Abstract
Tyrosinases are metalloenzymes belonging to the type-3 copper protein family which contain two copper ions in the active site. They are found in various prokaryotes as well as in plants, fungi, arthropods, and mammals and are responsible for pigmentation, wound healing, radiation protection, and primary immune response. Tyrosinases perform two sequential enzymatic reactions: hydroxylation of monophenols and oxidation of diphenols to form quinones which polymerize spontaneously to melanin. Two other members of this family are catechol oxidases, which are prevalent mainly in plants and perform only the second oxidation step, and hemocyanins, which lack enzymatic activity and are oxygen carriers. In the last decade, several structures of plant and bacterial tyrosinases were determined, some with substrates or inhibitors, highlighting features and residues which are important for copper uptake and catalysis. This review summarizes the updated information on structure–function correlations in tyrosinases along with comparison to other type-3 copper proteins.
Keywords: tyrosinase, catechol oxidase, hemocyanin, copper, type-3 copper proteins, X-ray structure
Introduction
Tyrosinases (EC 1.14.18.1) are copper-containing enzymes which exist in all life domains.1 They can be found in various prokaryotes as well as in plants, fungi, arthropods, and mammals. Tyrosinases are the catalysts in mammals responsible for the formation of melanin in skin and hair color, as well as browning in fruit and vegetables following cell damage. In plants, sponges, and many invertebrates, tyrosinases are important for wound healing and primary immune responses; in arthropods, they play a role in sclerotization, and in bacteria, tyrosinases protect DNA from UV damage.1–4
Using molecular oxygen, tyrosinase performs two sequential enzymatic reactions: the ortho-hydroxylation of monophenols (monophenolase or cresolase activity) and the oxidation of o-diphenols (diphenolase or catecholase activity) to the corresponding quinones. The reactive quinones then polymerize spontaneously to melanins.5 Because of their ability to oxidize small phenolic molecules as well as tyrosine residues in peptides and proteins, tyrosinases have found many applications in biotechnology, such as bioremediation, dye production, and biopolymer cross-linking.2
Tyrosinases belong to the “type-3 copper” family along with catechol oxidases (CO)—which perform only diphenolase activity—and hemocyanins (Hc)—which are oxygen carriers from the hemolymph of molluscs and arthropods.6,7 The members of this family have a conserved active site of six histidine residues, which are located in a four helical bundle, coordinating the two copper ions (CuA and CuB).6 Different spectroscopic and mechanistic studies have confirmed that the spectroscopic properties of the copper ions are similar in all type-3 copper proteins suggesting that the differences in activity may be attributed to the architecture and substrate accessibility of the different members.4–6 Although the catalytic mechanism of tyrosinases was intensively studied, and structural data of CO has been known since 1998,8 some mechanistic aspects are still a subject of debate. Several new structures of type-3 copper enzymes have been published in the past decade enabling new insights into structure–function links (Table I). This review will focus mainly on recently published works, emphasizing structural and functional aspects of tyrosinases and their similarity and discrepancy to other type-3 copper proteins.
Table I.
Structural Data, Conserved Residues of Catalytic Importance, and Activity Classification of Type-3 Copper Proteins
| Protein | Organism | PDB | Thioether bond | Placeholder | Blocker residue | Conserved Met | Conserved water | Conserved Glu/Asn | Activitya | References |
|---|---|---|---|---|---|---|---|---|---|---|
| Tyrosinase (TyrSc) | Streptomyces castaneoglobisporus | 1WX2b | No | Tyr98 (caddie) | Gly204 | Met201 | HOH431 | Glu182 Asn191 | ++ | Matoba et al., (2006)16 |
| Tyrosinase (TyrBm) | Bacillus megaterium | 3NM8b | No | No | Val218 | Met215 | HOH376 | Glu195 Asn205 | ++ | Sendovski et al., (2010)17 |
| Tyrosinase (PPOVv) | Vitis vinifera | 2P3X | Yes | Cleaved | Phe259 | Met256 | No | Glu235 Asn240 | ++ | Virador et al., (2010)55 |
| Tyrosinase (abPPO3) | Agaricus biporus | 2Y9Xb | Yes | Cleaved | Val283 | Met280 | HOH2095 | Glu256 Asn260 | ++ | Ismaya et al., (2011)44 |
| Tyrosinase (abPPO4) | Agaricus biporus | 4OUA | Yes | Phe454 | Ala270 | Met267 | HOH527 | Glu248 Asp252 | ++ | Mauracher et al., (2014)31 |
| Tyrosinase (TyrAo) | Asperugillus oryzae | 3W6Qb | Yes | Phe513 | Val359 | Met356 | HOH1103 | Glu325 Asn329 | ++ | Fujieda et al., (2013)32 |
| Catechol oxidase (IbCO) | Ipomoea batatas | 1BT3b | Yes | Cleaved | Phe261 | Met258 | HOH1007 | Glu236 Ile241 | + | Klabunde et al., (1998)8 |
| Catechol oxidase (AoCO4) | Asperugillus oryzae | 4J3Pb | No | No | Val299 | Met291 | HOH2198 | Gln273 Gly285 | + | Hakulinen et al., (2013)26 |
| Pro-phenoloxidase (proPOMc) | Manduca sexta | 3HHS | No | Phe85/Phe88 | Ser393/Glu395 | Met390 | HOH781 HOH700 | Glu351/353 Asn367/369 | ++ | Li et al., (2009)51 |
| Pro-phenoloxidase (proPOc) | Marsupenaeus japonicus | 3WKY | No | Phe72 | Val384 | Met381 | HOH831 | Glu343 Asn358 | ++ | Masuda et al., (2014)46 |
| Hemocyanin (Lobster Hc) | Panulirus interruptus | 1HCYb | No | Phe75 | Phe371 | Met368 | HOH779 | Glu329 Asn345 | + | Volbeda et al., (1989)47 |
| Hemocyanin (Crab Hc) | Limulus polyphemus | 1NOLb | No | Phe49 | Thr351 | Met348 | HOH662 | Glu309 Asn325 | ++ | Magnus et al., (1993)7 |
| Hemocyanin (Octopus Hc) | Octopus | 1JS8 | Yes | Leu2830 | Leu2689 | Met2686 | HOH436 | Glu2668 Asn2672 | ++ | Cuff et al., (1998)42 |
| Hemocyanin (Rapana Hc) | Rapana thomasiana | 1LNL | Yes | Leu343 | Leu199 | Met196 | No water molecules in the structure | Glu178 Asn182 | + | Perbandt et al., (2003)43 |
(+) denotes diphenolase activity only, (++) denotes mono- and diphenolase activity.
