Abstract
Biosensors for signaling molecules allow the study of physiological processes by bringing together the fields of protein engineering, fluorescence imaging, and cell biology. Construction of genetically encoded biosensors generally relies on the availability of a binding “core” that is both specific and stable, which can then be combined with fluorescent molecules to create a sensor. However, binding proteins with the desired properties are often not available in nature and substantial improvement to sensors can be required, particularly with regard to their durability. Ancestral protein reconstruction is a powerful protein-engineering tool able to generate highly stable and functional proteins. In this work, we sought to establish the utility of ancestral protein reconstruction to biosensor development, beginning with the construction of an l-arginine biosensor. l-arginine, as the immediate precursor to nitric oxide, is an important molecule in many physiological contexts including brain function. Using a combination of ancestral reconstruction and circular permutation, we constructed a Förster resonance energy transfer (FRET) biosensor for l-arginine (cpFLIPR). cpFLIPR displays high sensitivity and specificity, with a Kd of ∼14 µM and a maximal dynamic range of 35%. Importantly, cpFLIPR was highly robust, enabling accurate l-arginine measurement at physiological temperatures. We established that cpFLIPR is compatible with two-photon excitation fluorescence microscopy and report l-arginine concentrations in brain tissue.
Keywords: ancestral protein reconstruction, protein engineering, biosensor, fluorescence, neurobiology, Förster resonance energy transfer (FRET), nitric oxide
Introduction
Many amino acids have been shown to be mediators of cellular signaling and modulation.1,2 l-arginine specifically has been identified as the major substrate for the nitric oxide synthase (NOS) allowing for biosynthesis of nitric oxide (NO), a critical signaling molecule in the cardiovascular system, in the immune response, and important for neuronal function and plasticity.3,4 Because of the involvement of NO in many physiological processes, its role in signaling has been a pharmacological target in a variety of diseases, for example, NO donors as treatment for vascular diseases.5 In the brain, alterations of NO signaling have been in implicated in the pathophysiology of, among others, Alzheimer's disease,6,7 Huntington's disease,8 and epilepsy.9 Its rate of synthesis depends on a number of factors including the concentration of l-arginine,10 implying that changes of l-arginine concentration are relevant in disease. Indeed, brain l-arginine levels are affected by various relevant conditions including anxiety,11 pain,12 substance abuse,13 and Alzheimer's disease.14 A commonly used method to quantitatively investigate changes of l-arginine concentration is microdialysis.11–13 However, while microdialysis has been instrumental in gaining fundamental insights into l-arginine signaling it lacks the required spatial resolution for investigating changes of cellular distribution of l-arginine or localized l-arginine release or depletion. This is typically the domain of optical sensors which allow the experimenter to investigate concentrations and their changes non-invasively in space and time.
Optical sensors for amino acids generated so far typically rely on a molecular recognition event such as those found with the Fluorescent Indicator Protein (FLIP) sensors that exploit ligand mediated conformational changes of their binding core, usually a Periplasmic Binding Protein (PBP), to generate an observable change in fluorescence intensity.15,16 These binding proteins exist in two primary conformations; ligand free (open) and ligand bound (closed),17 previously likened to a “Venus fly trap” binding module.18 Existing PBP based sensors include those modified with fluorescent dyes to create fluorescence intensity optical sensors (EOS) and those using fluorescent proteins to construct Förster Resonance Energy Transfer (FRET) based biosensors capable of in vivo measurements.15,16,19,20
Extant, naturally occurring proteins often lack the properties that we require for medical or industrial applications and thus require substantial engineering. To be useful as a biosensor, the binding protein must be specific for its ligand. However, it is equally important that it can be expressed in high yield and maintains structural integrity throughout the course of the planned experiments. While proteins from thermophilic organisms could potentially fulfill these requirements, there are often other impediments related to their use. In this instance, PBPs from thermophilic organisms with affinity for l-arginine have been reported, but each is unsuitable for different reasons: the l-arginine binding protein from Thermotoga maritime has a domain swapped structure that complicates addition of fluorescent proteins in sensor construction,21 while thermophilic PBPs from Thermus thermophiles and Geobacillus stearothermophilus have low specificity for l-arginine, that is, they also bind other amino acids with high affinity.22,23 Existing PBP-based FRET sensors for l-arginine are compromised by suboptimal affinity for l-arginine (QBP sensor24) or sensitivity to physiological temperatures (ArtJ20), which could result in quantitative errors. We therefore sought to develop a novel strategy to generate stable and specific fluorescent reporters for l-arginine that can be used in complex tissue under physiological conditions.
Ancestral protein reconstruction (APR) is a technique that has been shown consistently to produce thermostable proteins.25–27 APR involves reconstruction of the sequence of an ancestral protein using statistical phylogenetic methods, allowing the ancestral protein to be characterized experimentally.28 The thermostability of ancestral proteins is somewhat controversial, variously being ascribed to the thermophilicity of ancient organisms from which they putatively originate,25 or through artifacts resulting from biases introduced by the maximum likelihood framework that can be used to infer ancestral sequences.29 As the polar amino acid PBP family is ancient, with homologs distributed throughout all domains of life,30 we expected that ancestral polar amino acid PBPs would also be thermostable and therefore provide useful scaffolds for biosensor engineering.
In this work we demonstrate that wild-type binding cores are poorly suited to measurement at physiological temperatures due to their low thermal stability, resulting in compromised experimental reliability. Use of an ancestrally reconstructed core allowed for the creation of cpFLIPR, a FRET sensor capable of reliable quantification of l-arginine concentrations in organized brain tissue at near-physiological temperature.
Results
Ancestral protein reconstruction
Maximum likelihood methods were used to infer a phylogeny of the polar amino acid PBP family from an alignment of 133 protein sequences and to reconstruct the sequences of ancestral nodes in this phylogeny (Fig. 1). Of the sequences used, 46 were bacterial homologs of proteins with high affinity for l-arginine, including lysine-/arginine-/ornithine-binding protein (ArgT), arginine-binding protein (ArtJ), and histidine-binding protein (HisJ), with the remainder being bacterial homologs of glutamine-binding protein (GlnH), aspartate-/glutamate-binding protein (GltI), cystine-binding protein (FliY), and cysteine-binding protein (CjaA). The maximum likelihood phylogeny reproduced general relationships between these proteins that have been inferred previously,31 such as the close relationship between the glutamine-binding protein and lysine-/arginine-/ornithine-/histidine-binding protein subfamilies.
Figure 1.
