Abstract
The use of banked human tissue, obtained with informed consent after elective surgical procedures, represents a powerful model for understanding underlying mechanisms of diseases or therapeutic interventions and for establishing prognostic markers. However, donated tissues typically have varying times of warm ischaemia in situ due to blood arrest or cold ischaemia due to procurement and transportation. Hence, before using these tissues, it is important to carry out pre-analytical studies to ensure that they are representative of the in vivo state. In particular, tissues of the gastrointestinal tract have been thought to have low RNA stability. Therefore, this study aimed to determine if extended warm or cold ischaemia times and snap-freezing or banking in RNA stabilization solution affects RNA integrity or gene expression in human ileum mucosa. In short, ileum mucosa was collected for up to 1.5 h and 6 h of simulated warm or cold ischaemia respectively. Subsequently, RNA integrity and gene expressions were determined. It was found that RNA integrity remained high over the course of warm and cold ischaemia examined and there were in general no significant differences between snap-freezing and banking in RNA stabilization solution. Following the same trend, there were in general no significant changes in gene expressions measured (MYC, HIF1α, CDX, HMOX1 and IL1β). In conclusion, RNA in the ileum mucosa is maintained at a high integrity and has stable gene expression over the examined time course of warm or cold ischaemia when banked in RNA stabilization solution or snap-frozen in liquid nitrogen. As the average warm and cold ischaemia times imposed by surgery and the process of tissue banking are shorter than the time period examined in this study, human ileum mucosa samples collected after surgeries could be used for gene expression studies.
Introduction
Comparative quantification of gene expression, through real-time quantitative polymerase chain reaction (RT-qPCR) or gene expression microarrays, is essential for understanding the molecular bases of diseases. For such studies, human tissue banks such as the one at the Surgical Clinic in the Hospital of the University of Munich, provide powerful models by banking leftover tissue in excess of what is required for diagnostic tests by the pathologists. For these tissues to provide relevant experimental results, the banked tissue must be of a high quality representative of the in vivo state. Unlike the use of animal models, human tissue is typically obtained from resected tissues from elective surgeries after obtaining informed consent from the donor. As such, there will be varying warm and cold ischaemia times associated with each tissue dependent on the blood arrest time during the operation or the transport time from the operation suite to the pathologist and finally to the tissue bank [1].
Studies have been undertaken by various investigators to examine the effect of temperature, time and banking method (snap-freezing/ banking in RNA stabilization solution) on RNA integrity and gene expression in a variety of tissues (S1 and S2 Tables). For the gut in particular, investigators have examined RNA quality and gene expression in the duodenum [2] and colon [3, 4]. However, to our knowledge, the effects of the above conditions on ileum mucosa have not been examined. This is of interest as previous studies have shown that RNA was least stable in tissues of the gastrointestinal tract [2, 5] possibly due to the presence of ribonucleases (RNases), which play a role in epithelial host defence [6–9].
For ileum collected in this hospital, the average warm ischaemia time (37°C) was 12 ± 13 min and the average cold ischaemia time (on ice) was 22 ± 15 min with values expressed as means ± standard deviation (N = 52). Therefore, this study aimed to examine the effects of extended warm or cold ischaemia time and banking method (snap-freezing in liquid nitrogen or banking in RNA stabilization solution) on RNA integrity and gene expression.
Materials and Methods
Human ileum collection
The tissues and data used in this study were provided by the Biobank (http://www.klinikum.uni-muenchen.de/Chirurgische-Klinik-und-Poliklinik-Grosshadern/de/0800-gewebebank/index.html) located in the Hospital of the University of Munich, which operates in accordance with the European Union-compliant ethical and legal framework of the Human Tissue and Cell Research (HTCR) Foundation (http://www.htcr.org) [10]. The process of tissue collection included obtaining written informed consent from all 6 donors. This framework has also been approved by the ethics commission of the Faculty of Medicine in the University of Munich and the Bavarian State Medical Association.
From each donor with colon carcinoma, a piece of terminal ileum was collected in the operation room during right hemicolectomy by the staff from the Biobank. During the surgical procedure, the terminal ileum part of the preparation was dissected out after the detachment of the branches of the middle colic artery and the right colic artery and before the complete resection of the whole preparation. The terminal ileum was dissected as follows. While the vascular structure remains intact, a 5 cm length of terminal ileum was isolated between two staplers. The vascular pedicle still supplying this part of the ileum was then clamped using an Overholt and the isolated ileum was then immediately separated and put on a back table for the preparation of ileum mucosa samples by the staff from the Biobank. The surgeon then completes the surgical intervention and removal of the remaining hemicolectomy preparation. During this procedure, no additional ileum tissue had to be resected as only the piece of terminal ileum, which is part of the hemicolectomy preparation, was separated in situ by slightly adapting surgical procedures with no effect on the therapeutic intervention.
