Abstract
It is now well established that prokaryotic cells assemble diverse proteins into dynamic cytoskeletal filaments that perform essential cellular functions. Although most of the filaments assemble on their own to form higher order structures, growing evidence suggests that there are a number of prokaryotic proteins that polymerise only in the presence of a matrix such as DNA, lipid membrane or even another filament. Matrix-assisted filament systems are frequently nucleotide dependent and cytomotive but rarely considered as part of the bacterial cytoskeleton. Here, we categorise this family of filament-forming systems as collaborative filaments and introduce a simple nomenclature. Collaborative filaments are frequent in both eukaryotes and prokaryotes and are involved in vital cellular processes including chromosome segregation, DNA repair and maintenance, gene silencing and cytokinesis to mention a few. In this review, we highlight common principles underlying collaborative filaments and correlate these with known functions.
Keywords: actin/tubulin cytoskeleton, bacterial cytoskeleton, cytomotive, DNA/membrane-assisted filaments, matrix-assisted filaments
Introduction
During the last two decades, it has been firmly established that prokaryotes, archaea and bacteria, like eukaryotes, possess a diverse set of highly dynamic “cytoskeletal elements” that control vital cellular processes such as cell division, DNA segregation and cell shape maintenance. In the absence of molecular motors, formation of nucleotide-driven, dynamic filaments is an effective way to exert force, induce curvature, push and pull cargos and maintain shape and, hence, these have been classed as cytomotive filaments (Löwe & Amos, 2009; Wickstead & Gull, 2011; Pilhofer & Jensen, 2013). Non-cytomotive (non-dynamic) filament systems exist in prokaryotes too, and they tend to fulfil more static, scaffolding roles (Ausmees et al, 2003; Lin & Thanbichler, 2013).
Based on structural similarities and sequence homology, prokaryotic cytoskeletal elements can be primarily grouped into four major classes: tubulin-like, actin-like, coiled coil-containing and others that are not homologous to any of the previous classes (Löwe & Amos, 2009; Lin & Thanbichler, 2013).
Amongst those filament classes exists a subset of conserved filament systems that do not always polymerise on their own but form higher order oligomeric structures in the presence of a matrix or scaffold such as DNA, lipid membrane bilayer or even another filament (Dunn et al, 1982; Leonard et al, 2005; Hui et al, 2010; Oliva et al, 2010; Szwedziak et al, 2012; van den Ent et al, 2014). These assisted filament-forming systems are often dynamic and nucleotide dependent and rely on surface composition/topology for polymerisation.
Because principles and properties are shared between matrix-assisted filaments and those that copolymerise with another filament, we now categorise these filaments as “collaborative”.
Collaborative filaments are equally common in eukaryotes, and as part of this review, we list a small number of examples from all kingdoms of life. Although prevalent in both eukaryotic and prokaryotic systems, collaborative filament systems are often not considered as “cytoskeletal elements”, further highlighting the imprecise nature of this term, which originated from historical observations of the three major filament classes in eukaryotes. In fact, with a growing list of complex filament systems in bacteria, the very definition of a cytoskeleton is problematic for highly dynamic, cytomotive filaments. Hence, the following two pairs of antonyms can be defined: cytomotive versus cytoskeletal and independent versus collaborative filaments. From the examples below, it will become clear that all four combinations exist.
A simple filament nomenclature
Since ever more filament systems with new architectures are being discovered, we propose to introduce a nomenclature to be able to describe the various filament architectures in short and unambiguous terms (Table1). All known filament systems, including complex ones such as microtubules and the most complex membrane:MinCD:FtsZ filament system (Ghosal et al, 2014), can be described using this nomenclature system. Superscript N denotes polymerisation, and nucleotide binding for cytomotive filaments is indicated by subscript, with NXP standing for nucleotide that will be hydrolysed by the filament to produce cytomotive behaviour. Colon denotes binding of subunits to each other, or a matrix or scaffold, and brackets are used to group these until no ambiguity remains, for example in the case of alternating copolymers such as MinCD or microtubules. Finally, at the start, the number of protofilaments is given as a number with subscript P or A denoting parallel or antiparallel architectures, as found amongst the actin-like proteins, for example.
Table 1.
Cytoskeletal, cytomotive, independent and collaborative filaments: systematic nomenclature and examples
| AN | Cytoskeletal filament | IF, crescentin |
| ANXPN | Cytomotive filament driven by nucleotide NXP | Single FtsZ protofilament |
| 2P(ANXP)N | 2 parallel protofilaments of cytomotive filament | Actin, ParM |
| 4P(ANXP)N | 4 parallel protofilaments of cytomotive filaments | TubZ |
| 5P(ANXP:BNXP)N | 5 parallel protofilaments of cytomotive copolymer | BtubAB (mini-microtubule) |
| 13P(ANXP:BNXP)N | 13 parallel protofilaments of cytomotive copolymer | Microtubule |
| AN:DNA | Collaborative filament on DNA | Dan filaments on DNA |
| ANXPN:DNA | Cytomotive, collaborative filament on DNA | ParA, RecA on DNA |
| AN:M | Collaborative filament on membrane | DivIVA on membrane |
| ANXPN:M | Cytomotive, collaborative filament on membrane | FtsA on membrane |
| 2A(ANXP)N:M | 2 antipar. protofil. of cytom. filament on membrane | MreB on membrane |
| (ANXPN:BNXPN):M | Two cytomo. filaments copolymerised on membrane | FtsZ:FtsA on membrane |
| (A:BNXP)N:M | Cytomotive, alternating copolymer on membrane | MinCD, septin on membrane |
| CNXPN:(A:BNXP)N:M | Above bound to another cytomotive filament | MinCD: FtsZ on membrane |
Superscript N denotes polymerisation of the monomer A and NXP stands for nucleotide (ATP/ADP/GTP/GDP etc) that may be hydrolysed by a cytomotive filament. Colon represents binding of subunits to each other or to a matrix or scaffold, and brackets are used to group these interactions until there is no ambiguity. The number of protofilaments is indicated at the very beginning with a subscript P or A implying parallel or antiparallel arrangement.