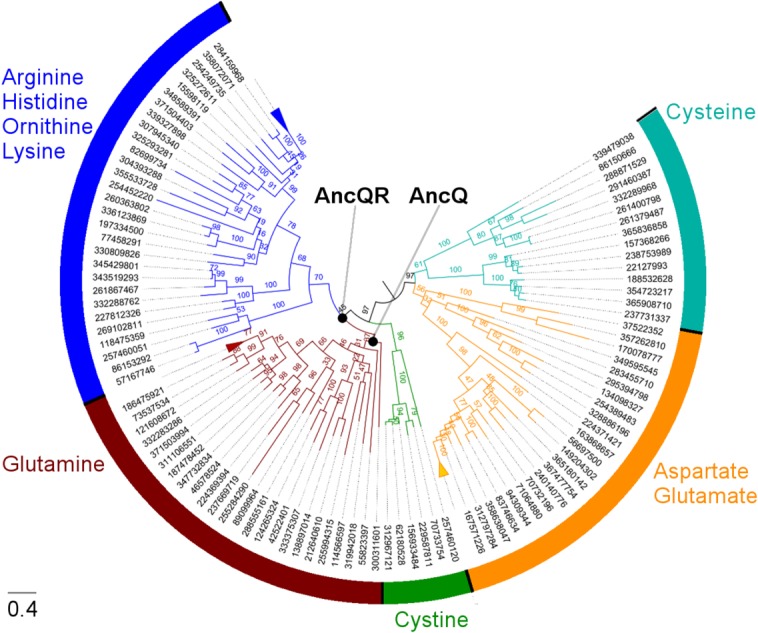
Reconstruction of ancestral polar amino acid PBPs. Midpoint-rooted maximum likelihood phylogeny of the amino acid-binding protein family used for reconstruction of ancestral sequences. Branches are drawn to scale and labeled with bootstrap values from 100 replicates. Sequences are identified by their GI number from the NCBI database, and are labeled with their putative binding specificity on the basis of functional annotation and homology to proteins of known function. The scale bar represents 0.4 substitutions per site. Three clades of closely related proteins are compressed for clarity.
Our primary aim in this work was not to investigate the evolution of these proteins but to generate a thermostable and functional scaffold that could be converted into a biosensor. To this end we investigated two ancestral nodes close to the divergence of the arginine-binding protein subfamily from the glutamine-binding protein subfamily (AncQ and AncQR; Fig. 1); we expected the corresponding ancestral proteins to be thermostable because their descendants (i.e. homologs of GlnH and ArgT) are found in phylogenetically diverse Gram-negative and Gram-positive bacteria, implying that proteins similar to AncQ and AncQR could have existed in ancient thermophilic bacteria.
Upon cloning of synthetic genes encoding the ancestral proteins followed by expression of the proteins in Escherichia coli, we found that AncQ and AncQR already had the desired properties for an l-arginine biosensor: high thermostability and sufficient specificity for l-arginine. Both ancestral proteins were expressed at very high levels in E. coli [Fig. 2(A)]. CD spectroscopy showed that both ancestral proteins were more stable than ArgT from Salmonella enterica (seArgT), which has a denaturation temperature (Tm) of 51.6°C [Fig. 2(B)], as well as other polar amino acid PBPs from mesophilic organisms.33 Indeed, AncQ is particularly stable, with a Tm exceeding 80°C [Fig. 2(B)]. Since preliminary screens performed by isothermal titration calorimetry (ITC) showed that AncQ and AncQR have similar binding specificity [Fig. 2(C)], we chose to use AncQ for construction of an l-arginine biosensor on the basis of its higher thermostability. Further characterization by ITC showed that AncQ binds l-arginine with high affinity (Kd = 5.0 µM); binding of other similar amino acids (l-glutamine, l-histidine, l-ornithine, and l-lysine) was low affinity (Kd >400 µM) [Fig. 2(D)], while binding of other proteinogenic amino acids was undetectable. The observed specificity of AncQ for l-arginine is higher than that of ArgT and comparable to that of ArtJ, both of which have been incorporated into l-arginine biosensors previously.20,34
Figure 2.
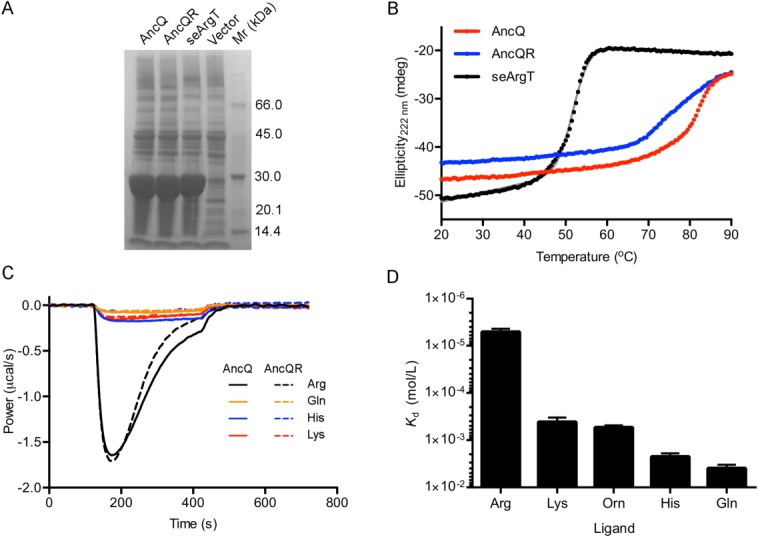
Characterization of the reconstructed ancestral proteins AncQ and AncQR. (A) SDS-PAGE gel showing overexpression of AncQ, AncQR, and seArgT in soluble fractions of E. coli crude cell lysates. Cultures were grown at 37°C in Terrific Broth media overnight. Samples were run on an ExpressPlus 4 to 20% PAGE gel (GenScript) and stained with Coomassie Blue. (B) Thermal unfolding of AncQ, AncQR and seArgT as monitored by CD spectroscopy. CD was measured at 222 nm. The Tm of seArgT was determined to be 51.6 ± 0.7°C (mean ± SD, n = 4) by fitting the data to a two-state denaturation model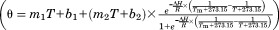 , where θ = CD (mdeg), T = temperature (°C), ΔH = enthalpy of unfolding (J/mol), R = gas constant (J/K/mol), Tm = denaturation temperature (°C), and θ = m1T + b1 and θ = m2T + b2 give the pretransition and post-transition baselines, respectively.32 (C) Baseline-corrected power traces resulting from continuous injection isothermal titration calorimetry experiments, illustrating binding of l-arginine, l-glutamine, l-histidine, and l-lysine by AncQ and AncQR. Amino acids (750 µM) were injected continuously into protein (50 µM) between t = 120 s and t = 420 s. Injection of other proteinogenic amino acids resulted in negligible deviations in power from baseline levels. (D) Binding specificity of AncQ. The Kd of AncQ for l-arginine, l-lysine, l-ornithine, l-histidine, and l-glutamine was determined by ITC. These experiments were performed in 50 mM Na2HPO4 pH 7.40, 100 mM NaCl buffer at 25°C. Error bars represent standard deviation of the mean from at least two separate titrations.
, where θ = CD (mdeg), T = temperature (°C), ΔH = enthalpy of unfolding (J/mol), R = gas constant (J/K/mol), Tm = denaturation temperature (°C), and θ = m1T + b1 and θ = m2T + b2 give the pretransition and post-transition baselines, respectively.32 (C) Baseline-corrected power traces resulting from continuous injection isothermal titration calorimetry experiments, illustrating binding of l-arginine, l-glutamine, l-histidine, and l-lysine by AncQ and AncQR. Amino acids (750 µM) were injected continuously into protein (50 µM) between t = 120 s and t = 420 s. Injection of other proteinogenic amino acids resulted in negligible deviations in power from baseline levels. (D) Binding specificity of AncQ. The Kd of AncQ for l-arginine, l-lysine, l-ornithine, l-histidine, and l-glutamine was determined by ITC. These experiments were performed in 50 mM Na2HPO4 pH 7.40, 100 mM NaCl buffer at 25°C. Error bars represent standard deviation of the mean from at least two separate titrations.
