Abstract
Histones and their modifications play an important role in the regulation of gene transcription. Numerous modifications, such as acetylation, phosphorylation, methylation, ubiquitination, and SUMOylation, have been described. These modifications almost always co-occur and thereby increase the combinatorial complexity of post-translational modification detection. The domains that recognize these histone modifications often occur in tandem in the context of larger proteins and complexes. The presence of multiple modifications can positively or negatively regulate the binding of these tandem domains, influencing downstream cellular function. Alternatively, these tandem domains can have novel functions from their independent parts. Here we summarize structural and functional information known about major tandem domains and their histone binding properties. An understanding of these interactions is key for the development of epigenetic therapy.
Keywords: histone, post-translational modifications, gene transcription, chromatin, protein modular domains
Introduction
One of the greatest scientific revelations in recent years is the discovery that as well as functioning as a structure for compacting the genome, chromatin is an active player in allowing access to it for gene transcription, editing, and repair. The basic structure of chromatin is the nucleosome, consisting of 147-bp of DNA coiled around dimers of each of the core histones: H2A, H2B, H3, and H4.1 Histones, H1 and H5, serve as linker proteins between nucleosomes for further compaction. The flexible and unstructured N and C-terminal tails that snake out from the central core of the nucleosome are subject to post-translational modifications. The wrapped DNA bases as well as the globular core of the nucleosome are also subject to modification by effector proteins. These modifications are important for epigenetic control of gene transcription in response to a variety of stimuli.2 The placement, detection, and removal of these histone and DNA modifications, or marks, is controlled by protein domains that are, respectively, “writers, readers, and erasers.3” The readout of these marks leads to multiple cellular processes, including transcription, cell differentiation, cell division, and apoptosis.4
A wide variety of post-translational modifications on histones have been found, including acetylation, methylation, phosphorylation, ubiquitination, SUMOylation, and crotonylation.5 In order to elicit downstream action, effector proteins must be able to bind histones in a modification-sensitive manner. The discovery of bromodomains as acetyl-lysine binding domains,6 PHD fingers as methyl-lysine binding domains,7,8 and the royal family of chromodomains, Tudor, and MBT domains9 illustrated how this may occur. These highly conserved structural domains make up larger proteins, which often assemble to form complexes and macromolecular machines that have the ability to expand chromatin to allow transcription or compact it to form silenced heterochromatin.10–12 In the last few years a number of structural and functional studies have helped us elucidate the details of the interactions of these individual effector domains with chromatin and other chromatin-interacting proteins.13 Genome-wide studies have also been performed to look at the breadth of post-translational modifications and correlations between their occurrence and cellular function.14,15 These studies have revealed that it is rare to find a correlation between a single histone modification and a given cellular output. This is perhaps due to the large degree of combinatorial complexity allowed by multiple, varying modifications on single histones and nucleosomes.16,17 The readout of multiple modifications often, but not always, requires two or more effector domains to work together. Clearly then to understand the relationship between a given set of histone modifications and their cellular function, it becomes necessary to study higher order arrangements of effector domains.18 This review seeks to describe the structure and function of paired chromatin associating domains, with a focus on pairs that have structural and biochemical information available (Table I). It is interesting to note with these domains that the whole can be different than the sum of two parts. For example, a paired PHD and bromodomain can often have different functions from simply simultaneous readout of methyl-lysine and acetyl-lysine. We focus on the molecular and cellular features of these tandem domains and the roles they have in epigenetic regulation.
Table I.
Tandem Domains, With Their Modifications Detected, Unique Properties and PDB IDs
| Protein | Tandem domain | Modification detected | Properties | PDB ID |
|---|---|---|---|---|
| TAF1 | Bromo-Bromo | H4K5/8/12/16ac | Simultaneous detection of two to four acetylated residues, located 25 Angstroms apart. | 1EQF, 3AAD |
| DPF3b | PHD-PHD | H3(1-9), H3K14ac, H4Nac | PHD1 binds H3K14ac while PHD2 binds the unmodified N-terminus of H3. PHD2 can also recognize an acetylated H4 N-terminus. | 2KWJ, 2KWN |
| CHD1 | CHD-CHD | H3K4me3 | Both chromodomains fuse to make a joint binding surface. | 2B2W |
| TRIM24 | PHD-Bromo | H3K4me0, H3K23ac | Simultaneous detection of H3K4me0 and H3K23ac. | 3O34, 3O34 |
| BPTF | PHD-Bromo | H3K4me3, H4Kac | Simultaneous intranucleosomal reading of H3K4me3 and H4Kac. | 2F6J |
| KAP1 | PHD-Bromo | None recognized | PHD functions as an intramolecular SUMO E3 ligase | 2RO1 |
| MLL1 | PHD-Bromo | H3K4me3 | Interacting domains acts as molecular switch to bind H3K4me3 and Cyp33 RRM simultaneously for gene repression. | 3LQJ, 2KU7 |
| UHRF1 | TTD-PHD | H3K9me3 and H3K4me0 | PBR of UHRF1 can block H3K9me3 binding. Binding of PI5P to PBR can relieve this block. | 4GY5, 3ASK, 3ASL |
| CBP/p300 | Bromo-RING-PHD-HAT | H3/4ac | RING and PHD domains can regulate HAT activity of adjacent HAT domain | 4N4F, 4BHW |
| ZMYND11 | Bromo-ZnF-PWWP | H3.3K36me3 | Shows preference for the histone variant H3.3. Binding interface extends across multiple domains. | 4N4I. 4NS5 |
Note: PDB, Protein Data Bank; SUMO, small ubiquitin-like modifier; PHD, plant homeodomain; TTD, tandem tudor domain; PBR, poly-basic region; PI5P, phosphatidylinositol-5-phosphate; HAT, histone acetyltransferase; RING, really interesting new gene; ZnF, zinc-finger; ac, acetylated; me0, unmethylated; me3, trimethylated.
Domains Repeated in Tandem
Taf1 (Bromodomain-bromodomain)
Over a dozen families of reader effector domains have been found in chromatin-associated proteins that are capable of binding to histones in a modification and sequence dependent manner. The same modifications, however, can occur in many different contexts. The histone h3 lysine 4 tri-methylation (H3K4me3) mark is typically present at the 5’ end of genes at levels that correlate with production of transcripts.19,20 However, it can be found with the H3K27me3 mark, which is typically associated with gene silencing, at genes poised for activation in embryonic stem (ES) cells, forming so-called “bivalent domains.21” The same histone mark occurring in different functional contexts can be distinguished by tandem domains, which can positively select for one mark while negatively selecting for another. In other cases, two different marks can lead to a positive combinatorial effect. This effect was first reported for TAF1 (formerly TAFII250), the largest subunit of the TFIID complex involved in assembly of the transcriptional machinery at promoters. Instead of recognition of a single acetyl-lysine by a bromodomain, the protein has tandem bromodomains that can bind multiple acetylated H4 histone tails. The crystal structure reported of the tandem bromodomains shows the domains, each arranged in a left-handed four-helix bundle topology, positioned adjacent to each other with the binding pockets separated by ∼25 angstroms, corresponding to distance of about 7 residues in the histone protein sequence. The N-terminal tail of H4 has lysine residues at positions 5, 8, 12, and 16, in accordance with the pocket distance. The tandem module binds much more strongly to the double mark H4K5acK12ac (Kd = 1.4 μM) than to the H4K16ac single mark (Kd ∼40 μM).22 Interestingly, bromodomain 1 of the mouse homolog of TAF1, Brdt, was recently shown to bind the H4K5acK8ac dual mark (Kd = 28 μM), but showed much weaker affinity for either mark individually.23 This illustrates that a single bromodomain can bind to a pair of acetyl-lysines or a pair of bromodomains can bind to four acetyl-lysines simultaneously. The structural information gives us a hint at the possible in vivo function of TAF1. Actively transcribed genes are generally hyper-acetylated at their promoter regions and therefore TAF1 can target the transcriptional machinery to these genes.