Why collaborative filaments?
Collaborative filaments utilise a matrix or scaffold for assembly, such as DNA, membrane or even another protein filament. The obvious question to ask is “why do these types of filaments use or even require a surface in contrast to independent filaments that spontaneously assemble to form higher order structures?”
For filaments to form, subunits have to come together to form a joint interface, burying hydrated surfaces. The energy gained from binding to each other has to overcompensate for the loss of hydration energy. For filaments with more than one protofilament (sometimes referred to as strands), lateral contacts between protofilaments exist in addition to the longitudinal contacts. For example, actin and ParM can be thought of as a double helical filament of two, parallel protofilaments: 2P(ParMAXP)N (Gayathri et al, 2012). For these filaments to form, longitudinal contacts along the protofilaments as well as sometimes much weaker lateral contacts between the protofilaments are required and both are critical for polymerisation (Fig 1B) (Alushin et al, 2014; Bharat et al, 2015; von der Ecken et al, 2015).
Figure 1. Schematic representation of cooperativity effects in different filament systems.
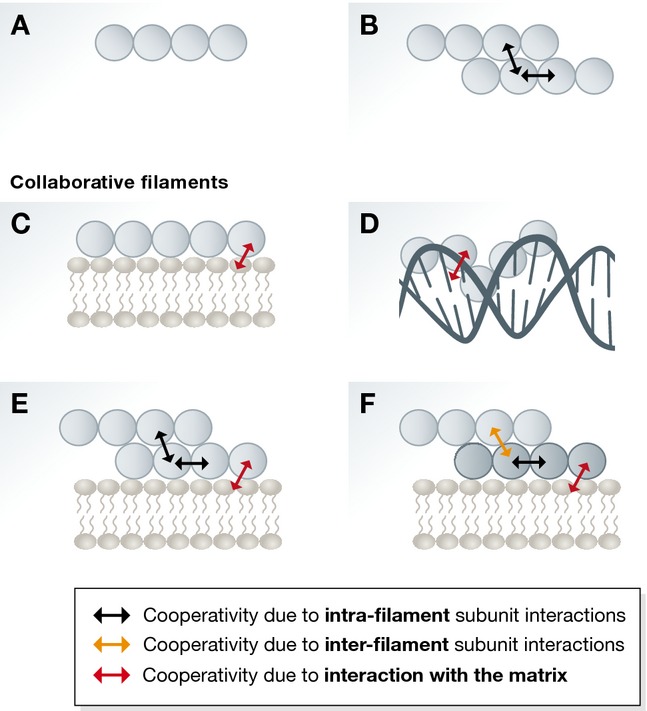
(A) In simple isodesmic assembly, there is no cooperativity effect. (B) For filaments with more than one protofilament (strand), assembly is enhanced by lateral contacts between subunits from different protofilaments. These intrafilament interactions (two-way black arrows) produce cooperativity. (C, D) An isodesmic filament system acquires cooperativity when it associates with a matrix such as membrane or DNA. The collaborative filaments could be considered as quasi-equivalent to multi-stranded filament systems since interactions with the matrix (indicated by two-way red arrow) are equivalent to lateral interactions within a multi-stranded filament. (E) When a multi-stranded filament associates with a matrix, in addition to the longitudinal and lateral interactions between subunits (indicated by two-way black arrows), there are also interactions with the matrix (indicated by two-way red arrow). All of these result in complex cooperative behaviour. (F) In an even more complex situation, two different multi-stranded filaments could copolymerise on a matrix. This leads to an even higher number of different types of interactions, and there could be cooperative effect within a multi-stranded filament of one type (indicated by two-way black arrows), within the other and between them (indicated by two-way orange arrows), and many of these interactions are facilitated by the interaction with the matrix (indicated by two-way red arrow).
Collaborative filaments instead form lateral bonds with a matrix, scaffold or a different type of protein filament.
In the case of a permanent matrix, such as lipid membrane or DNA, this has important consequences for the formation of the resulting collaborative filaments (Fig 1). Since the matrix provides binding energy as subunits directly bind to it laterally, filament formation may be restricted to the matrix surface, depending on the relative strengths of the longitudinal and lateral bonds formed. The matrix reduces the critical concentration for collaborative filament assembly by providing additional binding of subunits onto the surface and also by restricting subunit diffusion in two dimensions. In the case of linear, single-stranded filament assembly (also known as an isodesmic process) (Fig 1A), when the resulting filaments become collaborative (Fig 1C and D), they need to be considered as multi-stranded and acquire cooperativity and a critical concentration that includes the concentration of monomers and that of the surface (corresponding to the size of the surface, which translates into the number of available lateral binding sites). Cooperativity leads to a nonlinear relationship between monomer concentration and filament formation, simply by attaching the filament to a matrix. In the case of filaments that show cooperative assembly without matrix (Fig 1B), attaching them to a matrix will lead to significantly enhanced cooperativity (Fig 1E).
In the above instances, the matrix was considered to be a static support. However, studies in eukaryotes and prokaryotes have identified systems where one filament assembles on another pre-formed filament or polymerises together with another type of filament. In these cases, it can be considered that additional lateral interactions are formed between the subunits and because longitudinal binding events in the two or more filaments are different (Fig 1F), and may even have different subunit repeat lengths, very complex behaviour results. For example, the critical concentration becomes a function of two variables and this may result in very large cooperativity effects.
To make things even more interesting, various combinations of collaborative and cytomotive filaments and matrices exist, producing complex systems that may be tailored to specific functions.
Collaborative filaments on DNA
Protein filaments that form on DNA are common and have been found to form on either single- or double-stranded DNA. They are involved in indispensable cellular processes including DNA repair, DNA segregation, chromosome condensation and gene expression regulation (Dunn et al, 1982; Leonard et al, 2005; Lim et al, 2012b, 2013). Here, we discuss some of the well-studied nucleoprotein filaments as collaborative filament systems.
RecA filaments: DNA repair and maintenance (RecAAXPN:DNA)
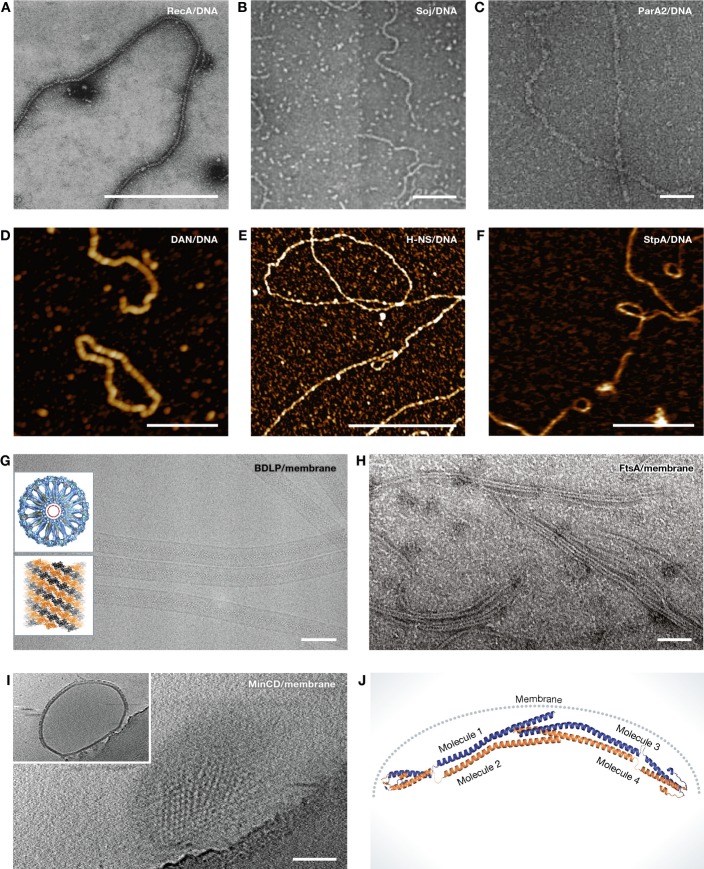
RecA (Rad51 in eukaryotes) is a DNA-dependent ATPase that forms filaments with single-stranded (ss) or double-stranded (ds) DNA (Fig 2A), and plays a central role in DNA damage repair and homologous recombination (Dunn et al, 1982; Ogawa et al, 1993). RecA-mediated homologous recombination occurs in three distinct steps. In the first step, RecA binds to ssDNA in the presence of ATP, in a highly cooperative manner with one RecA molecule per three to four nucleotides, and forms a right-handed helical nucleoprotein filament. In the second step, the RecA–ssDNA filament then aligns to a homologous dsDNA, and in the final step, the strand exchange reaction takes place coupled with RecA ATP hydrolysis (Lindsley & Cox, 1990). Once initial nucleation has occurred, more RecA subunits are added cooperatively in the 5′–3′ direction in an ATP-dependent manner. Although RecA/Rad51 filament assembly is more like an isodesmic process, interaction with DNA makes this process cooperative. While historically not thought of as cytomotive or cytoskeletal, a cytomotive role of RecA collaborative filaments has recently been discovered as RecA filaments were found to bridge the long distances between sister DNAs in E. coli during DNA double-stranded break repair (Lesterlin et al, 2014).
Figure 2. Different examples of collaborative filaments.
(A) RecA–ssDNA nucleoprotein filaments formed in the presence of ATP were visualised by negative staining electron microscopy (EM). Scale bar, 500 nm (reproduced with permission from Flory et al, 1984). (B) Electron micrographs of Soj–DNA collaborative filaments. These filaments are formed in the presence of ATP. Scale bar, 100 nm (reproduced with permission from Leonard et al, 2005). (C) ParA2–dsDNA nucleoprotein filaments formed in the presence of ATP and visualised by negative staining EM. Scale bar, 50 nm (reproduced with permission from Hui et al, 2010). (D) High-resolution atomic force microscopy (AFM) image showing Dan/DNA nucleoprotein filaments. Scale bar, 500 nm (reproduced with permission from Lim et al, 2013). (E) High-resolution AFM image of H-NS/DNA nucleoprotein filaments. Scale bar, 1 μm (reproduced with permission from Lim et al, 2012a). (F) H-NS paralogue StpA–DNA filaments imaged by AFM. Scale bar, 500 nm (reproduced with permission from Lim et al, 2012b). (G) Cryo-EM image of BDLP tubes. Insets showing cross section through three-dimensional reconstruction of a BDLP tube (looking along helical axis) and left-handed helical rise of BDLP filaments. Scale bar, 100 nm (reproduced with permission from Low et al, 2009). (H) Negatively stained electron micrographs of FtsA filaments formed on lipid monolayer. Scale bar, 50 nm (reproduced with permission from Szwedziak et al, 2012). (I) Electron cryotomography of MinCD copolymer-decorated liposomes. Shown is a surface view of a liposome. Inset showing additional layer formed by MinCD filaments on the liposome surface. Scale bar, 100 nm (reproduced with permission from Ghosal et al, 2014). (J) Composite model of B. subtilis DivIVA. DivIVA senses the curvature of the bacterial membrane and polymerises at specific locations (reproduced with permission from Oliva et al, 2010).