Construction of an l-arginine fluorescent sensor
AncQ was cloned into a specialized vector (pDOTS4), creating an enhanced cyan fluorescent protein (ECFP)-AncQ-Venus fluorescent protein (VFP) fusion construct, referred to henceforth as FLIPR (Fluorescent Indicator Protein for l-arginine) in keeping with the nomenclature developed by W. B. Frommer.16 Although ITC analysis showed that FLIPR retained the capacity to bind l-arginine, with a Kd of 25 µM (Table I) it did not exhibit a ligand mediated FRET response [Fig. 3(A)]. In the absence of a crystal structure of AncQ, we constructed a model using the PHYRE2 server35 [Fig. 3(B)]. Inspection of the homology model showed that the termini were located on the same lobe and therefore interfluorophore distance would not change between the open and closed conformations. Previous work by Okada et al.20 addresses this issue, increasing the dynamic range of the sensor through the use of circular permutation. The purpose of circular permutation is to place the N- and C-termini on separate lobes, so that changes in the relative positions of the fluorescent proteins are detectable through ligand-induced conformational change. In keeping with their methodology, the original termini were joined using a flexible linker peptide (Gly-Gly-Ser)4 and the second hinge strand of AncQ (residues 178–183) was removed. This yielded N and C termini in opposite lobes [Fig. 3(C)] and produced an active binding core, cpAncQ, with a Kd of 18 µM for l-arginine (Table I).
Table I.
Binding Affinities for AncQ, cpAncQ, FLIPR, and cpFLIPR as Determined by ITC
| Construct | Kd l-Arg (µM) | FRET ratio change |
|---|---|---|
| AncQ | 5.0 ± 0.7 | N/A |
| FLIPR | 28.8 ± 4.5 | 0 |
| cpAncQ | 18.5 ± 0.08 | N/A |
| cpFLIPR | 13.4 ± 0.38 | 0.35 |
Affinites were determined by incremental titration of 1250 µM l-arginine into 50 µM protein. These experiments were performed in triplicate in 50 mM Na2HPO4 pH 7.40, 100 mM NaCl buffer at 25°C.
Figure 3.
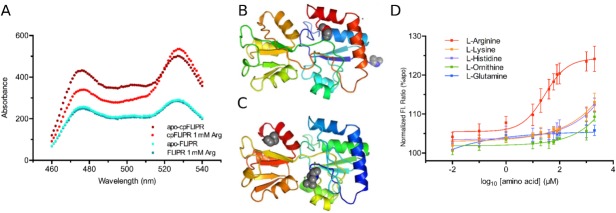
Characterization of FLIPR and cpFLIPR. A) Fluorescence spectra of FLIPR (turquoise and blue) and cpFLIPR (crimson and red) with peaks at 475 nm (CFP) and 525 nm (Venus) indicating change in fluorescence ratio upon addition of saturating ligand. FLIPR does not exhibit any change in ratio with addition of 1 mM
l-arginine, while cpFLIPR maximally shows a 35% change in ratio. B and C) PHYRE2 models of AncQ (B) and cpAncQ (C). N & C terminal regions are indicated by the blue and red secondary structures respectively, with spheres displaying the termini. The circular permutation is illustrated by the removal of the second hinge and the relocation of the termini (C). D) Sigmoidal dose response curves for cpFLIPR with l-arginine (red), l-lysine (orange), l-histidine (purple), l-ornithine (green), and l-glutamine (blue). Values are the (525 nm/475 nm) fluorescence ratio as a percentage of the same ratio for the apoenzyme. Curves were fitted using the following equation.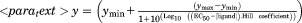 . Values are the mean ± standard error (n = 3).
. Values are the mean ± standard error (n = 3).
Next, the cpAncQ core was fused to the ECFP-VFP FRET pair yielding cpFLIPR. The new construct contained an N-terminal biotin tag to allow for binding to streptavidin. Fluorescence assays indicated a maximal fluorescence ratio change of 35% (excitation 433 nm, emission 525 nm/475 nm) at saturating levels of l-arginine compared with baseline [Fig. 3(A)]. We then conducted ITC analysis which showed that cpFLIPR binds l-arginine with a Kd of 13.4 µM. In comparison to current l-arginine sensors, this is a 130-fold improvement in Kd relative to the QBP variant24 and has an affinity in the relevant physiological range similar to that of cpArtJ.20,36 cpFLIPR appears to be specific for l-arginine; titration of the amino acids l-lysine, l-histidine, l-glutamine, and l-ornithine [Fig. 3(D)] did not show a significant change in fluorescence ratio at concentrations up to 500 µM.
Thermal stability of cpFLIPR and related sensors
To determine the relative stability of cpFLIPR in comparison to the current sensor cpArtJ, the fluorescence ratios of cpArtJ and cpFLIPR were monitored as a function of temperature [Fig. 4(A)], both with saturating concentrations of l-arginine. Construction of the circularly permuted sensor resulted in a loss of stability of both ArtJ and AncQ. The observed fluorescence ratio displayed a sigmoidal decay and yielded an effective Tm of 33°C and 50°C for cpArtJ and cpFLIPR, respectively, showing cpFLIPR was significantly more stable after sensor construction. The same experiments were performed with the ligand bound sensor to account for the potential stabilizing effects of bound ligand.37 An increase in stability of approximately 7°C was observed for both sensors, with the Tm increasing to 40°C and 56°C for cpArtJ and cpFLIPR, respectively. The thermal denaturation curve for cpArtJ showed a 15% increase (cpFLIPR only changed 4%) in ratio with temperature over the range of 25 to 35°C before quickly decreasing, indicating a strong temperature dependence of the CFP-VFP fluorescence ratio. These results suggest that cpArtJ is unable to accurately measure l-arginine concentrations at temperatures in excess of 30°C, while cpFLIPR can retain function to over 45°C.
Figure 4.