DPF3b (PHD-PHD)
While most bromodomains are known to bind acetyl-lysine, the plant homeodomain (PHD) finger is less predictable in its interaction with chromatin. The PHD fingers of AIRE24 and BHC8025 interact with unmodified H3K4 (H3K4me0), while most other reported PHD fingers bind to methylated histones.26 Recently, a distinct tandem PHD finger of DPF3b was shown to bind H3K14ac, a modification typically reserved for recognition by bromodomains.27 DPF3b functions in association with the BAF chromatin-remodeling complex to initiate transcription during muscle and heart development.28 An NMR structure–function study showed that PHD1 of DPF3b binds to H3K14ac, while PHD2 binds the H3R2-K4 region on the same tail [Fig. 1(A)]. The tandem PHD fingers bind to unmodified H3 peptide with Kd = 2.3 μM and a fourfold increase in affinity occurs when binding to H3K14ac (Kd = 0.5 μM). The same study also showed binding to an acetylated N-terminus of H4 (H4Nac) with Kd = 7.4 μM. PHD2 uses the same mechanism as other PHD fingers that bind to H3K4me0: a set of hydrophobic and acidic residues in the binding pocket. PHD1, however, uses a binding pocket on the opposite side of the domain that does not coincide with the residues typically used to bind H3R2 and H3K4. Interestingly, acetylation or methylation at H3K4 leads to a 15- or 20-fold reduction in affinity due to steric clashes with PHD2, illustrating how, as mentioned above, histone modifications can lead to negative selection with a tandem domain. DPF3b from transfected C2C12 myoblasts showed a decrease in binding to H3K4me3 peptides as compared to H3K14ac, in agreement with binding assays. Binding of DPF3b to H3K14ac provides recruitment of a chromatin-remodeling complex to target genes for pre-initiation of transcription. The placement of the H3K4me3 mark leads to dissociation of DPF3b and association of the transcriptional machinery for initiation and activation of transcription important for development.
Figure 1.
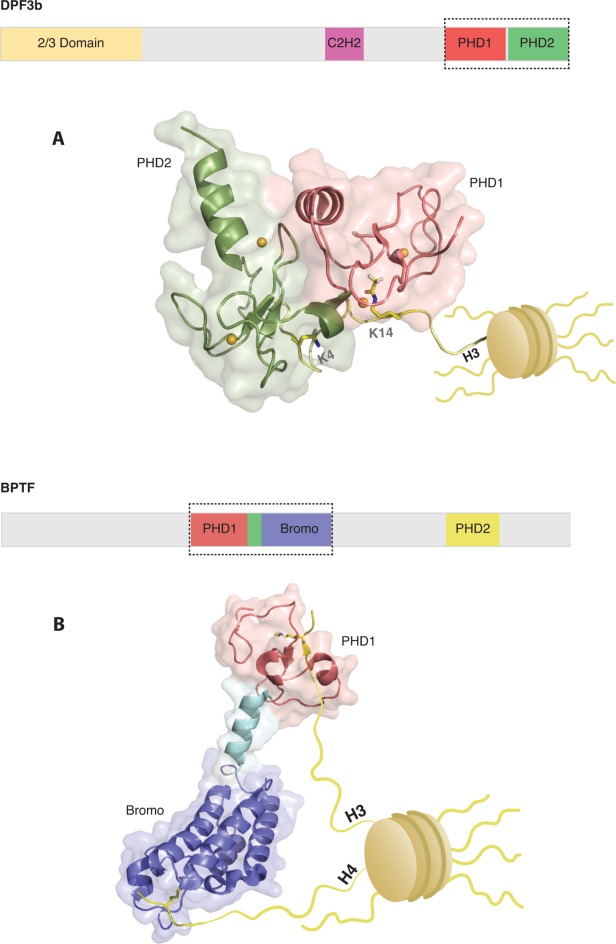
Reader domains can bind multiple marks on one, or multiple tails, simultaneously (A) Top: Domain architecture of full-length DPF3b. Bottom: The C-terminal tandem PHD fingers of DPF3b, PHD1 (red) and PHD2 (green), can recognize multiple marks on a single histone H3 tail. PHD1 of DPF3b recognizes H3K14ac while PHD2 simultaneously recognizes the unmodified N-terminus of H3. The outlined surface view illustrates the interaction surface between both PHD fingers. Chelated zinc atoms are shown as gold spheres. The unmodified H3K4me0 and H3K14ac marks are shown as sticks. (B) Top: Domain architecture of full-length BPTF. Bottom: PHD1 of BPTF (red) is in tandem with a bromodomain (blue), separated by a linker (aqua). PHD2 is not shown. The bromodomain recognizes H4K16ac while PHD1 simultaneously recognizes H3K4me3. The linker between the domains forms a rigid helix, preventing a direct interaction between PHD1 and the bromodomain. The histone marks H3K4me3 and H4K16ac are shown as sticks.
MOZ (PHD-PHD)
The monocytic leukemia zinc finger protein (MOZ, KAT6A, MYST3) is a MYST family histone acetyltransferase (HAT) that plays an essential role in a number of developmental processes, including acting as a co-activator for transcription factors specific for hematopoiesis.29 It also plays an important role in leukemia pathogenesis, present as fusion partners with CBP,30 p300,31 and TIF232 after chromosomal translocations. MOZ typically partners with ING5, EAF6, and BRPF1, BRPF2, or BRPF3 and acts as a catalytic subunit in a tetrameric complex in vivo.33 The structural basis for its targeting to specific genetic loci was recently discovered.34 Qiu et al. found that MOZ contains atypical tandem PHD fingers that can simultaneously bind H3R2me0 and H3K14ac, in similar fashion to DPF3b. The PHD fingers bind unmodified H3(1-18) with Kd = 64.9 μM and an approximate three-fold increase in binding occurs when H3 is acetylated at K14 (Kd = 23.3 μM). The structural mechanism for acetylated lysine recognition is quite similar to that of DPF3b, with an analogous hydrophobic pocket. However, DPF3b utilizes D263 and R289 to hydrogen bond with the carbonyl and amide moieties of the modification while MOZ uses S210 and N235. The mutations S210D and N235R maintain H3K14ac binding, reinforcing the conserved mechanism. However, there are some global changes in structure with functional consequences. The loop that precedes the first β strand of the MOZ PHD2 is longer than the analogous loop in DPF3b PHD2, which makes it possible to bind H3K4me3 (Kd = 286.5 μM). In follow up to their structural studies, Qiu et al. were able to demonstrate MOZ recruitment to the HOXA9 promoter, and not the HOXB1, HOXC5, and HOXD12 promoters. Introduction of point mutations S210A and D282A, which abolished histone binding, was able to significantly reduce promoter localization of MOZ. Also, the mutants reduced H3K14 acetylation at the HOXA9 promoter and HOXA9 mRNA levels approximately two-fold, indicating that the tandem PHD fingers are important for targeting of MOZ acetyltransferase activity. This result may explain aberrant targeting of MOZ fusion proteins to important hematopoietic regulators in AML.