ParA filament: DNA segregation (ParAAXPN:DNA)
ParAB operons are found on many plasmids and on some bacterial chromosomes and are involved in the segregation of DNA by an enigmatic mechanism (Ringgaard et al, 2009; Ptacin et al, 2010; Lim et al, 2014; Vecchiarelli et al, 2014). ParAs have been shown to form ATP-dependent dimers that non-specifically interact with DNA (Fig 2B and C) (Leonard et al, 2005). ParB from the same par locus stimulates the ATPase activity of ParA and disrupts the ParA–DNA interaction (Leonard et al, 2005). Although some of the earlier studies suggested that ParAs alone form filaments, several more recent studies failed to reproduce these and concluded that ParAs only form filaments on a matrix, and hence, they are collaborative (Suefuji et al, 2002; Ivanov & Mizuuchi, 2010; Ptacin et al, 2010; Vecchiarelli et al, 2010; Lim et al, 2014). Studies with the chromosomal ParA homologue Soj from Thermus thermophilus revealed that it binds to DNA in a nucleotide-dependent manner as a dimer and forms collaborative filaments (Fig 2B). Binding of Soj to DNA is non-specific but cooperative (Leonard et al, 2005; Hester & Lutkenhaus, 2007). A very similar example is Vibrio cholerae ParA2 that also forms helical filaments on double-stranded DNA (Fig 2C) (Hui et al, 2010). It has been proposed that the formation of the ParA–DNA collaborative filament is essential for DNA movement during cell division but the actual mechanism remains elusive, although several models have been proposed (Ringgaard et al, 2009; Howard & Gerdes, 2010; Ptacin et al, 2010; Vecchiarelli et al, 2010).
Dan filaments: DNA condensation (DanN:DNA)
Dan (DNA-binding protein under anaerobic conditions) is a DNA-associated transcription factor that regulates the expression of tartrate dehydratase enzyme by controlling the expression of the ttd operon (Lim et al, 2013). The concentration of Dan in E. coli under normal conditions is low, and Dan binds to the nucleoid in a non-specific manner with a somewhat higher affinity for GTTNATT sequences (Teramoto et al, 2010). Specific binding only occurs when Dan is bound to L-tartrate. However, under anaerobic conditions, the copy number of Dan increases more than 100-fold. At this high concentration, Dan binds to DNA cooperatively and forms a rigid collaborative filament that causes DNA compaction into complex higher order structures and reduces accessibility (Fig 2D) (Lim et al, 2013). Although how the Dan/DNA nucleoprotein works in vivo remains unclear, based on the experimental evidence in vitro, it has been suggested that these nucleoprotein filaments play a critical role in DNA protection and gene regulation. Cooperativity of the formation of Dan filaments, caused by their collaborative nature, is used to switch between highly ordered filaments and Dan's function as a non-polymerised, highly specific transcription factor.
H-NS and StpA filaments: gene silencing/regulation (H-NSN:DNA)
Heat-stable nucleoid-structuring proteins (H-NS) are a class of nucleoid-associated transcriptional repressors that regulate ∼5% of E. coli genes (Lim et al, 2012a). Like many other nucleoid-associated proteins, H-NS has been shown to form either stiff collaborative filaments or DNA bridges (Fig 2E), depending on the presence of divalent cations (Liu et al, 2010). Mechanistic insight into how H-NS might cause gene silencing came from atomic force microscopy studies. It has been suggested that H-NS first non-specifically nucleates at the regulatory region of the target gene. Subsequently, more subunits are added cooperatively onto DNA to form a long, stiff and continuous collaborative filament that sterically blocks RNA polymerase activity (Liu et al, 2010). The response regulator gene SsrB removes DNA-bound H-NS subunits and releases inhibition (Walthers et al, 2011).
StpA, a paralogue of H-NS, also exhibits very similar gene silencing activity by forming a nucleoprotein filament (Fig 2F). However, in contrast to H-NS, StpA filaments seem to interact with DNA causing DNA bridges (Dame et al, 2005). Therefore, StpA simultaneously induces DNA stiffening and compaction. The mechanism of StpA filament assembly seems to be very similar to H-NS, through initial nucleation followed by cooperative assembly.
Collaborative filaments on lipid membranes
Collaborative filaments also form on lipid membranes, and as for all collaborative filaments, membrane modulates polymerisation by increasing cooperativity and reducing critical concentration. In contrast to DNA, which offers sequence-specific binding, membrane is less defined, although head group-specific binding and membrane curvature provide specificity in some cases that distinguish between different membranes in cells.
Dynamin-like proteins (BDLP1GXP:BDLP2GXP)N:M
Dynamins are a class of mechanochemical enzymes found in eukaryotes as well as in some prokaryotes and are involved in the production and sensing of membrane curvature. The dynamin family of GTPases includes classical dynamins, dynamin-like proteins (DLPs), Mx proteins, OPA, mitofusins and GBPs (Low & Löwe, 2006; Bramkamp, 2012). Dynamins form GTP-dependent cooperative filaments on lipid surfaces and at least some act by constriction (Fig 2G). Low et al (2009) showed medium-resolution cryoEM data of a bacterial dynamin-like protein bound to lipid, constricting the lipid membrane into a very small tube. Collaborative assembly ensures that filament formation and GTP hydrolysis are restricted to membranes. Some dynamin systems are complex because in addition to lateral interactions with the membrane, filaments also bind to their neighbours, forming a helix that changes pitch and diameter to constrict the lipid tube inside (Stowell et al, 1999; Low & Löwe, 2010).