Several structures were deposited in PDB.
The Overall Structure of Tyrosinases
The overall structure of tyrosinase can be divided into three domains: the central domain, the N-terminal domain, and the C-terminal domain.9 The central domain, which is composed of six conserved histidine residues, contains the CuA and CuB oxidizing ions. Although, it is the most conserved domain among tyrosinases, CO and Hc (Fig. 1), the CuB site exhibits higher conservation than CuA.9–11 Biochemical investigations of tyrosinases from different species have shown the involvement of the conserved histidine residues in copper binding.11–14
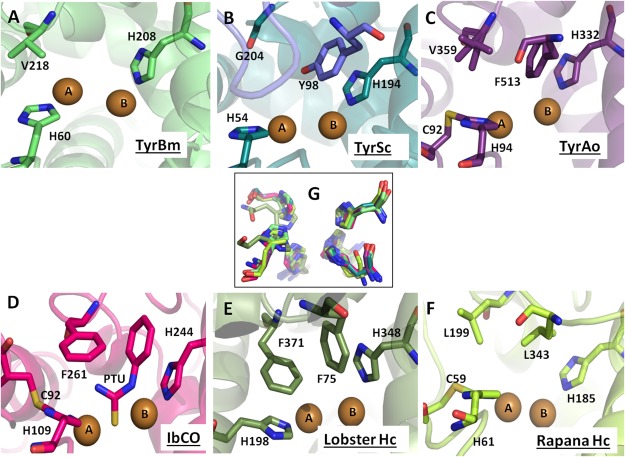
Figure 1. The active site of type-3 copper enzymes.
The active sites of type-3 copper proteins are shown in cartoon, active site residues are shown in sticks, and copper atoms are presented as brown spheres. (A) TyrBm active site and its blocker residue Val218 is shown in lime green. (B) The active site of TyrSc and its blocker residue Gly204 is shown in deep teal while the placeholder Tyr98 from the caddie protein is in blue. (C) The active site of TyrAo including Cys92 and His94 (thioether bond), a blocker residue Val359, and a placeholder Phe513 is presented in purple. (D) The active site of IbCO containing a thioehter bond between Cys92 and His109, a placeholder Phe261, and an inhibitor PTU is shown in pink. (E) Lobster Hc active site is presented in green with Phe371 and Phe75 as the blocker residue and a placeholder, respectively. (F) Rapana Hc active site is shown in lemon-green, containing a thioehter bond between Cys59 and His61, while Leu199 and Leu343 are the blocker residue and a placeholder, respectively. (G) Superposition of active site histidine residues of all type-3 copper proteins mentioned above. All figures were generated using PyMOL (http://www.pymol.org). Full names, PDB accession codes, and references are listed in Table I.
An unusual thioether bond formed between the second histidine residue coordinating CuA and an adjacent cysteine residue was found in CO of sweet potato (IbCO), tyrosinase from Aspergilus oryzae (TyrAo), and mollusc Hc (Octopus and Rapana Hc) but not in tyrosinases from Streptomyces castaneoglobisporus (TyrSc), Bacillus megaterium (TyrBm), mouse, humans, and arthropodan Hc (Lobster and Crab Hc) (Fig. 1).15–17 It was proposed that the role of the thioether bond is to stabilize the second histidine residue of the active site, which is located on a flexible loop, in contrast to the five other active-site hisitidine residues which are situated on a stable α-helical region.18
The N-terminal domain is a transit peptide which determines the final location of the enzyme and then undergoes proteolytic removal.14,19,20 In plant tyrosinases, the N-terminal peptide directs the enzyme to the chloroplast and, in human and mouse, it was suggested to be involved in melanosome transfer.21 Fungal tyrosinases are cytoplasmic enzymes thus do not contain a transit peptide, although in some cases they are associated with the cell wall.18 In bacterial tyrosinases, a TAT signal peptide which is responsible for proteins secretion was identified at the N-terminal domain in Verrucomicrobium spinosum and Streptomyces.22,23
The latent state of tyrosinase (pro-tyrosinase) is composed of the central domain and the C-terminal domain. The latter domain blocks the entrance into the active site by means of a “placeholder” residue which penetrates the active site similarly to the substrate or an inhibitor [Table I; Fig. 1(B–D)]. In vivo activation of pro-tyrosinases occurs by the proteolytic cleavage of the C-terminal domain, while in vitro activation might be achieved by inducing strong conformational changes using detergents.20 An analogous C-terminal domain is found in CO and in molluscs Hc [Table I; Fig. 1(F)], while in arthropods Hc, the N-terminal domain blocks the active site [Table I; Fig. 1(E)].24 Similar to tyrosinases, CO and several Hc are activated by the removal of the blocking domain.24 TyrSc does not contain the C-terminal domain, but its active site is covered by a placeholder provided from an associated “caddie” protein [Table I; Fig. 2(B)].16 In TyrBm, the C-terminal domain does not exist but in contrast to TyrSc, the active site is exposed and the enzyme is always found in its active form [Fig. 2(A)].17 Recently, a new group of short tyrosinase-like proteins was discovered and these proteins do not have a C-terminal domain.25 However, the active site of CO from Asperugillus oryzae (AoCO4), which belongs to this group, is not exposed to the surface, in contrast to TyrBm.26
Figure 2. Oxidation states of tyrosinase.