NSD family (PHD-C5HCH)
Another recent study35 found that the NSD (nuclear receptor SET domain-containing) family of H3K36me2 methyltransferases contains tandem C-terminal PHD-C5HCH domains. The three family members, NSD1, NSD2 (also known as MMSET), and NSD3 (also known as WHSC1L1) contain five PHD domains and it is this fifth domain that forms a tandem module with the C5HCH domain, which is a PHD-like domain. The NSD3 tandem domain is able to simultaneously bind H3K4me0 and K3K9me3 (Kd = 290 μM). Interestingly, the NSD2 tandem domain shows a preference for unmodified H3, whereas the NSD1 tandem domain did not show binding to histones at all. While the proteins are quite homologous,36 they are implicated in different diseases. Mutations in NSD1 cause Sotos syndrome,37 overexpression of NSD2 by the t(4;14) translocation is the cause for 15% of multiple myelomas,38 and both NSD1 and NSD3 can be fused to NUP98 to cause AML.39,40 Future studies will reveal whether differential targeting of the tandem PHD-C5HCH domain contributes to their varying disease phenotypes. Development of small molecules to target these domains may allow selective modulation of methyltransferase activity within the NSD family.
Chd1 (Chromodomain-chromodomain)
While the aforementioned examples illustrated the ability of tandem domains to bind multiple marks on histone tails, there are numerous examples in which tandem domains cooperate to bind a single mark. In addition to the PHD finger, a homologous sequence was found in the D. melanogaster heterochromatin protein-1 (HP-1) and Polycomb, termed the chromatin organization modifier domain, or chromodomain, that is capable of binding methyl-lysine. A tandem chromodomain is present in CHD1, part of the chromo helicase DNA-binding (CHD) proteins, which regulate ATP-dependent nucleosome assembly. The tandem chromodomains in human CHD1 cooperate to bind a single H3K4me3 mark (Kd = 5 μM).41 The crystal structure shows that both chromodomains join to make a continuous surface, with the H3 binding site occurring at an acidic surface bridging the two domains.
Simultaneous Readout by Different Domains
Trim24 (PHD-bromodomain)
While the same structural domains occur as repeats as in the above examples, there are many instances in which different domains are combined in tandem. An important example of this in which cooperative binding occurs is the tandem C-terminal PHD-bromodomain segment of TRIM24 (also called TIF-1α). TRIM24 is part of the TRIM/RBCC protein family, which is characterized by an N-terminal tripartite motif composed of a RING domain, B-box zinc fingers, and a coiled-coil region. These proteins also contain variable C-terminal domains, including tandem PHD-bromodomains.42 A recent structure–function study showed that the TRIM24 PHD finger targets unmodified H3K4me0, with similar structural characteristics to BHC80.43 The bromodomain binds to multiple H3 or H4 acetyllysine residues, with strongest affinity for H3K23ac. Isothermal titration calorimetry (ITC)-based binding assays showed that the PHD-Bromo tandem bound H3(1-15)K4me0 with a Kd of 8.6 μM and H3(13-23)K23ac with a Kd of 8.8 μM. Structural analysis showed that the H3K4 and H3K23ac peptides aligned in the same direction on the surface of the molecule when bound, suggesting that the tandem domains may be able to bind both peptides at the same time. Also, the distance between the Cα of H3K4 and H3K23 and the distance between the binding pockets on the domains is ∼30 Angstroms. In agreement, ITC studies showed binding to H3(1-33)K4K23ac peptide showed 90-fold higher affinity (Kd = 0.096 μM) as compared to binding to H3(1-15)K4 and H3(13-32)K23ac. While the structural study showed tandem binding to the H3K4me0 and acetylated lysine marks, there is no common interpretation for the presence of these marks in chromatin biology. However, there is a precedent in the co-regulation of ER-α by TRIM24, as interactions between the protein and nuclear receptors are ligand-dependent and because ligand-activated estrogen response elements (EREs) are independent of H3K4 methylation.44–46 Chromation immunoprecipitation (ChIP) showed, in agreement with structural studies, that TRIM24 interacts with ERα and chromatin lacking H3K4 methylation, but enriched for lysine acetylation in response to estrogen in MCF7 breast cancer cells. Depletion of TRIM24 by shRNA in tumor-derived breast cancer cells led to reduced survival and proliferation. In correlation, overexpression of TRIM24 in tissue samples from a non-metastatic breast cancer cohort correlated with poor patient survival, independent of ER status.43 In addition to this study, it was found that TRIM24 is a target of chromosomal translocation to form an oncogenic fusion protein in acute promyelocytic leukemia,47 papillary thyroid carcinoma,48 and myeloproliferative syndrome.49 This example illustrates the importance that studying tandem domains can have in discovery of novel epigenetic therapeutics.50–52
Trim33 (PHD-bromodomain)
Another member of the TRIM family, TRIM33, is a Smad-binding protein also with a C-terminal tandem PHD-bromodomain. A recent study implicated its histone readout capabilities in switching of master regulators of stem cell differentiation from poised to active states.53 It was previously found that TRIM33-deficient mice die early in development, with impairments in mesoderm formation and phenotypes suggestive of misregulated nodal (part of the TGF-β family) signaling.54 Furthermore, conditional deletion of TRIM33 in premalignant pancreatic progenitor cells yields a similar phenotype to that of SMAD4 knockout, implicating it in TGF-β family signaling.55,56 A recent study described the role of TRIM33 histone binding in nodal-dependent activation of the master regulator genes GSC and MIXL1.53 Xi et al. discovered that nodal signaling in embryoid bodies promoted formation of a TRIM33-Smad2/3 complex, which is targeted to the GSC and MIXL1 promoters to facilitate mesendoderm formation. They were also able to resolve the structural basis for this promoter localization. The tandem PHD-bromodomain recognizes H3K4me0 and H3K9me3 (through the PHD finger) and H3K18ac (through the bromodomain). The tandem domains were able to bind the H3K9me3K18ac dual peptide mark with greater affinity (Kd = 0.06 μM) than the sum of the affinities of the individual H3K9me3 mark (Kd = 0.20 μM) and H3K18ac mark (Kd = 0.21 μM), indicating a greater combinatorial effect in histone binding. Interestingly, the classical asparagine in the binding pocket of most bromodomains, which typically engages in hydrogen bonding with the acetyllysine, as in TRIM24, does not do so in TRIM33 as it is 8.2 Angstroms away. In functional agreement with their structural studies, TRIM33 was co-localized with H3K9me3 and H3K18ac at the GSC and MIXL1 promoters by ChIP-qPCR. Mononucleosome ChIP also showed that the modifications were present on the same nucleosomes. Regions rich in H3K9me3 are typically bound by the HP1 proteins,57 mediated by their chromodomains.9,12 This is thought to mediate condensation into heterochromatin.58 Xi et al. found that TRIM33 could outcompete HP1γ on peptides with both H3K9me3 and H3K18ac, but not H3K9me3 alone. Based on their structural and functional analysis, the authors posit that the GSC and MIXL1 loci are in a poised state with bound HP1γ prior to nodal activation. After activation by nodal, TRIM33 is able to cooperate with Smad2/3 to boot off HP1γ, bind to the GSC and MIXL1 promoters, and recruit HATs to promote gene activation and subsequently, cellular differentiation.
