Abstract
Laccase (p-diphenol:dioxygen oxidoreductase, EC 1.10.3.2) is a member of the multicopper oxidases and catalyzes the one-electron oxidation of a wide range of substrates, coupled with the reduction of oxygen to water. It is widely distributed in bacteria, fungi, plants and insects. Laccases are encoded by multigene family, and have been characterized mostly from fungi till now, with abundant industrial applications in pulp and paper, textile, food industries, organic synthesis, bioremediation and nanobiotechnology, while limited researches have been performed in plants, and no application has been reported. Plant laccases share the common molecular architecture and reaction mechanism with fungal ones, despite of difference in redox potential and pH optima. Plant laccases are implicated in lignin biosynthesis since genetic evidence was derived from the Arabidopsis LAC4 and LAC17. Manipulation of plant laccases has been considered as a promising and innovative strategy in plant biomass engineering for desirable lignin content and/or composition, since lignin is the major recalcitrant component to saccharification in biofuel production from lignocellulose, and therefore directly limits the fermentation yields. Moreover, plant laccases have been reported to be involved in wound healing, maintenance of cell wall structure and integrity, and plant responses to environmental stresses. Here, we summarize the properties and functions of plant laccase, and discuss the potential of biotechnological application, thus providing a new insight into plant laccase, an old enzyme with a promising beginning in lignocellulose biofuel production.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-015-0331-y) contains supplementary material, which is available to authorized users.
Keywords: Laccase, Lignin, Lignocellulose, Biofuel, Genetic engineering
Background
As a renewable, environmental-friendly and economical resource, biofuel has received much attention in recent years. After the “first generation” biofuel derived from starch and sugar-based raw materials like maize, sugarcane and sugar beet, the “second generation” biofuel production based on lignocellulosic biomass is emerging as a new promising industry [1]. Generally, the process of the second generation biofuel production includes pretreatment for liberation of polysaccharides, release of monomeric sugars based on enzymatic hydrolysis, fermentation and subsequential distillation. Feedstock for lignocellulosic biofuel production can be non-edible energy crops (Miscanthus, switchgrass, sweet sorghum, etc.), agricultural and forest residues, as well as municipal and industrial wastes, which are usually economically feasible, noncompeting with food supplies, and can be produced in large quantities [2].
The major component of the lignocellulosic biomass is the plant cell wall, a heterogeneous complex mainly consisting of cellulose, hemicellulose and lignin [1]. In the conversion process, lignin has been identified as the primary recalcitrant component to saccharification, since it tends to adsorb cellulolytic enzymes and restrict cellulose release, also the degradation byproducts can inhibit the activity of cellulolytic enzymes [3, 4]. As lignin directly limits the fermentation yield, its removal by pretreatment or modification prior to enzymatic hydrolysis and fermentation would be advantageous. Genetic engineering of plant cell wall has been considered as a promising strategy for desirable lignin content and/or composition towards higher biofuel production [5]. Manipulation of laccase, which catalyzes monolignols oxidation and polymerization in plant lignin synthesis, would be a great approach for plant biomass engineering [6, 7].
Laccase (p-diphenol:dioxygen oxidoreductase, EC 1.10.3.2), a family of copper-containing polyphenol oxidases, belongs to the multicopper oxidases (MCOs). Though first identified in the exudates of the Japanese lacquer tree Rhus vernicifera, studies on laccase have been performed predominantly in fungi in the past decades. And a great many reviews concerning fungal laccase have been published with emphasis on the general characteristics [8–13], catalytic properties [14–16], biological functions [17–19], industrial applications [20–26], and laccase engineering for higher catalytic activity and stability [27–29]. Compared to the extensive studies on fungal laccases, the research of plant laccases is just emerging in the past several years. This paper presents an overview of the current knowledge of plant laccases in aspects of the essential characteristics, gene expression pattern and regulation, genetic view of plant laccases in lignin biosynthesis, and divergent functions. Finally, the potential applications of plant laccase in energy plant engineering and phytoremediation are discussed.
Laccase: different sources with common structure but distinct properties
Typically, laccase contains four copper ions classified by different spectral and electronic paramagnetic resonance (EPR) properties: a mononuclear “blue” copper ion (Cu1) at the T1 site which confers the typical blue color to the protein, and a trinuclear copper cluster at the T2/T3 site, consisting of one T2 copper ion (Cu2) and two T3 copper ions (Cu3) [8, 9, 30]. Each of the copper ions plays a dissimilar role in the one-electron oxidation of various substrates, such as ortho- and para-diphenols, aminophenols, polyphenols, polyamines and aryl diamines, concomitantly with the full reduction of molecular oxygen to water (Fig. 1) [14, 25, 31]. Specifically, T1 is the site where substrate oxidation takes place while the reduction of oxygen and release of water occurs at the T2/T3 site [32, 33].
Fig. 1.
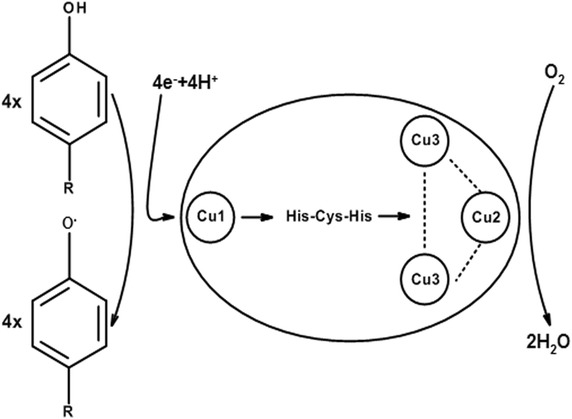
Schematic diagram of laccase catalytic cycle, adapted from Baldrian [10]. The laccase substrate is oxidized by Cu1, involving the loss of a single electron and the formation of a free radical, which may undergo further oxidation or nonenzymatic hydration, disproportionation and polymerization. Reduction of Cu1 is rate-limiting in the catalytic process. The extracted electron is transferred through highly conserved His-Cys-His motif to the trinuclear copper cluster, where molecular oxygen is reduced to water. The reduction of one molecule of oxygen to two molecules of water is accompanied by oxidation of four molecules of substrate
Laccase is widely distributed in plants, insects and microbes (including bacteria and fungi), and is encoded by multigene family, for example, the model plant Arabidopsis genome encodes 17 laccases, and the number of identified laccase genes in the Trametes villosa fungus and the insect Nephotettix cincticeps is 5 and 3, respectively [34–36]. Fungal laccases have been comprehensively identified from ascomycetes, deuteromycetes, and basidiomycetes especially the white-rot basidiomycetes, and are involved in lignin degradation, morphogenesis, pathogenesis and stress defence [8–10]. Genomic screening in bacteria has found widespread occurrence of laccases as well, although sequence and structure characterization were not widely reported [37–39]. The best-studied bacteria laccase so far is CotA in Bacillus subtilis, which participates in pigment biosynthesis and protection against UV light [40]. Moreover, laccase has been identified in archaea, such as the LccA from halophilic archaeon Haloferax volcanii, a highly thermostable and salt/solvent-tolerant laccase [21, 41]. In addition to microbial laccases, insect laccases have been continually identified in Monochamus alternatus, Bombyx mori, Apis mellifera, etc., which are mainly associated with cuticle sclerotization of insects [39, 42–44]. And to date, laccases have also been widely identified from plants besides Anacardiaceae, such as loblolly pine (Pinus taeda), sycamore maple (Acer pseudoplatanus), tobacco (Nicotiana tabacum), poplar (Populus trichocarpa), yellow poplar (Liridendron tulipifera), ryegrass (Lolium perenne) [45–50], etc. Moreover, in-depth research has been reported in Arabidopsis thaliana, maize (Zea mays), rice (Oryza Sativa), sugarcane (Saccharum officinarum), Brassica napus and Brachypodium distachyon [7, 51–55].
Laccases from plants, fungi, insects and bacteria are clustered separately in phylogenetic tree, although all of them have conserved copper binding amino acids, including ten histidines, one cysteine and an axial ligand of methionine, leucine or phenylalanine (Fig. 2). The axial ligand can roughly indicate the redox potential of a given laccase: a coordinating methionine for low potential whereas a non-coordinating leucine or phenylalanine for middle and high potential, respectively, but there are exceptions [14, 30, 56]. Generally, plant and insect laccases have lower redox potential. Microbial laccases fall into all three groups, among which the bacteria laccases belong to the low-redox-potential group, the basidiomycete white-rot fungi laccases mainly constitute the high-redox-potential group, and the majority of ascomycete and some basidiomycete fungi laccases are attached to the middle-redox-potential group [27, 29].
Fig. 2.
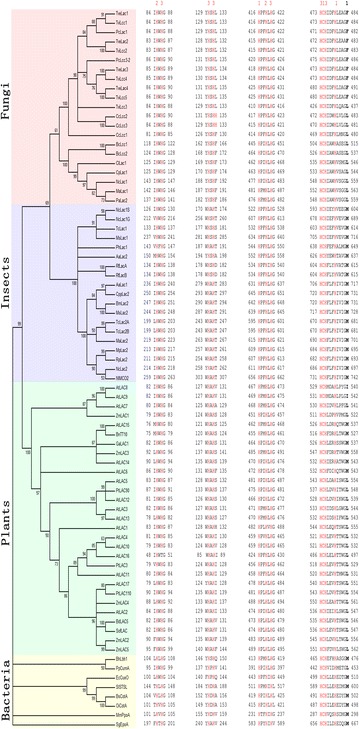
Phylogenetic analysis of representative laccases from plants, fungi, insects and bacteria. Laccases are encoded by multigene family, for example, the Arabidopsis genome encodes 17 laccases, and the number of identified laccase genes in Trametes villosa and Nephotettix cincticeps is 5 and 3, respectively. Laccases from plants, fungi, insects and bacteria are clustered separately in the phylogenetic tree for each taxon, but share conserved copper binding sites. The amino acids potentially involved in copper binding include ten histidines and one cysteine (in red), with numbers 1, 2 and 3 corresponding to the Cu1, Cu2 and Cu3 ions. An axial ligand of methionine, leucine, or phenylalanine (in bold) was indicated. The bootstrap consensus tree was constructed by MEGA with the Neighbor-Joining method. Database accession numbers of laccase sequences are listed in Additional file 1: Table S1
General enzymology characteristics of plant laccase
Plant laccases are glycoproteins with higher carbohydrate content (20–45 %) than fungal laccases (10–25 %), which has been reported to be responsible for copper retention, enzyme stability and activity. The molecules usually consist of 500–600 amino acids and weigh approximately 60–130 kDa, while the isoelectric point (pI) values range from 7.0 to 9.6 (Fig. 3a) [8, 56]. Most plant laccases are secreted proteins, with a few exceptions predicted to be located in mitochondria.
Fig. 3.
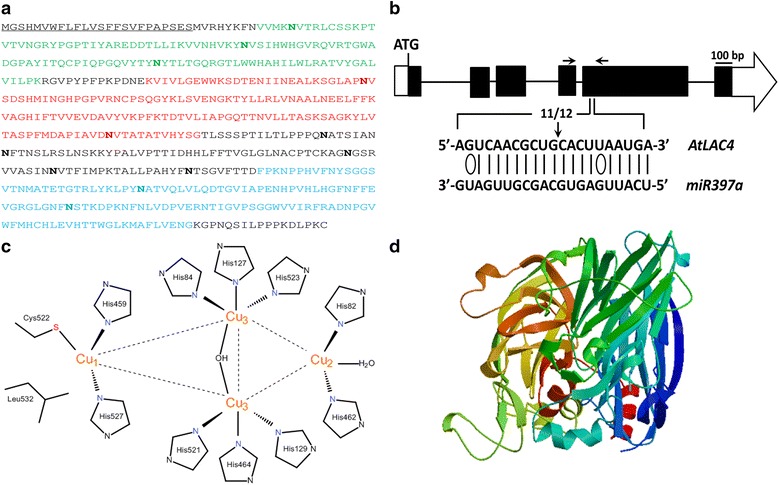
Sequence and structure of AtLAC4 from A. thalina. a The amino acid sequence of AtLAC4. The protein consists of 558 amino acids and weighs 61.5 kDa, with theoretical pI value of 9.31. It has a signal peptide (underlined) at the N-terminal predicted by SignalP 4.1 Server [98] and three conserved Cu-oxidase domains (colored in red, blue and green, respectively), according to Pfam [99]. Twelve asparagines predicted to be N-glycosylated by NetNGlyc 1.0 Server [100] were indicated in bold. b MiR397a mediated cleavage of AtLAC4, adapted from Abdel-Ghany et al. [63]. The black boxes represent exons and the horizontal lines represent introns. The white box represents 5′-UTR, while the white arrow represents 3′-UTR. Vertical arrow indicates the 5′ termini of miRNA-guided cleavage products, with the frequency of clones shown. Watson–Crick pairing (solid lines) and G:U wobble pairing (ellipse) between AtLAC4 target sequence and the complementary miRNA397a sequence are indicated. c The view of ligands at the copper center of AtLAC4. Cu1 is coordinated with two histidines, one cysteine and one leucine, Cu2 is coordinated by another two histidines and one H2O ligand, while six histidines coordinate the Cu3 pair in a symmetrical manner, with a bridging OH ligand. d Three-dimensional structure of AtLAC4 predicted by SWISS-MODEL [101]
Although, the active sites of plant laccases share the common molecular architecture (Fig. 3d) and reaction mechanism with fungal ones, they have lower redox potential and different pH requirements. The lower redox potential is due to a methionine or leucine rather than phenylalanine residue at the T1 site of plant laccases (Figs. 2, 3c) [9]. The pH optima greatly differs between laccases of plant and fungus origin, as the optimal pH for plant laccases is around physiological range of 7.0–10.0 while a lower acidic pH is optimal for the fungal ones [57]. Additionally, altered microenvironment at the active site was also predicted for both of the laccases [58, 59]. These characteristics may partly account for the different functions of plant and fungal laccase, one catalyzes lignin biosynthesis while the other is responsible for degradation, respectively. Further research is required to distinguish other differences.
Regulation of expression and subcellular localization of plant laccase
Plant laccases are expressed diversely in different tissues at various developmental stages. Tissue-specific expression patterns have been reported in Arabidopsis, such as LAC4 specifically expressed in interfascicular fibers, vascular bundles and seed coat columella, LAC7 in hydathodes and root hairs, LAC8 in pollen grains and phloem, LAC15 in seed coat cell walls, and LAC17 in interfascicular fibers [34, 60]. And in sugarcane, SofLAC has been detected to be preferentially expressed in sclerenchymatic bundles and parenchymatic cells surrounding the vascular bundles of young internodes [51]. The tissue-specific expression profiles were also detected for eight of the P. taeda laccases [61].
There are multiple cis-elements in promoter sequences of Arabidopsis laccases, indicating potential roles of transcription factors in regulating laccase expression [34]. MYB58, for instance, one of the SND1-regulated MYB transcription factors, is able to activate the expression of LAC4 gene directly [62]. In addition, 15 of the 17 laccases from Arabidopsis (except LAC6 and LAC14) contain copper response elements in promoter regions, suggesting the expression of these genes may be responsive to Cu levels [34]. Recently, miRNAs have also been reported to target laccases and function in the regulation of laccase expression at post-transcriptional level. Several laccases were potentially regulated by miRNAs under Cu deficient conditions in Arabidopsis, among which miR408 was predicted to target the coding sequences of LAC3, LAC12 and the 5′-UTR of LAC13; miR397 was predicted to target the fifth exon of LAC2, LAC4 (Fig. 3b) and LAC17; while miR857 was predicted to target the first exon of LAC7. Furthermore, the expression of these miRNAs was negatively correlated with that of the laccase targets [63]. A recent study found the involvement of miR397b in regulating both lignin content and seed number in Arabidopsis via modulating LAC4 [6]. Similar mechanisms have also been revealed in P. trichocarpa and O. Sativa, where Ptr-miR397a and OsmiR397 perform as negative regulators of PtrLACs and OsLAC, respectively [7, 64].
The expression of certain plant laccases are responsive to environmental stresses, based on large-scale sequencing analysis or expression detection. For example, the transcript level of a laccase gene was induced in tomato roots treated with 170 mM NaCl [65]. Similar increase of ZmLAC1 transcripts was observed in maize primary roots treated with varied NaCl concentrations [52]. Another study also detected sharp increase of laccase transcripts responding to lead (Pb) stress in maize, which was suggested to account in part for Pb hyperaccumulation in the line 178 [66].
Most plant laccases are predicted to be localized in the apoplast, on account of the presence of N-terminal signal peptide (Fig. 3a) which directs the protein into the secretory pathway. Though, limited experimental data have been gathered regarding the subcellular location of plant laccases. It has been reported in Arabidopsis that LAC4 and LAC17 are located in the secondary cell walls throughout protoxylem tracheary element differentiation [67]. And in B. distachyon, BdLAC5 and BdLAC6 were detected in the apoplasm in lignified interfascicular fibers [55]. The exception worth noting is that AtLAC15 was reported to be observed within the vacuole lumen instead of the cell wall, which may be related to its function [68].
Genetic evidence for the involvement of plant laccase in lignin biosynthesis
Being contrary to the ligninolytic activity of fungal laccases, plant laccases are involved in lignin polymerization [69, 70]. Lignin, the second most abundant biopolymer after cellulose, accounts for approximately 30 % of the organic carbon in the biosphere and has been essential in the evolutionary adaptation of plants from aquatic environment to land [71]. Lignin is derived primarily from three hydroxycinnamyl alcohol monomers: coniferyl, p-coumaryl and sinapyl alcohols (termed monolignols) (Fig. 4a), which are synthesized in the cytosol and thereafter is exported into the cell wall for endwise polymerization and incorporation into guaiacyl (G), p-hydroxyphenyl (H) and syringyl (S) lignin units, respectively (Fig. 4b) [72, 73]. The oxidation of monolignol molecules is thought to be catalyzed by one or more peroxidases; however, increasing evidences have indicated the involvement of laccase in this process [74, 75].
Fig. 4.
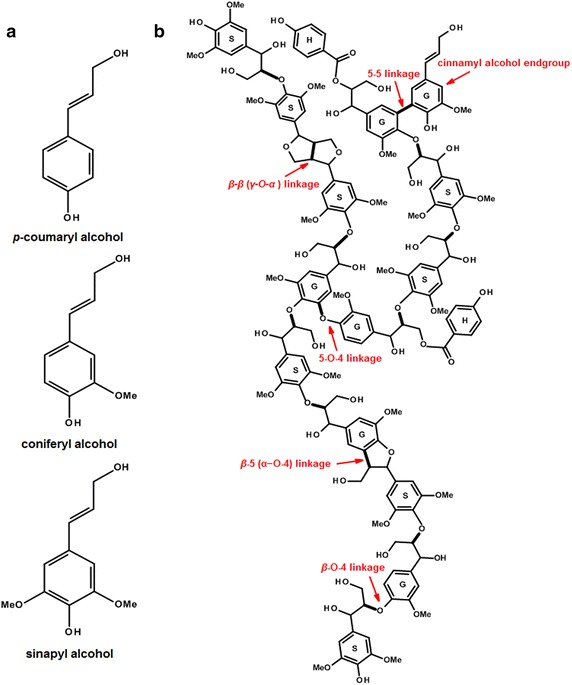
Monolignols and a lignin polymer model. a Primary lignin monomers: p-coumaryl alcohol, coniferyl alcohol and sinapyl alcohol. The three monolignols differ in the degree of methoxylation, and are catalyzed by peroxidase and/or laccases to form the corresponding p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) lignin unit, respectively. b A polymer model depicting the general feature of lignin. The model is constructed by Accelrys Draw 4.2 with two H units, six G units and eight S units, showing the main linkage types. The bolded bonds are formed in the radical coupling reactions. This is only a model, and do not imply any primary structure or composition of plant lignin
In vitro experiments were performed in P. taeda and A. pseudoplatanus as early as 20 years ago to associate laccase with lignification [45, 76]. Later in P. trichocarpa, three independent antisense poplar lines, lac3AS, lac90AS, and lac110AS were generated, but no significant alteration in lignin content and composition was observed. However, lac3AS exhibited a two- to threefold increase in total soluble phenolic content and a dramatic alteration of xylem fiber cell walls. The results indicated that LAC3 is essential for normal cell wall structure and integrity of poplar xylem fibers [77]. Chemical component analysis of the Arabidopsis lac15 mutant seeds revealed nearly 30 % decrease in extractable lignin content compared with wild-type seeds [78]. This was the first direct evidence for possible role of laccase in lignin synthesis.
Genetic evidence was not achieved until the year 2011 when Berthet et al. produced two double mutants of lac4-1lac17 and lac4-2lac17, the lignin content of which were 20 and 40 % lower than those of the control, respectively, while the single mutants lac4 and lac17 had moderately reduced lignin levels. Complementation of lac17 with LAC17 restored the mutant to a normal lignin profile. This study provided the first in vivo evidence that both of LAC4 and LAC17 contribute to the constitutive lignification of Arabidopsis stems [60]. Moreover, simultaneous disruption of LAC11 along with LAC4 and LAC17 almost completely abolished lignin deposition in lac4lac11lac17 triple mutant, causing severe plant growth defect, whereas the casparian strip develops normally through the activity of peroxidase, suggesting that laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis [79]. Another research in sugarcane discovered that the expression of SofLAC restored the lignin content but not the lignin composition in complemented Arabidopsis lac17 mutant, suggesting that SofLAC participates in lignification in sugarcane [51]. These findings indicate that genetic engineering of lignin-specific laccases is a potentially innovative and promising tool for fine-tuning lignin content and/or composition.
Diverse functions of plant laccase
Plant laccases are also involved in varieties of biological processes such as wound healing [17, 80], iron metabolism [81], and maintenance of cell wall structure and integrity [77]. It has been reported that mutants of the Arabidopsis laccases exhibit multiple phenotypes, suggesting different roles performed by them. For example, lac2 showed compromised root elongation under PEG-induced dehydration conditions; lac8 flowered earlier than wild-type plants, and lac15 showed an altered seed color [82].
Certain laccase members are predicted to take part in the polymerization of phenolic compounds. The Arabidopsis lac15 mutant has 59 % increase in soluble proanthocyanidin or condensed tannin in seeds [78]. Similar roles have been found for genes BnTT10, PtLAC3, and ZmLAC3, functioning in B. napus, P. trichocarpa and Z. maize, respectively [54, 77, 83].
Additionally, laccases are indicated to participate in plant response to environmental stresses, based on the findings that the expression of laccase was regulated by abiotic stresses [52, 65, 66]. Moreover, overexpression of a putative laccase gene from rice, OsChI1, increased the tolerance of transgenic Arabidopsis to drought and salinity stress [84]. The involvement of laccase in adversity stress response seems to be common, implying the potential of breeding energy crops on marginal lands, whereas further investigations are still needed to unravel the molecular mechanisms.
Application of plant laccase in energy plant improvement
So far, only fungi and a few bacteria laccases have been utilized in industry [24, 37]. Fungi laccases with high redox potential are particularly useful than other ones in application, attributing to a broader range of substrates as well as the expanding utilization of mature laccase mediator system [85, 86]. Indeed, fungi laccases have been widely applied in pulp and paper, textile, food industries, organic synthesis, bioremediation and nanobiotechnology [23, 26].
Moreover, studies on enzyme immobilization have recently focused on laccase, mainly referred to fungi laccase, aiming to optimize good performance in industrial and large-scale applications. Various methods such as physical coupling (entrapment, encapsulation) and chemical interactions (adsorption, covalent binding and self-immobilization) have been developed [87–89]. Immobilization of laccase offers preserved enzymatic activity, improved storage and operational stability, and fine reusability for enzyme applications in the industrial process.
As for plant laccase, however, no example from industrial application has been reported yet. On the other hand, manipulation of laccase in planta may be a bold attempt for desirable lignin content and/or composition towards optimum plant biomass. This distinguishes largely from the way of removing lignin by ligninolytic enzymes (fungal laccase, together with manganese peroxidase, lignin peroxidase, and versatile peroxidase), since the former strategy takes aim at acquisition of optimal plant biomass fit directly for biofuel production.
The chemical composition, more specifically, the lignocellulosic matrix of plant species can affect the potential biofuel yield, according to a batch anaerobic digestion tests of 41 energy crops, in which 80 % of the sample variation on biofuel yield can be explained through lignin [90]. In wheat and rice, the H unit proportion of lignin has been revealed to be one of the dominant factors positively determining biomass digestibility [91]. Similarly in sweet sorghum, the G-monomer has been detected to have negative effects on biomass enzymatic digestibility and ethanol fermentation [92]. Hence, modification of lignin composition can be potentially employed for higher biomass saccharification in bioenergy crops. On the other hand, a compensatory mechanism between lignin and cellulose has been observed in transgenic aspen (Populus tremuloides Michx.) and the Arabidopsis eli1 mutants, since repression of lignin biosynthesis promotes cellulose accumulation and reduced cellulose synthesis invokes lignification, respectively [93, 94]. From the perspective of energy utilization, increased cellulose content promises lower input but more output, providing directions and goals for energy plants improvement.
Previous studies have shown the inhibition of genes involved in monolignol biosynthesis, including phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), caffeic acid (5-hydroxyconiferaldehyde) O-methyltransferase (COMT), and (hydroxy)cinnamoyl CoA reductase (CCR), could successfully alter lignin profiles. However, unfavorable phenotypes such as collapsed xylem and dwarf stature, decreased pollen viability, and altered leaf and flower morphology often accompanied the desirable changes [95]. Plant laccases catalyze the last step of monolignols oxidation and polymerization in lignin synthesis; furthermore, certain plant laccase members participated in specific lignin unit deposition, such as the Arabidopsis LAC17 involved in the deposition of G lignin unit in the interfascicular fibers [60]. It is theoretically feasible to manipulate laccase gene expression for modifying lignin content and composition. The BdLAC5-misregulated Bd4442 mutant in B. distachyon has been reported to show 10 % decreased Klason lignin content and modification of the S/G ratio, while the mutant showed higher saccharification efficiency. The results provided clear evidence that laccases are promising targets to alleviate the recalcitrance of grass lignocelluloses [55]. On the other hand, due to the functional redundancy of laccase members, indirect modification through the upstream miRNA engineering would be more efficient as one miRNA could target several laccases. It has been reported that transgenic Arabidopsis plants overexpressing miR397b have reduced lignin deposition as well as enlarged seeds and increased seed yield [6]. In rice, overexpression of OsmiR397 enlarges grain size and promotes panicle branching, leading to an increase in overall grain yield of up to 25 % in a field trial [7]. Both of the studies indicated that regulation of laccase by miRNA have the potential for plant biomass engineering with less lignin and combined high yield properties.
To sum up, the application of plant laccase in biomass engineering through in planta manipulation of laccase/miRNA promises a bright development prospect, although no practice case has ever been reported in energy plant.
Perspectives
So far, researches of plant laccase have been focused on cloning, bioinformatics analysis, and verification of the function of a single laccase gene. Besides, functional studies of plant laccase have been mainly carried out in model plants Arabidopsis and rice. Laccase is encoded by multigene family and has divergent functions. Therefore, the identification of laccase family members in different plant species and the evolutionary analysis should be essential for better understanding of their functions. Additionally, more information is required to determine the precise effect of laccase manipulation on biofuel production from lignocellulose, along with technical challenges in commercialization to be solved in the future.
In bioenergy production, tissue-specific promoters should be employed to achieve optimal laccase gene expression without negative effects on growth of transgenic energy plants, in order for maximized ethanol production and economic benefits. For example, it has been demonstrated in canola (Brassica napus L.) that the expression of AtMYB32xs::IPT could delay leaf senescence and improve seed yield, with no penalty on growth or phenology, which is clearly superior to transgenic plants carrying IPT genes driven by strong constitutive promoter, SAG12 promoter, or SARK promoter [96].
Besides, the fact that certain members of plant laccases are involved in phenolic compounds oxidation imply the possibility of applying plant laccase for in vitro reaction or for industry in areas such as environmental pollution control, food industry, biosensors, textile industry, pharmaceutical industry, and in organic synthesis. Future studies on enzyme immobilization need pay attention to plant laccases as well.
The enhanced secretion of ex planta laccase may be of great value in phytoremediation of small organic pollutants. It has been reported in Arabidopsis that the overexpression of LAC1, a laccase gene specifically expressed in the roots of Gossypium arboretum, resulted in increased secretory laccase activity and enhanced resistance to several phenolic allelochemicals and 2,4,6-trichlorophenol (TCP) [97]. Therefore, ex planta phytoremediation, such as transgenic plants secreting extracellular laccases, can be another strategy for remediation of environmental contaminants.
Conclusion
Plant laccase is an important oxidoreductase and shares the common molecular architecture and reaction mechanism with fungal laccase. However, it has lower redox potential and different pH requirement, which may partly account for its function in lignin biosynthesis, being contrary to the ligninolytic activity of fungal laccase. Plant laccase is encoded by multigene family, the tissue-specific expression of which may be regulated by transcription factors, miRNAs, and environmental stresses. Laccases are involved in various biological processes and plant response to environmental stresses. Most plant laccases are predicted to be localized in apoplast, in accordance with their functional involvement in lignin biosynthesis. Furthermore, laccase has been considered as a potential target for fine-tuning of plant lignin content and/or composition to alleviate the recalcitrance of lignocellulose, which promises a bright future for energy plant improvement in biofuel production.
Authors’ contributions
JHW drafted the manuscript. JJF and YXL helped draft and revise the manuscript. SC and WTJ helped revise the manuscript. YXL and SZL initiated the review. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by National High Technology Research and Development Program of China (Grant No. 2012AA101805), and International Scientific and Technological Cooperation Program (Grant No. 2013DFA60470).
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests.
Abbreviations
- 4CL
4-coumarate:CoA ligase
- C4H
cinnamate 4-hydroxylase
- CCR
(hydroxy)cinnamoyl CoA reductase
- COMT
caffeic acid (5-hydroxyconiferaldehyde) O-methyltransferase
- EPR
electronic paramagnetic resonance
- MCOs
multicopper oxidases
- pI
isoelectric point
- PAL
phenylalanine ammonia lyase
- TCP
2,4,6-trichlorophenol
Additional file
Additional file 1: Table S1. Database accession numbers of laccase sequences for phylogenetic analysis.
Contributor Information
Jinhui Wang, Email: wangjinhui@ibcas.ac.cn.
Juanjuan Feng, Email: jjfeng@ibcas.ac.cn.
Weitao Jia, Email: jwtswu@sina.com.
Sandra Chang, Email: sandrahchang@gmail.com.
Shizhong Li, Email: szli@mail.tsinghua.edu.cn.
Yinxin Li, Email: yxli@ibcas.ac.cn.
References
- 1.Chang MCY. Harnessing energy from plant biomass. Curr Opin Chem Biol. 2007;11(6):677–684. doi: 10.1016/j.cbpa.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 2.Cheng JJ, Timilsina GR. Status and barriers of advanced biofuel technologies: a review. Renew Energ. 2011;36(12):3541–3549. doi: 10.1016/j.renene.2011.04.031. [DOI] [Google Scholar]
- 3.van der Pol EC, Bakker RR, Baets P, Eggink G. By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for (bio)chemicals and fuels. Appl Microbiol Biot. 2014;98(23):9579–9593. doi: 10.1007/s00253-014-6158-9. [DOI] [PubMed] [Google Scholar]
- 4.Zeng YN, Zhao S, Yang SH, Ding SY. Lignin plays a negative role in the biochemical process for producing lignocellulosic biofuels. Curr Opin Biotech. 2014;27:38–45. doi: 10.1016/j.copbio.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Sticklen M. Plant genetic engineering to improve biomass characteristics for biofuels. Curr Opin Biotech. 2006;17(3):315–319. doi: 10.1016/j.copbio.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Wang CY, Zhang SC, Yu Y, Luo YC, Liu Q, Ju CL, et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol J. 2014;12(8):1132–1142. doi: 10.1111/pbi.12222. [DOI] [PubMed] [Google Scholar]
- 7.Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol. 2013;31(9):848–852. doi: 10.1038/nbt.2646. [DOI] [PubMed] [Google Scholar]
- 8.Giardina P, Faraco V, Pezzella C, Piscitelli A, Vanhulle S, Sannia G. Laccases: a never-ending story. Cell Mol Life Sci. 2010;67(3):369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morozova OV, Shumakovich GP, Gorbacheva MA, Shleev SV, Yaropolov AI. “Blue” laccases. Biochemistry (Moscow). 2007;72(10):1136–1150. doi: 10.1134/S0006297907100112. [DOI] [PubMed] [Google Scholar]
- 10.Baldrian P. Fungal laccases—occurrence and properties. FEMS Microbiol Rev. 2006;30(2):215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- 11.Hakulinen N, Rouvinen J. Three-dimensional structures of laccases. Cell Mol Life Sci. 2015;72(5):857–868. doi: 10.1007/s00018-014-1827-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claus H. Laccases: structure, reactions, distribution. Micron. 2004;35(1–2):93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Kumar SVS, Phale PS, Durani S, Wangikar PP. Combined sequence and structure analysis of the fungal laccase family. Biotechnol Bioeng. 2003;83(4):386–394. doi: 10.1002/bit.10681. [DOI] [PubMed] [Google Scholar]
- 14.Mot AC, Silaghi-Dumitrescu R. Laccases: complex architectures for one-electron oxidations. Biochemistry (Moscow). 2012;77(12):1395–1407. doi: 10.1134/S0006297912120085. [DOI] [PubMed] [Google Scholar]
- 15.Wherland S, Farver O, Pecht I. Multicopper oxidases: intramolecular electron transfer and O2 reduction. J Biol Inorg Chem. 2014;19(4–5):541–554. doi: 10.1007/s00775-013-1080-7. [DOI] [PubMed] [Google Scholar]
- 16.Jones SM, Solomon EI. Electron transfer and reaction mechanism of laccases. Cell Mol Life Sci. 2015;72(5):869–883. doi: 10.1007/s00018-014-1826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayer AM, Staples RC. Laccase: new functions for an old enzyme. Phytochemistry. 2002;60(6):551–565. doi: 10.1016/S0031-9422(02)00171-1. [DOI] [PubMed] [Google Scholar]
- 18.Sharma KK, Kuhad RC. Laccase: enzyme revisited and function redefined. Indian J Microbiol. 2008;48(3):309–316. doi: 10.1007/s12088-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munk L, Sitarz AK, Kalyani DC, Mikkelsen JD, Meyer AS. Can laccases catalyze bond cleavage in lignin? Biotechnol Adv. 2015;33(1):13–24. doi: 10.1016/j.biotechadv.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 20.Arora DS, Sharma RK. Ligninolytic fungal laccases and their biotechnological applications. Appl Biochem Biotech. 2010;160(6):1760–1788. doi: 10.1007/s12010-009-8676-y. [DOI] [PubMed] [Google Scholar]
- 21.Kunamneni A, Plou FJ, Ballesteros A, Alcalde M. Laccases and their applications: a patent review. Recent Pat Biotech. 2008;2(1):15. doi: 10.2174/187220808783330965. [DOI] [PubMed] [Google Scholar]
- 22.Kudanga T, Le Roes-Hill M. Laccase applications in biofuels production: current status and future prospects. Appl Microbiol Biot. 2014;98(15):6525–6542. doi: 10.1007/s00253-014-5810-8. [DOI] [PubMed] [Google Scholar]
- 23.Kudanga T, Nyanhongo GS, Guebitz GM, Burton S. Potential applications of laccase-mediated coupling and grafting reactions: a review. Enzym Microb Tech. 2011;48(3):195–208. doi: 10.1016/j.enzmictec.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Jeon JR, Baldrian P, Murugesan K, Chang YS. Laccase-catalysed oxidations of naturally occurring phenols: from in vivo biosynthetic pathways to green synthetic applications. Microb Biotechnol. 2012;5(3):318–332. doi: 10.1111/j.1751-7915.2011.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Viswanath B, Rajesh B, Janardhan A, Kumar AP, Narasimha G. Fungal laccases and their applications in bioremediation. Enzym Res. 2014;2014:21. doi: 10.1155/2014/163242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couto SR, Herrera JLT. Industrial and biotechnological applications of laccases: a review. Biotechnol Adv. 2006;24(5):500–513. doi: 10.1016/j.biotechadv.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Mate DM, Alcalde M. Laccase engineering: from rational design to directed evolution. Biotechnol Adv. 2014;33(1):16. doi: 10.1016/j.biotechadv.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 28.Rodgers CJ, Blanford CF, Giddens SR, Skamnioti P, Armstrong FA, Gurr SJ. Designer laccases: a vogue for high-potential fungal enzymes? Trends Biotechnol. 2010;28(2):63–72. doi: 10.1016/j.tibtech.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Pardo I, Camarero S. Laccase engineering by rational and evolutionary design. Cell Mol Life Sci. 2015;72(5):897–910. doi: 10.1007/s00018-014-1824-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi UN, Singh P, Pandey VP, Kumar A. Structure-function relationship among bacterial, fungal and plant laccases. J Mol Catal B Enzym. 2011;68(2):117–128. doi: 10.1016/j.molcatb.2010.11.002. [DOI] [Google Scholar]
- 31.Ullrich R, Hofrichter M. Enzymatic hydroxylation of aromatic compounds. Cell Mol Life Sci. 2007;64(3):271–293. doi: 10.1007/s00018-007-6362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riva S. Laccases: blue enzymes for green chemistry. Trends Biotechnol. 2006;24(5):219–226. doi: 10.1016/j.tibtech.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 33.Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96(7):2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 34.Turlapati PV, Kim KW, Davin LB, Lewis NG. The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s) Planta. 2011;233(3):439–470. doi: 10.1007/s00425-010-1298-3. [DOI] [PubMed] [Google Scholar]
- 35.Yaver DS, Xu F, Golightly EJ, Brown KM, Brown SH, Rey MW, et al. Purification, characterization, molecular cloning, and expression of two laccase genes from the white rot basidiomycete Trametes villosa. Appl Environ Microb. 1996;62(3):834–841. doi: 10.1128/aem.62.3.834-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hattori M, Tsuchihara K, Noda H, Konishi H, Tamura Y, Shinoda T, et al. Molecular characterization and expression of laccase genes in the salivary glands of the green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae) Insect Biochem Molec. 2010;40(4):331–338. doi: 10.1016/j.ibmb.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 37.Sharma P, Goel R, Capalash N. Bacterial laccases. World J Microb Biot. 2007;23(6):823–832. doi: 10.1007/s11274-006-9305-3. [DOI] [Google Scholar]
- 38.Martins LO, Durao P, Brissos V, Lindley PF. Laccases of prokaryotic origin: enzymes at the interface of protein science and protein technology. Cell Mol Life Sci. 2015;72(5):911–922. doi: 10.1007/s00018-014-1822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shraddha, Ravi S, Simran S, Mohit K, Ajay K. Laccase: microbial sources, production, purification, and potential biotechnological applications. Enzym Res. 2011;2011:11. [DOI] [PMC free article] [PubMed]
- 40.Hullo MF, Moszer I, Danchin A, Martin-Verstraete I. CotA of Bacillus subtilis is a copper-dependent laccase. J Bacteriol. 2001;183(18):5426–5430. doi: 10.1128/JB.183.18.5426-5430.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uthandi S, Saad B, Humbard MA, Maupin-Furlow JA. LccA, an archaeal laccase secreted as a highly stable glycoprotein into the extracellular medium by Haloferax volcanii. Appl Environ Microb. 2010;76(3):733–743. doi: 10.1128/AEM.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu BL, Shen WF, Liu Y, Weng HB, He LH, Mu JJ, et al. Cloning and RNAi-mediated functional characterization of MaLac2 of the pine sawyer, Monochamus alternatus. Insect Mol Biol. 2008;17(3):303–312. doi: 10.1111/j.1365-2583.2008.00803.x. [DOI] [PubMed] [Google Scholar]
- 43.Yatsu J, Asano T. Cuticle laccase of the silkworm, Bombyx mori: purification, gene identification and presence of its inactive precursor in the cuticle. Insect Biochem Molec. 2009;39(4):254–262. doi: 10.1016/j.ibmb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Elias-Neto M, Soares MPM, Simoes ZLP, Hartfelder K, Bitondi MMG. Developmental characterization, function and regulation of a Laccase2 encoding gene in the honey bee, Apis mellifera (Hymenoptera, Apinae) Insect Biochem Molec. 2010;40(3):241–251. doi: 10.1016/j.ibmb.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 45.Bao W, Omalley DM, Whetten R, Sederoff RR. A laccase associated with lignification in loblolly pine xylem. Science. 1993;260(5108):672–674. doi: 10.1126/science.260.5108.672. [DOI] [PubMed] [Google Scholar]
- 46.Gavnholt B, Larsen K, Rasmussen SK. Isolation and characterisation of laccase cDNAs from meristematic and stem tissues of ryegrass (Lolium perenne) Plant Sci. 2002;162(6):873–885. doi: 10.1016/S0168-9452(02)00035-3. [DOI] [Google Scholar]
- 47.Kiefer-Meyer MC, Gomord V, O’Connell A, Halpin C, Faye L. Cloning and sequence analysis of laccase-encoding cDNA clones from tobacco. Gene. 1996;178(1–2):205–207. doi: 10.1016/0378-1119(96)00381-2. [DOI] [PubMed] [Google Scholar]
- 48.Lafayette PR, Eriksson KEL, Dean JFD. Nucleotide sequence of a cDNA clone encoding an acidic laccase from sycamore maple (Acer Pseudoplatanus L) Plant Physiol. 1995;107(2):667–668. doi: 10.1104/pp.107.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.LaFayette PR, Eriksson KEL, Dean JFD. Characterization and heterologous expression of laccase cDNAs from xylem tissues of yellow-poplar (Liriodendron tulipifera) Plant Mol Biol. 1999;40(1):23–35. doi: 10.1023/A:1026437406859. [DOI] [PubMed] [Google Scholar]
- 50.Ranocha P, McDougall G, Hawkins S, Sterjiades R, Borderies G, Stewart D, et al. Biochemical characterization, molecular cloning and expression of laccases—a divergent gene family—in poplar. Eur J Biochem. 1999;259(1–2):485–495. doi: 10.1046/j.1432-1327.1999.00061.x. [DOI] [PubMed] [Google Scholar]
- 51.Cesarino I, Araujo P, Mayer JLS, Vicentini R, Berthet S, Demedts B, et al. Expression of SofLAC, a new laccase in sugarcane, restores lignin content but not S:G ratio of Arabidopsis lac17 mutant. J Exp Bot. 2013;64(6):1769–1781. doi: 10.1093/jxb/ert045. [DOI] [PubMed] [Google Scholar]
- 52.Liang MX, Haroldsen V, Cai XN, Wu YJ. Expression of a putative laccase gene, ZmLAC1, in maize primary roots under stress. Plant Cell Environ. 2006;29(5):746–753. doi: 10.1111/j.1365-3040.2005.01435.x. [DOI] [PubMed] [Google Scholar]
- 53.McCaig BC, Meagher RB, Dean JFD. Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta. 2005;221(5):619–636. doi: 10.1007/s00425-004-1472-6. [DOI] [PubMed] [Google Scholar]
- 54.Zhang K, Lu K, Qu CM, Liang Y, Wang R, Chai YR, et al. Gene silencing of BnTT10 family genes causes retarded pigmentation and lignin reduction in the seed coat of Brassica napus. PLoS One. 2013;8(4):e61247. [DOI] [PMC free article] [PubMed]
- 55.Wang Y, Bouchabké-Coussa O, Lebris P, Antelme S, Soulhat C, Gineau E, et al. LACCASE 5 is required for lignification of the Brachypodium distachyon culm. Plant Physiol. 2015;168:13. doi: 10.1104/pp.114.255489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solomon EI, Heppner DE, Johnston EM, Ginsbach JW, Cirera J, Qayyum M, et al. Copper active sites in biology. Chem Rev. 2014;114(7):3659–3853. doi: 10.1021/cr400327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Madhavi V, Lele SS. Laccase: properties and applications. Bioresources. 2009;4(4):1694–1717. [Google Scholar]
- 58.Awasthi M, Jaiswal N, Singh S, Pandey VP, Dwivedi UN. Molecular docking and dynamics simulation analyses unraveling the differential enzymatic catalysis by plant and fungal laccases with respect to lignin biosynthesis and degradation. J Biomol Struct Dyn. 2015;33(9):1835–49. [DOI] [PubMed]
- 59.Hakulinen N, Kiiskinen LL, Kruus K, Saloheimo M, Paananen A, Koivula A, et al. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat Struct Biol. 2002;9(8):601–605. doi: 10.1038/nsb823. [DOI] [PubMed] [Google Scholar]
- 60.Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cezard L, Le Bris P, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23(3):1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato Y, Bao WL, Sederoff R, Whetten R. Molecular cloning and expression of eight laccase cDNAs in loblolly pine (Pinus taeda) J Plant Res. 2001;114(1114):147–155. doi: 10.1007/PL00013978. [DOI] [Google Scholar]
- 62.Zhou JL, Lee CH, Zhong RQ, Ye ZH. MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell. 2009;21(1):248–266. doi: 10.1105/tpc.108.063321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem. 2008;283(23):15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu SF, Li QZ, Wei HR, Chang MJ, Tunlaya-Anukit S, Kim H, et al. Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA. 2013;110(26):10848–10853. doi: 10.1073/pnas.1308936110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei JZ, Tirajoh A, Effendy J, Plant AL. Characterization of salt-induced changes in gene expression in tomato (Lycopersicon esculentum) roots and the role played by abscisic acid. Plant Sci. 2000;159(1):135–148. doi: 10.1016/S0168-9452(00)00344-7. [DOI] [PubMed] [Google Scholar]
- 66.Shen Y, Zhang YZ, Chen J, Lin HJ, Zhao MJ, Peng HW, et al. Genome expression profile analysis reveals important transcripts in maize roots responding to the stress of heavy metal Pb. Physiol Plant. 2013;147(3):270–282. doi: 10.1111/j.1399-3054.2012.01670.x. [DOI] [PubMed] [Google Scholar]
- 67.Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, et al. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol. 2014;166(2):798–807. doi: 10.1104/pp.114.245597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pang YZ, Cheng XF, Huhman DV, Ma JY, Peel GJ, Yonekura-Sakakibara K, et al. Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta. 2013;238(1):139–154. doi: 10.1007/s00425-013-1879-z. [DOI] [PubMed] [Google Scholar]
- 69.Eggert C, Temp U, Eriksson KEL. Laccase is essential for lignin degradation by the white-rot fungus Pycnoporus cinnabarinus. FEBS Lett. 1997;407(1):89–92. doi: 10.1016/S0014-5793(97)00301-3. [DOI] [PubMed] [Google Scholar]
- 70.Schuetz M, Smith R, Ellis B. Xylem tissue specification, patterning, and differentiation mechanisms. J Exp Bot. 2013;64(1):11–31. doi: 10.1093/jxb/ers287. [DOI] [PubMed] [Google Scholar]