FtsA filament: cell division (FtsAAXPN:M)
FtsA is a diverged bacterial actin homologue that directly interacts with the bacterial tubulin homologue FtsZ and anchors the nascent FtsZ ring to the cytoplasmic membrane (Pichoff & Lutkenhaus, 2005). FtsA interacts with the membrane through its C-terminal amphipathic helix. For a long time, it was incorrectly thought that FtsA does not polymerise, despite early reports using Streptococcus pneumonia FtsA (Lara et al, 2005). However, when T. maritima FtsA was applied to a lipid monolayer, it readily formed filaments and sheets, suggesting collaborative assembly (Fig 2H) (Szwedziak et al, 2012). These filaments showed a very similar repeat distance compared to what was seen in filaments deduced from a crystal structure (Szwedziak et al, 2012). Since non-polymerising FtsA mutants show a severe cell division defect, it was concluded that polymerisation of FtsA is indispensable for its function. Because in cells the number of FtsA molecules is low at ∼200 copies per E. coli cell (Wang & Gayda, 1992), it is expected that polymerisation will be limited to collaborative assembly on the membrane.
MinCD filaments: cell division inhibitor (MinC2:MinD2 AXP)N:M
In Gram-negative bacteria, the MinCDE septum site selection system comprising of the continuously oscillating proteins MinC, MinD and MinE prevents abnormal polar division. MinD is a Walker A cytoskeletal ATPase (WACA) that recruits MinC, the inhibitor of FtsZ activity, to the membrane and, together, they constitute the active inhibitor complex that prevents polar FtsZ-ring assembly (Hu et al, 2003; Lutkenhaus, 2007). Recently, MinC and MinD have been shown to form alternating copolymeric filaments that bind and decorate membrane (following the linear pattern in a protofilament of -MinC2-MinD2-) (Fig 2I) (Ghosal et al, 2014). In cells, the MinC concentration is 40 times lower than that of FtsZ (Dai & Lutkenhaus, 1992; Szeto et al, 2001). It has been proposed that MinCD copolymers inhibit FtsZ-ring assembly either by disrupting interfilament interactions or by affecting structural integrity of FtsZ filaments (Ghosal et al, 2014). Either way, substoichiometric amounts of MinCD could exert a strong inhibitory effect on FtsZ-ring assembly. Collaborative assembly ensures that filaments only form on membrane and enable polymerisation despite the low MinC concentration. The remaining free MinD is thought to engage in the oscillation process with MinE, its ATPase activator.
DivIVA filaments: septum site selection/cell wall addition (DivIVAN:M)
DivIVA is a coiled coil-containing peripheral membrane protein in Gram-positive bacteria. It has two primary roles: organising cell growth and coordinating polarity (Meniche et al, 2014; Sieger & Bramkamp, 2014). In B. subtilis, DivIVA has been shown to consistently localise at the cell poles and it also recruits the MinCD cell division inhibitor complex to protect cell poles from aberrant division (Edwards & Errington, 1997; Marston et al, 1998). A recent study in mycobacteria suggested that polar localisation of DivIVA is also essential for polar addition of new cell wall (Meniche et al, 2014). DivIVA has been shown to form bone-shaped oligomers that upon longer incubation form a two-dimensional network (Stahlberg et al, 2004). It has been suggested that in vivo, DivIVA senses the negative curvature at the cell poles through its N-terminal domain (Oliva et al, 2010) and forms a polar 2D network that recruits MinCD cell division inhibitors and guides polar localisation. Collaborative assembly with the membrane seems to cause the polar localisation, in which a curved membrane surface will preferentially bind the polymers (Fig 2J), since more binding sites can be satisfied in this way (Lenarcic et al, 2009; van Baarle et al, 2013).
Collaborative filaments involving more than one filament
From the above, it becomes clear that collaborative filaments are used to restrict polymerisation to certain areas, to be able to regulate their nucleation and elongation through cooperativity and to exert physical force on the matrix, or to cover it.
Interestingly, some collaborative filaments contain a second type of filamentous protein, with separate and different longitudinal interactions (Figs 1F and 3). This leads to an even greater number of interactions, and cooperativity may exist within a multi-stranded filament of one type, within the other and between them, as facilitated by lateral interactions between the two filaments.
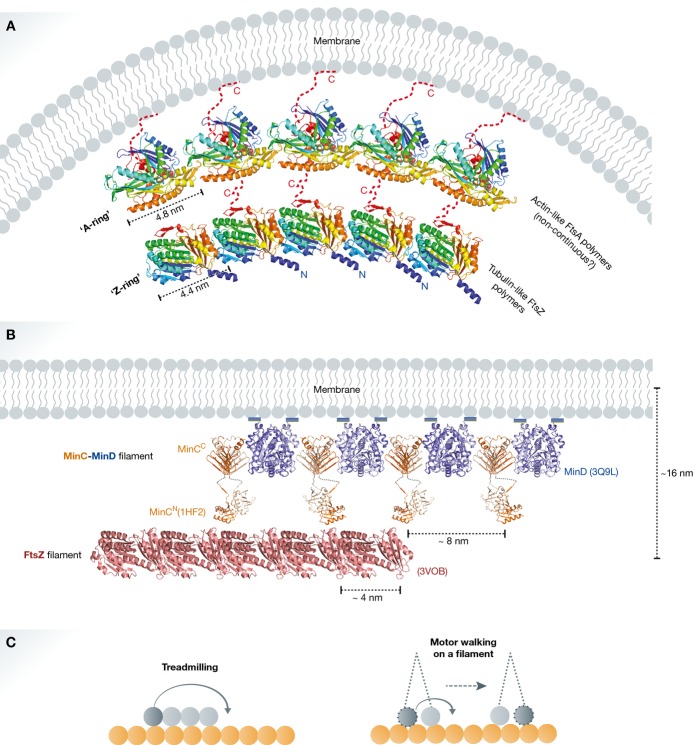
Figure 3. Two-protein collaborative filaments.