Uhrf1 (tandem Tudor-PHD)
There is also crosstalk between modes of epigenetic regulation, such as DNA methylation and histone modification. The nuclear protein, Ubiquitin-like with PHD and RING finger domains 1 (UHRF1), also known as ICB90 and NP95 in mouse, is a multi-domain protein that is required for the maintenance of CpG DNA methylation.59 It contains an SRA domain, which binds hemimethylated DNA,60 a tandem tudor domain (TTD), which mediates recognition of H3K9me3,61,62 and a PHD finger, which reads the unmodified N-terminal portion of the H3 tail.63–65 The TTD is only separated from the PHD finger domain by 15 residues and so structures were solved of the domains in tandem.66,67 The TTD-PHD linker was found to bind between the two Tudor domains and the PHD domain was able to combinatorially enhance TTD binding to H3K9me3 ∼6 fold. Interestingly, based on the structure of the TTD-PHD in complex with H3, it was thought that the isolated TTD-H3K9me3 structure had an incorrect mode of N-terminal H3 recognition because the isolated structure showed binding of the N-terminal H3 in the same location as the TTD-PHD linker. However, a new study shows that there is dynamic interplay between the TTD-PHD and the linker between the SRA and RING domains.68 In this study, a polybasic region (PBR) between the SRA and RING domains can bind in between the two Tudor domains, displacing the TTD-PHD linker and blocking TTD function. Binding of phosphatidylinositol-5-phosphate (PI5P) to the PBR can relieve this block and allow the TTD to bind H3K9me3. When the TTD is blocked, the PHD finger is free to bind to the unmodified N-terminal H3 tail. This study addresses why UHRF1 may localize to pericentromeric heterochromatin69 as well as have a role in gene repression in euchromatic regions.70 Structural studies on the full-length protein will elucidate the roles that the multiple domains and linker regions have in gene regulation.
Zmynd11 (PHD-Bromodomain-PWWP)
In addition to the canonical set of histone proteins, there exist histone variants. H3 can exist as multiple variants, including H3.1, H3.2, and H3.3.71,72 A recent study by Wen et al. demonstrated the effect of selective binding of a histone reader to histone variant H3.373. They found that ZMYND11, a candidate tumor suppressor, specifically recognizes H3K36me3 on H3.3, a specific mark of transcriptional elongation. ZMYND11, also known as BS69, can act as a co-repressor of E1A and transcription factors such as c-Myb and ETS-2. It contains multiple histone reader modules, such as a PHD finger, a bromodomain, and a PWWP domain. Wen et al. found specific binding of the bromodomain-PWWP module to H3.3K36me3 peptides (Kd = 56 μM). This was a gain of affinity of ∼8 fold as compared to binding to H3.1K36me3 peptides (Kd = 431 μM). The crystal structure of the ZMYND11 bromodomain-PWWP module elucidated a previously uncharacterized zinc-finger in between the two domains [Fig. 2(A)]. The bromodomain was found to vary from typical bromodomains, and several structural features suggest it may not bind to acetylated lysine. The histone variant H3.3 contains a serine at position 31 (S31), whereas H3.1 has A31. Residues from the bromodomain, zinc finger, and PWWP domain, all contribute to recognition of the motif centered at S31, illustrating integrated recognition of H3.3 by the three domains. In agreement with biochemical and structural studies, ZMYND11 was predominantly found throughout gene bodies and co-localized to H3.3 and H3K36me3 by ChIP-seq. Binding of ZMYND11 was also drastically reduced upon knockdown of SETD2, the major H3K36me3 methyltransferase. ZMYND11 was found to be both a repressor and activator, but a specific repressor of oncogenes, as seen by RNA-seq on ZMYND11 depleted cells. Accordingly, RNA Polymerase II (Pol II) was found to be increased at the 3’ end of ZMYND11-repressed genes in ZMYND11 depleted cells, suggesting that ZMYND11 may repress its target genes by restricting elongation by Pol II. Furthermore, a recent functional study showed that ZMYND11 interacts with regulators of RNA splicing, including EFTUD2. By antagonizing EFTUD2 through physical interaction, ZMYND11 can promote intron retention, connecting H3.3K36 methylation to RNA splicing events.74
Figure 2.
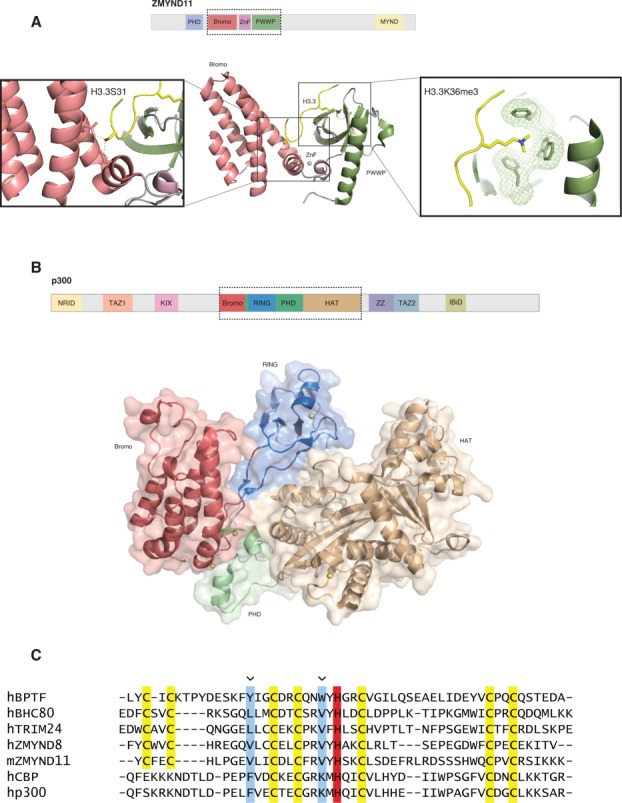
Structural studies reveal hidden zinc fingers in between tandem modules. (A) Top: Domain architecture of full-length ZMYND11. Bottom: Shown are the tandem bromodomain (red), zinc finger (purple), and PWWP (green) domains of ZMYND11. The histone H3 tail is shown in yellow. Visible in the left panel is the recognition of histone H3.3 specific residue S31 (H3.3S31). Recognition of this residue imparts the H3.3 specific binding of ZMYND11. The panel on the right illustrates binding of H3.3K36me3 by the “aromatic cage” of the PWWP domain. The “aromatic cage” is visible as mesh-covered sticks and H3.3K36me3 is visible as yellow sticks. (B) Top: Domain architecture of full-length p300. Bottom: Shown is the tandem bromodomain (red), RING (blue), PHD (green), and HAT (brown) module of P300. The RING domain inserts into the PHD domain. The PHD domain regulates catalytic activity of the adjacent HAT domain. (C) Sequence alignment shows that the PHD fingers of ZMYND8 and ZMYND11 have similar features to the H3K4me0 binding TRIM24 and BHC80 rather than the H3K4me3 binding BPTF. CBP and p300 lack the determinants for canonical H3K4 recognition. The zinc chelating cysteines are highlighted in yellow, the zinc chelating histidine is highlighted in red, the binding pocket residues are highlighted in blue and indicated with an arrow.
