Abstract
Odorant receptors (ORs) are the largest subfamily within class A G protein-coupled receptors (GPCRs). No experimental structural data of any OR is available to date and atomic-level insights are likely to be obtained by means of molecular modeling. In this article, we critically align sequences of ORs with those GPCRs for which a structure is available. Here, an alignment consistent with available site-directed mutagenesis data on various ORs is proposed. Using this alignment, the choice of the template is deemed rather minor for identifying residues that constitute the wall of the binding cavity or those involved in G protein recognition.
Keywords: odorant receptor, sequence, structure, binding cavity, olfactory
Introduction
Odorant molecules are perceived by mammals through extraordinary subtle mechanisms, notably involving odorant receptors (ORs).1 In human, the family of genes coding for ORs is one of the largest, as it represents more than 2% of our genome. At the protein level, ORs account for more than 4% of our proteome and constitute the largest subfamily of class A (or Rhodopsin like) G protein-coupled receptors (GPCRs). GPCRs are seven-transmembrane domain (7 TM) proteins that transmit extracellular signals across the plasma membrane. Although structures of some class A members have been experimentally solved, no experimental structure is to date available for any OR. For now, molecular modeling appears as the only way to propose atomic-level mechanisms of either ligand selectivity or receptor activation for these proteins on a structural basis. Models can either be made ab initio or based on sequence homology with respect to known experimental structures.2,3 In both cases, sequence alignment between the candidate receptor and the experimentally determined templates is undoubtedly the crucial step.
Within the motifs that represent hallmarks of class A GPCRs, most are shared by ORs,4 suggesting rather similar activation mechanism upon ligand binding and similar signal transduction. It follows that templates available for now may be sufficiently adapted to recover trustable OR models. Nevertheless, ORs conserved motifs are either broader or different than those observed in class A GPCRs. These motifs within OR sequences are as follows, with those shared by nonolfactory class A GPCRs written in bold:
GN in Trans-Membrane domain 1 (TM1),
LHxPMYFFLxxLSxxD in TM2,
MAYD(E)RYVAICxPLxY in TM3,
SY in TM5,
KAFSTCxSH in TM6,
PxLNPxIYSLNR in TM7.
Although TM1, 2, 3, and TM7 motifs are sufficiently conserved to lead to unambiguous alignments, TM4, 5, and 6 cases are more subtle and require additional data, ideally brought by experiments. An accurate sequence alignment will provide extremely useful information on residues forming the binding cavity or involved in receptor activation. Based on a thorough alignment and analysis of conservation thresholds between mouse and human ORs, such information was inferred and allowed identifying residues that contribute to ligand binding.5 In this article, we revisit and update this data by recapitulating available experimental results published so far. We combine information gained by sequence alignments and in vitro data using site-directed mutagenesis to provide an optimal sequence alignment consistent with experiment. In a second step, we use this alignment to assess the choice of the template for building a representative OR and to confirm that site-directed mutagenesis data can be interpreted on a structural basis using this model.
Results
Olfactory and nonolfactory GPCRs alignment
Alignments of TM1, TM2, and TM3 sequences are straightforward as the conserved motifs in each of these TM domains are clearly identified between ORs and available GPCR structures. Figure 1 recapitulates the alignment for ORs with available site-directed mutagenesis data. In TM1, the typical class A GPCR “GN” motif is conserved at 90 and 99% within human and mouse ORs, respectively.14,15 Here, residue N is referenced as N1.50, according to the Ballesteros-Weinstein notation.16 In TM2, the PMY motif found in ORs has no equivalence in any other class A GPCRs but the highly conserved LSxxD in ORs is straightforward to align with the highly conserved GPCR LAxAD (D2.50) motif. The alignment of TM3 is the easiest case because of the presence of both the D(E)RY motif (R3.50) involved in the activation of all class A GPCRs, and the cysteine residue C3.25 involved in the cysteine bridge with the extracellular loop 2 (ECL2). Within TM4, the tryptophan residue (W4.50) strongly conserved in nonolfactory GPCRs is also present in ORs, with conservation of 58% and 50% within human and mouse ORs, respectively. This residue provides a good anchoring point for fitting TM4 sequences of ORs and nonolfactory GPCRs. Before considering TM5 and TM6, we focus on TM7, where the NPxxY (P7.50) motif is conserved in all class A GPCRs making easy the alignment of TM7. In TM5, the highly conserved proline (P5.50) in class A GPCRs16 is moderately represented in ORs (conservation of 39% and 37%, in human and mouse ORs, respectively). However, the tyrosine residue of the “SY” (Y5.58) motif is strongly conserved in both GPCR subfamilies (100% and 93% in mouse and human ORs, respectively). Taking this tyrosine residue as a reference assesses the accurate alignment of TM5 and remains consistent with of the position of the proline residue (P5.50) between OR and sequences associated to available X-ray structures.