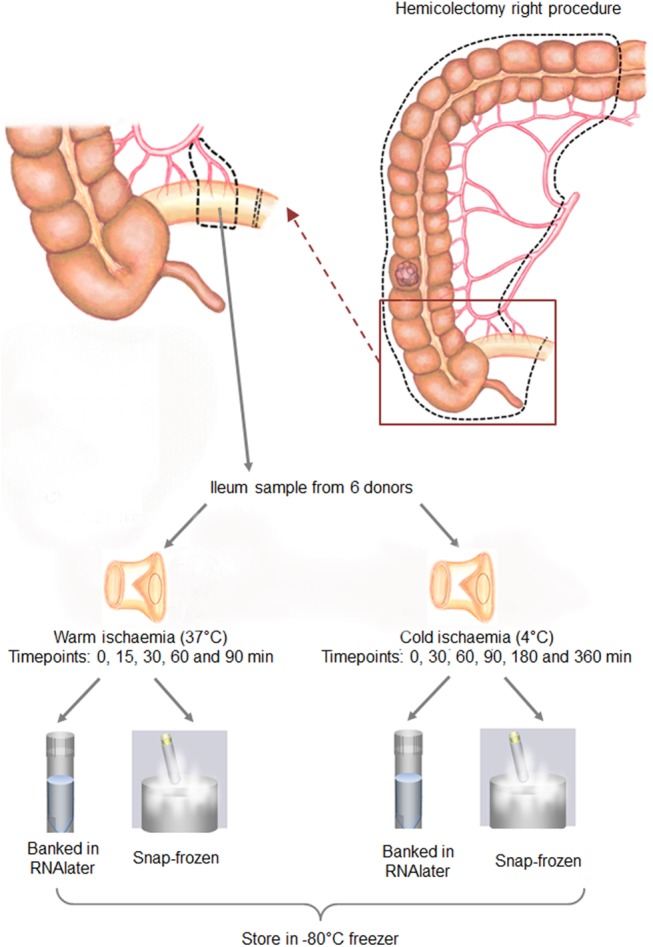
When the staff from the Biobank obtained the terminal ileum in the operation room, warm and cold ischaemia times remained less than 1 minute. As such, samples collected immediately were labeled as “time 0”. The whole ileum mucosa collection procedure is shown in Fig 1. In short, the piece of tissue collected from each donor was halved with one piece placed at 37°C to simulate warm ischemia (for 15, 30, 60 and 90 min) and the other at 4°C to simulate cold ischemia (for 30 min, 60 min, 3h and 6 h) respectively. At each time point for warm or cold ischaemia, two 30 mg pieces of ileum mucosa were sampled, one was snap-frozen and the other was banked using RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) according to manufacturer’s instructions.
Fig 1. Experimental plan for the collection of human ileum mucosa specimens from hemicolectomy right surgeries to examine the effects of warm or cold ischaemia and different processing methods used before banking of the tissues at -80°C.
H & E staining of ileum sections
FFPE ileum samples were sectioned at 6 μm thickness. These sections were stained according to the instructions for use that came with the Mayer’s haematoxylin solution (Merck, Hessen, Germany) with some modifications. The modifications were that the slides were rinsed in 1% HCl in 70% ethanol for 2 seconds after haematoxylin staining and a dip in distilled water was done after each rinse in running tap water before the next step.
Extraction and quantification of RNA from the ileum mucosa
The extraction of RNA from the ileum mucosa was performed using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. In brief, 30 mg of ileum mucosa was homogenised in the provided lysis buffer using the TissueLyser LT bead mill (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The homogenate was then transferred into a RNeasy spin column, which binds RNA using a silica-membrane. In order to remove any potential DNA contamination, an on-column DNA digestion step was done using the RNase-free DNase Set (Qiagen, Hilden, Germany). The bound RNA was finally eluted from the spin column in 30 μl of RNase free water.
Assessment of nucleic acid purity was done using the NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, USA). To quantify RNA, the Quant-iT Ribogreen RNA Assay Kit (Life Technologies, Darmstadt, Germany) was used according to the manufacturer’s protocol. RNA was quantitated with a fluorescent nucleic acid stain that can be detected using the Filtermax F3 microplate reader (Molecular Devices, Biberach an der Riss, Germany) operating with excitation / emission wavelengths of 480 nm / 520 nm respectively.
Determination of the RNA integrity
To determine the RNA integrity, 200 ng of RNA from each sample was loaded on the RNA 6000 Nano Chip (Agilent Technologies, Waldbronn, Germany) according to the manufacturer’s protocol. The RNA integrity was assessed with the Agilent 2100 Bioanalyzer system, which generates a RNA Integrity Number (RIN) [11].
Complementary DNA synthesis
RNA (1 μg) was transcribed into complementary DNA (cDNA) in a final volume of 20 μl using the SuperScript VILO cDNA Synthesis Kit (Life Technologies, Darmstadt, Germany) according to the manufacturer’s protocol. The synthesis of cDNA was primed by using random oligodeoxyribonucleotides. The newly synthesised cDNA was diluted 10 times for subsequent use in RT-qPCR. Both diluted and undiluted cDNA were stored at -20°C until further use.
Real-time-qPCR
The RT-qPCRs were performed using a StepOnePlus Real-Time PCR System (Life Technologies, Darmstadt, Germany) with TaqMan Gene Expression Assays. All primers, probes and TaqMan Fast Advanced Master Mix were purchased from Life Technologies (Darmstadt, Germany). The primers and probes used in this study are described in Table 1. Each reaction had a final volume of 20 μl and was set up as follow: 10 μl TaqMan Master Mix, 1 μl primer/probe mix (0.9 μM for the forward and reverse PCR primers and 0.25 μM for the probe), 4 μl diluted cDNA, 5 μl DNase/RNase free water. Each sample was loaded in duplicate and a negative control with all the components stated above except the cDNA was included on each reaction plate. Every reaction plate was repeated once again to ensure accuracy of results. The loaded MicroAmp Fast Optical 96-well Reaction Plate (Life Technologies, Darmstadt, Germany) was then centrifuged for 2 min at 1200 rpm prior to RT-qPCR. The thermal cycling method was set up according to the manufacturer’s protocol. Prior to gene expression analyses of the genes of interest (MYC, Hif1α, CDX2, HMOX1 and IL1β), a screening was done to determine appropriate reference genes from a panel (HPRT1, GUSB, PSMB6, RPL13, TBP) using geNorm [12]. The relative gene expression was determined using the 2-ΔΔC T method with the inclusion of a normalisation factor obtained by geNorm [12].