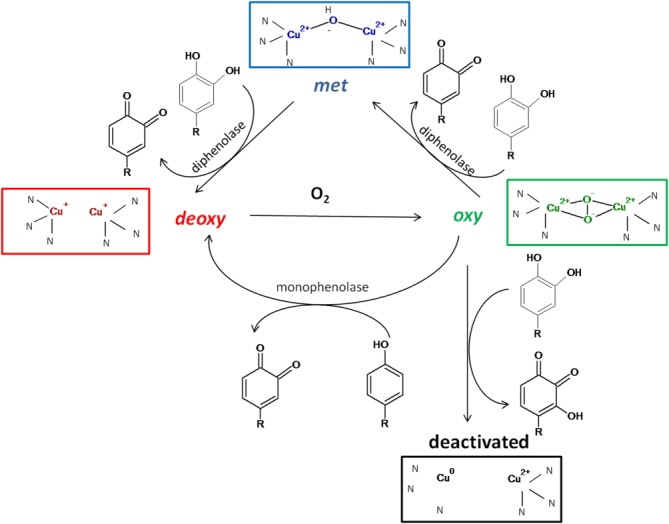
Four discrete states of the active site of tyrosinase are presented based on Ramsden and Riley (2014).5
Catalytic Mechanism
The properties of tyrosinase are determined by four oxidation states of its active site and were described in detail by Ramsden et al. (Fig. 2).5 Native tyrosinase is mostly found in the met state in which a hydroxyl ion is ligated by two Cu(II). In contrast to catechols, phenols cannot be oxidized by this form of tyrosinase. Catechol oxidation by the met state results in tyrosinase reduction to the deoxy state, in which both copper ions are reduced to Cu(I). The deoxy state binds oxygen to become the oxy state, in which two oxygen molecules in the peroxy configuration are ligated by two Cu(II). Deoxy tyrosinase oxidizes phenols and catechols to o-quinones, by different oxidative mechanisms. Catechol oxidation by a phenolic oxidative mechanism, results in reduction of copper to Cu0 and tyrosinase deactivation (suicide inactivation).4,5,27,28
Copper in the Binding Site
By comparing the oxygenated and deoxygenated structures of Crab Hc, it was observed that upon oxygenation, the distance between the copper ions decreased while the position of the histidine residues remained unchanged.7 Similar results were observed in the structure of IbCO and TyrSc.8,16 These observations indicate that during catalysis, both the oxidation state of Cu and its location are changed.
Structure determination of TyrSc in complex with a caddie protein revealed that while CuB is structurally stable in the active site, CuA and its ligating histidine residue His54 are flexible [Figs. 1(B) and 3(A)]. The authors suggested that flexibility of CuA and His54 derived from the fact that His54 is not restricted by a thioether bond and this flexibility might have functional implications.16 Using structural and biochemical data, Matoba et al., proposed a mechanism for copper transport in TyrSc.29 The authors showed that Cu is transferred from the caddie protein through His82, Met84, and His97 to the flexible His54 of TyrSc, which changes its conformation to coordinate the copper in the active site [Fig. 3(A)]. When copper reaches the active site, it first occupies the stable CuB position and then the CuA position.
Figure 3. Copper incorporation pathway.

A) Copper transport from the caddie protein (blue color) to TyrSc (deep teal). B) The active site and second shell residues of TyrBm participating in copper transport. The active site and the second shell residues of TyrBm participating in copper transport. (C) The conserved motif of Cys residues participating in copper transport in TyrAo. (D) Two positions of CuB in abPPO4. Full names, PDB codes and references are listed in Table I.
Copper plasticity was also observed in the structure of TyrBm.17 CuA showed flexibility over a range of 2Å concomitant with its coordinating residue, His60. In contrast to TyrSc, in the TyrBm structure CuB was not observed at all.17 The fact that TyrBm does not possess a caddie protein suggests a different mechanism for copper incorporation. Kanteev et al. have proposed that CuA and CuB enter the active site through different supporting residues.30 CuA transport is facilitated by two methionine residues Met61 and Met184 [Fig. 3(B)]. These residues transfer copper ions to the flexible His60, which in a similar way to His54 in TyrSc, positions the copper ion in the active site. On the other hand, it was proposed that CuB enters the active site through conserved Asn205 and Phe197, while the role of Asn205 is to stabilize His204, allowing efficient coordination of CuB in the active site [Fig. 3(B)].30
In the structures of Agaricus biporus tyrosinase (abPPO4), copper flexibility was observed as well. However in contrast to TyrBm and TyrSc, CuA was structurally stable but CuB exhibited flexibility and an additional conformation, next to a fourth histidine residue His282 [Fig. 3(D)].31 It is possible that CuB flexibility in abPPO4 is a result of the replacement of a conserved Asn with Asp252 (Table I). This Asp residue does not form polar interactions with His251which coordinates CuB in the active site; consequently, CuB is flexible in the active site [Fig. 3(B vs. D)].