Figure 1.
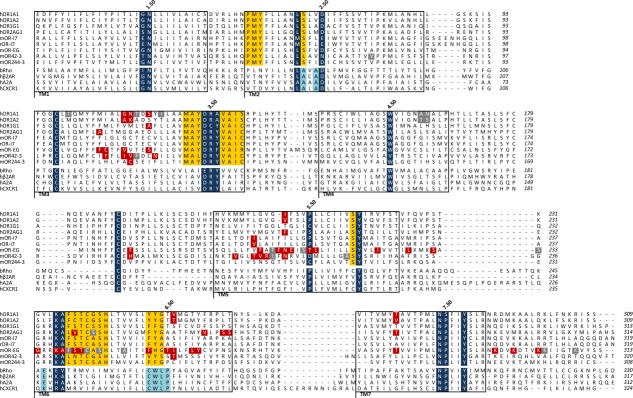
Alignment of ORs with some G protein-coupled receptors (GPCRs). Only ORs for which site-directed mutagenesis combined to molecular modeling was available are considered. Residues commonly conserved between ORs and non-OR GPCRs (dark blue), specific to ORs only (yellow), and specific to non-OR GPCRs only (light blue) are identified. Residues which experimentally modify the OR response upon odorant stimulation are shown in red, while those which do not change the OR response are in gray. Each transmembrane (TM) domain is boxed and the Ballesteros-Weinstein numbering scheme is indicated for Class-A GPCR. An alternative numbering scheme is proposed for the TM5 and TM6 of OR, which takes into account for highly conserved residues within these TMs (orange, italics). Site-directed mutagenesis data are reported for the Human (h) OR1A1 and hOR1A2,6 hOR1G1,7 hOR2AG1,8 Rat (r) and Mouse (m) I7,9 mOR-EG,10,11 mOR42-3,12 and mOR244-3.13 OR sequences are aligned with sequences of Bovine Rhodopsin (bRho), human β2-adrenergic (hβ2AR), human Adenosine-2A (hA2A), and human Chemokine-1 (CXCR1) receptors.
TM6 is even much trickier, as this TM lacks the CWxP (P6.50) motif considered as the TM6 hallmark of class A GPCRs. In TM6, ORs sequences show a highly conserved KAFSTCxSH motif for which the equivalence with nonolfactory GPCRs is not obvious. A “KA” motif can, however, be identified in nonolfactory GPCRs, and a 29% conserved proline in human ORs is aligned with the P6.50, assessing our alignment.
Intra and extra-cellular loops are also of importance for the function of a receptor. Here, we notably focus on ECL2 since it is involved in ligand binding and receptor structure. A disulfide bridge between ECL2 and C3.25 at the top of TM3 is common to all class A GPCRs. In ORs, three cysteines are present in ECL2 domain and one at the top of TM3, suggesting the presence of two disulfide bridges. Indeed, in addition to the canonical S-S bridge (between C973.25 and C179ECL2), identification of an additional S–S bridge within ECL2 (between C169ECL2 and C189ECL2) was characterized by mass spectrometry in hOR1D2.17 Forcing the alignment of the canonical cysteine bridge between ORs and nonolfactory GPCRs (C973.25-C179ECL2) provides a crucial data for the optimal alignment of ECL2.
This sequence alignment does not contain any gap within TM domains. The only gaps are set within loop sequences, consistent with a larger sequence and structure variability within loops with respect to the bundle.18 Based on the alignment of Figure 1, we next address the choice of template used for building a structural model consistent with site-directed mutagenesis data.