Table 1. Description of the TaqMan Gene Expression Assays used.
| Gene | Accession number | Exon | Amplicon length (bp) | Dye /Quencher | Assay number |
|---|---|---|---|---|---|
| HPRT1 (Reference gene) | NM_000194.2 | 1–2 | 72 | VIC / MGB | Hs01003267_m1 |
| GUSB (Reference gene) | NM_000181.3 | 8–9 | 96 | VIC / MGB | Hs00939627_m1 |
| PSMB6 (Reference gene) | NM_001270481.1 | 2–3 | 93 | VIC / MGB | Hs00382586_m1 |
| RPL13 (Reference gene) | NM_000977.3 | 6–6 | 137 | VIC / MGB | Hs00744303_s1 |
| TBP (Reference gene) | NM_001172085.1 | 2–3 | 91 | VIC / MGB | Hs00427620_m1 |
| MYC (Gene of interest) | NM_002467.4 | 1–2 | 87 | FAM / MGB | Hs00905030_m1 |
| Hif1α (Gene of interest) | NM_001530.3 | 1–2 | 62 | FAM / MGB | Hs00936371_m1 |
| CDX2 (Gene of interest) | NM_001265.4 | 2–3 | 81 | FAM / MGB | Hs01078080_m1 |
| HMOX1 (Gene of interest) | NM_002133.2 | 3–4 | 82 | FAM / MGB | Hs01110250_m1 |
| IL1β (Gene of interest) | NM_000576.2 | 3–4 | 91 | FAM / MGB | Hs01555410_m1 |
Statistical analyses
The data are represented as means ± standard error of the mean (SEM). For MYC, Hif1α and CDX2 gene expressions obtained from 6 donors (Fig 2), statistical analyses were performed using a two-way ANOVA followed by an unprotected Fisher's Least Significant Difference (LSD) test when the two-way ANOVA showed significant changes. For Fig 3 and HMOX1 and IL1β gene expressions in Fig 2 with different donor numbers, two-way ANOVAs were done with subsequent post tests performed using the Bonferroni method. Following statistical analyses, values were considered significantly different when the p value was less than 0.05. The statistical analyses were done using version 6 of Prism.
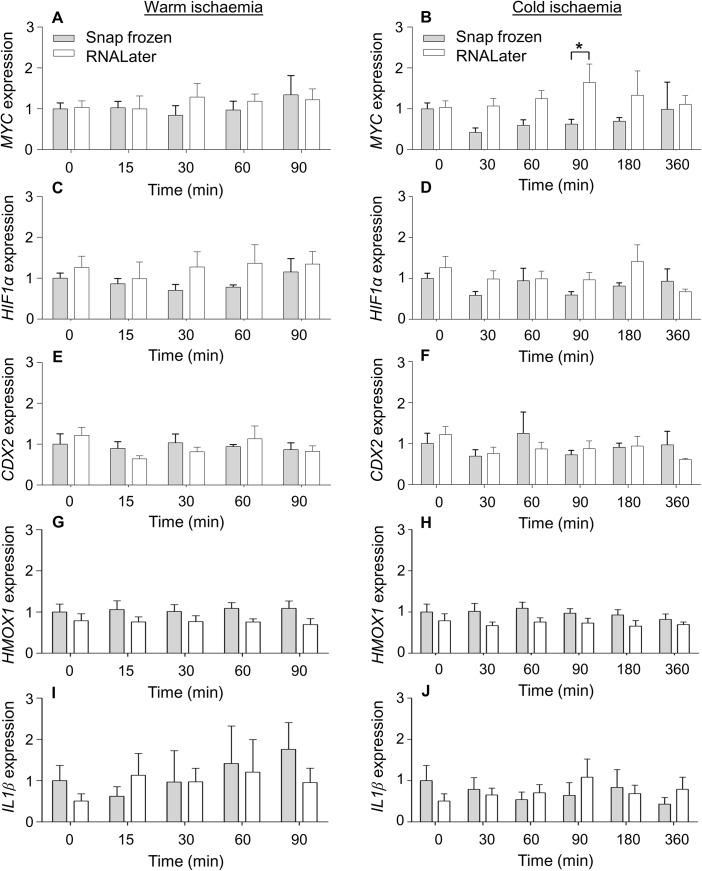
Fig 2. Normalised gene expressions of MYC (A, B), HIF1α (C, D), CDX2 (E, F), HMOX1 (G, H) or IL1β (I, J) after a time course of warm or cold ischaemia.
Values represent means ± SEM. All values were N = 6 unless otherwise stated. The following values were N = 5; HMOX1 expression at 0 and 90 min warm ischaemia (RNAlater) and 0 min cold ischaemia (RNAlater), IL1β expression at 90 min warm ischaemia (RNAlater) and 30 min cold ischaemia (snap frozen). The following values were N = 4; IL1β expression at 0 min warm or cold ischaemia (RNAlater). Four technical replicates were done per donor for each data point. *Significantly different from corresponding snap-frozen condition, p < 0.05.