Zmynd8 (PHD-Bromodomain-PWWP)
Interestingly, the crystal structure of the tandem PHD finger, bromodomain, and PWWP domain for the related ZMYND8 (also known as Prkcbp1) was also recently solved (PDB ID 4COS). This structure differs from that of the solved ZMYND11 structures due to the presence of the PHD finger, which makes contacts with αAZ and αB of the bromodomain. The PHD does not have the canonical aromatic cage of H3K4me3 binding PHD fingers [Fig. 2(C)]. However, it may contain a combination of hydrophobic and acidic residues that are characteristic of unmethylated-lysine binding PHD fingers, such as TRIM24 and BHC80, which bind H3K4me0. Wen et al. did not see binding of a PHD-containing construct to unmodified H3 peptides, which may indicate another level of recognition, or perhaps binding to another substrate.
BPTF (PHD-bromodomain)
All of the previous examples have focused on recognition of multiple post-translational marks on the same histone tail. Are there units of recognition that extend beyond a single tail with interesting cellular outputs? A recent study sought to answer this question by investigating the tandem PHD-bromodomain of BPTF, part of NURF, an ISWI-containing ATP-dependent chromatin remodeling complex.75 BPTF is an important part of NURF, essential for regulating chromatin structure during development.76 Previous structural work determined that the PHD finger of BPTF preferentially bound to H3K4me3 (Kd ∼ 2.7 μM), while the targets of the bromodomain were unknown.77 Ruthenburg et al. determined that the bromodomain showed binding to H4K12ac, H4K16ac, and H4K20ac by ITC.75 Furthermore, they found that a binding enhancement by bivalent targeting only for H3K4me3 in combination with H4K16ac and not H3K12ac, despite the promiscuity of the bromodomain determined by ITC. They found that the tandem domain pair was able to bind the two marks simultaneously in an intra-nucleosomal context, but not an inter-nucleosomal context [Fig. 1(B)]. In accordance with biophysical studies, ChIP experiments showed that BPTF co-localized with H3K4me3 and H4K16ac at the HOXA9 gene locus in HEK293 cells. To detect whether the co-occurrence of these marks occurred in cells, mononucleosomes were isolated by sucrose-gradient fractionation of MNase fragmented chromatin from HEK293 nuclei. The simultaneous presence of H3K4me3 and H4K16ac was indeed confirmed. Pull-downs with these purified mononucleosomes confirmed binding to BPTF as well, with no detectable binding to H3K12ac.75
Atypical Functions for Tandem Domains
Trim28 (PHD-bromodomain)
BPTF and TRIM24 both contain tandem PHD fingers and bromodomains that independently recognize histone modifications and provide higher order recognition of histone modifications. There are other functions, however, for PHD fingers and bromodomains in tandem that are independent of histone tail reading functions. The protein KAP1 (TIF-1β, TRIM28) is a corepressor for the KRAB domain-containing zinc finger proteins, a family of transcriptional silencers. Similar to TRIM24, its N-terminal RING domain, B-box zinc fingers, and coiled-coil domains are responsible for KRAB domain binding.78 The C-terminal HP1-binding domain and tandem PHD-bromodomain are required for gene silencing. The tandem PHD-bromodomain is responsible for the recruitment of the H3K9 histone methyltransferase (HMTase) SETDB1 and NuRD protein CHD3 to the promoters of KRAB regulated genes.79,80 Ivanov et al. found that the PHD domain of KAP1 acts in a similar way to the RING finger of the PIAS small ubiquitin-like modifier (SUMO) E3 ligases.81 This leads to intra-molecular SUMOylation and subsequently, KRAB-KAP1-mediated gene repression. A structure–function study showed that the tandem PHD-bromodomain adopted a different structure from that of BPTF, in which the tandem domains are connected by a rigid-three turn α-helix, separating the domains by ∼20 angstroms.82 Despite conservation of their individual structural folds, the PHD finger and bromodomain are not able to bind methyl-lysine and acetyl-lysine, respectively. The conserved tyrosine/asparagine pair present in bromodomain binding sites has been substituted by leucine/threonine, possessing drastically different properties.83 While most bromodomains contain an amphipathic helix αZ, the corresponding region in KAP1 contains a stretch of hydrophobic amino acids that form the anchor for the tandem domain interface. Analysis of the tandem domain and its interaction with the E2 ligase UBC9 by 1H-15N HSQC NMR showed major chemical shift perturbations localized to the tandem domain interface, indicating sites of UBC9 binding.82 Binding of UBC9 to the tandem domains led to autoSUMOylation on the bromodomain, where the major modification sites, K779 and K804, are located. ChIP studies showed that KAP1 binding led to a recruitment of SETDB1 and an increase in H3K9me3, a heterochromatin mark, in the promoter region of a reporter gene. Recruitment of SETDB1 was abrogated with transfection of lysine mutants, which were not capable of being SUMOylated.81
Mll1 (PHD-bromodomain)