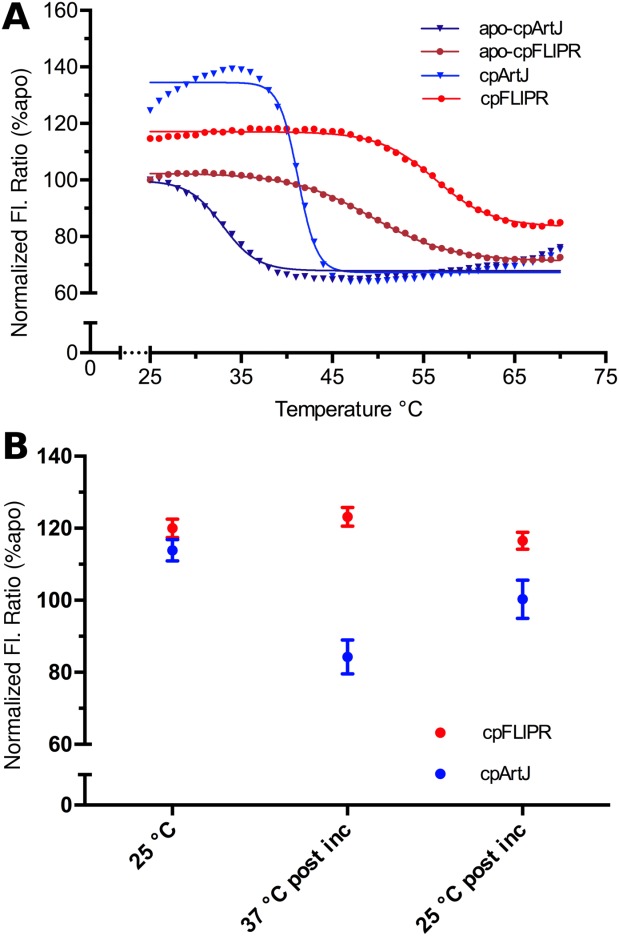
Comparison of stability between cpFLIPR and contemporary sensor cpArtJ (20). A) Fluorescence ratio (%, relative to apo) as a function of temperature. cpArtJ (apo: dark blue, 1 mM Arg: light blue) indicates a strong temperature dependence and lower stability than cpFLIPR (apo: crimson, 1 mM Arg: red). The data were fitted using a Boltzmann function (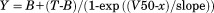 where B = Y min, T = Y max, V50 = Y mid. B) Ratio changes of cpArtJ (blue) and cpFLIPR (red) upon incubation at 37°C for 45 min normalized (as percentages) against the aposensor ratio. Pre incubation measurements were conducted as separate experiments. cpArtJ displays a diminished VFP/CFP ratio at 37°C postincubation and this change is irreversible as shown by the postincubation 25°C ratio, which is equivalent to that of the aposensor. All changes are statistically significant (P < 0.05) as determined by a t-test (preincubation relative to postincubation measurements) and a paired t-test (postincubation measurements).
where B = Y min, T = Y max, V50 = Y mid. B) Ratio changes of cpArtJ (blue) and cpFLIPR (red) upon incubation at 37°C for 45 min normalized (as percentages) against the aposensor ratio. Pre incubation measurements were conducted as separate experiments. cpArtJ displays a diminished VFP/CFP ratio at 37°C postincubation and this change is irreversible as shown by the postincubation 25°C ratio, which is equivalent to that of the aposensor. All changes are statistically significant (P < 0.05) as determined by a t-test (preincubation relative to postincubation measurements) and a paired t-test (postincubation measurements).
To investigate the kinetic stability of these sensors, under equilibrium conditions, they were incubated at 37°C for 45 min, to simulate a typical short experiment at physiological temperature. In the case of cpFLIPR, the VFP/CFP ratio change observed upon ligand binding at 25°C (120%) did not significantly change when the temperature was increased [Fig. 4(A)] or after 45 min of incubation [123%; Fig. 4(B)]. After cooling to 25°C, the VFP/CFP ratio remained almost identical to that preincubation (116%; t-test P < 0.05, n = 3). In contrast, the VFP/CFP ratio change observed upon ligand binding at 25°C for cpArtJ (113%) did change when the temperature was increased [Fig. 4(A)], and after 45 min at 37°C the sensor had lost all sensitivity, giving a negative ratio change (84% of apo cpArtJ). When the sensor was cooled to 25°C after the 37°C incubation, the VFP/CFP ratio was identical to that of unbound cpArtJ; that is, no change from ligand binding [100%, paired t-test P < 0.05, n = 3; Fig. 4(B)]. This experiment demonstrated that the negative effect of temperature upon cpArtJ is irreversible and likely a result of denaturation of the binding core. These data together with the melting curves indicate a significant increase in thermal and kinetic stability for cpFLIPR resulting in superior readout reliability.
cpFLIPR in two-photon excitation (2PE) fluorescence microscopy
2PE fluorescence microscopy is a powerful technique for imaging in living tissue in vivo and in situ. To test whether cpFLIPR is compatible with this method we imaged the sensor at an excitation wavelength of 800 nm in a meniscus under the objective and titrated increasing concentrations of l-arginine. Under these conditions, cpFLIPR had a Kd of 14 µM and a 7.2% change in fluorescence ratio [Fig. 5(A)].
Figure 5.
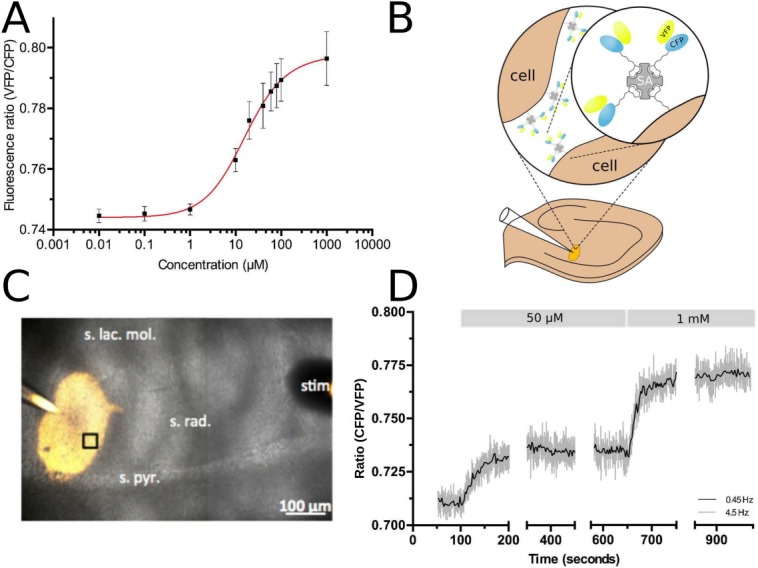
Characterization and in situ testing of cpFLIPR using two-photon excitation (2PE) fluorescence microscopy. A) Calibration curve of cpFLIPR using 2PE shows a dose-dependent VFP/CFP fluorescence ratio change. Fitting the relationship between fluorescence ratio R and l-arginine concentration c with a Hill equation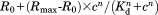 obtains a Kd of 14.04 ± 3.8 µM, n = 3. B) Schematic representation of sensor immobilization in brain tissue. Biotinylated cpFLIPR was conjugated to streptavidin (SA, gray) and then puffed into brain slices preincubated with EZ-link Biotin leading to its attachment to cell surfaces (brown). C) Illustrates the puff of cpFLIPR (yellow) into the striatum radiatum of the CA1 region of a rat hippocampal slice at a depth of ∼70 µm. The highlighted region was subsequently imaged at a depth of 59 µm. D) cpFLIPR responses to application of l-arginine in situ demonstrate its capability to measure steady state conditions and sensitivity to changes in ligand concentration in organized brain tissues. Fluorescence ratios at rest and after application of 50 µM and 1 mM of l-arginine were used to calculate the resting l-arginine concentration in acute brain slices.
obtains a Kd of 14.04 ± 3.8 µM, n = 3. B) Schematic representation of sensor immobilization in brain tissue. Biotinylated cpFLIPR was conjugated to streptavidin (SA, gray) and then puffed into brain slices preincubated with EZ-link Biotin leading to its attachment to cell surfaces (brown). C) Illustrates the puff of cpFLIPR (yellow) into the striatum radiatum of the CA1 region of a rat hippocampal slice at a depth of ∼70 µm. The highlighted region was subsequently imaged at a depth of 59 µm. D) cpFLIPR responses to application of l-arginine in situ demonstrate its capability to measure steady state conditions and sensitivity to changes in ligand concentration in organized brain tissues. Fluorescence ratios at rest and after application of 50 µM and 1 mM of l-arginine were used to calculate the resting l-arginine concentration in acute brain slices.