Structural investigations of tyrosinase from Aspergillus oryzae (TyrAo) have shown that the thioether bond between His94 and Cys92 are formed only in the presence of copper in the active site, while replacing Cys92 with Ala significantly decreased copper binding.32 In addition, a conserved motif of C522XXC525, similar to that found in copper chaperons, was observed in the C-terminal domain of TyrAo [Fig. 3(C)]. The replacement of these Cys residues to Ala also resulted in lower copper binding. It was suggested that the role of the C-terminal domain in TyrAo is not only to control enzymatic activity but also to facilitate copper transport into the active site, similar to chaperons.32 The conserved motifs of the Cys residues were also observed in Octopus Hc and IbCO, suggesting the existence of a similar mechanism of copper incorporation in these proteins.32 The role of Cys97 was also analyzed in aurone synthase from Coreopsis grandiflora (cgAUS1). However, in contrast to TyrAo, in cgAUS1, the replacement of Cys97 to Ala affected protein folding and hindered its activity, but did not have any effect on copper binding.13
Although little is known about copper transport in type-3 copper proteins, it seems that each protein has a different mechanism for copper incorporation depending on its structure and surroundings. Tyrosinases from mammalian and yeast origin receive copper from chaperons and transporters, which use Cys and Met motifs for copper incorporation.33,34 Others like TyrAo and TyrSc implement chaperon-like motifs in the C-terminal domains or by the aid of a caddie protein. Proteins similar to TyrBm which do not have a C-terminal domain or a caddie protein incorporate copper through Met and His residues found on their surface.
Although spectroscopic studies have shown that the active site behavior of type-3 copper proteins is similar during the catalytic cycle, the observed variations in copper plasticity are most likely due to the fact that different proteins have different mechanisms of copper incorporation and regulation.6
A Conserved Methionine Residue
Conserved methionine motifs are often found in copper transporters and chaperons, and are responsible for copper transport.35 Recently, it was shown that methionine interactions with aromatic amino acid residues stabilize the protein structure.36 In addition, the sulfur atom in methionine residues is easily oxidized to MetO or irreversibly MetO2.35,37 Several studies have shown that mutating a conserved Met374 in mammalian tyrosinase results in an inactive protein.38 This methionine (M215 in TyrBm) is highly conserved in type-3 copper proteins, and is located 3.4-5Å away from the second coordinating CuB histidine residue [Table I and Fig. 4(A)]. Using a very sensitive electrochemical method, it was shown that H2O2 is produced during tyrosinase activity which is then utilized as a cofactor in o-quinone generation.39 Furthermore, while low H2O2 concentrations activated tyrosinase activity, high H2O2 concentrations prevented its activity and subsequently hair melanogenesis.39,40 Computational analysis suggested that elevated H2O2 concentrations resulted in formation of a Met374O complex, which disturbs the orientation of the imidazole ring of His367 (the second coordinating CuB histidine residue).40 In this orientation, His367 cannot properly orient substrate entrance, which results in activity loss.6,16,40,41 Apparently, the role of this conserved methionine is to stabilize the orientation of the active site histidine residue, enabling tyrosinase activity. However, it is also possible that during the catalytic reaction, the generated H2O2 is trapped by this conserved methionine to prevent its harmful effect on the cell and to control tyrosinase activity and the rate of melanogenesis.39
Figure 4. Second shell residues of TyrBm.

(A) A conserved methionine in a distance of 3.9Å from the second histidine residue coordinating CuB is important for stabilizing this His residue and enabling tyrosinase activity. (B) A structurally conserved water molecule participating in substrate deprotonation is ligated by conserved Glu196 and Asn205 residues. See Table I for respective residues in other type-3 copper proteins.
Active Site Shielding
The active site of tyrosinases as well as most type-3 copper proteins is covered by a “placeholder” provided by an additional protein domain (Table I; Fig. 1).9,23 Usually, the placeholder is a bulky aromatic residue such as Phe or Tyr but a Leu residue may also act as a placeholder as in Octopus and Rapana Hc [Table I; Fig. 1(F)].7,16,31,32,42,43 The placeholder is located above the active site and is oriented parallel to the second coordinating CuB histidine residue, in similar orientation to the synthetic PTU inhibitor in the structure of IbCO [Fig. 1(D)] or tropolone inhibitor in the structure of tyrosinase from Agaricus biporus (abPPO3) (Table I).8,44 Since in most cases enzymatic activity is possible only after the removal of the placeholder, it was suggested that its role is to control enzymatic activity and to prevent an undesirable oxidation of phenolic compounds. TyrSc, in contrast to other known type-3 copper proteins, dissociates from its caddie protein without proteolytic cleavage or detergent use, a fact that enabled investigations of the placeholder.16,29 Replacing Tyr98 of the caddie protein with a Phe residue resulted in decreased activity. Determining the crystal structures of TyrSc variants, Matoba et al. suggested that Tyr98 forms a hydrogen bond network with structurally important water molecules which facilitate copper binding [Fig. 1(B)].16,29 Thus, in addition to the suggested role of the placeholder to control enzymatic activity, this position might also be important for protein maturation.
An additional position which covers the active site is located above the CuA and is mentioned in the literature as a blocker residue (Table I; Fig. 1).19 A bulky Phe261 at this position in IbCO was proposed to hinder the monophenoalse activity in CO. It was suggested that monophenol substrates bind to CuA and diphenol substrates to CuB.5,14 Furthermore, directed evolution on TyrBm showed that when the access to CuB is hindered by mutation R209H, the diphenolase activity decreases while the monophenolase activity increases.45 In addition, the comparison between a Marsupenaeus japonicus prophenoloxidase (POc) and lobster Hc (Table I) revealed that although the structures of both enzymes are very similar, only POc exhibited mono and diphenolase activity after SDS activation, while Hc was not active at all. The difference between those enzymes was attributed to the presence of a blocker residue Phe371in crustacean Hc in contrast to Val384 in POc.46,47
In contrast to previous observations, when a blocker residue Val218 in TyrBm was replaced with a bulky Phe, the monophenolase activity surprisingly increased and the diphenolase activity decreased [Fig. 1(A)].48 The crystal structure of variant V218F exhibited a conformational change of Phe218, which allowed the monophenoalse activity to occur, while the decrease in dipehnolase activity was attributed to the suicide inactivation due to the incorrect orientation of a diphenolic substrate.27,28,41 Moreover, when a blocker residue Phe273 was replaced to Ala in cgAUS, the enzyme lost its activity.13
It is evident, therefore, that the blocker residue participates in substrate orientation; however, while in some enzymes its role is extremely important, in others, it is negligible.