Another example in which loss of histone reading capabilities by a tandem domain has led to alternative functionality is MLL1, mixed lineage leukemia 1. MLL1 is essential for embryonic development and hematopoiesis Rearrangement of MLL1, located on chromosome 11q23, is responsible for a variety of leukemias, including acute lymphoblastic leukemia (ALL) and acute myelogenous leukemia (AML).84,85 In fact, the most frequent translocations in ALL are t(4;11) and t(11;19), associated with expression of MLL-AF4 and MLL-ENL, respectively, and a pro-B cell or mixed lineage phenotype.85 The protein is a member of the trithorax group of chromatin modifiers that antagonize the polycomb group of proteins to regulate gene expression in development.86,87 It contains a C-terminal H3K4 HKMTase SET domain as well as a tandem PHD-bromodomain and FYRN and FYRC domains.88 A recent study, which solved the structure of the PHD-bromodomain segment, showed that the two domains form an integrated structure, unlike seen in BPTF.89 Notably, the linker segment between the domains contained a proline (P1629) in the cis position, facilitating a compact, globular fold between the two domains with the αZ helix of the bromodomain anchoring the interaction. Despite the structural similarities to KAP1, however, the PHD finger showed binding to H3K4me3 (Kd = 4.3 μM) and H3K4me2 (Kd = 6.9 μM). The PHD finger alone showed much weaker binding to the histone marks as compared to the tandem domains. The bromodomain, in similar fashion to KAP1, was unable to bind acetylated histone peptides, perhaps due to the substitution of the conserved asparagine in the binding pocket to aspartate. It was previously found that the PHD finger adjacent to the bromodomain specifically interacts with the suppressor protein CyP33, a cyclophilin containing an N-terminal RNA recognition motif (RRM) and C-terminal peptidyl prolyl isomerase (PPIase) domain, and accordingly leads to an increase in HDAC1 binding to the MLL1 repression domain.90,91 Wang et al. found an interaction between the Cyp33 RRM domain and the MLL1 tandem PHD-bromodomain, but only in the presence of the CyP33 PPIase domain. Further studies showed that the PPIase domain facilitated a cis-trans isomerization of P1629 of MLL1, uncovering an interaction surface on the PHD domain for interaction with CyP33 RRM. The PHD domain was simultaneously able to bind RRM and H3K4me2/3. In agreement with structural studies, ChiP with anti-acetyl H3 antibodies showed a decrease in acetylation at HOXA9 and HOXC8, known MLL1 targets, when wild-type CyP33 was expressed in 293T cells. However, transfection with mutant CyP33, that showed no interaction with MLL1, showed no decrease in acetylation. This is perhaps due to the diminished Cyp33 dependent recruitment of HDAC1 to MLL target genes.89
P300/CBP (Bromodomain-RING-PHD)
CBP and p300 are a pair of closely related HATs that are crucial for coactivation of a number of genes.92 They are large, multi-modular proteins that contain several defined domains, including the bromodomain, PHD finger, CH1 (Taz1), KIX, CH3 (ZZ-Taz2), and HAT domains.92 The HAT domain, responsible for HAT activity, is flanked by the bromodomain and PHD finger. A recent study identified the structure of the bromodomain-PHD finger-HAT tandem module from p300,93 while another identified the bromodomain-PHD finger from CBP.94 The crystal structure of p300 revealed a discontinuous PHD finger, in which a RING domain was inserted just after the bromodomain [Fig. 2(B)], which the CBP structure was able to confirm. The bromodomain, RING domain, PHD finger, and HAT domain form a compact unit in which the RING domain displays contact with the HAT substrate-binding site. Mutations and deletions of the RING and PHD fingers lead to increased HAT autoacetylation and p53 acetylation, indicating a negative regulatory role for these domains on HAT activity. Furthermore, mutations present in Rubenstein-Taybi syndrome, a developmental condition that leads to learning difficulties and unusual facial features, lead to retention of HAT activity but increased autoacetylation as well.93,95,96 Interestingly, both structural studies could not find a histone binding partner for the PHD finger, despite extensive experiments with peptide arrays and NMR titration experiments. This is perhaps due to the absence of key aromatic residues that are present in the binding pocket of PHD fingers that interact with a histone substrate [Fig. 2(C)]. The PHD finger may instead play a structural and regulatory role in histone acetylation and HAT autoacetylation. It cannot be ruled out, however, that the PHD finger has an as-of-yet unidentified non-histone binding partner.
Future Work
While our understanding of the recognition of single and double marks has greatly improved, our knowledge of how more than two marks function in control of gene transcription in chromatin is limited. For example, simultaneous readers of enhancer marks (H3K4me1/2 and H3K27ac/me3) and bivalent promoter marks (H3K4me3/H3K27me3), have yet to be uncovered.97,98 Moving forward, it will be necessary to look at the structure and function of complexes that contain more than two consecutive domains to uncover these functions. For example, the trithorax family protein, ASH1L, has a region with a consecutive bromodomain, PHD finger, and bromodomain adjacent homology (BAH) domain, found to bind HDAC1.99 The leukocyte specific protein, SP140L, is important in pathogenesis of acute promyelocytic leukemia (APL) and viral infection and contains a region with consecutive SAND domain, PHD finger, and bromodomain.100 These two proteins also lack the conserved asparagine residue found to be essential for acetyl-lysine binding in most bromodomains, suggesting novel functions as performed by KAP1 and MLL1.
Structural biology, in combination with cell biology, has greatly increased our understanding of how epigenetics impacts normal and pathologic cell function. By understanding the molecular mechanisms by which histones are modified and recognized, we can begin to develop smart epigenetic therapies to target changes in pathologic protein function, such as the way the kinase field was revolutionized with the discovery of the bcr-abl specific imatinib to treat chronic myelogenous leukemia (CML).101 Recently, specific inhibitors of bromodomains were found for cancer therapy and male contraception.102,103 We are pursuing further studies of consecutive domain function and hope to apply this knowledge to rational drug design for epigenetic dysfunction in human disease.
Glossary
- BAH
bromodomain adjacent homology
- CHD
chromo helicase DNA-binding
- CML
chronic myelogenous leukemia
- ES
embryonic stem
- HAT
histone acetyltransferase
- HP-1
heterochromatin protein-1
- PHD
plant homeodomain
- RRM
RNA recognition motif
- TTD
tandem tudor domain
References
- 1.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis C. Translating the histone code. Science (New York, NY) 2001;293:1074. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Sims RJ, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- 5.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, Lu Z, Ye Z, Zhu Q, Wysocka J, Ye Y, Khochbin S, Ren B, Zhao Y. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 7.Li HT, Ilin S, Wang WK, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pena PV, Davrazou F, Shi XB, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ing2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED. Structure of the hp1 chromodomain bound to histone h3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 10.Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Ann Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs SA, Khorasanizadeh S. Structure of hp1 chromodomain bound to a lysine 9-methylated histone h3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 13.Seet BT, Dikic I, Zhou M-M, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 14.Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, Linder-Basso D, Plachetka A, Shanower G, Tolstorukov MY, Luquette LJ, Xi R, Jung YL, Park RW, Bishop EP, Canfield TP, Sandstrom R, Thurman RE, MacAlpine DM, Stamatoyannopoulos JA, Kellis M, Elgin SCR, Kuroda MI, Pirrotta V, Karpen GH, Park PJ. Comprehensive analysis of the chromatin landscape in drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Ann Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 17.Lee J-S, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Patel DJ. Combinatorial readout of dual histone modifications by paired Chromatin-associated modules. J Biol Chem. 2011;286:18363–18368. doi: 10.1074/jbc.R111.219139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human tafii250 double bromodomain module. Science (New York, NY) 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 23.Morinière J, Rousseaux S, Steuerwald U, Soler-López M, Curtet S, Vitte A-L, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Müller CW, Petosa C. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- 24.Chakravarty S, Zeng L, Zhou M-M. Structure and site-specific recognition of histone h3 by the PHD finger of human autoimmune regulator. Struct/Fold Design. 2009;17:670–679. doi: 10.1016/j.str.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone h3 lysine 4 links bhc80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez R, Zhou M-M. The PHD finger: a versatile epigenome reader. Trend Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng L, Zhang Q, Li S, Plotnikov AN, Walsh MJ, Zhou M-M. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature. 2010;466:258–262. doi: 10.1038/nature09139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange M, Kaynak B, Forster UB, Tönjes M, Fischer JJ, Grimm C, Schlesinger J, Just S, Dunkel I, Krueger T, Mebus S, Lehrach H, Lurz R, Gobom J, Rottbauer W, Abdelilah-Seyfried S, Sperling S. Regulation of muscle development by dpf3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Gene Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas T, Corcoran LM, Gugasyan R, Dixon MP, Brodnicki T, Nutt SL, Metcalf D, Voss AK. Monocytic leukemia zinc finger protein is essential for the development of long-term reconstituting hematopoietic stem cells. Gene Dev. 2006;20:1175–1186. doi: 10.1101/gad.1382606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borrow J, Stanton VP, Andresen JM, Becher R, Behm FG, Chaganti RSK, Civin CI, Disteche C, Dube I, Frischauf AM, Horsman D, Mitelman F, Volinia S, Watmore AE, Housman DE. The translocation t(8;l6)(p11, p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 31.Chaffanet M, Gressin L, Preudhomme C, Soenen-Cornu V, Birnbaum D, Pebusque MJ. MOZ is fused to p300 in an acute monocytic leukemia with t(8;22) Gene Chromosome Canc. 2000;28:138–144. doi: 10.1002/(sici)1098-2264(200006)28:2<138::aid-gcc2>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Carapeti M, Aguiar RCT, Goldman JM, Cross NCP. A novel fusion between MOZ and the nuclear receptor coactivator tif2 in acute myeloid leukemia. Blood. 1998;91:3127–3133. [PubMed] [Google Scholar]
- 33.Ullah M, Pelletier N, Xiao L, Zhao SP, Wang K, Degerny C, Tahmasebi S, Cayrou C, Doyon Y, Goh SL, Champagne N, Cote J, Yang XJ. Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol Cell Biol. 2008;28:6828–6843. doi: 10.1128/MCB.01297-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiu Y, Liu L, Zhao C, Han CC, Li FD, Zhang JH, Wang Y, Li GH, Mei YD, Wu MA, Wu JH, Shi YY. Combinatorial readout of unmodified h3r2 and acetylated h3k14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for hoxa9 transcription. Gene Dev. 2012;26:1376–1391. doi: 10.1101/gad.188359.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He C, Li FD, Zhang JH, Wu JH, Shi YY. The methyltransferase nsd3 has Chromatin-binding motifs, PHD5-C5HCH, that are distinct from other NSD (nuclear receptor SET domain) family members in their histone h3 recognition. J Biol Chem. 2013;288:4692–4703. doi: 10.1074/jbc.M112.426148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P. nsd3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics. 2001;74:79–88. doi: 10.1006/geno.2001.6524. [DOI] [PubMed] [Google Scholar]
- 37.Kurotaki N, Imaizumi K, Harada N, Masuno M, Kondoh T, Nagai T, Ohashi H, Naritomi K, Tsukahara M, Makita Y, Sugimoto T, Sonoda T, Hasegawa T, Chinen Y, Tomita H, Kinoshita A, Mizuguchi T, Yoshiura K, Ohta T, Kishino T, Fukushima Y, Niikawa N, Matsumoto N. Haploinsufficiency of nsd1 causes sotos syndrome. Nat Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 38.Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, Larratt LM, Mant MJ, Reiman T, Belch AR, Pilarski LM. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–4069. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosati nup98 is fused to the nsd3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) (vol 99, pg 3857, 2001) Blood. 2002;100:1132–1132. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 40.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-nsd1 links h3k36 methylation to Hox-a gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–U134. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 41.Flanagan JF, Mi L-Z, Chruszcz M, Cymborowski M, Clines KL, Kim Y, Minor W, Rastinejad F, Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone h3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 42.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ e3 ubiquitin ligases. BioEssays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 43.Tsai W-W, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai C-Y, Shi X, Schwarzer D, Plunkett W, Aronow B, Gozani O, Fischle W, Hung M-C, Patel DJ, Barton MC. trim24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren W-J, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung W-K, Liu ET, Wei C-L, Cheung E, Ruan Y. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Bassets I, Kwon Y-S, Telese F, Prefontaine GG, Hutt KR, Cheng CS, Ju B-G, Ohgi KA, Wang J, Escoubet-Lozach L, Rose DW, Glass CK, Fu X-D, Rosenfeld MG. Histone Methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thénot S, Henriquet C, Rochefort H, Cavaillès V. Differential interaction of nuclear receptors with the putative human transcriptional coactivator hTIF1. J Biol Chem. 1997;272:12062–12068. doi: 10.1074/jbc.272.18.12062. [DOI] [PubMed] [Google Scholar]
- 47.Pandolfi PP, Zhong S, Delva L, Rachez C, Cenciarelli C, Gandini D, Zhang H, Kalantry S, Freedman LP. A RA-dependent, tumour-growth suppressive transcription complex is the target of the PML-RAR|[alpha]| and t18 oncoproteins. Nat Genet. 1999;23:287–295. doi: 10.1038/15463. [DOI] [PubMed] [Google Scholar]
- 48.Klugbauer S, Rabes H. The transcription coactivator htif1 and a related protein are fused to the RET receptor tyrosine kinase in childhood papillary thyroid carcinomas. Oncogene. 1999;18 doi: 10.1038/sj.onc.1202824. [DOI] [PubMed] [Google Scholar]
- 49.Belloni E, Trubia M, Gasparini P, Micucci C, Tapinassi C, Confalonieri S, Nuciforo P, Martino B, Lo-Coco F, Pelicci PG, Di Fiore PP. 8p11 myeloproliferative syndrome with a novel t(7;8) translocation leading to fusion of theFGFR1 andTIF1 genes. Genes, Chromosomes Cancer. 2005;42:320–325. doi: 10.1002/gcc.20144. [DOI] [PubMed] [Google Scholar]
- 50.Brown R, Strathdee G. Epigenomics and epigenetic therapy of cancer. Trend Mol Med. 2002;8:S43–S48. doi: 10.1016/s1471-4914(02)02314-6. [DOI] [PubMed] [Google Scholar]