We next tested cpFLIPR in 300 µm thick acute slices of rat hippocampus. To this end, we adopted a labeling procedure that anchors fluorescent proteins in the extracellular space.38 Brain slices were biotinylated using a commercially available biotinylation agent. With the help of its biotin tag attached to the CFP moiety [Fig. 5(B)], cpFLIPR (20–40 µM, in PBS) was bound to streptavidin (2.5–5 µM) and then injected with a patch pipette ∼70 µm deep into the stratum radiatum of the CA1 region of the hippocampus by a mild pressure pulse [see Fig. 5(C) for an illustration]. This resulted in a bright labeling [Fig. 5(C)] that was stable for the duration of a typical experiment. Over 15 min, the ratio of VFP and CFP fluorescence intensities did not change significantly (+0.36 ± 0.99%, P = 0.75, one-population Student's t-test). To test if this labeling procedure impairs slice viability we recorded field responses to Schaffer collateral stimulation from the stratum radiatum of the CA1 region in biotinylated and control slices. No significant changes of fiber volley amplitudes, field response slopes or paired-pulse ratios were detected (n = 8, all groups, P > 0.2 throughout, unpaired Student's t-tests) indicating that axonal, post-, and presynaptic function are not affected. We then used cpFLIPR to measure the extracellular resting concentration of l-arginine in brain slices. After an initial recording to obtain a resting VFP/CFP fluorescence ratio, increasing concentrations of l-arginine were added to the extracellular solution. The sensor responded to 50 µM and 1 mM l-arginine with a change in VFP/CFP fluorescence ratio of 3.4 ± 0.4% and 8.7 ± 0.9% respectively [n = 3; Fig. 5(D)]. Using the calibration obtained in free solution (Kd and n) and the VFP/CFP fluorescence ratios obtained from baseline, 50 µM and 1 mM measurements in the slice, respectively, we estimated a resting l-arginine concentration of 16.8 ± 4.6 µM (n = 4) in the extracellular space of rat brain. To the best of our knowledge, this is the first measurement of extracellular resting l-arginine concentrations in intact neuropil, and is close to values measured in rat hippocampus and human cerebrospinal fluid of 5 to 20 µM.36,39,40
Discussion
In this study we describe the use of ancestral protein reconstruction to generate a thermostable binding core that allowed for the construction of a FRET sensor capable of specifically detecting l-arginine in complex tissue such as rat brain slices at physiological temperature. This work shows that ancestral protein reconstruction allows us to circumvent this problem of low resistance to thermal denaturation inherent in many contemporary sensors derived from mesophilic proteins. Whereas thermostable l-arginine-binding proteins derived from thermophilic organisms have been reported, other properties are less desirable, such as the domain-swapped dimeric structure of TmArgBP21 or the low specificity of TTC0807 and GsArtJ.22,23 The increased thermostability of the ancestral core compared with contemporary proteins gave AncQ a higher tolerance to manipulations such as circular permutation, which has previously been shown to decrease ligand affinity20 and thermodynamic stability.41,42 This destabilization is often attributed to the strain introduced by the linking of the original termini.42 Specifically, in the case of cpArtJ, the destabilizing effects of circular permutation resulted in a strong temperature dependence of FRET and thereby impeded data reliability, whereas cpFLIPR upon circular permutation retained stable function at temperatures in excess of the physiological range and even increased dynamic range (Fig. 4).
Immobilization of cpFLIPR in the extracellular space of acute rat brain slices using its biotin tag resulted in a stable and bright label for several hours, while faithfully reporting l-arginine concentration in the extracellular space of the tissue upon application of the amino acid. Using our calibration data in free solution and the fluorescence ratios obtained for varying concentrations of l-arginine washed onto the slice, we calculate a resting concentration of ∼17 µM in the extracellular space of the rat brain. This is in good agreement with values measured in rat hippocampus and human cerebrospinal fluid of 5 to 20 µM36,39,40 indicating that cpFLIPR can be used to measure physiologically relevant extracellular concentrations of l-arginine in intact brain tissue. This result also suggests that homeostatic mechanisms maintaining extracellular l-arginine concentrations could be preserved in acute slices, similar to other relevant transmitters like GABA43 or NMDA receptor co-agonists44 cpFLIPR therefore represents a non-invasive tool to study the cellular processes that release, remove and/or deplete extracellular l-arginine11–14 at high spatial and temporal resolution. Since the anchoring of cpFLIPR to surface membranes is not specific for brain tissue the use of cpFLIPR is not limited to brain or reduced preparations like tissue slices. Indeed, its suitability for two-photon excitation at 800 nm should also prove useful for in vivo imaging deep in the intact tissue.
In summary, we show that cpFLIPR can be used in experimental approaches that involve both single- and two photon fluorescence excitation, therefore rendering it versatile for various experimental setups ranging from in vitro to in situ and possibly in vivo conditions. The successful application of this sensor supports evidence that circular permutation is a useful and generally applicable technique for improving dynamic range of PBP-based sensors,20 particularly when deriving the binding core through reconstruction of ancestral proteins.
Materials and Methods
Phylogenetic analysis
Bacterial homologs of GltI (NCBI GI number: 15800369), GlnH (15800563), ArgT (309702622), HisJ (15802856), and FliY (312967121) from Escherichia coli, and CjaA from Campylobacter jejuni (86150666) were collected from the NCBI database of nonredundant protein sequences using the BLAST server. Sequences were divided into subfamilies and aligned within their subfamily using MUSCLE version 3.8.3145 and the resulting alignments were combined by profile-profile alignment in MUSCLE. Model selection for the phylogenetic analysis was done using PROTTEST version 2.4 46,47 on the basis of the Akaike Information Criterion, which was calculated for BIONJ trees inferred using various substitution models. This analysis supported use of the LG substitution matrix, with rate heterogeneity modeled using the discrete-gamma model with four rate categories, a fraction of invariant sites fixed at 0.3%, and equilibrium amino acid frequencies estimated from the data. Unrooted phylogenetic trees were calculated using the maximum likelihood method implemented in PhyML version 3.047 and bootstrapped with 100 replicates. Ancestral protein sequences were reconstructed using the empirical Bayes method implemented in PAML version 4.5.48 The posterior probability distribution at each site for each ancestral node was also calculated using PAML. Putative N-terminal signal sequences and artifactual insertions caused by gaps in the multiple sequence alignment were removed manually. Sequences corresponding to a putative ancestor of the l-arginine and l-glutamine PBPs, as well as a putative ancestor of only the l-glutamine family were selected for more detailed analysis. The rationale for including the ancestor of l-glutamine binding proteins was that the precise binding specificities of these potential ancestors could not be reliably predicted given their evolutionary separation from contemporary proteins. These were the only ancestors screened in this work, that is, no dysfunctional ancestors were generated.