Substrate Deprotonation
Different studies have shown that a proton has to be removed from the monophenol substrate prior to hydroxylation.49,50 Several mechanisms have been proposed for the substrate deprotonation scenario.50,51 However, recent QM/MM studies have shown that the movement of the proton to a structurally conserved water molecule and then to a conserved glutamic acid is the most energetically feasible mechanism.52 Goldfeder et al. proposed that in TyrBm, this water molecule is activated by two conserved residues Glu195 and Asn205, and acts as a base abstracting the proton from monophenols [Fig. 4(B)].53 This conserved water molecule is found in all type-3 copper protein structures known to date (Table I).
Substrate Orientation in the Binding Site
The existence of two catalytic activities in the active site of tyrosinase in contrast to only diphenolase activity of CO together with the presence of a bulky Phe residue above CuA leads to the suggestion that monophenols bind to CuA while diphenols bind to CuB [Fig. 1(D)].5,14 However, recently determined TyrBm crystal structures with monophenol and diphenol substrates in the active site disproved this assumption. It was shown that tyrosine and l-dopa were similarly oriented toward CuA through π–π interactions with the second coordinating CuB histidine residue, His208 (similar to the orientation of the placeholder).42,49 The carboxyl tail of the substrates formed polar contacts with Arg209, which was found to be important for TyrBm activity (Fig. 5).45 Moreover, it was observed that in order to allow substrate entrance, the TyrBm blocker residue V218 moves 1.4 Å, confirming its role in substrate orientation.53
Figure 5. Similar orientation of tyrosine and l-Dopa in the active site of TyrBm.
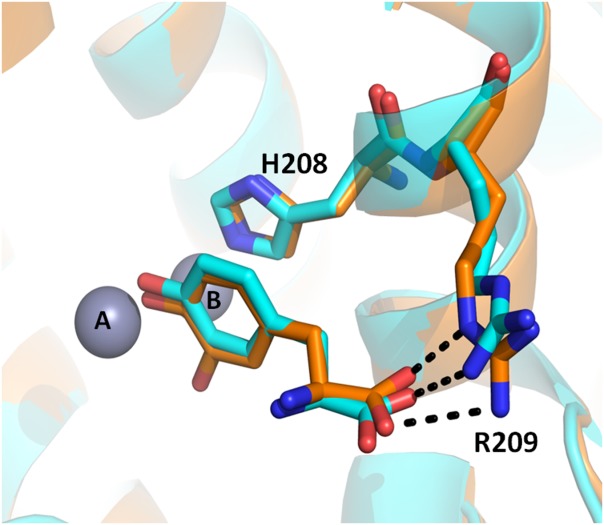
Superposition of the TyrBm structure with tyrosine (cyan, PDB accession code 4P6R) and the structure with l-Dopa (orange, PDB accession code 4P6S), showing similar orientation of monophenols and diphenols substrates in the active site of tyrosinases.53 The carboxyl side chains of the substrates form hydrogen bonds with Arg209. Zinc ions, replacing Cu ions in the crystal structure, are presented as grey spheres.
Monophenolase versus diphenolase Activity
In light of the new discovery regarding the similar binding of monophenolic and diphenolic substrates in the active site (Fig. 5), the explanation to the lack of monophenolase activity in CO was revised.53 Based on the previously suggested catalytic mechanism by Decker et al. and Deeth and Dietrich,49,50 it was suggested that the hydroxylation of monophenol substrates on the o-position occurs via an electrophilic substitution mechanism and is mediated by the rotation of three histidine residues coordinating CuA, followed by butterfly distortion of the Cu2O2 and consequent substrate reorientation. According to Goldfeder et al., this reorientation is not possible in CO in which the active site movement is restricted by a thioether bond together with a blocker residue Phe above CuA [Fig. 1(D)].53 In contrast, the hydroxylation of diphenol substrates does not involve an electrophilic attack, consequently might not require the reorganization and rotation steps, therefore, can occur in a partially blocked active site of CO. However, recent crystal structures and biochemical investigations of CO from Aspergillus oryzae (AoCO4) and polyphenol oxidase from Vitis vinifera (PPOVv) challenge the proposed assumption regarding the lack of monophenolase activity in CO.26,54,55
AoCO4 does not contain a thioether bond or a bulky Phe blocker residue (Table I), which may restrict active site reorganization nevertheless; it does not possess a monophenolase activity. In addition, its diphenolase activity is 100-fold lower than that of IbCO.26 Structural examination of AoCO4 revealed that instead of two conserved Glu and Asn residues, which were proposed to be important for substrate deprotonation and monophenolase activity [Fig. 4(B)], this protein possesses Gln237 and Gly285, respectively (Table I). Interestingly, IbCO which also does not possess monophenolase activity has Ile241 instead of Asn at the respective position to Gly285. Furthermore, although the active site of AoCO4 is not hindered by a blocker residue, it is not exposed to the surface and accessibility to the active site is restricted by external loops. Perhaps, the absence of monophenolase activity in AoCO4 can be attributed to the lack of conserved Asn and Glu residues, while the relatively low diphenolase activity might be a result of the limited active site accessibility.