Three-dimensional structure and comparison with experimental data
Here, we analyze the accuracy of the alignment by translating it into atomic-level models. Five models of the human OR1G1 are built either with Modeller19 using different receptor structures as templates (Bovine Rhodopsin, Human β2-adrenergic, Human Chemokine-1, and a combination of them three) or by means of the ab initio GEnSeMBLE (GPCR Ensemble of Structures in Membrane BiLayer Environment) complete sampling method.3,20,21
Figure 2 gathers information inferred from these models. Focusing on the helical TM domains, all structures are similar with Cα Root Mean Square deviations (RMSd) lower than 3 Å [see Fig. 2(C)] between pairs of models, at the exception of that based on the chemokine receptor. The latter exhibits a RMSd value of ∼6 Å with respect to other structures. The main difference when using the Chemokine receptor template appears for TM1, TM2, and TM7 which show a small deviation with respect to other templates. This difference has however a small influence on the position of residues lining the binding cavity. Focusing on eight of them (1043.32, 1083.36, 2025.42, 2065.46, 2526.48, 2566.52, 2606.56, and 2797.42, vide infra), we compute a Cα RMSd of 3.2 Å between the multitemplate model and that build with the chemokine receptor. Importantly, despite these tertiary structure weak dissimilarities, all models exhibit similar secondary folds. Furthermore, residues that constitute the wall of the binding cavity and those involved in the signaling pathway through a contact with the G protein appear to be located in the same regions.22,23 As observed in all class A GPCRs, the canonical binding site is made up by residues belonging to TM3, TM5, TM6, and TM7.5 Inspection of TM3 3D-structure shows that side-chains of residues 1093.37, 1083.36, 1053.33, and 1043.32 participate to the binding cavity. This is consistent with a modification of the odorant response when tested in mutants expressed in vitro (Fig. 1). In the models, residue 1123.40 is located under the binding cavity. Its non-synonymous mutation is consistent with a general decrease of the OR response to odorants in hOR1G1 (Ala → Ser),24 mOR-EG (Ser → Ala or Val),10,11, and hOR1A1 (Ser → Ala).6
Figure 2.
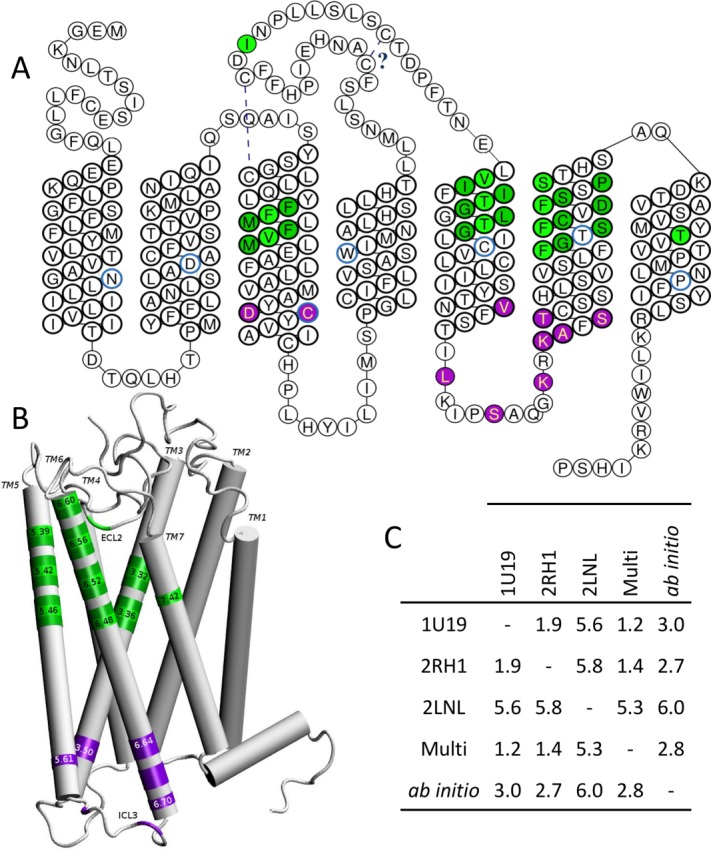
Residues governing the function of mammalian ORs projected onto the sequence and the structure of hOR1G1. A, snakeplot of the OR sequence with residues involved in odorant contact in green and those involved in the OR activation through a contact with the G Protein in purple. Residues in light green will be strongly in contact with the odorant, those in dark green contribute to the wall of the binding cavity. Number 50 residue of the Ballesteros-Weinstein notation are circled in blue. The cysteine bridges are also indicated. B, position of important residues on the structure of the receptor, with some Ballesteros-Weinstein notations. C, C-α positions Root Mean Square deviation (in Å) between models build using Bovine Rhodopsin (PDB ID: 1U19), β2-adrenergic (PDB ID: 2RH1), Chemokine-1 (PDB ID: 2LNL) receptor, or a multitemplate (Multi) of the three receptors cited above, or an ab initio model (See Supporting Information for PDB structure of each model).
TM4 would contribute to lining the binding cavity through one or two residues located at the top of the helix. Mutations at these positions (4.55 and 4.56) however do not affect responsiveness of the receptor,6 suggesting that this contribution is deemed rather minor.
Amino-acids belonging to TM5 largely contribute to define the binding cavity. Side-chains of residues 1995.39, 2025.42, 2065.46 point inward the cavity, consistent with a modification of the response to odorants upon mutation on mOR-EG10,11 and mOR42-3 in vitro.12 In mOR-EG, mutations at residues located deeper into the structure (5.50 and 5.51) also affected responsiveness of the receptor when stimulated by odorants. They would rather contribute to stabilize the receptor since they correspond to positions within the sequence showing a larger conservation (Pro at ∼40% at position 5.50, Phe/Leu at 64% at 5.51, and Ile at ∼85% at 5.61) than hypervariable residues found within the cavity.5 The main contribution of TM6 to the function of the receptor stems not only from residues within the binding cavity but also from others involved in the activation. The highly conserved aromatic residue at position 6.48 (Y/F252 is conserved at ∼95%) is located at the bottom of the binding cavity. One, two, and three helix turns above, residues 2556.51−2566.52, 2596.55−2606.56, and 2636.59−2646.60 are pointing to the cavity. These positions are in line with in vitro data on mOR-EG,10,11 mOR42-3,12 hOR2AG1,8 hOR1A1, and hOR1A2, where the response of the receptor upon odorant stimulation is modified by mutations at these positions.6 Deeper into the intracellular part, the “KAFSTCASH” is likely to take part in the contact with the G protein upon activation, as shown on mOR-EG.22 The contribution of TM7 to the binding pocket is mostly coming from residue 2797.42, consistent with its impact on ligand recognition on several ORs in vitro.6,8,10
Conclusion
We have built an alignment of mammalian Odorant Receptor sequences that recapitulates available experimental data obtained by site-directed mutagenesis. More particularly, the debatable alignment of TM5 and TM6 are now consistent with data provided by several other studies. The effect of the template in the case of homology-based approaches is deemed rather minor if one is interested in identifying residues that belong to the binding cavity or those potentially involved in the coupling of a G protein to the OR. These data provide a robust starting point for initiating mechanistic or structural studies involving odorant receptor and their complexes with ligands.
Materials and Methods
The alignment was performed with Jalview.25 Sequences have been firstly aligned with ClustalW before manual adjustments. Tools of GPCRDB have been used to obtain a snakeplot. Three-dimensional models have been built either with Modeller19 by homology modeling using a mono- or multitemplate (Bovine Rhodopsin PDB ID: 1U19, Human β2-adrenergic PDB ID: 2RH1 and Human Chemokine-1 PDB ID: 2LNL) or by an ab initio protocol with the GEnSeMBLE (GPCR Ensemble of Structures in Membrane BiLayer Environment) complete sampling method.20 Visual analysis, images, and RMSd calculations have been performed with VMD.26
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supporting Information.
References
- 1.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 2.Yarnitzky T, Levit A, Niv MY. Homology modeling of G-protein-coupled receptors with X-ray structures on the rise. Curr Opin Drug Discov Devel. 2010;13:317–325. [PubMed] [Google Scholar]
- 3.Charlier L, Topin J, de March CA, Lai PC, Crasto CJ, Golebiowski J. Molecular modelling of odorant/olfactory receptor complexes. In: Crasto CJ, editor. Olfactory receptors. New York: Humana Press; 2013. pp. 53–65. [DOI] [PubMed] [Google Scholar]
- 4.Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- 5.Man O, Gilad Y, Lancet D. Prediction of the odorant binding site of olfactory receptor proteins by human–mouse comparisons. Protein Sci. 2004;13:240–254. doi: 10.1110/ps.03296404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmiedeberg K, Shirokova E, Weber H-P, Schilling B, Meyerhof W, Krautwurst D. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 2007;159:400–412. doi: 10.1016/j.jsb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Launay G, Sanz G, Pajot-Augy E, Gibrat J-F. Modeling of mammalian olfactory receptors and docking of odorants. Biophys Rev. 2012;4:255–269. doi: 10.1007/s12551-012-0080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelis L, Wolf S, Hatt H, Neuhaus EM, Gerwert K. Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angew Chem Int Ed. 2012;51:1274–1278. doi: 10.1002/anie.201103980. [DOI] [PubMed] [Google Scholar]
- 9.Krautwurst D, Yau K-W, Reed RR. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell. 1998;95:917–926. doi: 10.1016/s0092-8674(00)81716-x. [DOI] [PubMed] [Google Scholar]
- 10.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baud O, Etter S, Spreafico M, Bordoli L, Schwede T, Vogel H, Pick H. The mouse eugenol odorant receptor: structural and functional plasticity of a broadly tuned odorant binding pocket. Biochemistry. 2011;50:843–853. doi: 10.1021/bi1017396. [DOI] [PubMed] [Google Scholar]
- 12.Abaffy T, Malhotra A, Luetje CW. The molecular basis for ligand specificity in a mouse olfactory receptor: a network of functionally important residues. J Biol Chem. 2007;282:1216–1224. doi: 10.1074/jbc.M609355200. [DOI] [PubMed] [Google Scholar]
- 13.Sekharan S, Ertem MZ, Zhuang H, Block E, Matsunami H, Zhang R, Wei JN, Pan Y, Batista VS. QM/MM model of the mouse olfactory receptor mor244-3 validated by site-directed mutagenesis experiments. Biophys J. 2014;107:L5–L8. doi: 10.1016/j.bpj.2014.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zozulya S, Echeverri F, Nguyen T. The human olfactory receptor repertoire. Genome Biol. 2001;2:1–12. doi: 10.1186/gb-2001-2-6-research0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
- 16.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In: Stuart CS, editor. Methods in neurosciences. New York: Academic Press; 1995. pp. 366–428. [Google Scholar]
- 17.Cook BL, Steuerwald D, Kaiser L, Graveland-Bikker J, Vanberghem M, Berke AP, Herlihy K, Pick H, Vogel H, Zhang S. Large-scale production and study of a synthetic G protein-coupled receptor: human olfactory receptor 17-4. Proc Natl Acad Sci USA. 2009;106:11925–11930. doi: 10.1073/pnas.0811089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheatley M, Wootten D, Conner MT, Simms J, Kendrick R, Logan RT, Poyner DR, Barwell J. Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol. 2012;165:1688–1703. doi: 10.1111/j.1476-5381.2011.01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, Pieper U, Sali A. Current Protocols in Bioinformatics. New York: Wiley; 2006. Comparative protein structure modeling using modeller. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bray JK, Abrol R, Goddard WA, Trzaskowski B, Scott CE. SuperBiHelix method for predicting the pleiotropic ensemble of G-protein–coupled receptor conformations. Proc Natl Acad Sci USA. 2014;111:E72–E78. doi: 10.1073/pnas.1321233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim S-K, Goddard W., III Predicted 3D structures of olfactory receptors with details of odorant binding to OR1G1. J Comput Aided Mol Des. 2014;28:1–16. doi: 10.1007/s10822-014-9793-4. [DOI] [PubMed] [Google Scholar]
- 22.Kato A, Katada S, Touhara K. Amino acids involved in conformational dynamics and G protein coupling of an odorant receptor: targeting gain-of-function mutation. J Neurochem. 2008;107:1261–1270. doi: 10.1111/j.1471-4159.2008.05693.x. [DOI] [PubMed] [Google Scholar]
- 23.Kato A, Touhara K. Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell Mol Life Sci. 2009;66:3743–3753. doi: 10.1007/s00018-009-0111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Launay G, Teletchea S, Wade F, Pajot-Augy E, Gibrat JF, Sanz G. Automatic modeling of mammalian olfactory receptors and docking of odorants. Protein Eng Des Sel. 2012;25:377–386. doi: 10.1093/protein/gzs037. [DOI] [PubMed] [Google Scholar]
- 25.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.