Cloning
The protein sequences of ArgT from Salmonella enterica with signal peptide removed (seArgT; NCBI GI number 446677077), and the ancestral proteins AncQ and AncQR were back-translated, producing nucleotide sequences codon-optimized for expression in E. coli. The resulting genes were synthesized (GeneArt) and cloned into the NdeI/EcoRI site of the pETMCSIII plasmid49 using standard restriction-ligation methods. This allowed expression of the proteins with N-terminal hexahistidine tags under control of a T7 promoter. The cpAncQ gene was synthesized (GeneArt) and cloned into the same plasmid by Gibson assembly50 using in-house prepared reagents. The cpArtJ sequence was constructed based off the literature20 and synthesized by GeneArt for sensor cloning. Sensor constructs were cloned into a series of vector backbones denoted as “pDOTS#.” These utilize the vector system used previously16 (Addgene Plasmid #13537) containing a pRSET backbone with an N-terminal 6×HisTag. YbeJ was removed from the CFP-Venus cassette and replaced with a SapI linker designed around the method of golden gate cloning,51 whereby SapI (NEB) restriction sites (GCTCTTC/GAAGAGC) are used to clone in a gene of interest; this yielded the vector pDOTS4. All gene fragments were generated by PCR to have the reciprocal SapI sites at both the 5' and 3' ends. A single amino acid insertion at both ends occurs due to cloning (isoleucine at 5' and leucine at 3'). AncQ was cloned into pDOTS4 yielding FLIPR, while cpAncQ was cloned into pDOTS10 giving cpFLIPR. pDOTS10 contains the insertion of a biotin tag from the PinPoint™ Xa-1 Vector (Promega) in between the His tag and the first fluorescent protein. To obtain usable quantities of vector DNA, E. coli Top10 cells were transformed by electroporation with target construct and following successful transformation used to inoculate 10 mL Luria Broth (LB) media. The vector DNA was purified using a QIAprep Spin Miniprep Kit (Qiagen, Germany) following the manufacturer's protocol.
Protein expression and purification
Proteins were expressed in E. coli BL21 (DE3) cells, transformed by electroporation. AncQ, AncQR, and seArgT were grown in auto-induction media (per L: 20 g tryptone, 5 g yeast extract, 5 g NaCl, 3 g KH2HPO4, 6 g Na2HPO4, 5 mL glycerol, 2 g lactose, 0.5 g glucose, 100 mg ampicillin) at 37°C for 24 h, while cpAncQ was grown in Terrific Broth (TB) media with 100 mg/L ampicillin at 25 to 28°C to OD600 = 0.7, induced using 0.5 to 0.75 mM IPTG and incubated for 24 h. FLIP constructs were grown in auto-induction media at 25 to 28°C for 48 h from a single colony inoculation. Cells were harvested 48 h after induction and resuspended in buffer A (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole pH 7.4). The resuspended cells were lysed using a French Press or by sonication. The resultant lysate was centrifuged at 13,500 rpm for 45 min to sediment insoluble material. Soluble fractions were filtered and loaded onto a 5 mL Ni-NTA column (GE Life Sciences) equilibrated in buffer A. Purification was performed using an Akta Purifier FPLC. The protein was eluted in 100% buffer B (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole pH 7.4). Target protein-containing fractions (as determined by UV absorbance and SDS-PAGE) were pooled and exhaustively dialyzed in dialysis buffer (20 mM NaH2PO4, 100 mM NaCl, pH 7.4) to remove imidazole and small contaminant molecules. Purified samples were concentrated using a 10/30 kDa Amicron spin concentrator (Millipore).
Protein binding and thermostability characterization
Samples for ITC experiments were prepared in dialysis buffer except where otherwise indicated, and experiments were performed at 25°C. Preliminary ITC experiments to determine the binding specificity of AncQ and AncQR were performed using a VP-ITC calorimeter (MicroCal). AncQ and AncQR were screened for binding to 17 proteinogenic amino acids (all except Pro, Trp, and Tyr) by injection of 50 µL of 750 µM amino acid solution into 150 µL of 50 µM protein over 300 s. The signal was recorded for 300 s following the injection. The rest of the experiments were performed using a Nano-ITC low-volume instrument (TA Instruments). A typical experiment involved at least 15 2 µL injections of 1.25 mM ligand into 50 µM protein. Corrections for background heat effects were made using ligand-buffer control titrations. Data were analyzed in NanoAnalyze version 2.3.6 (TA Instruments); the association constant (Ka), binding enthalpy (ΔH), and where appropriate, stoichiometry (n) for each interaction were obtained by fitting the integrated data to the independent binding model.
The thermal stability of PBP binding cores was assessed by circular dichroism (CD) spectroscopy using a Chirascan™ spectropolarimeter (Applied Photophysics). Before CD experiments, proteins were refolded on-column to ensure the complete absence of bound ligands; briefly, this was done by loading the protein onto a 5 mL Ni-NTA column, washing the column with unfolding buffer (8M urea, 20 mM NaH2PO4, 500 mM NaCl pH 7.40), and refolding the protein by application of a gradient into buffer A over 75 min. The eluted proteins were then purified by size-exclusion chromatography on a HiLoad™ 26/600 Superdex™ 200 pg column (GE Life Sciences), eluting in dialysis buffer, to remove misfolded aggregates. Proteins were diluted to 0.5 mg/mL in dialysis buffer. The path length of the cuvette was 1 mm. CD at 222 nm was measured at 0.5°C intervals as the temperature was increased at 1°C/min from 20°C to 90°C.
Fluorescence spectra were recorded using the Cary Eclipse Varian spectrophotometer scan program with an excitation of 433/5 nm, measuring emissions from 460 to 540 nm with peaks registered at 475 and 525 nm for ECFP and Venus (VFP), respectively. Temperature dependent measurements were obtained using an Applied Physics Chirascan™ fluorescence photomultiplier with 433/3 nm excitation and peak fluorescence at 483 and 533 nm. Proteins were used at a concentration of 50 µM in dialysis buffer with fluorescence measured at 1°C intervals as the temperature was increased at 0.3°C/min from 20°C to 90°C.
Fluorescent titrations we carried out using the Cary Eclipse spectrophotometer using an excitation of 433/5 nm, measuring emissions from 460 to 540 nm with peaks registered at 475 and 525 nm for ECFP and Venus (VFP) respectively. To a 50 µM protein sample with a volume of 975 µL, ligand concentration was increased incrementally from an initial concentration of 10 nM to a final concentration of 2 mM. Ratios were calculated as 525/475 nm.
Hippocampal slice preparation and biotinylation
Hippocampal slices were prepared as described previously.52 Briefly, 300 µm thick acute horizontal slices were obtained from 3- to 4-week-old Wistar rats in full compliance with national and institutional guidelines on animal experimentation. Slices were prepared in an ice-cold slicing solution containing (in mM): NaCl 60, sucrose 105, KCl 2.5, MgCl2 7, NaH2PO4 1.25, ascorbic acid 1.3, sodium pyruvate 3, NaHCO3 26, CaCl2 0.5, and glucose 10 (osmolarity 300–305 mOsm), and kept in the slicing solution at 34°C for 15 min before being stored at room temperature (RT, 21–23°C) in an extracellular solution containing (in mM) NaCl 131, KCl 2.5, MgSO4 1.3, NaH2PO4 1.25, NaHCO3 21, CaCl2 2, and glucose 10 (pH 7.35–7.45; osmolarity adjusted to 295–305 mOsm). For the first 45 min this solution was supplemented with 50 µM Sulfo-NHS EZ Link Biotin (Thermo Fisher). After a brief washing step, slices were kept in extracellular solution at RT until use. For in-slice titrations and recordings, slices were transferred to a submersion-type recording chamber and superperfused with extracellular solution at 34°C. All solutions were continuously bubbled with 95% O2/5% CO2. For injections of cpFLIPR into the tissue, patch clamp pipettes (2–4 MΩ) were backfilled with PBS to which 20 to 40 µM cpFLIPR and 2.5 to 5 µM streptavidin (Life Technologies) had been added. The pipette was inserted ∼70 µm deep into the stratum radiatum of the CA1 region of the hippocampus. cpFLIPR was injected by applying mild pressure to the back end of the pipette.
Extracellular recordings of neuronal activity from acute slices were performed as documented previously.52 Briefly, hippocampal CA3 to CA1 synapses were stimulated by brief current pulses (100 µs, stimulus intensity 20, 40, 80, 160, and 320 µA) to Schaffer collaterals delivered by a concentric bipolar electrode. Double stimuli separated by 25, 50, 100, 200, and 400 ms were used to test paired-pulse behavior. Neuronal field responses were recorded by an extracellular electrode filled with extracellular solution connected to a Multiclamp 700B amplifier and stored for offline analysis.
Two-photon excitation fluorescence microscopy
We used a FV10MP imaging system (Olympus) optically linked to a femtosecond pulse laser Vision S (Coherent, λ = 800 nm) and equipped with a 25× (NA 1.05) objective (Olympus). The laser power was adjusted to ∼3 mW under the objective for meniscus imaging. For imaging in slices, laser power was adjusted to ∼6 mW under the objective which corresponds to an estimated intensity of ∼3 mW at 50 µm depth in the slice where measurements were taken. cpFLIPR CFP and Venus fluorescence signals were collected with photomultiplier tubes connected to a single photon counting board (Picoharp, Picoquant). Their arrival times were recorded using Symphotime software (Picoquant). Offline analysis was performed using Origin (OriginLab) and custom written scripts in Matlab (Mathworks). The ratio of Venus and CFP fluorescence was calculated from the number of photons detected by the respective detectors in time bins of 222 ms.
References
- 1.Coruzzi G, Bush DR. Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 2001;125:61–64. doi: 10.1104/pp.125.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meijer AJ, Dubbelhuis PF. Amino acid signalling and the integration of metabolism. Biochem Biophys Res Commun. 2004;313:397–403. doi: 10.1016/j.bbrc.2003.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Gensert JM, Ratan RR. The metabolic coupling of arginine metabolism to nitric oxide generation by astrocytes. Antioxidants Redox Signal. 2006;8:919–928. doi: 10.1089/ars.2006.8.919. [DOI] [PubMed] [Google Scholar]
- 4.Prast H, Philippu A. Nitric oxide as modulator of neuronal function. Prog Neurobiol. 2001;64:51–68. doi: 10.1016/s0301-0082(00)00044-7. [DOI] [PubMed] [Google Scholar]
- 5.Miller M, Megson I. Recent developments in nitric oxide donor drugs. Br J Pharmacol. 2007;151:305–321. doi: 10.1038/sj.bjp.0707224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molokanova E, Akhtar MW, Sanz-Blasco S, Tu S, Piña-Crespo JC, McKercher SR, Lipton SA. Differential effects of synaptic and extrasynaptic NMDA receptors on Aβ-induced nitric oxide production in cerebrocortical neurons. J Neurosci. 2014;34:5023–5028. doi: 10.1523/JNEUROSCI.2907-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Rowan MJ, Anwyl R. β-amyloid-mediated inhibition of NMDA receptor-dependent long-term potentiation induction involves activation of microglia and stimulation of inducible nitric oxide synthase and superoxide. J Neurosci. 2004;24:6049–6056. doi: 10.1523/JNEUROSCI.0233-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deckel AW. Nitric oxide and nitric oxide synthase in Huntington's disease. J Neurosci Research. 2001;64:99–107. doi: 10.1002/jnr.1057. [DOI] [PubMed] [Google Scholar]
- 9.Rundfeldt C, Koch R, Richter A, Mevissen M, Gerecke U, Löscher W. Dose-dependent anticonvulsant and proconvulsant effects of nitric oxide synthase inhibitors on seizure threshold in a cortical stimulation model in rats. Eur J Pharmacol. 1995;274:73–81. doi: 10.1016/0014-2999(94)00711-f. [DOI] [PubMed] [Google Scholar]
- 10.Bode-Böger SM, Scalera F, Ignarro LJ. The L-arginine paradox: importance of the L-arginine/asymmetrical dimethylarginine ratio. Pharmacol Therapeut. 2007;114:295–306. doi: 10.1016/j.pharmthera.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Bosch OJ, Sartori SB, Singewald N, Neumann ID. Extracellular amino acid levels in the paraventricular nucleus and the central amygdala in high-and low-anxiety dams rats during maternal aggression: regulation by oxytocin. Research report. Stress. 2007;10:261–270. doi: 10.1080/10253890701223197. [DOI] [PubMed] [Google Scholar]
- 12.Silva E, Hernandez L, Contreras Q, Guerrero F, Alba G. Noxious stimulation increases glutamate and arginine in the periaqueductal gray matter in rats: a microdialysis study. Pain. 2000;87:131–135. doi: 10.1016/S0304-3959(00)00275-X. [DOI] [PubMed] [Google Scholar]
- 13.Lallemand F, Ward RJ, Dravolina O, De Witte P. Nicotine-induced changes of glutamate and arginine in naive and chronically alcoholized rats: an in vivo microdialysis study. Brain Res. 2006;1111:48–60. doi: 10.1016/j.brainres.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 14.Tarbit I, Perry E, Perry R, Blessed G, Tomlinson B. Hippocampal free amino acids in Alzheimer's disease. J Neurochem. 1980;35:1246–1249. doi: 10.1111/j.1471-4159.1980.tb07883.x. [DOI] [PubMed] [Google Scholar]
- 15.Hires SA, Zhu Y, Tsien RY. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc Natl Acad Sci USA. 2008;105:4411–4416. doi: 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okumoto S, Looger LL, Micheva KD, Reimer RJ, Smith SJ, Frommer WB. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc Natl Acad Sci USA. 2005;102:8740–8745. doi: 10.1073/pnas.0503274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dwyer MA, Hellinga HW. Periplasmic binding proteins: a versatile superfamily for protein engineering. Curr Opin Struct Biol. 2004;14:495–504. doi: 10.1016/j.sbi.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 19.Namiki S, Sakamoto H, Iinuma S, Iino M, Hirose K. Optical glutamate sensor for spatiotemporal analysis of synaptic transmission. Eur J Neurosci. 2007;25:2249–2259. doi: 10.1111/j.1460-9568.2007.05511.x. [DOI] [PubMed] [Google Scholar]
- 20.Okada S, Ota K, Ito T. Circular permutation of ligand-binding module improves dynamic range of genetically encoded FRET-based nanosensor. Protein Sci. 2009;18:2518–2527. doi: 10.1002/pro.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruggiero A, Dattelbaum JD, Staiano M, Berisio R, D'Auria S, Vitagliano L. A loose domain swapping organization confers a remarkable stability to the dimeric structure of the arginine binding protein from Thermotoga maritima. Plos One. 2014;9:11. doi: 10.1371/journal.pone.0096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanemaru Y, Hasebe F, Tomita T, Kuzuyama T, Nishiyama M. Two ATP-binding cassette transporters involved in (S)−2-aminoethyl-cysteine uptake in Thermus thermophilus. J Bacteriology. 2013;195:3845–3853. doi: 10.1128/JB.00202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vahedi-Faridi A, Eckey V, Scheffel F, Alings C, Landmesser H, Schneider E, Saenger W. Crystal structures and mutational analysis of the arginine-, lysine-, histidine-binding protein ArtJ from Geobacillus stearothermophilus. Implications for interactions of ArtJ with its cognate ATP-binding cassette transporter, Art(MP) J Mol Biol. 2008;375:448–459. doi: 10.1016/j.jmb.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 24.Bogner M, Ludewig U. Visualization of arginine influx into plant cells using a specific FRET-sensor. J Fluor. 2007;17:350–360. doi: 10.1007/s10895-007-0192-2. [DOI] [PubMed] [Google Scholar]
- 25.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. 2008;451:U702–U704. doi: 10.1038/nature06510. Nature. [DOI] [PubMed] [Google Scholar]
- 26.Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425:285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- 27.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of precambrian beta-lactamases. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 28.Thornton JW. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat Rev Genet. 2004;5:366–375. doi: 10.1038/nrg1324. [DOI] [PubMed] [Google Scholar]
- 29.Williams PD, Pollock DD, Blackburne BP, Goldstein RA. Assessing the accuracy of ancestral protein reconstruction methods. PLoS Comput Biol. 2006;2:598–605. doi: 10.1371/journal.pcbi.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felder CB, Graul RC, Lee AY, Merkle HP, Sadee W. The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci. 1999;1:E2. doi: 10.1208/ps010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berntsson RPA, Smits SHJ, Schmitt L, Slotboom DJ, Poolman B. A structural classification of substrate-binding proteins. FEBS Lett. 2010;584:2606–2617. doi: 10.1016/j.febslet.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 32.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreimer DI, Malak H, Lakowicz JR, Trakhanov S, Villar E, Shnyrov VL. Thermodynamics and dynamics of histidine-binding protein, the water-soluble receptor of histidine permease. Implications for the transport of high and low affinity ligands. Eur J Biochem. 2000;4252:4242–4252. doi: 10.1046/j.1432-1033.2000.01470.x. [DOI] [PubMed] [Google Scholar]
- 34.Ausili A, Staiano M, Dattelbaum J, Varriale A, Capo A, D'Auria S. Periplasmic binding proteins in thermophiles: characterization and potential application of an arginine-binding protein from Thermotoga maritima: a brief thermo-story. Life. 2013;3:149–160. doi: 10.3390/life3010149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 36.Armengou A, Hurtado O, Leira R, Obón M, Pascual C, Moro MA, Lizasoain I, Castillo J, Dávalos A. l-Arginine levels in blood as a marker of nitric oxide–mediated brain damage in acute stroke: a clinical and experimental study. J Cereb Blood Flow Metab. 2003;23:978–984. doi: 10.1097/01.WCB.0000080651.64357.C6. [DOI] [PubMed] [Google Scholar]
- 37.Waldron TT, Murphy KP. Stabilization of proteins by ligand binding: application to drug screening and determination of unfolding energetics. Biochemistry. 2003;42:5058–5064. doi: 10.1021/bi034212v. [DOI] [PubMed] [Google Scholar]
- 38.Okubo Y, Sekiya H, Namiki S, Sakamoto H, Iinuma S, Yamasaki M, Watanabe M, Hirose K, Iino M. Imaging extrasynaptic glutamate dynamics in the brain. Proc Natl Acad Sci USA. 2010;107:6526–6531. doi: 10.1073/pnas.0913154107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardingham N, Dachtler J, Fox K. The role of nitric oxide in pre-synaptic plasticity and homeostasis. Front Cell Neurosci. 2013;7:1–19. doi: 10.3389/fncel.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lerma J, Herranz AS, Herreras O, Abraira V, Del RRM. In vivo determination of extracellular concentration of amino acids in the rat hippocampus. A method based on brain dialysis and computerized analysis. Brain Res. 1986;384:145–155. doi: 10.1016/0006-8993(86)91230-8. [DOI] [PubMed] [Google Scholar]
- 41.Beernink PT, Yang YR, Graf R, King DS, Shah SS, Schachman HK. Random circular permutation leading to chain disruption within and near α helices in the catalytic chains of aspartate transcarbamoylase: effects on assembly, stability, and function. Protein Sci. 2001;10:528–537. doi: 10.1110/ps.39001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinemann U, Hahn M. Circular permutation of polypeptide chains: implications for protein folding and stability. Prog Biophys Mol Biol. 1995;64:121–143. doi: 10.1016/0079-6107(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 43.Semyanov A, Walker MC, Kullmann DM. GABA uptake regulates cortical excitability via cell type–specific tonic inhibition. Nat Neurosci. 2003;6:484–490. doi: 10.1038/nn1043. [DOI] [PubMed] [Google Scholar]
- 44.Henneberger C, Papouin T, Oliet SH, Rusakov DA. Long-term potentiation depends on release of D-serine from astrocytes. Nature. 2010;463:232–236. doi: 10.1038/nature08673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 47.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Systematic Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 48.Yang ZH. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 49.Neylon C, Brown SE, Kralicek AV, Miles CS, Love CA, Dixon NE. Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: a model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry. 2000;39:11989–11999. doi: 10.1021/bi001174w. [DOI] [PubMed] [Google Scholar]
- 50.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Meth. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 51.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS One. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henneberger C, Rusakov DA. Monitoring local synaptic activity with astrocytic patch pipettes. Nat Protoc. 2012;7:2171–2179. doi: 10.1038/nprot.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]