In contrast to AoCO4, PPOVv exhibits both monophenolase and diphenolase activities, but contains a thioether bond together with a bulky Phe259 above CuA, similar to IbCO [Table I and Fig. 1(D)].54,55 However, in contrast to IbCO, PPOVv contains a conserved Asn240 residue (Table I). Decker and co-workers have shown that Phe259 in PPOVv exhibits similar flexibility to Phe218 in TyrBm variant V218F and its presence together with a thioether bond does not hinder monophenolase activity but may cause the slow hydroxylation rate.41,54
Alignment of tyrosinase and CO sequences using ConSurf (http://consurf.tau.ac.il/) revealed that while the Glu residue is highly conserved among the enzymes, Asn is conserved mostly among tyrosinases. Interestingly, these conserved residues were also found in Hc structures. It is thus suggested that the presence of conserved Glu and Asn next to the active site is pivotal for monophenolase activity of type-3 copper enzymes.
Although several mechanisms have been proposed to account for the differences between the catalytic activity of CO and tyrosinase, none of them succeed to enclose all the components in this puzzle. It seems that for monophenolase activity to occur, the protein active site has to possess flexibility to allow reorganization and substrate rotation.50,53 Moreover, substrate deprotonation is an obligatory step in monophenolase activity, which is most likely facilitated by activation of a conserved water molecule by the action of conserved Glu and Asn residues. Still, further biochemical and structural investigations are required to clarify this assumption. The full impact of the conserved Glu and Asn residues are poorly understood and elucidating their role will undoubtedly shed more light on this conundrum.
To conclude, although type-3 copper proteins have been intensively investigated over the past 20 years, further structural, computational, and biochemical studies are essential for understanding the functional differences among this class of proteins.
References
- 1.Claus H, Decker H. Bacterial tyrosinases. Syst Appl Microbiol. 2006;29:3–14. doi: 10.1016/j.syapm.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Faccio G, Kruus K, Saloheimo M, Thöny-Meyer L. Bacterial tyrosinases and their applications. Process Biochem. 2012;47:1749–1760. [Google Scholar]
- 3.Fairhead M, Thony-Meyer L. Bacterial tyrosinases: old enzymes with new relevance to biotechnology. New Biotechnol. 2012;29:183–191. doi: 10.1016/j.nbt.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-Ferrer A, Rodriguez-Lopez JN, Garcia-Canovas F, Garcia-Carmona F. Tyrosinase: a comprehensive review of its mechanism. Biochim Biophys Acta. 1995;22:1–11. doi: 10.1016/0167-4838(94)00204-t. [DOI] [PubMed] [Google Scholar]
- 5.Ramsden CA, Riley PA. Tyrosinase: the four oxidation states of the active site and their relevance to enzymatic activation, oxidation and inactivation. Biorg Med Chem. 2014;22:2388–2395. doi: 10.1016/j.bmc.2014.02.048. [DOI] [PubMed] [Google Scholar]
- 6.Decker H, Schweikardt T, Tuczek F. The first crystal structure of tyrosinase: all questions answered? Angew Chem Int Ed. 2006;45:4546–4550. doi: 10.1002/anie.200601255. [DOI] [PubMed] [Google Scholar]
- 7.Magnus KA, Hazes B, Ton-That H, Bonaventura C, Bonaventura J, Hol WG. Crystallographic analysis of oxygenated and deoxygenated states of arthropod hemocyanin shows unusual differences. Proteins. 1994;19:302–309. doi: 10.1002/prot.340190405. [DOI] [PubMed] [Google Scholar]
- 8.Klabunde T, Eicken C, Sacchettini JC, Krebs B. Crystal structure of a plant catechol oxidase containing a dicopper center. Nat Struct Mol Biol. 1998;5:1084–1090. doi: 10.1038/4193. [DOI] [PubMed] [Google Scholar]
- 9.van Gelder CWG, Flurkey WH, Wichers HJ. Sequence and structural features of plant and fungal tyrosinases. Phytochemistry. 1997;45:1309–1323. doi: 10.1016/s0031-9422(97)00186-6. [DOI] [PubMed] [Google Scholar]
- 10.Tran L, Taylor J, Constabel C. The polyphenol oxidase gene family in land plants: lineage-specific duplication and expansion. BMC Genomics. 2012;13:395. doi: 10.1186/1471-2164-13-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schweikardt T, Olivares C, Solano F, Jaenicke E, Garcia-Borron JC, Decker H. A three-dimensional model of mammalian tyrosinase active site accounting for loss of function mutations. Pigment Cell Res. 2007;20:394–401. doi: 10.1111/j.1600-0749.2007.00405.x. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura M, Nakajima T, Ohba Y, Yamauchi S, Lee BR, Ichishima E. Identification of copper ligands in Aspergillus oryzae tyrosinase by site-directed mutagenesis. Biochem J. 2000;350:537–545. [PMC free article] [PubMed] [Google Scholar]
- 13.Kaintz C, Mayer RL, Jirsa F, Halbwirth H, Rompel A. Site-directed mutagenesis around the CuA site of a polyphenol oxidase from Coreopsis grandiflora (cgAUS1) FEBS Lett. 2015;589:789–797. doi: 10.1016/j.febslet.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olivares C, Garcia-Borron JC, Solano F. Identification of active site residues involved in metal cofactor binding and stereospecific substrate recognition in mammalian tyrosinase. Implications to the catalytic cycle. Biochemistry. 2002;41:679–686. doi: 10.1021/bi011535n. [DOI] [PubMed] [Google Scholar]