(A) FtsA is a bacterial actin homologue that recruits the nascent FtsZ ring to the cytoplasmic membrane. This is an example of a two-protein collaborative filament system on a matrix. There is a mismatch of repeat distances between FtsA and FtsZ polymers (∼5 nm and ∼4 nm, respectively). The mismatch causes filament curvature upon collaborative assembly (modified from Szwedziak et al, 2012). (B) MinCD cell division inhibitors form alternating copolymeric filaments on the membrane and interact with FtsZ filaments preventing polar cell division. This is another example of a two-protein collaborative filament system on a matrix. Here, the repeat distance of MinCD filaments (∼8 nm) is in multiples of the FtsZ filament repeat distance (∼4 nm). Hence, very little or no curvature is generated. This system shows cooperativity allowing MinC to attack FtsZ filaments over FtsZ monomers; thus, substoichiometric amounts of MinC can regulate Z-ring assembly (reproduced with permission from Ghosal et al, 2014). (C) Comparison between a two-filament collaborative filament system, where one cytomotive filament treadmills on another relatively stable filament (i) and a molecular motor walking on a filament (ii). If the movement of the lagging motor head to the front in the direction of the movement is assumed as treadmilling of a very short filament, consisting of only two subunits, then both systems have similar working principles. This postulation does not account for motor-to-motor communication, filament-induced nucleotide hydrolysis and the torque generation.
Heterogeneous collaborative filaments provide another interesting property: the ability to create curvature since the repeat distances (from subunit to subunit along the two or more longitudinal axes) within the different types of filaments may be different. When such filaments polymerise together or bind laterally to each other, curvature or even helical appearance will be induced. Importantly, homotopic, multi-stranded filaments made from identical subunits will not be able to induce curvature this way, it is a unique property facilitated by collaborative assembly of two different filaments.
FtsA–FtsZ two-filament collaborative filaments: cell division (FtsZGXPN:FtsAAXPN):M
During bacterial cell division, the cytokinetic FtsZ ring is tethered to the membrane by FtsA, which itself most likely forms short polymers as discussed. This process is collaborative but there is a mismatch of repeat distances between FtsA and FtsZ filaments (∼5 and ∼4 nm, respectively) (Fig 3A) (Szwedziak et al, 2012). The mismatch causes bending of filaments upon collaborative assembly. Indeed, in vitro reconstitution of the FtsA–FtsZ interaction on liposome surfaces showed that they spontaneously form collaborative filaments on the membrane and induce negative curvature, leading to membrane constriction (Szwedziak et al, 2014). Two-protein collaborative filament systems with a mismatch in subunit repeat lead to force generation without an active bending mechanism in either of the filaments. Cytomotive properties of the participating filaments may be used to disassemble the structure, completing the force-generating cycle.
MinCD–FtsZ two-filament collaborative filaments: cell division inhibition FtsZGXPN:(MinC2:MinD2 AXP)N:M
Another example of a two-protein collaborative filament system is the MinCD copolymeric filament and FtsZ filament interaction (Fig 3B). Recently, reconstitution of MinCD–FtsZ interactions on liposome surfaces suggested that the MinCD–FtsZ interaction is cooperative (Ghosal et al, 2014). In contrast to the previously described FtsA–FtsZ interaction, the subunit repeat distances of MinCD (∼8 nm) and FtsZ (∼4 nm) filaments match in multiples. The result is very little bending, and maybe this is not so surprising as the system is not meant to exert mechanical bending force. Avidity, which is a consequence of the extreme cooperativity between two filaments binding to each other with very many potential lateral interactions linking the two, is probably the main reason why MinCD filaments exist as they make it possible for the cell division inhibitor MinC to only attack FtsZ filaments over free FtsZ monomers. Interestingly, a somewhat similar mechanism has also been proposed for the inhibition of FtsZ-ring assembly by nucleoid occlusion factor SlmA. One of the published models suggests SlmA forms collaborative filaments on DNA and disorient FtsZ filaments affecting their lateral interactions that are needed for productive constriction and cell division (Cho et al, 2011; Tonthat et al, 2013).
Two-protein collaborative filaments in eukaryotes
Several examples of two-protein collaborative filament systems have been reported in eukaryotic cells. For example, septins have been shown to guide the directionality of the microtubule plus end during epithelial polarity establishment (Bowen et al, 2011). During this process, septin filaments polymerise along the length of microtubules and regulate microtubule dynamics and organisation. Interestingly, the septin–microtubule two-protein collaborative filament system is somewhat reminiscent of MinCD–FtsZ in bacteria. MinCD collaborative filaments are distantly related to the heteromeric septin filaments, and FtsZ is the bacterial tubulin homologue. Therefore, it has been proposed that septins and MinD might have a common evolutionary origin.
Another example of two-protein collaborative filament formation is the interaction between septins and actin filaments. Septins and F-actin are central components of many eukaryotic cytokinetic rings. Recent studies have demonstrated that septin rods copolymerise with F-actin and cause bundling as well as bending of F-actin filaments (Mavrakis et al, 2014). Septin-induced bending of F-actin filaments is mostly caused by a mismatch in repeat distance between septin and F-actin filaments, analogous to the FtsA–FtsZ interaction discussed above.
Conclusion
From the above, it is clear that filament architectures and mechanisms of assembly range from the simple isodesmic to extremely complex two-protein matrix-associated collaborative filament systems, and examples of such complex collaborative assemblies are probably fairly common as they are important components of large, self-assembling systems. The addition of a lateral binding partner to all, or a subset of subunits along the filament, either through a neighbouring matrix or indeed another filament creates opportunities for complex and emerging properties of the resulting systems.
