Abstract
At their C-termini, cytosolic Hsp70s have an EEVD tetrapeptide that interacts with J-protein co-chaperones of the B, but not A, class. This interaction is required for partnering with yeast B-type J-proteins in protein folding. Here we report conservation of this feature. Human B-type J-proteins also have a stringent EEVD requirement. Human A-type J-proteins function less well than their yeast orthologs with Hsp70ΔEEVD. Changes in the Zinc binding domain, a domain absent in B-type J-proteins, overcomes this partial EEVD dependence. Our results suggest that the structurally similar A- and B-class J-proteins of the cytosol have evolved conserved, yet distinct, features that enhance specialized functionality of Hsp70 machinery.
Introduction
Ubiquitous Hsp70 molecular chaperones play central roles in many cellular processes, from protein folding to protein translocation across membranes [1-4]. Regardless of the specific cellular function in which an Hsp70 is participating, it functions with a J-protein co-chaperone [5]. All J-proteins have a highly conserved J-domain, which serves to stimulate the ATPase activity of its partner Hsp70. This stimulation serves to stabilize Hsp70's interaction with a client protein, by driving allosteric conformational changes in the Hsp70 peptide binding domain (PDB) that trap the client in the peptide-binding cleft [6].
Typically, the most abundant J-proteins, regardless of the cellular compartment in which they reside, are of the Class A (e.g. cytosolic Ydj1 and DnaJA2, in Saccharomyces cerevisiae and Homo sapiens, respectively) and Class B (e.g. cytosolic Sis1 and DnaJB1, in S. cerevisiae and H. sapiens, respectively) types [5, 7]. Class A and B are sometimes called Class I and Class II, respectively. Even though J-proteins are often structurally diverse outside of the J-domain, Class A J-proteins and a subset of Class B J-proteins have many similarities. These include, in addition to the N-terminal J-domain, an adjacent glycine-rich region, two structurally similar ß-barrel domains (CTD1 and CTD2) containing the client binding cleft, and a C-terminal dimerization domain.
However, despite the significant similarities, these Class A and Class B J-proteins have significant differences. For example, Class A J-proteins have a Zinc binding domain (ZnBD) protruding from CTD1, that consists of two Zinc centers [8]. In addition, Class B proteins of the cytosol have a binding site in CTD1 for the conserved EEVD tetrapeptide found at the extreme C-terminus of cytosolic Hsp70s, called EEVD(Hsp70) throughout [9, 10]. Recent analysis of Sis1 showed that the Sis1:EEVD(Hsp70) interaction is required for Sis1 to function in in vitro protein folding assays, as indicated by the ability of the yeast Hsp70 Ssa1 lacking its EEVD motif (Ssa1ΔEEVD) to refold denatured luciferase when partnering with wild-type Ydj1, but not with wild-type Sis1 [11].
EEVD(Hsp70) also interacts with a number of “targeting factors” that, for example, serve to target Hsp70 clients to the Hsp90 chaperone system and the mitochondrial outer membrane [12-16]. These results suggested that this interaction might play a physiological role, such as minimizing the entry of Class B bound clients into these pathways. Thus, it is of interest to know whether these fundamental characteristics have been conserved. Therefore, we set out to compare the ability of human orthologs of Sis1 and Ydj1, DnaJB1 and DnaJA2, respectively, to function with Hsp70s lacking their EEVD motifs. We found that wild-type DnaJB1, like wild-type Sis1, stringently required EEVD(Hsp70). Furthermore, human DnaJA2 performed less well than its yeast ortholog Ydj1 in protein refolding. These functional differences were traced, at least in part, to sequence differences in the Zn binding domain of the two proteins, suggesting that the Zn binding domain may have diverged, reflecting the evolving complexities of multicellular organisms.
Materials and Methods
Protein purification
Ydj1 and Hsp104 were purified from Escherichia coli and Ssa1 was purified from yeast, as described [11]. The templates for Hsp70A1A (Hsp72), DnaJB1, DnaJA2 and Apg2 constructs were obtained from Addgene (Cambridge, MA) cloned into pMAL-His-TEV [17] and expressed in Rosetta 2 (DE3) pLys E coli cells. DnaJB1, DnaJA2 and Apg2 were purified in a manner similar to that described for Ydj1/Sis1 [11], using Ni chromatography and subsequent removal of MBP-His tag with TEV protease. Hsp72 was purified in a similar manner with an additional ATP agarose chromatography step after TEV cleavage. Hsp72ΔEEVD and variant J-protein plasmids were constructed using the QuickChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). DnaJA2Zn-Xdj1 and DnaJA2Zn-Ydj1 were constructed by replacing the ZnBD of DnaJA2 (143-209) with that of Xdj1 (159-235) and Ydj1 (143-208), respectively.