- 15.Decker H, Schweikardt T, Nillius D, Salzbrunn U, Jaenicke E, Tuczek F. Similar enzyme activation and catalysis in hemocyanins and tyrosinases. Gene. 2007;398:183–191. doi: 10.1016/j.gene.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 16.Matoba Y, Kumagai T, Yamamoto A, Yoshitsu H, Sugiyama M. Crystallographic evidence that the dinuclear copper center of tyrosinase is flexible during catalysis. J Biol Chem. 2006;281:8981–8990. doi: 10.1074/jbc.M509785200. [DOI] [PubMed] [Google Scholar]
- 17.Sendovski M, Kanteev M, Ben-Yosef Adir SV, Fishman NA. First structures of an active bacterial tyrosinase reveal copper plasticity. J Mol Biol. 2011;405:227–237. doi: 10.1016/j.jmb.2010.10.048. [DOI] [PubMed] [Google Scholar]
- 18.Marusek CM, Trobaugh NM, Flurkey WH, Inlow JK. Comparative analysis of polyphenol oxidase from plant and fungal species. J Inorg Biochem. 2006;100:108–123. doi: 10.1016/j.jinorgbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Kaintz C, Mauracher SG, Rompel A. Type-3 copper proteins: Recent advances on polyphenol oxidases. In: Christo ZC, editor. Advances in Protein Chemistry and Structural Biology. Vol. 97. Academic Press; 2014. pp. 1–35. [DOI] [PubMed] [Google Scholar]
- 20.Mayer AM. Polyphenol oxidases in plants and fungi: Going places? A review. Phytochemistry. 2006;67:2318–2331. doi: 10.1016/j.phytochem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Lerch K. Protein and active-site structure of tyrosinase. Prog Clin Biol Res. 1988;256:85–98. [PubMed] [Google Scholar]
- 22.Schaerlaekens K, Schierová M, Lammertyn E, Geukens N, Anné J, Van Mellaert L. Twin-arginine translocation pathway in Streptomyces lividans. J Bacteriol. 2001;183:6727–6732. doi: 10.1128/JB.183.23.6727-6732.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fairhead M, Thöny-Meyer L. Role of the C-terminal extension in a bacterial tyrosinase. Febs J. 2010;277:2083–2095. doi: 10.1111/j.1742-4658.2010.07621.x. [DOI] [PubMed] [Google Scholar]
- 24.Flurkey WH, Inlow JK. Proteolytic processing of polyphenol oxidase from plants and fungi. J Inorg Biochem. 2008;102:2160–2170. doi: 10.1016/j.jinorgbio.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 25.Gasparetti C, Faccio G, Arvas M, Buchert J, Saloheimo M, Kruus K. Discovery of a new tyrosinase-like enzyme family lacking a C-terminally processed domain: production and characterization of an Aspergillus oryzae catechol oxidase. Appl Microbiol Biotechnol. 2010;86:213–226. doi: 10.1007/s00253-009-2258-3. [DOI] [PubMed] [Google Scholar]
- 26.Hakulinen N, Gasparetti C, Kaljunen H, Kruus K, Rouvinen J. The crystal structure of an extracellular catechol oxidase from the ascomycete fungus Aspergillus oryzae. J Biol Inorg Chem. 2013;18:917–929. doi: 10.1007/s00775-013-1038-9. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz-Muñoz JL, Acosta-Motos JR, Garcia-Molina F, Varon R, Garcia-Ruíz PA, Tudela J, Garcia-Cánovas F, Rodríguez-López JN. Tyrosinase inactivation in its action on dopa. BBA-Proteins Proteom. 2010;1804:1467–1475. doi: 10.1016/j.bbapap.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Ramsden CA, Stratford MRL, Riley PA. The influence of catechol structure on the suicide-inactivation of tyrosinase. Org Biomol Chem. 2009;7:3388–3390. doi: 10.1039/b910500j. [DOI] [PubMed] [Google Scholar]
- 29.Matoba Y, Bando N, Oda K, Noda M, Higashikawa F, Kumagai T, Sugiyama M. A molecular mechanism for copper transportation to tyrosinase that is assisted by a metallochaperone caddie protein. J Biol Chem. 2011;286:30219–30231. doi: 10.1074/jbc.M111.256818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanteev M, Goldfeder M, Chojnacki M, Adir N, Fishman A. The mechanism of copper uptake by tyrosinase from Bacillus megaterium. JBIC Journal of Biological Inorganic Chemistry. 2013;18:895–903. doi: 10.1007/s00775-013-1034-0. [DOI] [PubMed] [Google Scholar]
- 31.Mauracher SG, Molitor C, Al-Oweini R, Kortz U, Rompel A. Latent and active abPPO4 mushroom tyrosinase cocrystallized with hexatungstotellurate(VI) in a single crystal. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:2301–2315. doi: 10.1107/S1399004714013777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fujieda N, Yabuta S, Ikeda T, Oyama T, Muraki N, Kurisu G, Itoh S. Crystal structures of copper-depleted and copper-bound fungal pro-tyrosinase: Insights into enogenous cysteine-dependent copper incorporation. J Biol Chem. 2013;288:22128–22140. doi: 10.1074/jbc.M113.477612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang N, Hebert DN. Tyrosinase maturation through the mammalian secretory pathway: bringing color to life. Pigment Cell Research. 2006;19:3–18. doi: 10.1111/j.1600-0749.2005.00288.x. [DOI] [PubMed] [Google Scholar]