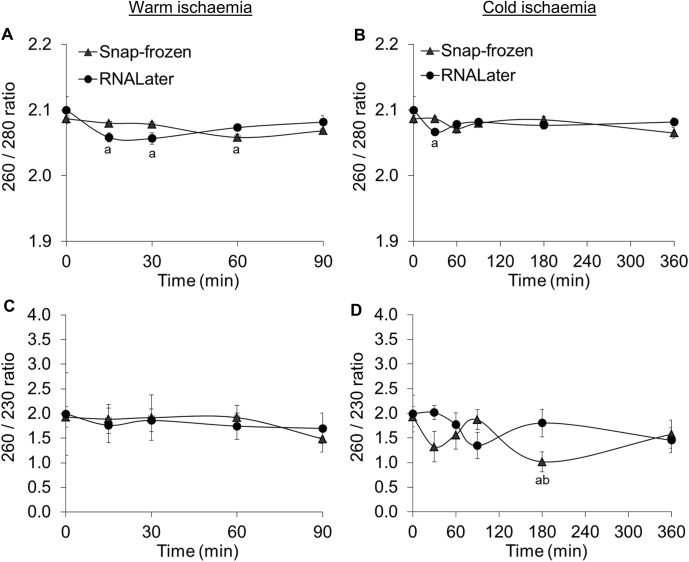
Fig 3. 260/ 280 and 260/ 230 ratios after a time course of warm (A, C) or cold (B, D) ischaemia.
Values represent means ± SEM. All values were N = 6 except for 3 values that were N = 5; 260/ 280 ratio for warm ischaemia at 30 min (snap frozen), 260/ 230 ratios for warm ischaemia at 0 min (RNAlater) and 30 min (snap frozen). aSignificantly different from corresponding 0 min condition, p < 0.05. bSignificantly different from corresponding 90 min condition, p < 0.05.
Results
Nucleic acid purity was sufficient for downstream experiments
260/280 ratios with values between 2 and 2.1 show that there is no protein contamination of the isolated RNA samples (Fig 3A and 3B). However, 260/ 230 ratios were below the range of 2 to 2.2, indicating that there may be residual chemical contamination from the RNA extraction procedure (Fig 3C and 3D). Thus, in order to avoid an overestimation of RNA concentration by spectrophotometric methods due to chemical contamination, RNA in solution was quantified using a fluorescent nucleic acid stain.
The ileum mucosa retains tissue and cellular structure
H & E staining showed that the epithelium of the mucosa remains intact and the tissue retains characteristic tissue and cellular structure over the time course of warm or cold ischaemia (Fig 4).
Fig 4. H & E staining of ileum section at 0 min (A), 90 min warm ischaemia (B) and 360 min cold ischaemia (C).
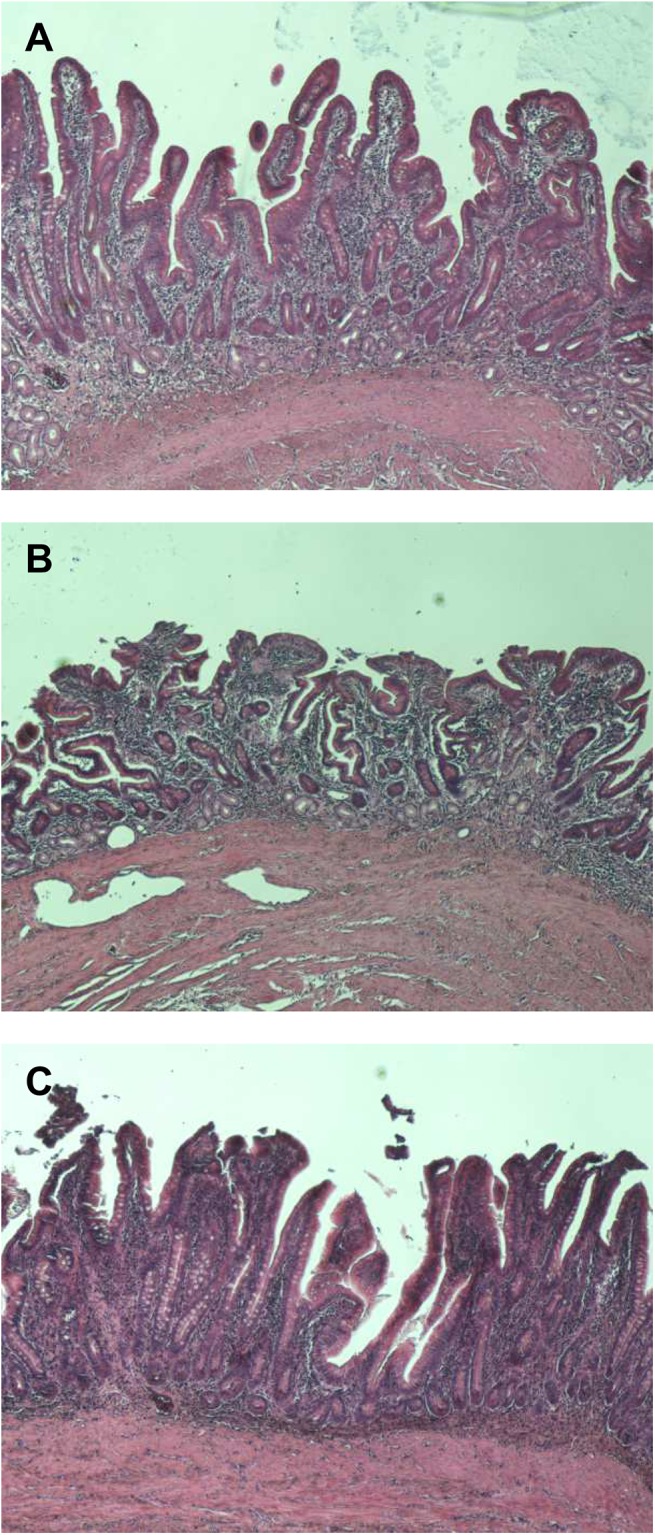
The images were acquired at x 40 magnification.