- 51.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 52.Zhang G, Sanchez R, Zhou M-M. Scaling the druggability landscape of human bromodomains, a new class of drug targets. J Med Chem. 2012;55:7342–7345. doi: 10.1021/jm3011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xi QR, Wang ZX, Zaromytidou AI, Zhang XHF, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, Studer L, Mark W, Patel DJ, Massague J. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morsut L, Yan KP, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, Dupont S, Losson R, Piccolo S. Negative control of smad activity by ectodermin/tif1 gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 55.Vincent DF, Yan KP, Treilleux I, Gay F, Arfi V, Kaniewsky B, Marie JC, Lepinasse F, Martel S, Goddard-Leon S, Iovanna JL, Dubus P, Garcia S, Puisieux A, Rimokh R, Bardeesy N, Scoazec JY, Losson R, Bartholin L. Inactivation of tif1 gamma cooperates with kras(G12D) to induce cystic tumors of the pancreas. Plos Genet. 2009;5 doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Gene Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon SH, Workman JL. The heterochromatin protein 1 (hp1) family: put away a bias toward hp1. Mol Cell. 2008;26:217–227. [PubMed] [Google Scholar]
- 58.Ruthenburg AJ, Li H, Patel DJD, Allis C. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. uhrf1 plays a role in maintaining DNA methylation in mammalian cells. Science (New York, NY) 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 60.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human uhrf1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- 61.Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, Wang YX, Bronner C, Chedin F, Arrowsmith CH, Dhe-Paganon S. Recognition of multivalent histone states associated with heterochromatin by uhrf1 protein. J Biol Chem. 2011;286:24300–24311. doi: 10.1074/jbc.M111.234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, Arrowsmith CH, Strahl BD. Association of uhrf1 with methylated h3k9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, Shi YG, Xu Y, Patel DJ, Shi Y. PHD finger recognition of unmodified histone h3r2 links uhrf1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of uhrf1 and insights into recognition of unmodified histone h3 arginine residue 2. Nat Publishing Group. 2011;21:1374–1378. doi: 10.1038/cr.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang C, Shen J, Yang Z, Chen P, Zhao B, Hu W, Lan W, Tong X, Wu H, Li G, Cao C. Structural basis for site-specific reading of unmodified r2 of histone h3 tail by uhrf1 PHD finger. Nat Publishing Group. 2011;21:1379–1382. doi: 10.1038/cr.2011.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, Ariyoshi M, Shirakawa M. Recognition of modification status on a histone h3 tail by linked histone reader modules of the epigenetic regulator uhrf1. Proc Natl Acad Sci. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng J, Yang Y, Fang J, Xiao J, Zhu T, Chen F, Wang P, Li Z, Yang H, Xu Y. Structural insight into coordinated recognition of trimethylated histone h3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of uhrf1 (Ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem. 2013;288:1329–1339. doi: 10.1074/jbc.M112.415398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gelato KA, Tauber M, Ong MS, Winter S, Hiragami-Hamada K, Sindlinger J, Lemak A, Bultsma Y, Houliston S, Schwarzer D, Divecha N, Arrowsmith CH, Fischle W. Accessibility of different histone H3-binding domains of uhrf1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol Cell. 2014;54:905–919. doi: 10.1016/j.molcel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 69.Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol Biol Cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim JK, Estève P-O, Jacobsen SE, Pradhan S. uhrf1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acid Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 72.Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 73.Wen H, Li Y, Xi Y, Jiang S, Stratton S, Peng D, Tanaka K, Ren Y, Xia Z, Wu J, Li B, Barton MC, Li W, Li H, Shi X. zmynd11 links histone h3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508:263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo R, Zheng L, Park JW, Lv R, Chen H, Jiao F, Xu W, Mu S, Wen H, Qiu J, Wang Z, Yang P, Wu F, Hui J, Fu X, Shi X, Shi YG, Xing Y, Lan F, Shi Y. Bs69/zmynd11 reads and connects histone h3.3 lysine 36 Trimethylation-decorated chromatin to regulated Pre-mRNA processing. Mol Cell. 2014;56:298–310. doi: 10.1016/j.molcel.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell. 2011;145:692–706. doi: 10.1016/j.cell.2011.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, Wu C, Allis CD. A PHD finger of NURF couples histone h3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 77.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Gene Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 79.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ. Setdb1: a novel KAP-1-associated histone h3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Gene Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schultz DC, Friedman JR, Rauscher FJ. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Gene Dev. 2001;15:428–443. doi: 10.1101/gad.869501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, Sadofsky MJ, Zhou M-M, Rauscher FJ., III PHD Domain-mediated e3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ, Zhou M-M. Structural insights into human kap1 PHD finger-bromodomain and its role in gene silencing. Nat Struct Mol Biol. 2008;15:626–633. doi: 10.1038/nsmb.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanchez R, Zhou M-M. The role of human bromodomains in chromatin biology and gene transcription. Curr Opin Drug Discov Dev. 2009;12:659–665. [PMC free article] [PubMed] [Google Scholar]
- 84.Ayton PM, Cleary ML. Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene. 2001;20:5695–5707. doi: 10.1038/sj.onc.1204639. [DOI] [PubMed] [Google Scholar]
- 85.Hess JL. MLL: a histone methyltransferase disrupted in leukemia. Trend Mol Med. 2004;10:500–507. doi: 10.1016/j.molmed.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 86.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009:1–12. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 87.Schuettengruber B, Martinez A-M, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 88.Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- 89.Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro isomerization in mll1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141:1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fair K, Anderson M, Bulanova E, Mi H, Tropschug M, Diaz MO. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol Cell Biol. 2001;21:3589–3597. doi: 10.1128/MCB.21.10.3589-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Xia Z-B, Anderson M, Diaz MO, Zeleznik-Le NJ. MLL repression domain interacts with histone deacetylases, the polycomb group proteins hpc2 and BMI-1, and the corepressor C-terminal-binding protein. Proc Natl Acad Sci U S A. 2003;100:8342–8347. doi: 10.1073/pnas.1436338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Gene Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 93.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–1046. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 94.Plotnikov AN, Yang S, Zhou TJ, Rusinova E, Frasca A, Zhou MM. Structural insights into Acetylated-histone h4 recognition by the Bromodomain-PHD finger module of human transcriptional coactivator CBP. Structure. 2014;22:353–360. doi: 10.1016/j.str.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rack JG, Lutter T, Kjaereng Bjerga GE, Guder C, Ehrhardt C, Varv S, Ziegler M, Aasland R. The PHD finger of p300 influences its ability to acetylate histone and non-histone targets. J Mol Biol. 2014;426:3960–3972. doi: 10.1016/j.jmb.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 96.Kalkhoven E, Roelfsema JH, Teunissen H, den Boer A, Ariyurek Y, Zantema A, Breuning MH, Hennekam RC, Peters DJ. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-taybi syndrome. Hum Mol Genet. 2003;12:441–450. doi: 10.1093/hmg/ddg039. [DOI] [PubMed] [Google Scholar]
- 97.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanaka Y, Nakayama Y, Taniguchi M, Kioussis D. Regulation of early T cell development by the PHD finger of histone lysine methyltransferase ash1. Biochem Biophys Res Commun. 2008;365:589–594. doi: 10.1016/j.bbrc.2007.10.159. [DOI] [PubMed] [Google Scholar]
- 100.Bloch DB, de la Monte SM, Guigaouri P, Filippov A, Bloch KD. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 101.Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the abl tyrosine kinase on the growth of Bcr-abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 102.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matzuk MM, McKeown MR, Filippakopoulos P, Li Q, Ma L, Agno JE, Lemieux ME, Picaud S, Yu RN, Qi J, Knapp S, Bradner JE. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]