- 35.Larson CA, Adams PL, Blair BG, Safaei R, Howell SB. The role of the methionines and histidines in the transmembrane domain of mammalian copper transporter 1 in the cellular cccumulation of cisplatin. Mol Pharmacol. 2010;78:333–339. doi: 10.1124/mol.110.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J Biol Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schöneich C, Williams TD, Squier TC. Loss of conformational stability in calmodulin upon methionine oxidation. Biophys J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spritz RA, Oh J, Fukai K, Holmes SA, Ho L, Chitayat D, France TD, Musarella MA, Orlow SJ, Schnur RE, et al. Novel mutations of the tyrosinase (TYR) gene in type I oculocutaneous albinism (OCA1) Human Mutat. 1997;10:171–174. doi: 10.1002/(SICI)1098-1004(1997)10:2<171::AID-HUMU11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 39.Mastore M, Kohler L, Nappi AJ. Production and utilization of hydrogen peroxide associated with melanogenesis and tyrosinase-mediated oxidations of DOPA and dopamine. Febs J. 2005;272:2407–2415. doi: 10.1111/j.1742-4658.2005.04661.x. [DOI] [PubMed] [Google Scholar]
- 40.Wood JM, Decker H, Hartmann H, Chavan B, Rokos H, Spencer JD, Hasse S, Thornton MJ, Shalbaf M, Paus R, et al. Senile hair graying: H2O2-mediated oxidative stress affects human hair color by blunting methionine sulfoxide repair. Faseb J. 2009;23:2065–2075. doi: 10.1096/fj.08-125435. [DOI] [PubMed] [Google Scholar]
- 41.Decker H, Tuczek F. Tyrosinase/catecholoxidase activity of hemocyanins: structural basis and molecular mechanism. Trends in Biochemical Sciences. 2000;25:392–397. doi: 10.1016/s0968-0004(00)01602-9. [DOI] [PubMed] [Google Scholar]
- 42.Cuff ME, Miller KI, van Holde KE, Hendrickson WA. Crystal structure of a functional unit from Octopus hemocyanin. J Mol Biol. 1998;278:855–870. doi: 10.1006/jmbi.1998.1647. [DOI] [PubMed] [Google Scholar]
- 43.Perbandt M, Guthohrlein EW, Rypniewski W, Idakieva K, Stoeva S, Voelter W, Genov N, Betzel C. The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity. Biochemistry. 2003;42:6341–6346. doi: 10.1021/bi020672x. [DOI] [PubMed] [Google Scholar]
- 44.Ismaya WT, Rozeboom HJ, Weijn A, Mes JJ, Fusetti F, Wichers HJ, Dijkstra BW. Crystal structure of Agaricus bisporus mushroom tyrosinase: identity of the tetramer subunits and interaction with tropolone. Biochemistry. 2011;50:5477–5486. doi: 10.1021/bi200395t. [DOI] [PubMed] [Google Scholar]
- 45.Shuster Ben-Yosef V, Sendovski M, Fishman A. Directed evolution of tyrosinase for enhanced monophenolase/diphenolase activity ratio. Enzyme and Microbial Technology. 2010;47:372–376. [Google Scholar]
- 46.Masuda T, Momoji K, Hirata T, Mikami B. The crystal structure of a crustacean prophenoloxidase provides a clue to understanding the functionality of the type-3 copper proteins. Febs J. 2014;281:2659–2673. doi: 10.1111/febs.12812. [DOI] [PubMed] [Google Scholar]
- 47.Volbeda A, Hol WG. Crystal structure of hexameric haemocyanin from Panulirus interruptus refined at 3.2 A resolution. J Mol Biol. 1989;209:249–279. doi: 10.1016/0022-2836(89)90276-3. [DOI] [PubMed] [Google Scholar]
- 48.Goldfeder M, Kanteev M, Adir N, Fishman A. Influencing the monophenolase/diphenolase activity ratio in tyrosinase. BBA-Proteins Proteom. 2013;3:629–633. doi: 10.1016/j.bbapap.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 49.Deeth RJ, Diedrich C. Structural and mechanistic insights into the oxy form of tyrosinase from molecular dynamics simulations. J Biol Inorg Chem. 2010;15:117–129. doi: 10.1007/s00775-009-0577-6. [DOI] [PubMed] [Google Scholar]
- 50.Rolff M, Schottenheim J, Decker H, Tuczek F. Copper-O2 reactivity of tyrosinase models towards external monophenolic substrates: molecular mechanism and comparison with the enzyme. Chem Soc Rev. 2011;40:4077–4098. doi: 10.1039/c0cs00202j. [DOI] [PubMed] [Google Scholar]
- 51.Li Y, Wang Y, Jiang H, Deng J. Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes. Proc Natl Acad Sci USA. 2009;106:17002–17006. doi: 10.1073/pnas.0906095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegbahn PE, Borowski T. Comparison of QM-only and QM/MM models for the mechanism of tyrosinase. Faraday Discuss. 2011;148:109–117. doi: 10.1039/c004378h. [DOI] [PubMed] [Google Scholar]
- 53.Goldfeder M, Kanteev M, Isaschar-Ovdat S, Adir N, Fishman A. Determination of tyrosinase substrate-binding modes reveals mechanistic differences between type-3 copper proteins. Nat Commun. 2014;5 doi: 10.1038/ncomms5505. DOI 10.1038/ncomms5505. [DOI] [PubMed] [Google Scholar]
- 54.Fronk P, Hartmann H, Bauer M, Solem E, Jaenicke E, Tenzer S, Decker H. Polyphenoloxidase from Riesling and Dornfelder wine grapes (Vitis vinifera) is a tyrosinase. Food Chem. 2015;183:49–57. doi: 10.1016/j.foodchem.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 55.Virador VM, Reyes Grajeda JP, Blanco-Labra A, Mendiola-Olaya E, Smith GM, Moreno A, Whitaker JR. Cloning, sequencing, purification, and crystal structure of Grenache (Vitis vinifera) polyphenol oxidase. J Agric Food Chem. 2010;58:1189–1201. doi: 10.1021/jf902939q. [DOI] [PubMed] [Google Scholar]