The ileum mucosa retains high RNA integrity after an extended period of warm and cold ischaemia
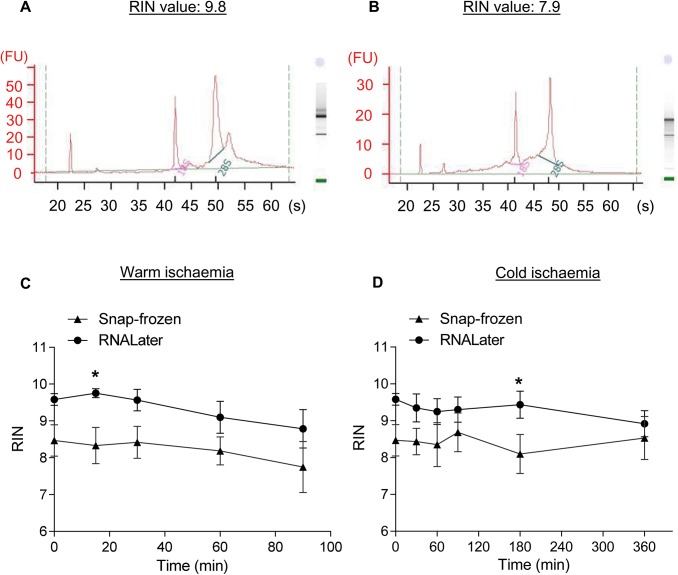
The RNA integrity of each sample was assessed using the RIN assigned by the Bioanalyzer. Electropherograms representing the highest (9.8) and lowest (7.9) average RIN obtained in this study are shown in Fig 5A and 5B. When RIN was plotted against warm ischaemia time (Fig 5C) or cold ischaemia time (Fig 5D), there were generally no significant differences between snap-freezing or using RNA stabilization solution to bank the ileum mucosae over the time course. There were significant 1.2-fold increases in RIN when the tissue was banked using RNA stabilization solution only after 15 minutes of warm ischemia and 180 minutes of cold ischemia.
Fig 5. A representative electropherogram is shown for the highest (A) and lowest (B) average RNA Integrity Number (RIN) obtained in this study. RIN of ileum mucosa specimens collected by snap-freezing in liquid nitrogen or banked using RNA stabilization solution after a time course of warm (C) or cold ischaemia (D).
Values represent means ± SEM with N = 6. *Significantly different from corresponding snap-frozen condition, p < 0.05.
Warm and cold ischaemia time does not affect gene expression in the ileum mucosa
Prior to gene expression analyses, suitable reference genes for this study had to be found. Therefore, 5 commonly used reference genes, HPRT1, GUSB, PSMB6, TBP and RPL13, were tested. GeNorm output indicated that RPL13 was the least stable reference gene (Fig 6A) and suggested that four reference genes, HPRT1, GUSB, PSMB6 and TBP, should be used to keep the pairwise variation less than 0.12 (Fig 6B). These four reference genes were sufficient as Vandesompele et al. [12] proposed that no additional reference gene is required when pairwise variations are less than 0.15.
Fig 6. Genorm analysis to determine (A) average expression stability values of the reference genes during stepwise exclusion of the least stable reference gene and (B) pairwise variation analysis between the normalization factors to determine the number of reference genes required for accurate normalization.
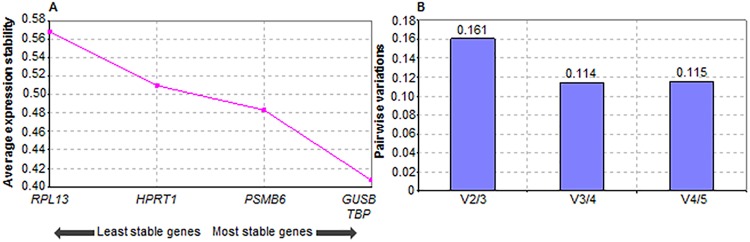
All values from 6 donors were used for the analysis with each gene expression assay done with 4 technical replicates.
To assess whether an extended period of warm or cold ischaemia affects gene expression in the ileum mucosa, the expressions of 5 genes of interest were examined. These genes include 4 inducible genes, which can respond to external stimuli by a rapid activation of gene expression and an intestine-specific gene (CDX2). In short, the 4 inducible genes examined included an immediate early gene (MYC), the hypoxia-inducible factor 1α (HIF1α), an inflammatory response gene (IL1β) and oxidative stress inducible gene (HMOX1). Real-time-qPCR analysis showed that none of these 5 genes showed significant differences in relative gene expression when compared across various time points for both warm and cold ischaemia (Fig 2). In addition, the processing of the ileum mucosa by snap-freezing or through the use of RNA stabilization solution for banking did not result in any significant differences except for a significant increase in the relative expression of MYC when banked using RNA stabilization solution after 90 min of cold ischemia by 2.6-fold (Fig 2B).