Luciferase refolding assay
Luciferase refolding assay for human chaperone proteins was carried out as described [18] with some modifications. Firefly luciferase (5 μM; Sigma-Aldrich, St. Louis, MO) was denatured for 30 min at 47°C in buffer G [100 mM KOAc, 20 mM Hepes-KOH, pH 7.5 and 5 mM Mg(OAc)2] containing 6M guanidinium hydrochloride. The denatured luciferase was diluted to give a final concentration of 30 nM, in refolding buffer [100 mM KOAc, 20 mM Hepes-KOH, pH 7.5, 5 mM Mg(OAc)2, 39 mM NaCl and 4 mM ATP] containing J-protein (DnaJB1 and DnaJA2 at 6 μM) and Hsp72 (3 μM) and incubated at 30°C for up to 150 min. The amounts of J-proteins and Hsp72 used were determined by optimization tests over a range of concentrations: J-proteins from 1 μM to 8 μM; Hsp72 from 0.5 μM to 4 μM. To measure refolding, 1 μl of the refolding mixture was first added to 24 μl of buffer G containing 2 mM DTT and 0.1 mg/ml of bovine serum albumin (BSA), and then 50 μl of luciferase assay system (Promega, Madison, WI). Measurements were taken in a BioTek synergy2 plate reader.
Luciferase refolding assay for DnaJA2/Ydj1 with Ssa1 (Fig. 2B) was carried out as described [11]. Briefly, luciferase was denatured for 1 hr 20 min at 30°C in buffer A [25 mM Hepes (pH 7.4), 50 mM KCl and 5 mM MgCl2] containing DTT (5 mM) and guanidinium hydrochloride (6M), and then, to give a final concentration of 30 nM, diluted in refolding buffer [buffer A containing 2 mM ATP]. The refolding mixture contained J-proteins (DnaJA2, 4 μM; Ydj1, 3.2 μM) and Ssa1 (2 μM for DnaJA2 and 1.6 μM for Ydj1). Luciferase refolding assay for DnaJB1/DnaJA2 with Hsp72, Apg2 and Hsp104 (Fig. 2C) was carried out as described above for human chaperone proteins except 6M urea at 30°C, rather than guanidinium hydrochloride at 47°C, was used for denaturation because guanidinium hydrochloride inhibits the activity of Hsp104. The refolding mixture contained J-proteins (8 μM), Hsp72 (4 μM), Apg2 (0.6 μM with DnaJB1; 0.4 μM with DnaJA2) and Hsp104 (1 μM).
Fig. 2.
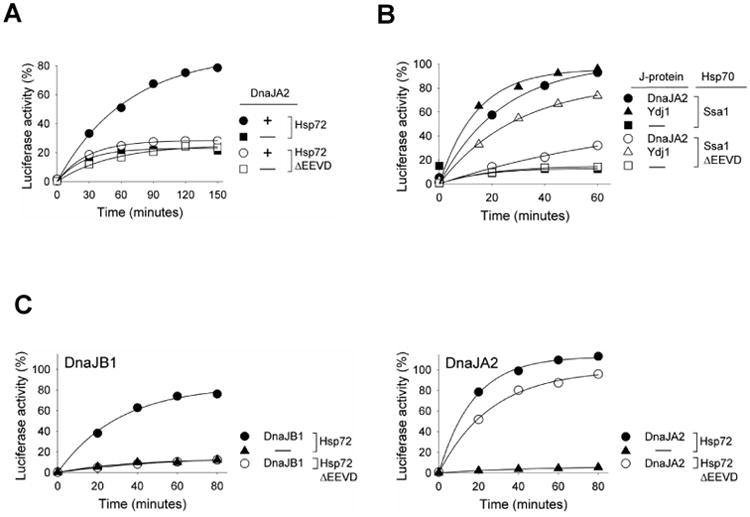
DnaJA2 is more dependent on EEVD(Hsp70) than Ydj1 in protein refolding. (A, B) 6M GuHCl denatured luciferase was diluted into refolding buffer containing (A) Hsp72/DnaJA2 and (B) Ssa1/DnaJA2 or Ydj1. Aliquots were taken at the indicated times and luciferase activity determined. Activity of non-denatured luciferase was set at 100%. (C) 6M urea denatured luciferase was diluted into refolding buffer containing Hsp72/DnaJB1or DnaJA2 in addition to the nucleotide exchange factor Apg2 and Hsp104. Aliquots were taken at the indicated times and luciferase activity was determined (non-denatured luciferase: 100%).
The numerical value of percent refolding for each figure is provided in Supplementary Figure 1.
Results
Class B J-protein dependence on EEVD(Hsp70) interaction
To compare the activities of yeast and human J-proteins, we first tested whether the protein refolding ability of human Class B J-protein DnaJB1 required the presence of the EEVD motif of human Hsp70A1A (called Hsp72), as did yeast Class B Sis1 when functioning with the yeast Hsp70 Ssa1. Refolding of ∼70% of denatured luciferase was observed in assays containing full-length Hsp72 partnering with DnaJB1 (Fig 1A, left panel). However, in the presence of Hsp72 lacking its C-terminal EEVD (called Hsp72ΔEEVD throughout) no activity was detected above the background levels found in the absence of J-protein (Fig 1A, right panel).
Fig. 1.
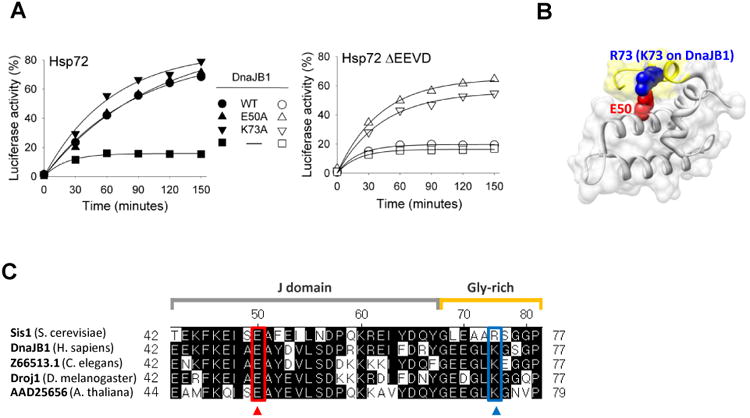
Dependence of DnaJB1 on EEVD(Hsp70) for protein refolding activity. (A) Luciferase was denatured in 6M GuHCl and diluted into refolding buffer containing Hsp72/DnaJB1 mixtures. Aliquots were removed at the indicated times and luciferase activity determined. Activity of non-denatured luciferase was set at 100%. (B) Molecular structure of S. cerevisiae Sis1 N-terminal 1-81 residues fragment (PDB ID: 4RWU), rendered using UCSF Chimera (San Francisco, CA). Side chains of E50 and R73 (K73 on DnaJB1) residues are shown in space-filled mode. J-domain and glycine-rich region are colored gray and yellow, respectively. (C) Alignment of selected regions of J-domain and glycine-rich region of DnaJB1 orthologs, performed using DNASTAR (Madison, WI). The two amino acids involved in internal interaction between J-domain and glycine-rich region of Sis1 of Saccharomyces cerevisiae are boxed (E50 [red] and R73 [blue], respectively).
In the case of Sis1, we previously found through structural analysis that E50 of the J-domain forms a salt bridge with R73 of the glycine-rich region (Fig 1B). In addition, alteration of either E50 or R73 to alanine overcame the dependence of Sis1 on EEVD(Hsp70) [11]. Sequence comparison revealed that the charge of these residues is conserved in Sis1 orthologs across species (Fig 1C). For example, residue 50 is also glutamic acid in DnaJB1. Position 73 is a lysine in human DnaJB1 and many other Sis1 orthologs. To assess whether the relief of the dependence on EEVD(Hsp70) is conserved, we individually altered E50 and K73 of DnaJB1 to alanine. These variants were purified and tested for their ability to partner with Hsp72 and Hsp72ΔEEVD in protein refolding. In reactions containing Hsp72ΔEEVD plus either DnaJB1E50A or DnaJB1K73A, approximately 65 and 55 % of denatured luciferase was refolded, respectively, to its active form. This substantial gain of activity with Hsp72ΔEEVD indicates that this fundamental principles of Class B J-protein dependence on EEVD(Hsp70) for functionality has been conserved.