- 71.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annu Rev Plant Biol. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 72.Miao YC, Liu CJ. ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci USA. 2010;107(52):22728–22733. doi: 10.1073/pnas.1007747108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W. Lignin biosynthesis and structure. Plant Physiol. 2010;153(3):895–905. doi: 10.1104/pp.110.155119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mechin V, Baumberger S, Pollet B, Lapierre C. Peroxidase activity can dictate the in vitro lignin dehydrogenative polymer structure. Phytochemistry. 2007;68(4):571–579. doi: 10.1016/j.phytochem.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 75.Morreel K, Ralph J, Kim H, Lu FC, Goeminne G, Ralph S, et al. Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol. 2004;136(3):3537–3549. doi: 10.1104/pp.104.049304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sterjiades R, Dean JFD, Eriksson KEL. Laccase from sycamore maple (Acer Pseudoplatanus) polymerizes monolignols. Plant Physiol. 1992;99(3):1162–1168. doi: 10.1104/pp.99.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, et al. Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiol. 2002;129(1):145–155. doi: 10.1104/pp.010988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liang MX, Davis E, Gardner D, Cai XN, Wu YJ. Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta. 2006;224(5):1185–1196. doi: 10.1007/s00425-006-0300-6. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Q, Nakashima J, Chen F, Yin YB, Fu CX, Yun JF, et al. LACCASE is necessary and nonredundant with PEROXIDASE for lignin polymerization during vascular development in Arabidopsis. Plant Cell. 2013;25(10):3976–3987. doi: 10.1105/tpc.113.117770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huttermann A, Mai C, Kharazipour A. Modification of lignin for the production of new compounded materials. Appl Microbiol Biot. 2001;55(4):387–394. doi: 10.1007/s002530000590. [DOI] [PubMed] [Google Scholar]
- 81.Hoopes JT, Dean JFD. Ferroxidase activity in a laccase-like multicopper oxidase from Liriodendron tulipifera. Plant Physiol Bioch. 2004;42(1):27–33. doi: 10.1016/j.plaphy.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 82.Cai XN, Davis EJ, Ballif J, Liang MX, Bushman E, Haroldsen V, et al. Mutant identification and characterization of the laccase gene family in Arabidopsis. J Exp Bot. 2006;57(11):2563–2569. doi: 10.1093/jxb/erl022. [DOI] [PubMed] [Google Scholar]
- 83.Caparros-Ruiz D, Fornale S, Civardi L, Puigdomenech P, Rigau J. Isolation and characterisation of a family of laccases in maize. Plant Sci. 2006;171(2):217–225. doi: 10.1016/j.plantsci.2006.03.007. [DOI] [Google Scholar]
- 84.Cho HY, Lee C, Hwang SG, Park YC, Lim HL, Jang CS. Overexpression of the OsChI1 gene, encoding a putative laccase precursor, increases tolerance to drought and salinity stress in transgenic Arabidopsis. Gene. 2014;552(1):98–105. doi: 10.1016/j.gene.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 85.Canas AI, Camarero S. Laccases and their natural mediators: biotechnological tools for sustainable eco-friendly processes. Biotechnol Adv. 2010;28(6):694–705. doi: 10.1016/j.biotechadv.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 86.Morozova OV, Shumakovich GP, Shleev SV, Yaropolov YI. Laccase-mediator systems and their applications: a review. Appl Biochem Microbiol. 2007;43(5):523–535. doi: 10.1134/S0003683807050055. [DOI] [PubMed] [Google Scholar]
- 87.Fernandez-Fernandez M, Sanroman MA, Moldes D. Recent developments and applications of immobilized laccase. Biotechnol Adv. 2013;31(8):1808–1825. doi: 10.1016/j.biotechadv.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 88.Champagne PP, Ramsay J. Reactive blue 19 decolouration by laccase immobilized on silica beads. Appl Microbiol Biot. 2007;77(4):819–823. doi: 10.1007/s00253-007-1208-1. [DOI] [PubMed] [Google Scholar]
- 89.Sheldon RA. Enzyme immobilization: the quest for optimum performance. Adv Synth Catal. 2007;349(8–9):1289–1307. doi: 10.1002/adsc.200700082. [DOI] [Google Scholar]
- 90.Dandikas V, Heuwinkel H, Lichti F, Drewes JE, Koch K. Correlation between biogas yield and chemical composition of energy crops. Bioresour Technol. 2014;174:316–320. doi: 10.1016/j.biortech.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 91.Wu ZL, Zhang ML, Wang LQ, Tu YY, Zhang J, Xie GS, et al. Biomass digestibility is predominantly affected by three factors of wall polymer features distinctive in wheat accessions and rice mutants. Biotechnol Biofuels. 2013;6(1):813. [DOI] [PMC free article] [PubMed]
- 92.Li M, Feng SQ, Wu LM, Li Y, Fan CF, Zhang R, et al. Sugar-rich sweet sorghum is distinctively affected by wall polymer features for biomass digestibility and ethanol fermentation in bagasse. Bioresour Technol. 2014;167:14–23. doi: 10.1016/j.biortech.2014.04.086. [DOI] [PubMed] [Google Scholar]
- 93.Cano-Delgado A, Penfield S, Smith C, Catley M, Bevan M. Reduced cellulose synthesis invokes lignification and defense responses in Arabidopsis thaliana. Plant J. 2003;34(3):351–362. doi: 10.1046/j.1365-313X.2003.01729.x. [DOI] [PubMed] [Google Scholar]
- 94.Hu WJ, Harding SA, Lung J, Popko JL, Ralph J, Stokke DD, et al. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol. 1999;17(8):808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- 95.Boudet AM, Kajita S, Grima-Pettenati J, Goffner D. Lignins and lignocellulosics: a better control of synthesis for new and improved uses. Trends Plant Sci. 2003;8(12):576–581. doi: 10.1016/j.tplants.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 96.Kant S, Burch D, Badenhorst P, Palanisamy R, Mason J, Spangenberg G. Regulated expression of a cytokinin biosynthesis gene IPT delays leaf senescence and improves yield under rainfed and irrigated conditions in canola (Brassica napus L.) PLoS One. 2015;10(1):18. doi: 10.1371/journal.pone.0116349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang GD, Li QJ, Luo B, Chen XY. Ex planta phytoremediation of trichlorophenol and phenolic allelochemicals via an engineered secretory laccase. Nat Biotechnol. 2004;22(7):893–897. doi: 10.1038/nbt982. [DOI] [PubMed] [Google Scholar]
- 98.SignalP 4.1 Server. 2015. http://www.cbs.dtu.dk/services/SignalP/. Accessed 8 July 2015.
- 99.Pfam. 2015. http://pfam.xfam.org/search. Accessed 8 July 2015.
- 100.NetNGlyc 1.0 Server. 2015. http://www.cbs.dtu.dk/services/NetNGlyc/. Accessed 8 July 2015.
- 101.SWISS-MODEL. 2015. http://swissmodel.expasy.org/. Accessed 8 July 2015.