Discussion
This study aimed to determine the effect of warm or cold ischaemia time on RNA integrity and gene expression in the ileum mucosa. Due to the nature of the surgery described in the method section above, ileum samples could be obtained in the operation room without warm ischaemia in situ. Ileum tissue was then sampled at various time-points after warm or cold ischaemia for comparison against the time 0 sample, which is a good representation of the in vivo condition (Fig 1).
RNA integrity is indicated by RIN, which is a number assigned by a software algorithm that ranges between 1 (totally degraded RNA) to 10 (fully intact RNA) [11]. Various publications have recommended minimum acceptable RINs after examining the relationship between RIN and the reliability of data obtained by downstream applications [13–18]. For RT-qPCR, Fleige et al. [14] stated that a RIN higher than 5 indicates good total RNA quality for amplification of PCR products up to 200 bp and a RIN higher than 8 indicates optimal total RNA for this application. However, Botling et al. [13] found that the critical RIN associated with potentially erroneous RT-qPCR expression values was below 8. This higher RIN requirement from Botling et al. [13] could be due to the use of oligo-dT primers for cDNA synthesis. For gene expression microarrays, authors have recommended RINs ≥ 6 [17], ≥7 [16, 18] or >5 (provided that genes that are short or have probe binding sites close to the 5’ end are excluded) [15]. In this study, the RINs (7.8±0.6–9.8±0.1) obtained from all time-points, temperatures or banking methods indicate that the RNA isolated from the ileum mucosa is of a very high quality. Thus, these RNA samples should be of a sufficient quality for downstream analyses using RT-qPCR or gene expression microarrays. It is interesting to note that such high RIN can be obtained despite the expression of RNases in the gut [6–9]. This could be due to tissue and cellular structure being maintained at these early stages (Fig 4), which would restrict exogeneous RNase access to intracellular RNA [3, 13].
With regard to the RIN, S1 Table summarises the effect of time, temperature and banking method on this number. In short, it has been found that the use of RNA stabilization solution generally results in higher RIN compared to snap-freezing [19–21]. Similarly, this study found that significantly higher RINs (1.2-fold) were obtained for 2 of the examined time-points when the samples were banked in RNA stabilization solution instead of being snap-frozen. In contrast, although the samples banked in RNA stabilization solution tended to have higher RINs, there were in general no significant differences between the 2 banking methods. As samples here were snap-frozen or placed in RNA stabilization solution at the same time using the same dissection technique, this discrepancy could be due to the nature of the banking method itself. Both methods preserve RNA through the prevention of RNase activity either by sub-zero temperatures or by precipitating RNases and RNA. However, for snap-frozen samples, enzymatic activity is only temporarily inactivated by the low temperature. If the sample is thawed, this process destroys tissue and cellular structure allowing RNases to commence degradation of RNA as the temperature rises [13, 18]. As such, great care must be taken when processing the sample for subsequent steps to prevent RNA degradation and to obtain comparable RNA quality.
Further, some authors have found that there were no significant differences in RIN over a time course in human colon cancers (up to 4 h) [22], rectal and distal sigmoid tumours (up to 2 h) [19], pancreatic tumours (up to 1 h) [16] or mouse skin (up to 1 h) [23] (S2 Table). However, Hong et al. [24] found that RIN in human colorectal cancers decreased with time (S1 Table). This study has results similar to the first group; RINs in human ileum mucosa were not significantly different for up to 3 h of warm ischaemia or 6 h of cold ischaemia. As stated above, RNA integrity appears to be maintained in intact tissue. It is possible that RIN can be decreased in tissues subjected to trauma during surgery or dissection [20] or in samples with necrosis [13].
For gene expression, there was only a significant increase in the relative expression of MYC after 90 minutes of cold ischaemia when banked using RNA stabilization solution. This result could be due to cold ischaemia triggering a transient overexpression of MYC at 90 minutes. However, this scenario is not very likely as a corresponding increase in MYC expression should have occurred in the snap-frozen samples. Instead, this increase could be due to an inadvertent sampling of cancerous tissue at this time-point, although the tissue obtained is from the resection margin, as it is known that colorectal carcinomas frequently have overexpression of MYC [25, 26]. Other than this significant value, there are in general no significant changes in gene expression. Similarly, the studies done on a wide variety of tissues show that although there were changes in gene expression with time and banking method, the majority of the genes examined did not have significantly different expression levels (S2 Table) [4, 19, 21, 23, 27–31]. Considering that many of the above studies were done with microarrays, which in many cases have a 3’ bias due to the requirement for mRNAs with a poly-A tail and subsequent hybridization to a probe that may bind at some distance from the 3’ end [5, 32], the gene expression profiles obtained were surprisingly stable. Thus, this can explain why gene expression in ileum mucosa is stable as RT-qPCR is more robust especially when random hexamers are used for reverse transcription of cDNA, amplicon lengths are short and suitable reference genes are chosen [14].
In conclusion, ileum mucosa has a high RNA integrity and stable gene expression over a time course of warm or cold ischaemia whether banked in RNA stabilization solution or snap-frozen in liquid nitrogen. Since the average warm and cold ischaemia times imposed by surgery and the process of tissue banking is shorter than what was examined in this study, human ileum mucosa samples could be used for gene expression studies. Nonetheless, it is important to check that one’s genes of interest are not in the small proportion of genes affected by such conditions before proceeding. As to whether snap-freezing or banking in RNA stabilization solution should be done, both methods offer different advantages and disadvantages. RNA stabilization solution offers ease of use compared to liquid nitrogen and is also more operator-friendly compared to snap-frozen samples, which should not be thawed at all during processing steps. On the flip side, snap-frozen samples offer more versatility as samples banked using RNA stabilization solution have been found to provide poor microscopic images due to a loss in tissue morphology [13, 33] and have been found to be unsuitable for immunohistochemistry [33].