DnaJA2 is less functional than Ydj1 in partnering with Hsp70ΔEEVD
We next assessed the ability of a human Class A J-protein to partner with human Hsp72 and Hsp72ΔEEVD. We chose DnaJA2 for analysis because it had previously been shown to function in refolding of denatured luciferase [18]. When DnaJA2 partnered with full-length Hsp72, 80% recovery of luciferase activity was obtained by 150 min. However, no activity above background levels was detected with Hsp72ΔEEVD (Fig 2A). This result was unexpected because the yeast orthologs of DnaJA2, Ydj1 and Xdj1 [19], function with Ssa1ΔEEVD only slightly less efficiently than with full-length Ssa1 [11].
To begin to understand the basis of this difference between yeast and human Class A J-proteins in their functioning with an Hsp70 partner lacking its C-terminal EEVD, we first tested whether DnaJA2 could function with yeast Ssa1 (Fig 2B). We found that DnaJA2 functioned efficiently with full-length Ssa1; 93% of luciferase activity was recovered in 60 min. However, activity with Ssa1ΔEEVD was minimal, reaching 32%, compared to 15% for the control reaction lacking J-protein. We next tested the EEVD dependence of DnaJA2 in a reaction that contained yeast Hsp104, as well as the human Hsp70 proteins (Fig 2C, right panel). Hsp104 is a disaggregase [20, 21] that facilitates Hsp70:J-protein driven refolding of luciferase denatured by urea because it solubilizes aggregates that may form under these conditions. DnaJA2 was nearly as active with Hsp72ΔEEVD as with full-length Hsp72, reaching close to 100% reactivation within 80 min.
We also tested whether DnaJB1 refolding activity was EEVD(Hsp70)-dependent in this assay to determine whether it, like yeast Sis1, is inactive with Ssa1ΔEEVD even in the presence of Hsp104. No activity above background was detected for DnaJB1 with Hsp72ΔEEVD, although it functioned effectively with full-length Hsp72 (Fig 2C, left panel). Together these results suggest that while yeast and human B class proteins have similar functional requirements for Hsp70 partnering, some differences exist between DnaJA2 and its ortholog in their efficiency of functioning with Hsp70 lacking its C-terminal EEVD.
Zn binding domain alterations overcome defect in refolding activity of DnaJA2 with Hsp70ΔEEVD
With a goal of better understanding this difference between the class A J-proteins of yeast (Ydj1 and Xdj1) and human (DnaJA2), we focused on the ZnBD, as it is the main feature that distinguishes Class A from Class B J-proteins, and has been shown to play critical roles in their function [22-24]. We constructed chimeric genes substituting the Zn binding domains of Ydj1 and Xdj1 for that of DnaJA2, generating DnaJA2Zn-Ydj1 and DnaJA2Zn-Xdj1 respectively. Both DnaJA2Zn-Ydj1 and DnaJA2Zn-Xdj1 were active, as they facilitated refolding of luciferase with full-length Hsp72 at least as efficiently as wild-type DnaJA2 (Fig 3). Furthermore, both chimeras were able to facilitate refolding with Hsp72ΔEEVD significantly better than native DnaJA2, with DnaJA2Zn-Ydj1 reaching 77% refolding. These results suggest that the yeast ZnBD fostered partnering with Hsp70 lacking its EEVD motif better than the human domain.
Fig. 3.
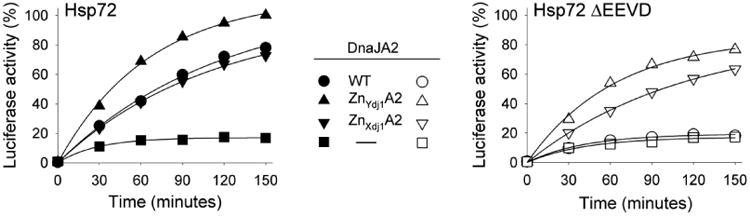
Substitution of Zn Binding Domain (ZnBD) of Ydj1/Xdj1 for that of DnaJA2 overcomes dependence of DnaJA2 on EEVD(Hsp70). Luciferase was denatured by treatment with 6M GuHCl and diluted into refolding buffer containing wild type or chimeric DnaJA2 and either Hsp72 or Hsp72 ΔEEVD. Aliquots were removed at the indicated times and enzymatic activity determined. Activity of non-denatured enzyme was set at 100%. DnaJA2Zn-Ydj1 (ZnYdj1A2) and DnaJA2Zn-Xdj1 (ZnXdj1A2).
We then compared the sequences of the ZnBD of yeast Class A J-proteins Xdj1 and Ydj1 with DnaJA2. The overall sequence identity of ZnBD of DnaJA2 with that of Xdj1 and Ydj1 is 25% and 47%, respectively (Fig 4A). We noted 5 positions in the alignment in which the yeast proteins were identical, but different in DnaJA2. We then made constructs to encode DnaJA2 variants having each residue changed to the one present in the yeast protein (i.e. S153K, A161S, A175G, S187D and E198P). In addition we changed residue A145 of DnaJA2 to E, as it is a charged residue in the yeast proteins. The variants functioned indistinguishably from wild-type DnaJA2 partnering with Hsp72, except S153K, which was inactive (Fig 4B). Like wild-type DnaJA2, two additional variants, A161S and E198P, were inactive with Hsp72ΔEEVD. However, three had substantial activity. A145E and S187D, which alter residues in Zn centers 1 and 2 (Fig 4C,D), respectively, facilitated refolding to a level of 70% in 150 min, while A175G, which is at the tip of the domain, had intermediate activity, reaching 50% in that time.
Fig. 4.
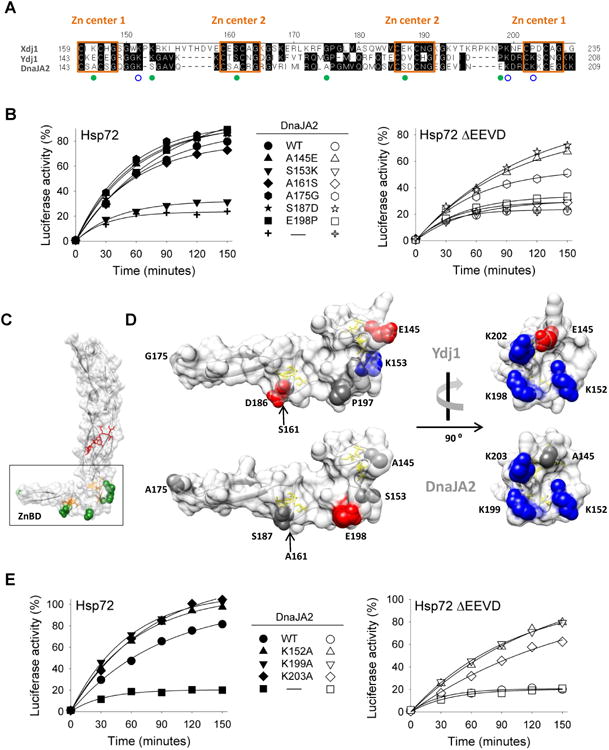
Substitution of conserved residues of ZnBD of Ydj1/Xdj1 partially overcomes EEVD dependence of DnaJA2. (A) Alignment of ZnBD of Xdj1, Ydj1 and DnaJA2, performed using DNASTAR (Madison, WI). Numbering at top is that of DnaJA2. Boxed residues indicate consensus CXXCXG sites comprising the indicated Zn centers. Filled green circles indicate the residues changed in DnaJA2 variants based on sequence conservation between Xdj1 and Ydj1; blue open-circles indicate lysine residues changed to alanine in DnaJA2 to explore the influence of charged amino acids on refolding activity. (B, E) Luciferase was denatured by treatment with 6M GuHCl and diluted into refolding buffer containing wild type or variant DnaJA2 and either Hsp72 or Hsp72ΔEEVD. Activity of non-denatured enzyme was set at 100%. (C) Molecular structure of S. cerevisiae Ydj1 ZnBD (boxed) and CTD1 and CTD2, using coordinates of Ydj1 C-terminus (PDB ID: 1NLT), generated using UCSF Chimera (San Francisco, CA)[33]. Cysteine residues of Zn centers 1 and 2 are shown in yellow (stick) and amino acids altered in DnaJA2 (filled green circles in A) are shown as green (space-filled). Peptide bound to CTD1 is shown in red. (D) Blow-up of the ZnBD. Structures of Ydj1 (top) and DnaJA2 (bottom, model structure). Cysteine residues of Zn centers 1 and 2 are shown in yellow (stick) and amino acids altered in DnaJA2 (marked with open circles in A) are shown as space-filled models; acidic side chains, red; basic side chains, blue; all others, gray. Left panels: ZnBD in the same orientation as shown in C. Right panel: rotated 90° about a vertical axis as indicated. Right panel also illustrates the relative location of additional residues altered in DnaJA2 (open blue circles in A). The model for DnaJA2 was generated using SWISS-MODEL [34].
A145E and S187D are of particular interest because both alter an uncharged residue to a negatively charged residue, suggesting that a charge change may play a role in the enhanced ability of the variants to function in refolding with Hsp72ΔEEVD We noted a set of positively charged residues near A145, in close proximity to Zn center 1. Therefore, to test the idea that overall charge may be of importance, we purified variants having each lysine, individually, changed to alanine. All three had higher than wild-type activity, with DnaJA2K152A and DnaJA2K199A being the most active, resulting in greater than 80% refolding when partnering with Hsp72ΔEEVD (Fig 4E). These results are consistent with charge playing a role in the ability of a Class A J-protein to function with Hsp70 lacking its EEVD.
Discussion
We found that human Class B J-protein DnaJB1 requires the EEVD motif at Hsp70's C-terminus to facilitate protein refolding, whether in a heterologous system partnering with yeast Hsp70 or in a homologous system partnering with human Hsp70. The conservation of this requirement between fungi and humans is consistent with the fact that both the yeast and human J-protein:Hsp70 pairs stably interact via the EEVD motif, as shown by structural and biochemical analyses [9, 10, 25, 26] and a previous report that DnaJB1 did not refold luciferase with Hsp72ΔEEVD [27]. That the same alterations at position 50 of the J-domain and 73 of the glycine-rich region have a positive effect on both the yeast [11] and human proteins suggests a conservation of the fundamental functional importance of interaction between these two domains in regulating Class B J-protein function in coordination with interaction of its CTD1 with Hsp70(EEVD). Although structural analysis of Sis1 revealed a salt bridge between E50 and R73 [11], the molecular basis of the restorative ability of these alterations is not yet understood. The relatively stable EEVD(Hsp70)/Class B J-protein interaction may overcome a constraint caused by the J-domain:glycine-rich region interaction, either by “simply” increasing the local concentration of the J-domain and/or by facilitating positioning of the J-domain such that it can most efficiently stimulate Hsp70's ATPase activity.
Regardless of the mechanism, since Hsp70's EEVD is known to be important for interaction with cellular proteins involved in protein folding and targeting pathways [12-16], the interplay amongst EEVD(Hsp70) interacting proteins likely has broader effects on cellular physiology. In the yeast system, disruption of the EEVD interaction of Hsp70 with Sis1 increases Sis1's ability to substitute for the Class A J-protein (Ydj1) in vivo [11]. This effect has been suggested to be due to more efficient transfer of Sis1-bound client to Hsp70s whose EEVD is already interacting with such targeting factors. A role in triaging Class B bound client trafficking is likely the case in the mammalian cytosol as well.
Overall, our results suggest that the Class A J-proteins do not have a stringent requirement for the presence of the EEVD of Hsp70 for partnering, as do the Class B J-proteins. However, we did observe that, unlike yeast orthologs Ydj1 and Xdj1, DnaJA2 function in luciferase refolding was essentially abolished when it was paired with Hsp72ΔEEVD. Evidence presented here indicates that the ZnBD is in good part responsible for this difference between the yeast and human Class A proteins. First, this lack of DnaJA2 function was reversed for chimeric proteins in which the ZnBD of either yeast ortholog was substituted for the native ZnBD. Secondly, the function of Hsp72ΔEEVD with DnaJA2 was enhanced by alterations within the ZnBD that more closely mimicked the properties of the yeast orthologs. One was a change of the relatively inflexible alanine (A175) within the terminal loop of the β-hairpin to glycine. Most, however, were changes that either introduced an acidic side chain (A145E, S187D) or removed a basic side chain (K152A, K199A, K203A).
The presence of the ZnBD in Class A and the interaction of EEVD(Hsp70) with Class B are the most obvious characteristics distinguishing these two classes of J-proteins. Therefore, considering the inherent negative charge of EEVD, it is intriguing that an increase in negative charge of the ZnBD enhanced the ability of DnaJA2 to function with Hsp72ΔEEVD. Such negative charges may enhance the functionality of the J-protein:Hsp70 partnership. However, although major advances regarding the ATP/ADP cycle of conformational changes have been made [6, 28], data is limited regarding the mechanistic role of either an intramolecular function of EEVD(Hsp70) or the ZnBD (Class A J-proteins). The results reported here clearly indicate that Hsp70ΔEEVD has functionality in protein folding, as alterations of J-proteins can increase activity to be nearly on par with wild-type in protein refolding assays. However, full activity is usually not achieved, suggesting, and consistent with previous results [27], that the EEVD has additional affects on Hsp70 activity independent of its specific J-protein partner. Existing data indicate that the Zn-binding domain plays an important role in the transfer of client proteins from a J-protein to Hsp70 [22, 23, 29]. Thus, charge:charge interactions may play a role in the correct, efficient positioning of Class A J-protein in its interaction with Hsp70, as is suggested for the positioning role of the interaction between EEVD(Hsp70) and Class B J-proteins.
Earlier results demonstrated that overexpression of Hsp72ΔEEVD was able to protect luciferase from heat inactivation in vivo as well as full-length Hsp72 [30]. While surprising at the time, because most experiments utilized Class B J-proteins as the canonical J-protein, and thus lack of functionality was expected, the observed result was likely due to lack of a stringent requirement of Class A J-proteins for Hsp70 to have a C-terminal EEVD. The myriad functional interacting partners of Hsp70s, either direct, such as nucleotide exchange factors [31, 32], or indirect such as other compensating chaperone systems [2], likely provides the explanation of this result. Clearly much remains to be known about the complexities of Hsp70 chaperone machineries in particular and functional interactions amongst chaperone networks in general.
Supplementary Material
HIGHLIGHTS.
-- Class A and B J-protein Hsp70 co-chaperones are structurally conserved
-- Human and yeast cytosolic Class B J-proteins require Hsp70 C-terminal EEVD for activity
-- Human Class A DnaJA2 is more dependent upon EEVD of Hsp70 than yeast homolog Ydj1
-- DnaJA2 reliance on EEVD of Hsp70 overcome by Ydj1-like changes to Zn Binding Domain
-- Conserved Class A J-proteins have evolved distinct features
Acknowledgments
We thank Szymon Ciesielski for useful discussions and help with modeling. This work was supported by National Institutes of Health grants GM31107 and GM27870 (E.A.C.)
Footnotes
Author Contribution: HYY and EAC conceived the study. HYY, TZ and EAC designed the experiments. HYY and TZ performed experiments. HYY, TZ and EAC analyzed the data and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Naturereviews Molecular cell biology. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annual review of biochemistry. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 3.Sontag EM, Vonk WI, Frydman J. Sorting out the trash: the spatial nature of eukaryotic protein quality control. Current opinion in cell biology. 2014;26:139–146. doi: 10.1016/j.ceb.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulz C, Schendzielorz A, Rehling P. Unlocking the presequence import pathway. Trends in cell biology. 2015;25:265–275. doi: 10.1016/j.tcb.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nature reviews Molecular cell biology. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends in biochemical sciences. 2013;38:507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Hageman J, Kampinga HH. Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell stress & chaperones. 2009;14:1–21. doi: 10.1007/s12192-008-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Qian X, Sha B. The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure. 2003;11:1475–1483. doi: 10.1016/j.str.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Li J, Wu Y, Qian X, Sha B. Crystal structure of yeast Sis1 peptide-binding fragment and Hsp70 Ssa1 C-terminal complex. The Biochemical journal. 2006;398:353–360. doi: 10.1042/BJ20060618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki H, Noguchi S, Arakawa H, Tokida T, Hashimoto M, Satow Y. Peptide-binding sites as revealed by the crystal structures of the human Hsp40 Hdj1 C-terminal domain in complex with the octapeptide from human Hsp70. Biochemistry. 2010;49:8577–8584. doi: 10.1021/bi100876n. [DOI] [PubMed] [Google Scholar]
- 11.Yu HY, Ziegelhoffer T, Osipiuk J, Ciesielski SJ, Baranowski M, Zhou M, Joachimiak A, Craig EA. Roles of intramolecular and intermolecular interactions in functional regulationof the Hsp70 J-protein co-chaperone Sis1. Journal of molecular biology. 2015;427:1632–1643. doi: 10.1016/j.jmb.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/s0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 13.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 14.Wu Y, Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nature structural & molecular biology. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- 15.Odunuga OO, Hornby JA, Bies C, Zimmermann R, Pugh DJ, Blatch GL. Tetratricopeptide repeat motif-mediated Hsc70-mSTI1 interaction. Molecular characterization of the critical contacts for successful binding and specificity. The Journal of biological chemistry. 2003;278:6896–6904. doi: 10.1074/jbc.M206867200. [DOI] [PubMed] [Google Scholar]
- 16.Alvira S, Cuellar J, Rohl A, Yamamoto S, Itoh H, Alfonso C, Rivas G, Buchner J, Valpuesta JM. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by Hop. Nature communications. 2014;5:5484. doi: 10.1038/ncomms6484. [DOI] [PubMed] [Google Scholar]
- 17.Meyer AE, Hoover LA, Craig EA. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. The Journal of biological chemistry. 2010;285:961–968. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzankov S, Wong MJ, Shi K, Nassif C, Young JC. Functional divergence between co-chaperones of Hsc70. The Journal of biological chemistry. 2008;283:27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahi C, Kominek J, Ziegelhoffer T, Yu HY, Baranowski M, Marszalek J, Craig EA. Sequential duplications of an ancient member of the DnaJ-family expanded the functional chaperone network in the eukaryotic cytosol. Molecular biology and evolution. 2013;30:985–998. doi: 10.1093/molbev/mst008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 21.Haslberger T, Bukau B, Mogk A. Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2010;88:63–75. doi: 10.1139/o09-118. [DOI] [PubMed] [Google Scholar]
- 22.Linke K, Wolfram T, Bussemer J, Jakob U. The roles of the two zinc binding sites in DnaJ. The Journal of biological chemistry. 2003;278:44457–44466. doi: 10.1074/jbc.M307491200. [DOI] [PubMed] [Google Scholar]
- 23.Fan CY, Ren HY, Lee P, Caplan AJ, Cyr DM. The type I Hsp40 zinc finger-like region is required for Hsp70 to capture non-native polypeptides from Ydj1. The Journal of biological chemistry. 2005;280:695–702. doi: 10.1074/jbc.M410645200. [DOI] [PubMed] [Google Scholar]
- 24.Banecki B, Liberek K, Wall D, Wawrzynow A, Georgopoulos C, Bertoli E, Tanfani F, Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. The Journal of biological chemistry. 1996;271:14840–14848. doi: 10.1074/jbc.271.25.14840. [DOI] [PubMed] [Google Scholar]
- 25.Aron R, Lopez N, Walter W, Craig EA, Johnson J. In vivo bipartite interaction between the Hsp40 Sis1 and Hsp70 in Saccharomyces cerevisiae. Genetics. 2005;169:1873–1882. doi: 10.1534/genetics.104.037242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian X, Hou W, Zhengang L, Sha B. Direct interactions between molecular chaperones heat-shock protein (Hsp) 70 and Hsp40: yeast Hsp70 Ssa1 binds the extreme C-terminal region of yeast Hsp40 Sis1. The Biochemical journal. 2002;361:27–34. doi: 10.1042/0264-6021:3610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. The EMBO journal. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. Journal of molecular biology. 2015;427:1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cyr DM, Ramos CH. Specification of Hsp70 function by Type I and Type II Hsp40. Sub-cellular biochemistry. 2015;78:91–102. doi: 10.1007/978-3-319-11731-7_4. [DOI] [PubMed] [Google Scholar]
- 30.Michels AA, Kanon B, Bensaude O, Kampinga HH. Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. The Journal of biological chemistry. 1999;274:36757–36763. doi: 10.1074/jbc.274.51.36757. [DOI] [PubMed] [Google Scholar]
- 31.Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. The Journal of biological chemistry. 2014;289:1402–1414. doi: 10.1074/jbc.M113.521997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracher A, Verghese J. GrpE, Hsp110/Grp170, HspBP1/Sil1 and BAG domain proteins: nucleotide exchange factors for Hsp70 molecular chaperones. Sub-cellular biochemistry. 2015;78:1–33. doi: 10.1007/978-3-319-11731-7_1. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Biasini M, Bienert S, Waterhouse A, Arnold G, Studer T, Schmidt F, Kiefer TG, Cassarino M, Bertoni M, Bordoli L, Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