The most obvious, but by no means only consequence is that collaborative filaments may be restricted in occurrence to the site of the matrix or scaffold they bind to.
The large number of additional, lateral binding sites sometimes creates large cooperativity effects that enable filaments to bind with extreme affinity to their partner, even if the individual binding energies, per subunit, are rather small. The simplest case of this is actually within homotopic, non-collaborative filaments, where it was found that lateral interactions holding the protofilaments together are surprisingly weak (Alushin et al, 2014; von der Ecken et al, 2015). Avidity enables the selection of filament binding over monomers, by a very large margin.
Perhaps more surprising are the consequences of geometry: collaborative filaments with two different filaments, and differing subunit repeat lengths create curvature. Additionally, on flat or straight matrices, collaborative filaments will cause bending or select for bent geometry under certain circumstances that involve additional interactions within the filament.
Finally, we would like to raise the enticing possibility that collaborative filaments acted as precursors for molecular motors (Fig 3Ci–ii), which so far have only been found in eukaryotes: processive motors walking along microtubules or actin filaments, mostly contain two ATPase heads of the myosin, kinesin or dynein type. Communication between the heads ensures that binding, release and finding of the next binding site for each head leads to movement. Now, if one considers the movement of the lagging motor head to the front, in the direction of the movement, as treadmilling of a very short filament consisting of only two subunits, then this could be considered a special situation of a collaborative filament. It could be described as one stable filament collaborating with another cytomotive filament, treadmilling. In fact, such data have recently been recorded in the case of the tubulin-like TubZRC system (Fink & Löwe, 2015, Fig S1 and Movie S3 therein). Thus, processive motor activity could arise by covalently linking two subunits of a collaborative cytomotive filament treadmilling on another, more stable filament. However, this postulation does not account for motor-to-motor communication, filament-induced nucleotide hydrolysis and subsequent torque generation as observed in modern motor proteins.
Acknowledgments
We would like to thank Linda A. Amos, Fusinita van den Ent and Gero Fink (all MRC-LMB) for helpful discussions and comments. This work was supported by the Medical Research Council (U105184326) and the Wellcome Trust (095514/Z/11/Z).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, Nogales E. High-resolution microtubule structures reveal the structural transitions in alphabeta-tubulin upon GTP hydrolysis. Cell. 2014;157:1117–1129. doi: 10.1016/j.cell.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Kuhn JR, Jacobs-Wagner C. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell. 2003;115:705–713. doi: 10.1016/s0092-8674(03)00935-8. [DOI] [PubMed] [Google Scholar]
- van Baarle S, Celik IN, Kaval KG, Bramkamp M, Hamoen LW, Halbedel S. Protein-protein interaction domains of Bacillus subtilis DivIVA. J Bacteriol. 2013;195:1012–1021. doi: 10.1128/JB.02171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat TA, Murshudov GN, Sachse C, Löwe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106–110. doi: 10.1038/nature14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen JR, Hwang D, Bai X, Roy D, Spiliotis ET. Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J Cell Biol. 2011;194:187–197. doi: 10.1083/jcb.201102076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramkamp M. Structure and function of bacterial dynamin-like proteins. Biol Chem. 2012;393:1203–1214. doi: 10.1515/hsz-2012-0185. [DOI] [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992;174:6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dame RT, Luijsterburg MS, Krin E, Bertin PN, Wagner R, Wuite GJ. DNA bridging: a property shared among H-NS-like proteins. J Bacteriol. 2005;187:1845–1848. doi: 10.1128/JB.187.5.1845-1848.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K, Chrysogelos S, Griffith J. Electron microscopic visualization of recA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982;28:757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Izore T, Bharat TA, Johnson CM, Löwe J. Bacterial actin MreB forms antiparallel double filaments. Elife. 2014;3:e02634. doi: 10.7554/eLife.02634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink G, Löwe J. Reconstitution of a prokaryotic minus end-tracking system using TubRC centromeric complexes and tubulin-like protein TubZ filaments. Proc Natl Acad Sci USA. 2015;112:E1845–E1850. doi: 10.1073/pnas.1423746112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory J, Tsang SS, Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc Natl Acad Sci USA. 1984;81:7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayathri P, Fujii T, Moller-Jensen J, van den Ent F, Namba K, Löwe J. A bipolar spindle of antiparallel ParM filaments drives bacterial plasmid segregation. Science. 2012;338:1334–1337. doi: 10.1126/science.1229091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Trambaiolo D, Amos LA, Löwe J. MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat Commun. 2014;5:5341. doi: 10.1038/ncomms6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester CM, Lutkenhaus J. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc Natl Acad Sci USA. 2007;104:20326–20331. doi: 10.1073/pnas.0705196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M, Gerdes K. What is the mechanism of ParA-mediated DNA movement? Mol Microb iol. 2010;78:9–12. doi: 10.1111/j.1365-2958.2010.07316.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Saez C, Lutkenhaus J. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. J Bacteriol. 2003;185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui MP, Galkin VE, Yu X, Stasiak AZ, Stasiak A, Waldor MK, Egelman EH. ParA2, a Vibrio cholerae chromosome partitioning protein, forms left-handed helical filaments on DNA. Proc Natl Acad Sci USA. 2010;107:4590–4595. doi: 10.1073/pnas.0913060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov V, Mizuuchi K. Multiple modes of interconverting dynamic pattern formation by bacterial cell division proteins. Proc Natl Acad Sci USA. 2010;107:8071–8078. doi: 10.1073/pnas.0911036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, Biton J, Vicente M, Mingorance J, Massidda O. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol. 2005;55:699–711. doi: 10.1111/j.1365-2958.2004.04432.x. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Löwe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer–a conserved biological switch. EMBO J. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesterlin C, Ball G, Schermelleh L, Sherratt DJ. RecA bundles mediate homology pairing between distant sisters during DNA break repair. Nature. 2014;506:249–253. doi: 10.1038/nature12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Lee SY, Kenney LJ, Yan J. Nucleoprotein filament formation is the structural basis for bacterial protein H-NS gene silencing. Sci Rep. 2012a;2:509. doi: 10.1038/srep00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Lee SY, Teramoto J, Ishihama A, Yan J. The nucleoid-associated protein Dan organizes chromosomal DNA through rigid nucleoprotein filament formation in E. coli during anoxia. Nucleic Acids Res. 2013;41:746–753. doi: 10.1093/nar/gks1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Whang YR, Kenney LJ, Yan J. Gene silencing H-NS paralogue StpA forms a rigid protein filament along DNA that blocks DNA accessibility. Nucleic Acids Res. 2012b;40:3316–3328. doi: 10.1093/nar/gkr1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. Elife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Thanbichler M. Nucleotide-independent cytoskeletal scaffolds in bacteria. Cytoskeleton (Hoboken) 2013;70:409–423. doi: 10.1002/cm.21126. [DOI] [PubMed] [Google Scholar]
- Lindsley JE, Cox MM. Assembly and disassembly of RecA protein filaments occur at opposite filament ends. Relationship to DNA strand exchange. J Biol Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- Liu Y, Chen H, Kenney LJ, Yan J. A divalent switch drives H-NS/DNA-binding conformations between stiffening and bridging modes. Genes Dev. 2010;24:339–344. doi: 10.1101/gad.1883510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Low HH, Löwe J. Dynamin architecture–from monomer to polymer. Curr Opin Struct Biol. 2010;20:791–798. doi: 10.1016/j.sbi.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Löwe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Amos LA. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int J Biochem Cell Biol. 2009;41:323–329. doi: 10.1016/j.biocel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu Rev Biochem. 2007;76:539–562. doi: 10.1146/annurev.biochem.75.103004.142652. [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis M, Azou-Gros Y, Tsai FC, Alvarado J, Bertin A, Iv F, Kress A, Brasselet S, Koenderink GH, Lecuit T. Septins promote F-actin ring formation by crosslinking actin filaments into curved bundles. Nat Cell Biol. 2014;16:322–334. doi: 10.1038/ncb2921. [DOI] [PubMed] [Google Scholar]
- Meniche X, Otten R, Siegrist MS, Baer CE, Murphy KC, Bertozzi CR, Sassetti CM. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA. 2014;111:E3243–E3251. doi: 10.1073/pnas.1402158111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, Veprintsev DB, Hamoen LW, Löwe J. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29:1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pilhofer M, Jensen GJ. The bacterial cytoskeleton: more than twisted filaments. Curr Opin Cell Biol. 2013;25:125–133. doi: 10.1016/j.ceb.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieger B, Bramkamp M. Interaction sites of DivIVA and RodA from Corynebacterium glutamicum. Front Microbiol. 2014;5:738. doi: 10.3389/fmicb.2014.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg H, Kutejova E, Muchova K, Gregorini M, Lustig A, Muller SA, Olivieri V, Engel A, Wilkinson AJ, Barak I. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol Microbiol. 2004;52:1281–1290. doi: 10.1111/j.1365-2958.2004.04074.x. [DOI] [PubMed] [Google Scholar]
- Stowell MH, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- Suefuji K, Valluzzi R, RayChaudhuri D. Dynamic assembly of MinD into filament bundles modulated by ATP, phospholipids, and MinE. Proc Natl Acad Sci USA. 2002;99:16776–16781. doi: 10.1073/pnas.262671699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, King GF. The dimerization function of MinC resides in a structurally autonomous C-terminal domain. J Bacteriol. 2001;183:6684–6687. doi: 10.1128/JB.183.22.6684-6687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Bharat TA, Tsim M, Löwe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. Elife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SM, Löwe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teramoto J, Yoshimura SH, Takeyasu K, Ishihama A. A novel nucleoid protein of Escherichia coli induced under anaerobiotic growth conditions. Nucleic Acids Res. 2010;38:3605–3618. doi: 10.1093/nar/gkq077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonthat NK, Milam SL, Chinnam N, Whitfill T, Margolin W, Schumacher MA. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci USA. 2013;110:10586–10591. doi: 10.1073/pnas.1221036110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Han YW, Tan X, Mizuuchi M, Ghirlando R, Biertumpfel C, Funnell BE, Mizuuchi K. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Seol Y, Neuman KC, Mizuuchi K. A moving ParA gradient on the nucleoid directs subcellular cargo transport via a chemophoresis force. Bioarchitecture. 2014;4:154–159. doi: 10.4161/19490992.2014.987581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walthers D, Li Y, Liu Y, Anand G, Yan J, Kenney LJ. Salmonella enterica response regulator SsrB relieves H-NS silencing by displacing H-NS bound in polymerization mode and directly activates transcription. J Biol Chem. 2011;286:1895–1902. doi: 10.1074/jbc.M110.164962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Gayda RC. Quantitative determination of FtsA at different growth rates in Escherichia coli using monoclonal antibodies. Mol Microbiol. 1992;6:2517–2524. doi: 10.1111/j.1365-2958.1992.tb01428.x. [DOI] [PubMed] [Google Scholar]
- Wickstead B, Gull K. The evolution of the cytoskeleton. J Cell Biol. 2011;194:513–525. doi: 10.1083/jcb.201102065. [DOI] [PMC free article] [PubMed] [Google Scholar]