Supporting Information
(DOCX)
(DOCX)
Acknowledgments
The authors would like to acknowledge Stefanie Schreiber, Sandra Mülek, Beatrice Rauter, Nadine Gesse, Ute Bossmanns and Sabrina Fröba for their help with tissue collection and bench work. Also, we would like to thank Edeltraud Hanesch for creating the illustrations in Fig 1. This study was supported by the non-profit state-controlled HTCR Foundation, which granted the necessary rights for the utilisation of the donor tissues and data.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was carried out with financial support from the German Federal Ministry of Education and Research (BMBF grant: M4 Munich Biotech Excellence Cluster Biobank Alliance, grant number 01EX1020B). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Thasler WE, Thasler RM, Schelcher C, Jauch KW. Biobanking for research in surgery: are surgeons in charge for advancing translational research or mere assistants in biomaterial and data preservation? Langenbeck's archives of surgery / Deutsche Gesellschaft fur Chirurgie. 2013;398(4):487–99. Epub 2013/02/23. 10.1007/s00423-013-1060-y . [DOI] [PubMed] [Google Scholar]
- 2. Ibberson D, Benes V, Muckenthaler MU, Castoldi M. RNA degradation compromises the reliability of microRNA expression profiling. BMC biotechnology. 2009;9:102 Epub 2009/12/23. 10.1186/1472-6750-9-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Micke P, Ohshima M, Tahmasebpoor S, Ren ZP, Ostman A, Ponten F, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Laboratory investigation; a journal of technical methods and pathology. 2006;86(2):202–11. Epub 2006/01/13. 10.1038/labinvest.3700372 . [DOI] [PubMed] [Google Scholar]
- 4. Spruessel A, Steimann G, Jung M, Lee SA, Carr T, Fentz AK, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques. 2004;36(6):1030–7. Epub 2004/06/24. . [DOI] [PubMed] [Google Scholar]
- 5. Lee J, Hever A, Willhite D, Zlotnik A, Hevezi P. Effects of RNA degradation on gene expression analysis of human postmortem tissues. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2005;19(10):1356–8. Epub 2005/06/16. 10.1096/fj.04-3552fje . [DOI] [PubMed] [Google Scholar]
- 6. Cho S, Zhang J. Zebrafish ribonucleases are bactericidal: implications for the origin of the vertebrate RNase A superfamily. Molecular biology and evolution. 2007;24(5):1259–68. Epub 2007/03/10. 10.1093/molbev/msm047 . [DOI] [PubMed] [Google Scholar]
- 7. Futami J, Tsushima Y, Murato Y, Tada H, Sasaki J, Seno M, et al. Tissue-specific expression of pancreatic-type RNases and RNase inhibitor in humans. DNA and cell biology. 1997;16(4):413–9. Epub 1997/04/01. . [DOI] [PubMed] [Google Scholar]
- 8. Harder J, Schroder JM. RNase 7, a novel innate immune defense antimicrobial protein of healthy human skin. The Journal of biological chemistry. 2002;277(48):46779–84. Epub 2002/09/24. 10.1074/jbc.M207587200 . [DOI] [PubMed] [Google Scholar]
- 9. Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature immunology. 2003;4(3):269–73. Epub 2003/01/28. 10.1038/ni888 . [DOI] [PubMed] [Google Scholar]
- 10. Thasler WE, Weiss TS, Schillhorn K, Stoll PT, Irrgang B, Jauch KW. Charitable State-Controlled Foundation Human Tissue and Cell Research: Ethic and Legal Aspects in the Supply of Surgically Removed Human Tissue For Research in the Academic and Commercial Sector in Germany. Cell and tissue banking. 2003;4(1):49–56. Epub 2004/07/17. 10.1023/A:1026392429112 . [DOI] [PubMed] [Google Scholar]
- 11. Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M, et al. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC molecular biology. 2006;7:3 Epub 2006/02/02. 10.1186/1471-2199-7-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology. 2002;3(7):RESEARCH0034 Epub 2002/08/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Botling J, Edlund K, Segersten U, Tahmasebpoor S, Engstrom M, Sundstrom M, et al. Impact of thawing on RNA integrity and gene expression analysis in fresh frozen tissue. Diagnostic molecular pathology: the American journal of surgical pathology, part B. 2009;18(1):44–52. Epub 2009/02/14. 10.1097/PDM.0b013e3181857e92 . [DOI] [PubMed] [Google Scholar]
- 14. Fleige S, Walf V, Huch S, Prgomet C, Sehm J, Pfaffl MW. Comparison of relative mRNA quantification models and the impact of RNA integrity in quantitative real-time RT-PCR. Biotechnology letters. 2006;28(19):1601–13. Epub 2006/08/11. 10.1007/s10529-006-9127-2 . [DOI] [PubMed] [Google Scholar]
