC-R (with the HindIII restriction site) (Table 5), and the pETDuet–PT7–budB–PT7–budC was constructed based on the pETDuet–PT7–budB.
Construction of plasmid pET28a–lysR–Pabc–budB–budC
The lysR, Pabc, budB and budC are sequentially clustered in one operon in the E. cloacae SDM. The fragments lysR–Pabc and budB–budC of E. cloacae SDM were amplified through PCR with the primer pairs lysR–Pabc-F(BglII)/lysR–Pabc-R(overlap) and budB–budC-F(overlap)/budB–budC-R(HindIII), respectively. Then the fragments lysR–Pabc and budB–budC of E. cloacae SDM were then ligated through gene splicing by overlap extension using the primers lysR–Pabc-F(BglII) and budB–budC-R(HindIII). Then fragment lysR–Pabc–budB–budC was ligated to the pEASY-Blunt vector to get pEASY-Blunt–lysR–Pabc–budB–budC. Then, pEASY-Blunt–lysR–Pabc–budB–budC was digested with BglII and HindIII, and the gel-purified lysR–Pabc–budB–budC fragment was ligated to the pET28a vector digested with the same restriction enzymes. The resulting plasmid was designated pET28a–lysR–Pabc–budB–budC.
Biocatalyst preparation and bioconversion conditions
LB medium supplemented with 100 μg/mL ampicillin or 50 μg/mL kanamycin was used to cultivate E. coli BL21 (pETDuet-1) and E. coli BL21 (pET28a–lysR–Pabc–budB–budC). The cells of the strains were grown for 12 h, then centrifuged at 13,000×g for 5 min, and washed twice with 67 mM phosphate buffer (pH 7.4). The E. coli BL21 (pETDuet–PT7–budB–PT7–budC) were grown in LB medium containing 100 μg/mL of ampicillin at 37 °C on a rotary shaker (180 rpm). The cultures were induced with 1 mM IPTG at an OD620 nm of 0.6 at 16 °C for about 10 h. The cells were harvested by centrifugation at 6000×g for 5 min at 4 °C and then washed twice with 67 mM phosphate buffer (pH 7.4). The cell pellets were resuspended in 67 mM phosphate buffer (pH 7.4) as biocatalysts for further bioconversion study.
(2S,3S)-2,3-BD production was carried out using 40.0 g/L of glucose as the substrate and 5.0 g DCW/L whole cells of the recombinant as the biocatalysts. 20 mL of mixture with 10 mM Fe3+ was reacted at 30 °C and 180 rpm in 50 mL flasks. pH was controlled at 7.0 by adding 10 M NaOH.
Enzyme activity assays
To measure enzyme activity, the cells of the different E. coli strains were resuspended in 67 mM phosphate buffer (pH 7.4) and disrupted with an ultrasonic cell-breaking apparatus (Xinzhi, Ningbo, China). Cell debris was removed through centrifugation at 13,000×g for 15 min. The resulting supernatants were used in the successive enzyme activity assays. All enzyme assays were performed at 30 °C with the proper enzyme in 67 mM phosphate buffer (pH 7.4).
Activity of ALS was measured by monitoring the conversion of pyruvate to α-acetolactate [36]. One unit of ALS activity was defined as the amount of enzyme that produced 1 μmol of α-acetolactate per minute.
ALDC activity was assayed by detecting the production of acetoin from α-acetolactate [37]. α-Acetolactate was prepared immediately before use of ethyl 2-acetoxy-2-methyl-acetoacetate, according to the protocol supplied by the manufacturer. One unit of ALDC activity was defined as the amount of protein that formed 1 μmol of acetoin per min.
meso-BDH activity was assayed by measuring the change in absorbance at 340 nm corresponding to the oxidation of NADH or reduction of NAD when diacetyl or acetoin was used as the substrate [38]. For the reduction reaction, 5 mM acetoin or diacetyl and 0.2 mM NADH were used for the enzyme assay, and 10 mM meso-2,3-BD and 1 mM NAD were used for the oxidation reactions. One unit of enzyme activity was defined as the amount of enzyme that consumed 1 μmol of NADH or produced 1 μmol of NADH per minute.
The protein concentration was determined by the Lowry procedure using bovine serum albumin as the standard [39].
Analytical methods
Samples were withdrawn periodically and centrifuged at 12,000×g for 10 min. The concentration of glucose was measured enzymatically by a bio-analyzer (SBA-40D, Shandong Academy of Sciences, China) after diluting to an appropriate concentration. The concentrations of 2,3-BD were analyzed by GC (Varian 3800) as described previously [17]. The GC system was equipped with a capillary GC column (AT. SE-54, inside diameter, 0.32 mm; length, 30 m, Chromatographic Technology Center, Lanzhou Institute of Chemical Physics, China). The ratio of the three stereoisomers of 2,3-BD was analyzed by GC (Agilent GC6820) using a fused silica capillary column (Supelco Beta DEXTM 120, inside diameter 0.25 mm; length 30 m) [40]. The stereoisomeric purity of (2S,3S)-2,3-BD was defined as (([S])/([M] + [S] + [R])) × 100 %, where [S], [M] and [R] represent the concentrations of (2S,3S)-2,3-BD, meso-2,3-BD and (2R,3R)-2,3-BD, respectively.
Authors’ contributions
CG, CM and PX participated in the design of the study. HC, BX and YW executed the experimental work. LL, XZ, XL and PL analyzed the data. CG, CM and PX contributed reagents and materials. CG, CM and PX wrote and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Chinese National Program for High Technology Research and Development (2011AA02A207 and 2012AA022104), the National Natural Science Foundation of China (31470164, 31400027 and J1103515), the Program for High Technology Research and Development of Shandong Province (2014GSF121030) and the Young Scholars Program of Shandong University (2015WLJH25).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- GC
gas chromatography
- HPLC
high-performance liquid chromatography
- 2,3-BD
butane-2,3-diol
- ALS
α-acetolactate synthase
- ALDC
α-acetolactate decarboxylase
- BDH
2,3-BD dehydrogenase
- NADH
reduced nicotinamide adenine dinucleotide
- NAD
nicotinamide adenine dinucleotide
Additional file
Additional file 1: Figure S1. Verification of recombinant vectors.Figure S2. Time course of byproducts production from glucose using whole cells of E. coli BL21 (pETDuet-PT7-bud B-PT7-bud C) with Fe3+ addition. Table S1. Cost analyses of various (2S,3S)-2,3-BD production processes.
Contributor Information
Haipei Chu, Email: chuhp@163.com.
Bo Xin, Email: xinbosdu@gmail.com.
Peihai Liu, Email: liuph_513@sina.com.
Yu Wang, Email: wang_yu@sjtu.edu.cn.
Lixiang Li, Email: lilixiang@sdu.edu.cn.
Xiuxiu Liu, Email: 1179796910@qq.com.
Xuan Zhang, Email: 809401655@qq.com.
Cuiqing Ma, Email: macq@sdu.edu.cn.
Ping Xu, Email: pingxu@sjtu.edu.cn.
Chao Gao, Email: jieerbu@sdu.edu.cn.
References
- 1.Celińska E, Grajek W. Biotechnological production of 2,3-butanediol—current state and prospects. Biotechnol Adv. 2009;27:715–725. doi: 10.1016/j.biotechadv.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Ji XJ, Huang H, Ouyang PK. Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv. 2011;29:351–364. doi: 10.1016/j.biotechadv.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Zeng AP, Sabra W. Microbial production of diols as platform chemicals: recent progresses. Curr Opin Biotechnol. 2011;22:749–757. doi: 10.1016/j.copbio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Ji XJ, Huang H, Du J, Zhu JG, Ren LJ, Hu N, et al. Enhanced 2,3-butanediol production by Klebsiella oxytoca using a two-stage agitation speed control strategy. Bioresour Technol. 2009;100:3410–3414. doi: 10.1016/j.biortech.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 5.Wang A, Xu Y, Ma C, Gao C, Li L, Wang Y, et al. Efficient 2,3-butanediol production from cassava powder by a crop-biomass-utilizer, Enterobacter cloacae subsp. dissolvens SDM. PLoS One. 2012;7:e40442. doi: 10.1371/journal.pone.0040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver JW, Machado IM, Yoneda H, Atsumi S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc Natl Acad Sci USA. 2013;110:1249–1254. doi: 10.1073/pnas.1213024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ji XJ, Xia ZF, Fu NH, Nie ZK, Shen MQ, Tian QQ, et al. Cofactor engineering through heterologous expression of an NADH oxidase and its impact on metabolic flux redistribution in Klebsiella pneumoniae. Biotechnol Biofuels. 2013;6:7. doi: 10.1186/1754-6834-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao Z, Wang X, Huang Y, Huo F, Zhu X, Xi L, et al. Thermophilic fermentation of acetoin and 2,3-butanediol by a novel Geobacillus strain. Biotechnol Biofuels. 2012;5:88. doi: 10.1186/1754-6834-5-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L, Sun J, Hao Y, Zhu J, Chu J, Wei D, et al. Microbial production of 2,3-butanediol by a surfactant (serrawettin)-deficient mutant of Serratia marcescens H30. J Ind Microbiol Biotechnol. 2010;37:857–862. doi: 10.1007/s10295-010-0733-6. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Zhang L, Li K, Wang Y, Gao C, Han B, et al. A newly isolated Bacillus licheniformis strain thermophilically produces 2,3-butanediol, a platform and fuel bio-chemical. Biotechnol Biofuels. 2013;6:123. doi: 10.1186/1754-6834-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie NZ, Li JX, Song LF, Hou JF, Guo L, Du QS, et al. Genome sequence of type strain Paenibacillus polymyxa DSM 365, a highly efficient producer of optically active (R,R)-2,3-butanediol. J Biotechnol. 2015;195:72–73. doi: 10.1016/j.jbiotec.2014.07.441. [DOI] [PubMed] [Google Scholar]
- 12.Adlakha N, Yazdani SS. Efficient production of (R,R)-2,3-butanediol from cellulosic hydrolysate using Paenibacillus polymyxa ICGEB2008. J Ind Microbiol Biotechnol. 2015;42:21–28. doi: 10.1007/s10295-014-1542-0. [DOI] [PubMed] [Google Scholar]
- 13.Dai JJ, Cheng JS, Liang YQ, Jiang T, Yuan YJ. Regulation of extracellular oxidoreduction potential enhanced (R,R)-2,3-butanediol production by Paenibacillus polymyxa CJX518. Bioresour Technol. 2014;167:433–440. doi: 10.1016/j.biortech.2014.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Ji XJ, Liu LG, Shen MQ, Nie ZK, Tong YJ, Huang H. Constructing a synthetic metabolic pathway in Escherichia coli to produce the enantiomerically pure (R,R)-2,3-butanediol. Biotechnol Bioeng. 2015;112:1056–1059. doi: 10.1002/bit.25512. [DOI] [PubMed] [Google Scholar]
- 15.Yan Y, Lee CC, Liao JC. Enantioselective synthesis of pure (R,R)-2,3-butanediol in Escherichia coli with stereospecific secondary alcohol dehydrogenases. Org Biomol Chem. 2009;7:3914–3917. doi: 10.1039/b913501d. [DOI] [PubMed] [Google Scholar]
- 16.Lian J, Chao R, Zhao H. Metabolic engineering of a Saccharomyces cerevisiae strain capable of simultaneously utilizing glucose and galactose to produce enantiopure (2R,3R)-butanediol. Metab Eng. 2014;23:92–99. doi: 10.1016/j.ymben.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Chu H, Gao C, Tao F, Zhou Z, Li K, et al. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab Eng. 2014;23:22–33. doi: 10.1016/j.ymben.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Li K, Wang Y, Chen C, Xu Y, Zhang L, et al. Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R,3R)-2,3-butanediol from lignocellulose-derived sugars. Metab Eng. 2015;28:19–27. doi: 10.1016/j.ymben.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 19.Qi G, Kang Y, Li L, Xiao A, Zhang S, Wen Z, et al. Deletion of meso-2,3-butanediol dehydrogenase gene budC for enhanced d-2,3-butanediol production in Bacillus licheniformis. Biotechnol Biofuels. 2014;7:16. doi: 10.1186/1754-6834-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Chen T, Zhao X, Chamu J. Metabolic engineering of thermophilic Bacillus licheniformis for chiral pure d-2,3-butanediol production. Biotechnol Bioeng. 2012;109:1610–1621. doi: 10.1002/bit.24427. [DOI] [PubMed] [Google Scholar]
- 21.Bai F, Dai L, Fan J, Truong N, Rao B, Zhang L, et al. Engineered Serratia marcescens for efficient (3R)-acetoin and (2R,3R)-2,3-butanediol production. J Ind Microbiol Biotechnol. 2015;42:779–786. doi: 10.1007/s10295-015-1598-5. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Qin J, Gao C, Hua D, Ma C, Li L, et al. Production of (2S,3S)-2,3-butanediol and (3S)-acetoin from glucose using resting cells of Klebsiella pneumonia and Bacillus subtilis. Bioresour Technol. 2011;102:10741–10744. doi: 10.1016/j.biortech.2011.08.110. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Li L, Ma C, Gao C, Tao F, Xu P. Engineering of cofactor regeneration enhances (2S,3S)-2,3-butanediol production from diacetyl. Sci Rep. 2013;3:2643. doi: 10.1038/srep02643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Wang Y, Zhang L, Ma C, Wang A, Tao F, et al. Biocatalytic production of (2S,3S)-2,3-butanediol from diacetyl using whole cells of engineered Escherichia coli. Bioresour Technol. 2012;115:111–116. doi: 10.1016/j.biortech.2011.08.097. [DOI] [PubMed] [Google Scholar]
- 25.Ui S, Takusagawa Y, Sato T, Ohtsuki T, Mimura A, Ohkuma M, et al. Production of l-2,3-butanediol by a new pathway constructed in Escherichia coli. Lett Appl Microbiol. 2004;39:533–537. doi: 10.1111/j.1472-765X.2004.01622.x. [DOI] [PubMed] [Google Scholar]
- 26.Ui S, Takusagawa Y, Ohtsuki T, Mimura A, Ohkuma M, Kudo T. Stereochemical applications of the expression of the l-2,3-butanediol dehydrogenase gene in Escherichia coli. Lett Appl Microbiol. 2001;32:93–98. doi: 10.1046/j.1472-765x.2001.00869.x. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Song Q, Yu M, Wang Y, Xiong B, Zhang Y, et al. Characterization of a stereospecific acetoin(diacetyl) reductase from Rhodococcus erythropolis WZ010 and its application for the synthesis of (2S,3S)-2,3-butanediol. Appl Microbiol Biotechnol. 2014;98:641–650. doi: 10.1007/s00253-013-4870-5. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Z, Lv C, Gao C, Qin J, Ma C, Liu Z, et al. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PLoS One. 2010;5:e8860. doi: 10.1371/journal.pone.0008860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jantama K, Polyiam P, Khunnonkwao P, Chan S, Sangproo M, Khor K, et al. Efficient reduction of the formation of by-products and improvement of production yield of 2,3-butanediol by a combined deletion of alcohol dehydrogenase, acetate kinase-phosphotransacetylase, and lactate dehydrogenase genes in metabolically engineered Klebsiella oxytoca in mineral salts medium. Metab Eng. 2015;30:16–26. doi: 10.1016/j.ymben.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 30.Jung MY, Mazumdar S, Shin SH, Yang KS, Lee J, Oh MK. Improvement of 2,3-butanediol yield in Klebsiella pneumoniae by deletion of the pyruvate formate-lyase gene. Appl Environ Microbiol. 2014;80:6195–6203. doi: 10.1128/AEM.02069-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JM, Hong WK, Lee SM, Heo SY, Jung YR, Kang IY, et al. Identification and characterization of a short-chain acyl dehydrogenase from Klebsiella pneumoniae and its application for high-level production of l-2,3-butanediol. J Ind Microbiol Biotechnol. 2014;41:1425–1433. doi: 10.1007/s10295-014-1483-7. [DOI] [PubMed] [Google Scholar]
- 32.Jiang LQ, Fang Z, Guo F, Yang LB. Production of 2,3-butanediol from acid hydrolysates of Jatropha hulls with Klebsiella oxytoca. Bioresour Technol. 2012;107:405–410. doi: 10.1016/j.biortech.2011.12.083. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Wei D, Shi J, Wang M, Hao J. Mechanism of 2,3-butanediol stereoisomer formation in Klebsiella pneumoniae. Appl Microbiol Biotechnol. 2014;98:4603–4613. doi: 10.1007/s00253-014-5526-9. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Tao F, Xu P. Glycerol dehydrogenase plays a dual role in glycerol metabolism and 2,3-butanediol formation in Klebsiella pneumoniae. J Biol Chem. 2014;289:6080–6090. doi: 10.1074/jbc.M113.525535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Zhang Y, Liu Q, Meng L, Hu M, Lv M, et al. Production of diacetyl by metabolically engineered Enterobacter cloacae. Sci Rep. 2015;5:9033. doi: 10.1038/srep09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stormer FC. 2,3-Butanediol biosynthetic system in Aerobacter aerogenes. Methods Enzymol. 1975;41:518–532. doi: 10.1016/S0076-6879(75)41108-9. [DOI] [PubMed] [Google Scholar]
- 37.Phalip V, Monnet C, Schmitt P, Renault P, Godon JJ, Diviès C. Purification and properties of the alpha-acetolactate decarboxylase from Lactococcus lactis subsp. lactis NCDO 2118. FEBS Lett. 1994;351:95–99. doi: 10.1016/0014-5793(94)00820-5. [DOI] [PubMed] [Google Scholar]
- 38.Zhang L, Xu Q, Zhan S, Li Y, Lin H, Sun S, et al. A new NAD(H)-dependent meso-2,3-butanediol dehydrogenase from an industrially potential strain Serratia marcescens H30. Appl Microbiol Biotechnol. 2014;98:1175–1184. doi: 10.1007/s00253-013-4959-x. [DOI] [PubMed] [Google Scholar]
- 39.Markwell MA, Haas SM, Bieber LL, Tolbert NE. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 40.Gao C, Zhang L, Xie Y, Hu C, Zhang Y, Li L, et al. Production of (3S)-acetoin from diacetyl by using stereoselective NADPH-dependent carbonyl reductase and glucose dehydrogenase. Bioresour Technol. 2013;137:111–115. doi: 10.1016/j.biortech.2013.02.115. [DOI] [PubMed] [Google Scholar]


