Abstract
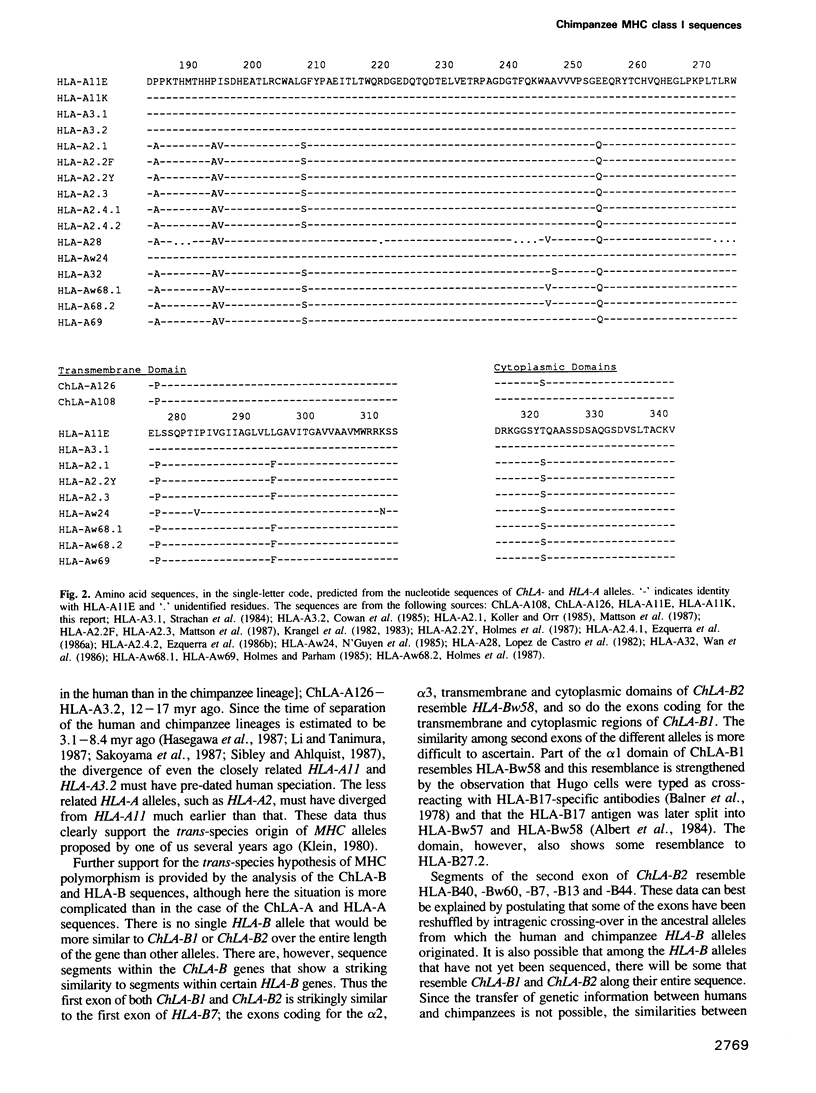
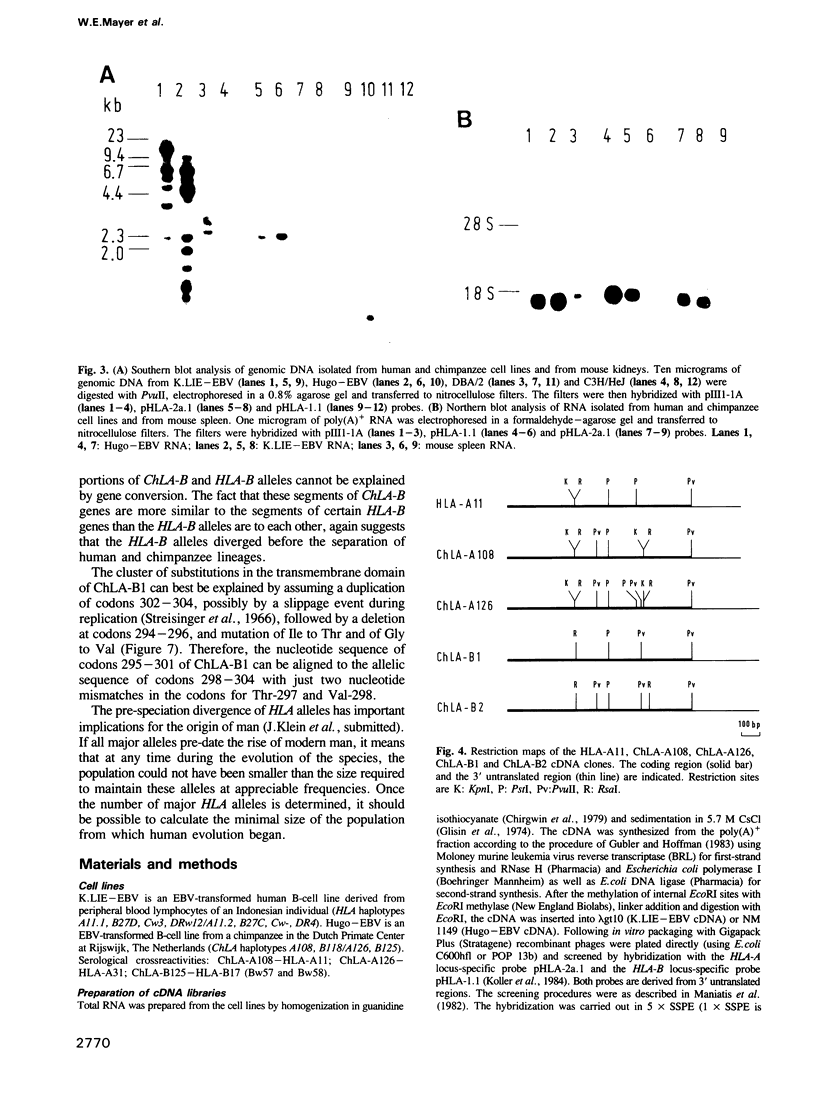
To obtain an insight into the evolutionary origin of the major histocompatibility complex (MHC) class I polymorphism, a cDNA library was prepared from a heterozygous chimpanzee cell line expressing MHC class I molecules crossreacting with allele-specific HLA-A11 antibodies. The library was screened with human class I locus-specific DNA probes, and clones encoding both alleles at the A and B loci have been identified and sequenced. In addition, the sequences of two HLA-A11 subtypes differing by a single nucleotide substitution have been obtained. The comparison of chimpanzee and human sequences revealed a close similarity (up to 98.5%). The chimpanzee A locus alleles showed greatest similarity to the human HLA-A11/A3 family of alleles, one of them being very close to HLA-A11. Similarly, segments of the ChLA-B alleles displayed greatest similarity to certain HLA-B alleles. The calculated evolutionary branch point for the A11-like alleles is 7 x 10(6) to 9 x 10(6) years, whereas the other A locus alleles diverged between 12 x 10(6) and 17 x 10(6) years ago. Since the human and chimpanzee lineages separated 5 x 10(6) to 7 x 10(6) years ago, our data support the notion that during evolution, MHC alleles are transmitted from one species to the next.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aparicio P., Rojo S., Jaraquemada D., López de Castro J. A. Fine specificity of HLA-B27 cellular allorecognition. HLA-B27f is a functional variant distinguishable by cytolytic T cell clones. J Immunol. 1987 Aug 1;139(3):837–841. [PubMed] [Google Scholar]
- Balner H., Gabb B. W., D'Amaro J., van Vreeswijk W., Visser T. P. Evidence for two linked loci controlling ther serologically defined leukocyte antigens of chimpanzees (ChL-A). Tissue Antigens. 1974;4(4):313–328. doi: 10.1111/j.1399-0039.1974.tb00258.x. [DOI] [PubMed] [Google Scholar]
- Balner H., van Vreeswijk W., Roger J. H., D'Amaro J. The major histocompatibility complex of chimpanzees: identification of several new antigens controlled by the A and B loci of ChLa. Tissue Antigens. 1978 Jul;12(1):1–18. [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987 Oct 8;329(6139):506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- Bjorkman P. J., Saper M. A., Samraoui B., Bennett W. S., Strominger J. L., Wiley D. C. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987 Oct 8;329(6139):512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Cowan E. P., Jelachich M. L., Biddison W. E., Coligan J. E. DNA sequence of HLA-A11: remarkable homology with HLA-A3 allows identification of residues involved in epitopes recognized by antibodies and T cells. Immunogenetics. 1987;25(4):241–250. doi: 10.1007/BF00404694. [DOI] [PubMed] [Google Scholar]
- Cowan E. P., Jordan B. R., Coligan J. E. Molecular cloning and DNA sequence analysis of genes encoding cytotoxic T lymphocyte-defined HLA-A3 subtypes: the E1 subtype. J Immunol. 1985 Oct;135(4):2835–2841. [PubMed] [Google Scholar]
- Ezquerra A., Bragado R., Vega M. A., Strominger J. L., Woody J., López de Castro J. A. Primary structure of papain-solubilized human histocompatibility antigen HLA-B27. Biochemistry. 1985 Mar 26;24(7):1733–1741. doi: 10.1021/bi00328a025. [DOI] [PubMed] [Google Scholar]
- Ezquerra A., Doménech N., van der Poel J., Strominger J. L., Vega M. A., López de Castro J. A. Molecular analysis of an HLA-A2 functional variant CLA defined by cytolytic T lymphocytes. J Immunol. 1986 Sep 1;137(5):1642–1649. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Frischauf A. M., Garoff H., Lehrach H. A subcloning strategy for DNA sequence analysis. Nucleic Acids Res. 1980 Dec 11;8(23):5541–5549. doi: 10.1093/nar/8.23.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumaki Y., Collins F., Kole R., Stoeckert C. J., Jr, Jagadeeswaran P., Duncan C. H., Weissman S. M., Jagadeeswaran P., Pan J., Forget B. G. Sequences of human repetitive DNA, non-alpha-globin genes, and major histocompatibility locus genes. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1079–1086. [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H., Yano T. Man's place in Hominoidea as inferred from molecular clocks of DNA. J Mol Evol. 1987;26(1-2):132–147. doi: 10.1007/BF02111287. [DOI] [PubMed] [Google Scholar]
- Hayashida H., Miyata T. Unusual evolutionary conservation and frequent DNA segment exchange in class I genes of the major histocompatibility complex. Proc Natl Acad Sci U S A. 1983 May;80(9):2671–2675. doi: 10.1073/pnas.80.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes N., Ennis P., Wan A. M., Denney D. W., Parham P. Multiple genetic mechanisms have contributed to the generation of the HLA-A2/A28 family of class I MHC molecules. J Immunol. 1987 Aug 1;139(3):936–941. [PubMed] [Google Scholar]
- Holmes N., Parham P. Exon shuffling in vivo can generate novel HLA class I molecules. EMBO J. 1985 Nov;4(11):2849–2854. doi: 10.1002/j.1460-2075.1985.tb04013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaulin C., Perrin A., Abastado J. P., Dumas B., Papamatheakis J., Kourilsky P. Polymorphism in mouse and human class I H-2 and HLA genes is not the result of random independent point mutations. Immunogenetics. 1985;22(5):453–470. doi: 10.1007/BF00418091. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ota T. On the stochastic model for estimation of mutational distance between homologous proteins. J Mol Evol. 1972 Dec 29;2(1):87–90. doi: 10.1007/BF01653945. [DOI] [PubMed] [Google Scholar]
- Klein J., Figueroa F. Evolution of the major histocompatibility complex. Crit Rev Immunol. 1986;6(4):295–386. [PubMed] [Google Scholar]
- Klein J. Origin of major histocompatibility complex polymorphism: the trans-species hypothesis. Hum Immunol. 1987 Jul;19(3):155–162. doi: 10.1016/0198-8859(87)90066-8. [DOI] [PubMed] [Google Scholar]
- Koller B. H., Orr H. T. Cloning and complete sequence of an HLA-A2 gene: analysis of two HLA-A alleles at the nucleotide level. J Immunol. 1985 Apr;134(4):2727–2733. [PubMed] [Google Scholar]
- Koller B. H., Sidwell B., DeMars R., Orr H. T. Isolation of HLA locus-specific DNA probes from the 3'-untranslated region. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5175–5178. doi: 10.1073/pnas.81.16.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottmann A. H., Seemann G. H., Guessow H. D., Roos M. H. DNA sequence of the coding region of the HLA-B44 gene. Immunogenetics. 1986;23(6):396–400. doi: 10.1007/BF00372673. [DOI] [PubMed] [Google Scholar]
- Krangel M. S., Biddison W. E., Strominger J. L. Comparative structural analysis of HLA-A2 antigens distinguishable by cytotoxic T lymphocytes. II. Variant DK1: evidence for a discrete CTL recognition region. J Immunol. 1983 Apr;130(4):1856–1862. [PubMed] [Google Scholar]
- Krangel M. S., Taketani S., Biddison W. E., Strong D. M., Strominger J. L. Comparative structural analysis of HLA-A2 antigens distinguishable by cytotoxic T lymphocytes: variants M7 and DR1. Biochemistry. 1982 Nov 23;21(24):6313–6321. doi: 10.1021/bi00267a042. [DOI] [PubMed] [Google Scholar]
- Lalanne J. L., Bregegere F., Delarbre C., Abastado J. P., Gachelin G., Kourilsky P. Comparison of nucleotide sequences of mRNAs belonging to the mouse H-2 multigene family. Nucleic Acids Res. 1982 Feb 11;10(3):1039–1049. doi: 10.1093/nar/10.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987 Mar 5;326(6108):93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- López de Castro J. A., Strominger J. L., Strong D. M., Orr H. T. Structure of crossreactive human histocompatibility antigens HLA-A28 and HLA-A2: possible implications for the generation of HLA polymorphism. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3813–3817. doi: 10.1073/pnas.79.12.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Castro J., Bragado R., Strong D. M., Strominger J. L. Primary structure of papain-solubilized human histocompatibility antigen HLA-B40 (-Bw60). An outline of alloantigenic determinants. Biochemistry. 1983 Aug 2;22(16):3961–3969. doi: 10.1021/bi00285a036. [DOI] [PubMed] [Google Scholar]
- Mattson D. H., Handy D. E., Bradley D. A., Coligan J. E., Cowan E. P., Biddison W. E. DNA sequences of the genes that encode the CTL-defined HLA-A2 variants M7 and DK1. Immunogenetics. 1987;26(3):190–192. doi: 10.1007/BF00365911. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCune J. M., Humphreys R. E., Yocum R. R., Strominger J. L. Enhanced representation of HL-A antigens on human lymphocytes after mitogenesis induced by phytohemagglutinin or Epstein-Barr virus. Proc Natl Acad Sci U S A. 1975 Aug;72(8):3206–3209. doi: 10.1073/pnas.72.8.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyada C. G., Klofelt C., Reyes A. A., McLaughlin-Taylor E., Wallace R. B. Evidence that polymorphism in the murine major histocompatibility complex may be generated by the assortment of subgene sequences. Proc Natl Acad Sci U S A. 1985 May;82(9):2890–2894. doi: 10.1073/pnas.82.9.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T., Nishida T. Nucleotide sequence divergence and functional constraint in mRNA evolution. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7328–7332. doi: 10.1073/pnas.77.12.7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- N'Guyen C., Sodoyer R., Trucy J., Strachan T., Jordan B. R. The HLA-AW24 gene: sequence, surroundings and comparison with the HLA-A2 and HLA-A3 genes. Immunogenetics. 1985;21(5):479–489. doi: 10.1007/BF00430931. [DOI] [PubMed] [Google Scholar]
- Neefjes J. J., Breur-Vriesendorp B. S., van Seventer G. A., Iványi P., Ploegh H. L. An improved biochemical method for the analysis of HLA-class I antigens. Definition of new HLA-class I subtypes. Hum Immunol. 1986 Jun;16(2):169–181. doi: 10.1016/0198-8859(86)90046-7. [DOI] [PubMed] [Google Scholar]
- Perler F., Efstratiadis A., Lomedico P., Gilbert W., Kolodner R., Dodgson J. The evolution of genes: the chicken preproinsulin gene. Cell. 1980 Jun;20(2):555–566. doi: 10.1016/0092-8674(80)90641-8. [DOI] [PubMed] [Google Scholar]
- Rojo S., Aparicio P., Choo S. Y., Hansen J. A., López de Castro J. A. Structural analysis of an HLA-B27 population variant, B27f. Multiple patterns of amino acid changes within a single polypeptide segment generate polymorphism in HLA-B27. J Immunol. 1987 Aug 1;139(3):831–836. [PubMed] [Google Scholar]
- Sakoyama Y., Hong K. J., Byun S. M., Hisajima H., Ueda S., Yaoita Y., Hayashida H., Miyata T., Honjo T. Nucleotide sequences of immunoglobulin epsilon genes of chimpanzee and orangutan: DNA molecular clock and hominoid evolution. Proc Natl Acad Sci U S A. 1987 Feb;84(4):1080–1084. doi: 10.1073/pnas.84.4.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C. G., Ahlquist J. E. DNA hybridization evidence of hominoid phylogeny: results from an expanded data set. J Mol Evol. 1987;26(1-2):99–121. doi: 10.1007/BF02111285. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steinmetz M., Hood L. Genes of the major histocompatibility complex in mouse and man. Science. 1983 Nov 18;222(4625):727–733. doi: 10.1126/science.6356354. [DOI] [PubMed] [Google Scholar]
- Strachan T., Sodoyer R., Damotte M., Jordan B. R. Complete nucleotide sequence of a functional class I HLA gene, HLA-A3: implications for the evolution of HLA genes. EMBO J. 1984 Apr;3(4):887–894. doi: 10.1002/j.1460-2075.1984.tb01901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Szöts H., Riethmüller G., Weiss E., Meo T. Complete sequence of HLA-B27 cDNA identified through the characterization of structural markers unique to the HLA-A, -B, and -C allelic series. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1428–1432. doi: 10.1073/pnas.83.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega M. A., Bragado R., Iványi P., Peláez J. L., López de Castro J. A. Molecular analysis of a functional subtype of HLA-B27. A possible evolutionary pathway for HLA-B27 polymorphism. J Immunol. 1986 Dec 1;137(11):3557–3565. [PubMed] [Google Scholar]
- Vega M. A., Ezquerra A., Rojo S., Aparicio P., Bragado R., López de Castro J. A. Structural analysis of an HLA-B27 functional variant: identification of residues that contribute to the specificity of recognition by cytolytic T lymphocytes. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7394–7398. doi: 10.1073/pnas.82.21.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega M. A., Wallace L., Rojo S., Bragado R., Aparicio P., López de Castro J. A. Delineation of functional sites in HLA-B27 antigens. Molecular analysis of HLA-B27 variant Wewak I defined by cytolytic T lymphocytes. J Immunol. 1985 Nov;135(5):3323–3332. [PubMed] [Google Scholar]
- Wan A. M., Ennis P., Parham P., Holmes N. The primary structure of HLA-A32 suggests a region involved in formation of the Bw4/Bw6 epitopes. J Immunol. 1986 Dec 1;137(11):3671–3674. [PubMed] [Google Scholar]
- Ways J. P., Coppin H. L., Parham P. The complete primary structure of HLA-Bw58. J Biol Chem. 1985 Oct 5;260(22):11924–11933. [PubMed] [Google Scholar]
- Ways J. P., Lawlor D. A., Wan A. M., Parham P. A transposable epitope of HLA-B7, B40 molecules. Immunogenetics. 1987;25(5):323–328. doi: 10.1007/BF00404425. [DOI] [PubMed] [Google Scholar]
- Weiss E. H., Kuon W., Dörner C., Lang M., Riethmüller G. Organization, sequence and expression of the HLA-B27 gene: a molecular approach to analyze HLA and disease associations. Immunobiology. 1985 Dec;170(5):367–380. doi: 10.1016/S0171-2985(85)80061-9. [DOI] [PubMed] [Google Scholar]
- Weiss E. H., Mellor A., Golden L., Fahrner K., Simpson E., Hurst J., Flavell R. A. The structure of a mutant H-2 gene suggests that the generation of polymorphism in H-2 genes may occur by gene conversion-like events. Nature. 1983 Feb 24;301(5902):671–674. doi: 10.1038/301671a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zemmour J., Ennis P. D., Parham P., Dupont B. Comparison of the structure of HLA-Bw47 to HLA-B13 and its relationship to 21-hydroxylase deficiency. Immunogenetics. 1988;27(4):281–287. doi: 10.1007/BF00376123. [DOI] [PubMed] [Google Scholar]