- 15. Opitz L, Salinas-Riester G, Grade M, Jung K, Jo P, Emons G, et al. Impact of RNA degradation on gene expression profiling. BMC medical genomics. 2010;3:36 Epub 2010/08/11. 10.1186/1755-8794-3-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rudloff U, Bhanot U, Gerald W, Klimstra DS, Jarnagin WR, Brennan MF, et al. Biobanking of human pancreas cancer tissue: impact of ex-vivo procurement times on RNA quality. Annals of surgical oncology. 2010;17(8):2229–36. Epub 2010/02/18. 10.1245/s10434-010-0959-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Strand C, Enell J, Hedenfalk I, Ferno M. RNA quality in frozen breast cancer samples and the influence on gene expression analysis—a comparison of three evaluation methods using microcapillary electrophoresis traces. BMC molecular biology. 2007;8:38 Epub 2007/05/24. 10.1186/1471-2199-8-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thompson KL, Pine PS, Rosenzweig BA, Turpaz Y, Retief J. Characterization of the effect of sample quality on high density oligonucleotide microarray data using progressively degraded rat liver RNA. BMC biotechnology. 2007;7:57 Epub 2007/09/15. 10.1186/1472-6750-7-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bray SE, Paulin FE, Fong SC, Baker L, Carey FA, Levison DA, et al. Gene expression in colorectal neoplasia: modifications induced by tissue ischaemic time and tissue handling protocol. Histopathology. 2010;56(2):240–50. Epub 2010/01/28. 10.1111/j.1365-2559.2009.03470.x . [DOI] [PubMed] [Google Scholar]
- 20. Lawson MH, Rassl DM, Cummings NM, Russell R, Morjaria JB, Brenton JD, et al. Tissue banking of diagnostic lung cancer biopsies for extraction of high quality RNA. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer. 2010;5(7):956–63. Epub 2010/06/01. 10.1097/JTO.0b013e3181ddbbe9 . [DOI] [PubMed] [Google Scholar]
- 21. Wolfe LM, Thiagarajan RD, Boscolo F, Tache V, Coleman RL, Kim J, et al. Banking placental tissue: an optimized collection procedure for genome-wide analysis of nucleic acids. Placenta. 2014;35(8):645–54. Epub 2014/06/22. 10.1016/j.placenta.2014.05.005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bao WG, Zhang X, Zhang JG, Zhou WJ, Bi TN, Wang JC, et al. Biobanking of fresh-frozen human colon tissues: impact of tissue ex-vivo ischemia times and storage periods on RNA quality. Annals of surgical oncology. 2013;20(5):1737–44. Epub 2012/06/20. 10.1245/s10434-012-2440-1 . [DOI] [PubMed] [Google Scholar]
- 23. Gopee NV, Howard PC. A time course study demonstrating RNA stability in postmortem skin. Experimental and molecular pathology. 2007;83(1):4–10. Epub 2006/12/19. 10.1016/j.yexmp.2006.11.001 . [DOI] [PubMed] [Google Scholar]
- 24. Hong SH, Baek HA, Jang KY, Chung MJ, Moon WS, Kang MJ, et al. Effects of delay in the snap freezing of colorectal cancer tissues on the quality of DNA and RNA. Journal of the Korean Society of Coloproctology. 2010;26(5):316–23. Epub 2010/12/15. 10.3393/jksc.2010.26.5.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maestro R, Viel A, Boiocchi M. Correlation between chromosome 5q deletions and different mechanisms of c-myc overexpression in human colorectal cancer. British journal of cancer. 1991;63(2):185–6. Epub 1991/02/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Viel A, Maestro R, Toffoli G, Grion G, Boiocchi M. c-myc overexpression is a tumor-specific phenomenon in a subset of human colorectal carcinomas. Journal of cancer research and clinical oncology. 1990;116(3):288–94. Epub 1990/01/01. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. De Cecco L, Musella V, Veneroni S, Cappelletti V, Bongarzone I, Callari M, et al. Impact of biospecimens handling on biomarker research in breast cancer. BMC cancer. 2009;9:409 Epub 2009/11/26. 10.1186/1471-2407-9-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin DW, Coleman IM, Hawley S, Huang CY, Dumpit R, Gifford D, et al. Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2006;24(23):3763–70. Epub 2006/07/11. 10.1200/JCO.2005.05.1458 . [DOI] [PubMed] [Google Scholar]
- 29. Schlomm T, Nakel E, Lubke A, Buness A, Chun FK, Steuber T, et al. Marked gene transcript level alterations occur early during radical prostatectomy. European urology. 2008;53(2):333–44. Epub 2007/04/24. 10.1016/j.eururo.2007.03.075 . [DOI] [PubMed] [Google Scholar]
- 30. Sewart S, Barraclough R, Rudland PS, West CR, Barraclough DL. Molecular analysis of a collection of clinical specimens stored at 4 degrees C as an alternative to snap-freezing. International journal of oncology. 2009;35(2):381–6. Epub 2009/07/07. . [PubMed] [Google Scholar]
- 31. Lee SM, Schelcher C, Gashi S, Schreiber S, Thasler RM, Jauch KW, et al. RNA stability in human liver: comparison of different processing times, temperatures and methods. Molecular biotechnology. 2013;53(1):1–8. Epub 2012/01/25. 10.1007/s12033-011-9493-4 . [DOI] [PubMed] [Google Scholar]
- 32. Almeida A, Paul Thiery J, Magdelenat H, Radvanyi F. Gene expression analysis by real-time reverse transcription polymerase chain reaction: influence of tissue handling. Analytical biochemistry. 2004;328(2):101–8. Epub 2004/04/29. 10.1016/j.ab.2004.02.004 . [DOI] [PubMed] [Google Scholar]
- 33. Hoffmann G, Ijzer J, Brinkhof B, Schotanus BA, van den Ingh TS, Penning LC, et al. Comparison of different methods to obtain and store liver biopsies for molecular and histological research. Comparative hepatology. 2009;8:3 Epub 2009/07/10. 10.1186/1476-5926-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper.