Abstract
PERK is serine/threonine kinase localized to the endoplasmic reticulum (ER) membrane. PERK is activated and contributes to cell survival in response to a variety of physiological stresses that affect protein quality control in the ER, such as hypoxia, glucose depravation, increased lipid biosynthesis, and increased protein translation. Pro-survival functions of PERK are triggered by such stresses, suggesting that development of small-molecule inhibitors of PERK may be efficacious in a variety of disease scenarios. Hence, we have conducted a detailed enzymatic characterization of the PERK kinase to develop a high-throughput-screening assay (HTS) that will permit the identification of small-molecule PERK inhibitors. In addition to establishing the Km of PERK for both its primary substrate, eIF2α, and for adenosine triphosphate, further mechanistic studies revealed that PERK targets its substrate via either a random/steady-state ordered mechanism. For HTS, we developed a time-resolved fluorescence resonance energy transfer–based assay that yielded a robust Z′ factor and percent coefficient of variation value, enabling the successful screening of 79,552 compounds. This approach yielded one compound that exhibited good in vitro and cellular activity. These results demonstrate the validity of this screen and represent starting points for drug discovery efforts.
Keywords: PERK, HTS, small-molecule inhibitors
Introduction
The accumulation of misfolded proteins in the lumen of the endoplasmic reticulum (ER) triggers the activation of a signaling pathway commonly referred to as the unfolded protein response (UPR). The UPR consists of a cascade of events leading to translation attenuation, transactivation of ER chaperone gene promoters, and activation of protein degradation pathways. The UPR is mediated by three primary signal transducer molecules that span the ER membrane: PKR-like ER kinase (PERK/PEK/EIF2AK3),1 inositol-requiring enzyme 1 (Ire1α and Ire1β),2 and transmembrane transcription factor ATF6.3 Together, PERK, Ire1, and ATF6 affect cell fate following ER stress by regulating expression of ER chaperones and components of the ER-associated degradation system,4,5 while PERK uniquely serves to reduce new protein synthesis.
PERK regulates protein translation via phosphorylation of eukaryotic translation initiation factor 2α (eIF2α). While phosphorylation of eIF2α reduces translation of many messenger RNAs (mRNAs), it concurrently leads to increased translation efficiency of select mRNAs such as those encoding the ATF4 transcription factor and the antiapoptotic proteins cIAP1/2.6 PERK activation contributes to cell survival via activation of ATF47 and Nrf28 transcription factors and through induction of growth arrest, the latter a consequence of inhibition of cyclin D1 translation.9
Because UPR and PERK activation are associated with a variety of disease phenotypes, including cancer, diabetes, and neurodegenerative and heart diseases,10 we were compelled to fully characterize the enzymatic properties of PERK and subsequently develop a robust assay for the identification of small-molecule PERK inhibitors. Herein we describe the steady-state enzymatic characterization of PERK and determine optimal assay conditions for high-throughput screening (HTS). Using this assay, we carried out an initial screen of approximately 80,000 compounds and describe the initial characterization of two compounds.
Materials and Methods
Chemicals
Adenosine triphosphate (ATP), magnesium chloride, sodium chloride, DMSO, HEPES, and β-actin were purchased from Sigma Chemicals (St. Louis, MO). Full-length human eIF2α-flag tagged protein (molecular mass of 37.1 kDa) was purchased from Blue Sky BioProducts (Worcester, MA). Human recombinant catalytic domain of PERK protein (amino acids 536–1116) conjugated with N-terminal GST tag (molecular mass 93.6 kDa) was purchased from Life Technologies (Carlsbad, CA). A truncated version of PERK containing the catalytic domain was used as it exhibited more robust activity, which benefits the establishment of an HTS assay. The selection of proteins used in all experiments in this study was based on quantity, purity, and activity with a focus on adaptation for a high-throughput screen.
[γ-P33]ATP was purchased from PerkinElmer (Boston, MA). Flag-d2 and p-eIF2α(S51)-Eu3+ antibodies were purchased from Cisbio Bioassays (Bedford, MA). Phospho-eIF2α(S51) and total eIF2α antibodies were obtained from Cell Signaling Technology (Danvers, MA). Multiscreen PH filtration plates were purchased from EMD-Millipore (Billerica, MA).
Compound Library
The Laboratory for Drug Discovery in Neurodegeneration (LDDN) compound library was used for the high-throughput assay. The LDDN library consists of 150,000 compounds purchased from multiple commercial vendors, as well as sets of proprietary compounds. It has been built with various computational filters to select compounds with an increased probability of oral bioavailability and blood-brain barrier (BBB) penetration, which includes calculations of polar surface area (physicochemical descriptor that strongly correlates with oral bioavailability and the ability to cross the BBB), Lipinski’s “rule of five,” and other desirability filters. A subset of the LDDN chemical library consisting of approximately 80,000 compounds was used and consists of Prestwick collection of Food and Drug Administration (FDA)–approved drugs, an National Institute of Neurological Disorders and Stroke (NINDS) collection of known bioactive compounds, purified compounds from natural sources, and peptides and small molecules purchased from Peakdale (Chapel-en-le-Frith, UK), Maybridge (Tintagel, UK), Cerep (Redmond, WA), ChemBridge (San Diego, CA), Bionet (Camelford, UK), Prestwick (Washington, DC), SPECS (Delft, The Netherlands) and Chemical Diversity Lab (San Diego, CA). Furthermore, they were chosen for the following additional properties: low proportions of compounds that contain known toxicophores, low proportion of compounds that contain reactive functional groups, and maximization of molecular diversity. In addition, the LDDN has collected proprietary compounds from various academic laboratories throughout the world and provided some of their own unique compounds synthesized by LDDN chemists.
Radiometric Kinase Reaction
For characterizing the enzyme kinetics and establishing optimal assay conditions, a radiometric assay was used. After optimization, the following reaction conditions were chosen: 20 mM HEPES (pH 7.2), 10 mM MgCl2, 150 mM NaCl and 0.01% Tween 20, and 1 µM ATP/1 µCi [γ-33P] ATP, 1 µM eIF2α with 8 nM PERK. Reactions were run for 90 min at room temperature and stopped by the addition of 75 mM phosphoric acid. Reaction mixtures were transferred to Multiscreen PH filtration plates, followed by extensive washes. Filter plates were dried and product formation was quantified using a scintillation counter. Background product formation was assessed in reactions run in the absence of PERK. Optimal enzyme concentration was determined as 8 nM, and the reaction progress curve of phospho-eIF2α formation was linear over at least 60 min (Suppl. Fig. S1).
Time-Resolved Fluorescence Resonance Energy Transfer Base Assay and HTS
For screening a small-molecule compound library, we developed a robust time-resolved fluorescence resonance energy transfer (TR-FRET) assay (Z′ > 0.7; % coefficient of variation [CV] of negative and positive controls <<10% and signal/background [S/B] ratio of 2.45) in 384-well white plates that measures direct phosphorylation of PERK kinase-specific substrate eIF2α with a phospho-specific antibody conjugated with fluorescent cryptate donor Europium (Eu3+). The TR-FRET kinase assay was conducted in buffer containing 20 mM HEPES (pH 7.2), 10 mM MgCl2, 150 mM NaCl, and 0.01% Tween 20. All kinase reaction reagents such as PERK, eIF2α, and ATP were prepared in this buffer. PERK enzyme (8 nM final concentration) was preincubated for 30 min at room temperature with the small molecules (10 µM final concentration) in a volume of 2 µL. After preincubation, 1 µL of the eIF2α/ATP mixture was added to initiate the kinase reaction (1 µM eIF2α substrate and 1 µM ATP final concentration) and incubated for 45 min at room temperature. The screen was run at the Km concentration of eIF2α and ATP, permitting the potential identification of all classes of inhibitors (noncompetitive and competitive). The enzymatic reaction was stopped by the addition of the detection reagents (p-eIF2α(S51)-Eu3+ and Flag-d2 antibodies), which were dissolved in homogeneous time-resolved fluorescence (HTRF) Transcreener buffer (Cisbio) containing 60 mM EDTA and incubated 60 min (final volume of 6 µL). The detection method is based on a phospho-specific antibody to eIF2α labeled with a long-life fluorescent cryptate donor Europium (Eu3+) and anti-Flag antibody labeled with HTRF acceptor fluorophore d2. Kinase reaction progress curves were linear for at least 60 min. An EnVision plate reader (PerkinElmer, Waltham, MA) was used to measure the FRET signal.
Data analysis
Km value for ATP was calculated based on the Michaelis-Menten equation. The Km value for eIF2α was analyzed by the Hill equation:
| (1) |
where v0 is the initial velocity, [E0] is the total enzyme concentration, kcat is the turnover number, [S] is the substrate concentration, S0 5 is the substrate concentration where v0 is half of the maximal velocity, and K is the dissociation constant for the enzyme-substrate inhibitory complex. Data were fit using SigmaPlot (Systat Software, San Jose, CA) or GraphPad Prism (GraphPad Software, La Jolla, CA) software. HTS data were loaded into ActivityBase software (IDBS, London, UK) for calculations and analysis.
Docking Studies
Glide 4.5 (Portland, OR) was used for all docking calculations of both ATP and LDN-0020506 into the structure of PERK (4G31). LDN-0020506 was docked first with an unbiased grid search over the whole receptor. The best inhibitor pose was then used to dock the ATP molecule. The induced fit docking protocol with a softened potential docking was performed to generate 20 initial poses. The softened potential docking consisted of scaling the van der Waals radii by 0.5 except in the event when alanine substitutions were introduced, in which case the receptor scaling was set to 0.7. In this case, residues of the hinge region, gatekeeper residue, and the allosteric pocket were mutated to alanine to enhance the hit rate of poses in the initial docking that are close to the correct answer, and the Glide hydrogen bond energy cutoff filter was decreased to −0.05 kcal/mol. This ensures that all retained poses contain at the very least one weak hydrogen bond with the receptor. Second, the Glide Coulomb-vdW energy cutoff filter was increased to 10 kcal/mol, enabling toleration of more steric clashes than in a normal docking run. Poses with a root mean squared deviation (RMSD) of less than 0.5 Å and a maximum atomic displacement of less than 1.2 Å were eliminated as redundant to increase diversity in the retained ligand poses. An inner grid box of 10 Å was used to fit the ligand center, and an outer box size of 20 Å was used. For each of the top 20 poses (with respect to GlideScore) from the initial softened potential docking step, a full cycle of protein refinement was performed. Prime uses the OPLS-AA (optimized potentials for liquid simulations-all atom representations) parameter and a surface Generalized Born implicit solvent model. First, a list was generated consisting of all residues having at least one atom within 5 Å of an atom in any of the 20 ligand poses. All side chains in the list underwent a conformational search and minimization. Three residues that were mutated to alanine in the initial docking stage were returned to their original identity prior to the search. After convergence to a low-energy solution, an additional minimization was performed allowing all residues in the list (backbone and side chain) and the ligand to be relaxed. The complexes were ranked by PrimeEnergy (molecular mechanics plus solvation), and those within 30 kcal/mol of the minimum energy structure were passed through for a final round of Glide docking and scoring.
Cell Culture Conditions
Mouse embryonic fibroblasts (MEFs) from wild-type mice were prepared as previously described11 and grown in Dulbecco’s modified Eagle’s medium (DMEM) (high-glucose formulation) with 4 mM L-glutamine, 10% (v/v) fetal bovine serum, 100 U/mL penicillin, 100 µg/mL streptomycin, and 55 mM β-mercaptoethanol.
Western Blot Analysis
Cells were lysed in EBC buffer (50 mM Tris [pH 8.0], 120 mM NaCl, 0.5% NP-40) supplemented with protease and phosphatase inhibitors (1 mM phenylmethylsulfonyl fluoride, 20 U/mL aprotinin, 0.4 mM NaF, 0.4 mM sodium orthovanadate, and 10 mM β-glycerophosphate). The lysates were run on 10% polyacrylamide gels and analyzed by the Western blot method. The following antibodies were used: phospho-eIF2α and eIF2α from Cell Signaling Technology.
Results
PERK Phosphorylation of eIF2α; Linearity of Reaction
We initially compared the enzymatic activity of full-length versus a truncated form of PERK wherein the transmembrane and ER-localized N-terminal domains were removed. The kinase activity of full-length PERK was significantly reduced relative to the truncated version of this enzyme (data not shown). We therefore chose to use the truncated version as it provides a more robust enzyme for the establishment of an HTS assay for the identification of inhibitors of the catalytic activity of the PERK enzyme. We used human PERK recombinant protein, which comprised the catalytic domain and an N-terminal GST epitope tag (Invitrogen, Carlsbad, CA). As a PERK substrate, we used full-length (amino acids 1–315) Flag-tagged eIF2α, a well-documented PERK substrate.9
We determined optimal buffer composition, including salt dependence, NaCl versus KCl and MgCl2 versus MnCl2 concentration, and pH of the buffer. Optimal phosphorylation of eIF2α was observed at a final concentration of 10 mM MgCl2. We subsequently assessed enzyme activity toward eIF2α at constant ionic strength with pH values ranging from 5 to 10 in 0.4-unit intervals, identifying an optimal pH of 7.2. After optimization of MgCl2 and pH, we evaluated the ionic strength of our buffer by titration with NaCl and KCl (Suppl. Fig. S2). For experiments presented in Supplemental Figure S2A – C, we used saturation conditions for the kinase reaction, which helped us develop optimal kinase buffer conditions.
On the basis of these experiments, we used the following buffer: 20 mM HEPES (pH 7.2), 10 mM MgCl2, 150 mM NaCl, and 0.01% Tween 20.
We established an enzyme concentration that provided linear kinase reaction kinetics over time (Suppl. Fig. S1). Based on these results, 8 nM PERK kinase, 1 µM ATP, and 1 µM eIF2α were selected.
PERK Uses a Random/Steady-State Ordered Kinetic Mechanism of Substrate Binding
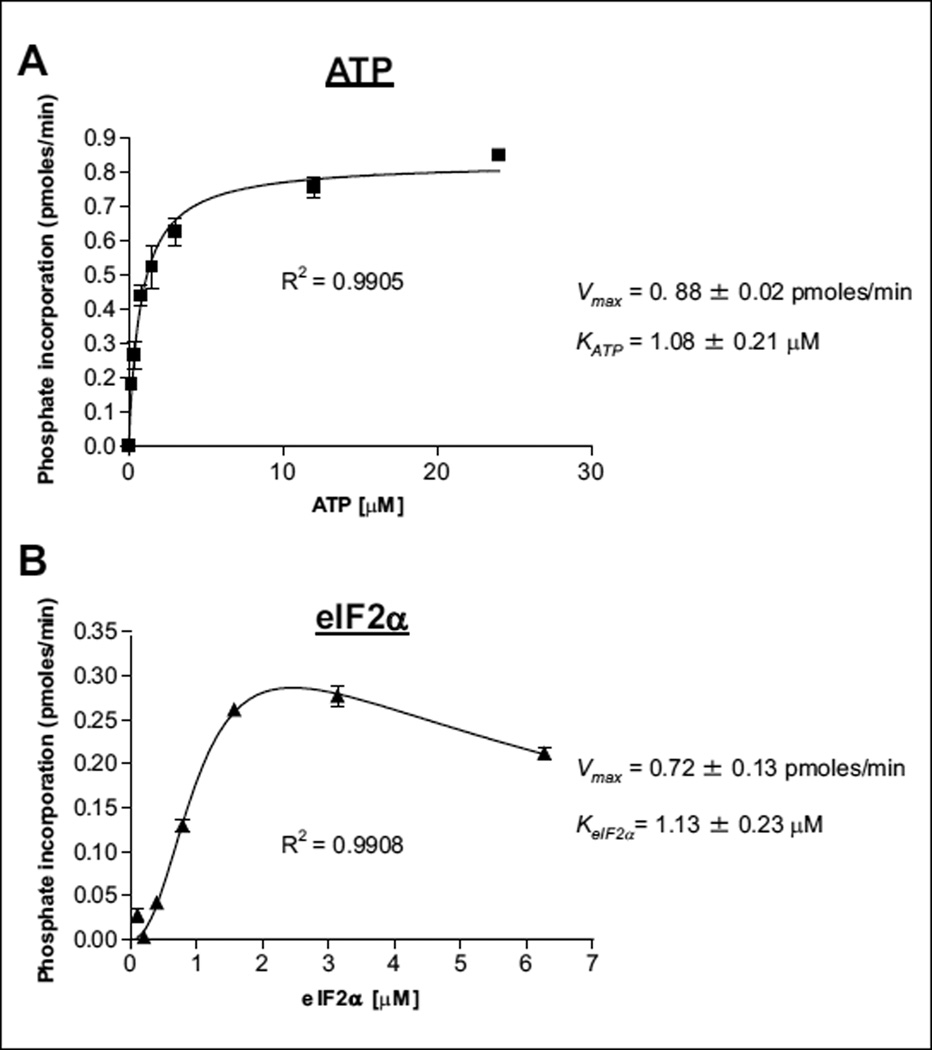
A radiometric assay was used to establish key kinetic parameters such as the Km values for eIF2α and ATP (Fig. 1), and to investigate the kinetic mechanism of this two-substrate reaction. When eIF2α was the variable substrate, two striking features were observed. First, strong substrate inhibition was observed at high eIF2α concentrations (Fig. 1B). Second, eIF2α bound to the enzyme cooperatively, as evidenced by the sigmoidal curve at low eIF2α concentrations with n = 2.2 ± 0.5 (Fig. 1B). The sigmoidal kinetics or the cooperative binding of eIF2α suggests dimer formation, which is consistent with evidence that PERK activation is dependent on dimerization.12 The crystal structures of PERK indicate that dimerization occurs through the N-lobes of the kinase domains.
Figure 1.
Kinetic characterization of PERK enzyme substrates. Initial velocity measurements were used for the (A) Km determination for adenosine triphosphate (ATP) and (B) Km determination for eIF2α by the radiometric kinase assay. In total, 8 nM PERK and 2 µM eIF2α was used for the ATP titration (A), 8-nM PERK, and 1 µM ATP was used for the eIF2α titration (B).
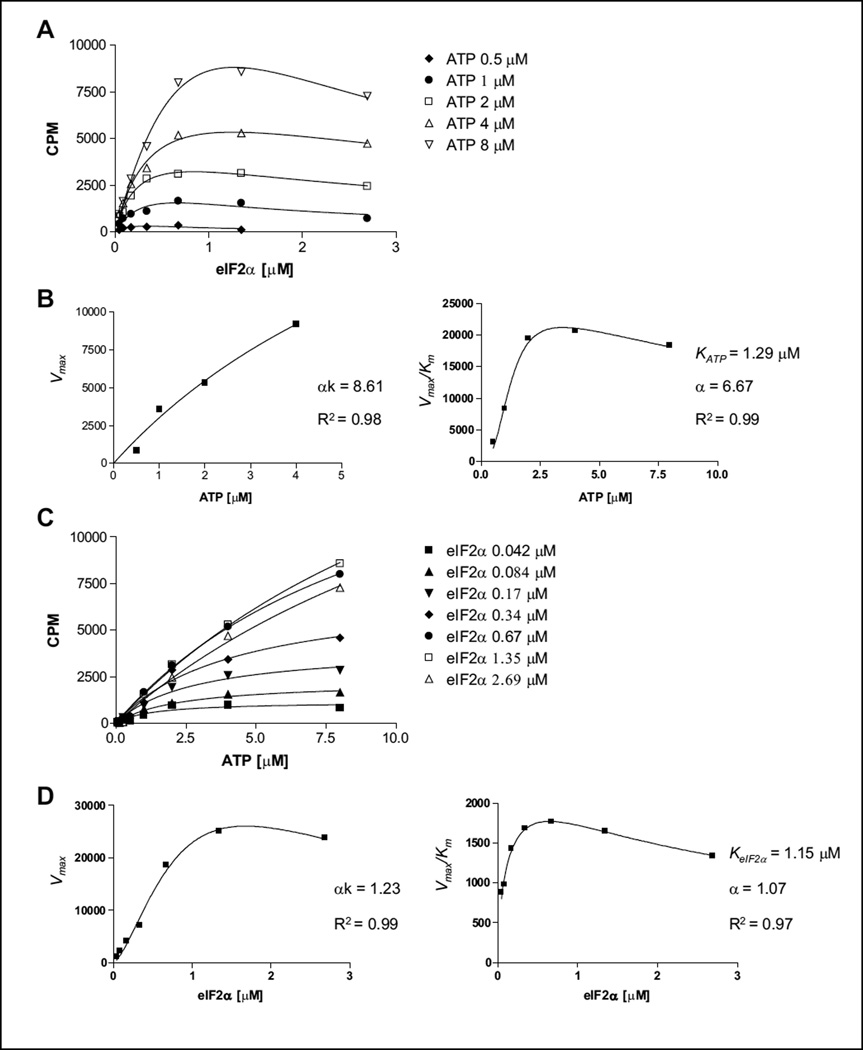
To determine the kinetic mechanism of PERK-catalyzed eIF2α phosphorylation, we conducted initial velocity analysis. Initial velocities were measured as a function of [eIF2α] (in the range of 0.042–3 µM) at various [ATP] (0.5–8 µM) (Fig. 2). Data were globally fitted to equations for three standard kinetic mechanisms: ping-pong, ordered, and random/steady-state ordered.13–15 The best fit was the random/steady-state ordered mechanism (data not shown). To avoid complicating this analysis, points reflecting eIF2α substrate inhibition were not included. To judge the data more carefully, therefore, we also analyzed the entire data set using the replot method and confirmed the kinetic mechanism based on the shape of the replots, as described previously.16,17 Briefly, when eIF2α was the variable substrate, each data set was analyzed by fitting the data to the equation reflecting both cooperative binding and substrate inhibition (as presented in Fig. 2A). Next, the replots of (Vmax)eIF2α versus [ATP] and (Vmax/Km) eIF2α versus [ATP] were constructed. Both (Vmax)eIF2α and (Vmax/Km) eIF2α were hyperbolically dependent on ATP concentration (Fig. 2B). When ATP was the variable substrate, simple kinetics were observed (Fig. 2C). Data were fitted to the basic Michaelis-Menten equation, and replots of (Vmax)ATP versus [eIF2α] and (Vmax/Km)ATP versus [eIF2α] were generated (Fig. 2D). Hyperbolic curves were observed in these two replots with a clear sign of substrate inhibition. The sigmoidal kinetics was reproduced in the replot of (Vmax)ATP versus [eIF2α] (Fig. 2D). The hyperbolic shape of all the four replots suggests that the reaction follows either a random or a steady-state ordered mechanism (Suppl. Fig. S3).14,15 Finally, the kinetic parameters of KeIF2α =, KATP =, and α = were determined from the replots, which are consistent with parameters calculated from global fitting. We chose 1 µM of each substrate (ATP and eIF2α), which is close to their Km values, as an optimal concentration for further experiments and a small-molecule compounds screen.
Figure 2.
Kinetic mechnism studies for PERK toward adenosine triphosphate (ATP) and eIF2α substrates. (A) Titration of ATP in the range of 0.5 to 8 µM versus an eIF2a concentration of 0.04 to 3 µM. From each ATP concentration plot, Vmax values of each reaction were calculated. (B) Determination of αk = 8.61 and α = 6.67 values. Based on the curve fit, we demonstrate that PERK kinase follows a random mechanism toward the ATP substrate. (C) Titration of eIF2α in the range of 0.04 to 3 µM versus an ATP concentration of 0.5 to 8 µM. From each eIF2α concentration plot, Vmax values of each reaction were calculated. (D) Determination of αk = 1.23 and α = 1.07 values. Based on the curve fit, we demonstrate that PERK kinase follows a random or steady-state ordered mechanism toward the eIF2α substrate. Experiments were repeated a minimum of three times. One representative experiment is shown.
Assay Development for HTS
For HTS development, DMSO sensitivity in the radiometric assay was assessed as all compounds in the library are solubilized in DMSO. DMSO tolerance was evaluated over a concentration range of 0.05% to 3.3% v/v. No significant change in eIF2α phosphorylation was observed (Suppl. Fig. S2D). The final concentration of DMSO for the HTS library screen was 0.6% v/v. After all kinase buffer conditions were optimized, reaction kinetics was assessed for linearity with respect to time over 60 minutes (Suppl. Fig. S2E).
Once the critical kinetic parameters for PERK-dependent phosphorylation of eIF2α were established such as kinase buffer (20 mM HEPES [pH 7.2], 10 mM MgCl2, 150 mM NaCl, and 0.01% Tween 20; 1 µM ATP [Fig. 1A]; 1 µM eIF2α [Fig. 1B] with 8 nM PERK [Suppl. Fig. S1]), a non-radiometric assay was developed for the HTS. From several alternative assay formats (including ATP/NADH-coupled; template-directed self-assembly kinase, HTRF transcreener ADP; data not shown), an HTRF assay was selected for development in a 384-well format. This assay is a TR-FRET– based technology from Cisbio that uses the principles of both TRF (time-resolved fluorescence) and FRET (fluorescence resonance energy transfer).18 This method enabled the assessment of substrate phosphorylation using an Eu3+ cryptate-labeled phospho-specific antibody (p-eIF2α(S51)-Eu3+) and anti-Flag antibody labeled with the d2 fluorophore, which recognizes the Flag epitope tag conjugated to eIF2α. In this system, a FRET signal is generated between both fluorophores (Eu3+ and d2) when the antibodies are both bound to eIF2α, following phosphorylation by PERK (Suppl. Fig. S4). Optimization was performed in a volume of 20 µL with 8 nM PERK, 1 µM eIF2α, and 1 µM ATP for 45 min, after which detection reagents were added and incubated for 1 h at room temperature. Titration of both antibodies revealed 83.3 ng of anti–Flag-d2 and 0.78 ng of anti–p-eIF2α(S51)-Eu3+ antibodies (Suppl. Fig. S5A) as optimal. Next, we further reduced the reaction volumes for HTS to 6 µL/reaction and detection reagents by a factor of 3.3, using 25 ng of anti–Flag-d2 antibodies (27.8 nM final concentration per reaction) and 0.24 ng of anti–p-eIF2α(S51)-Eu3+ antibodies (0.27 nM final concentration per reaction). The detection reagents were dissolved in HTRF Transcreener buffer containing 0.1% bovine serum albumin (BSA), 0.4 M potassium fluoride, and 60 mM EDTA (Cisbio). The final step prior to HTS was to reassess reaction linearity in the small volume. The reaction progress curve was still linear for least 60 min (Suppl. Fig. S5B).
With the reaction conditions finalized, the HTS was performed in a 384-well plate format, testing compounds at a final concentration of 10 µM. Each plate included no enzyme controls and full reaction in the absence of compound. Compounds were preincubated with 8 nM PERK kinase for 30 min to allow detection of any slowly binding compounds following which eIF2α and ATP substrates (1 µM final) were added to each well for an additional 45 min. HTS was run at the Km for PERK substrates (1 µM eIF2α and 1 µM ATP) for optimal inhibitor identification. Enzyme reactions were terminated by the addition of detection reagents (25 ng of anti–Flag-d2 antibodies and 0.24 ng of anti–p-eIF2α(S51)-Eu3+ antibodies) and incubated for 1 h. FRET signals were evaluated using the EnVision plate reader (PerkinElmer). The entire HTS was run at room temperature with the use of a Beckman (Brea, CA) NX liquid-handling system.
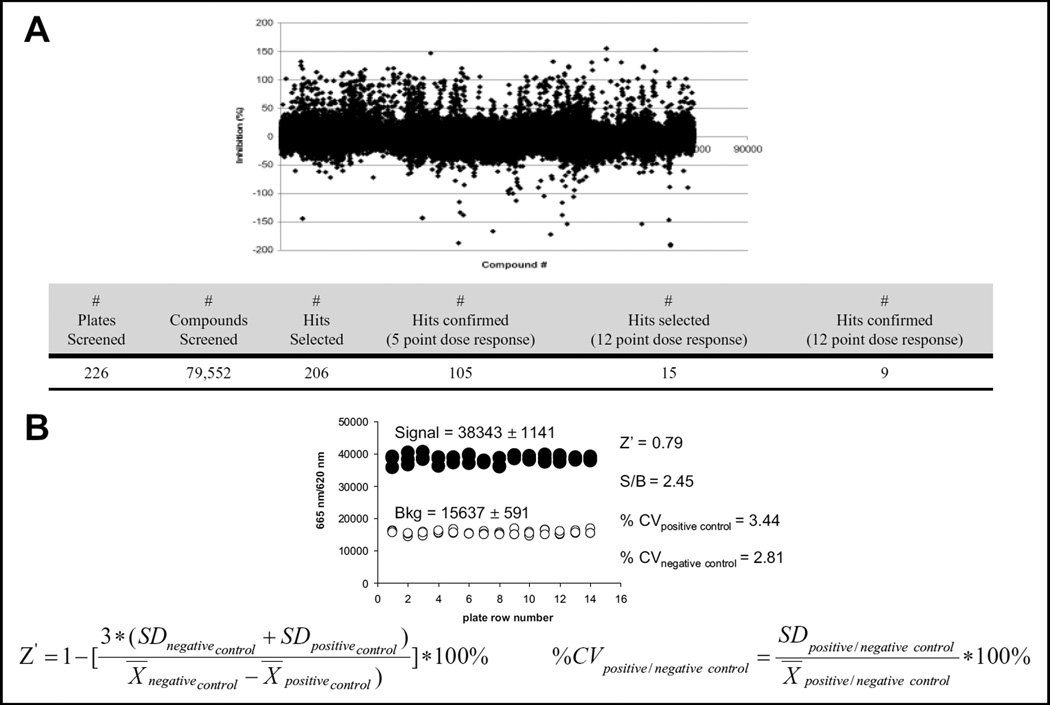
In total, 79,552 compounds were successfully screened. The observed FRET signals were converted to percent of inhibition for each screened compound (Fig. 3A). As a measure of assay robustness, Z′ factor, % CV values of negative and positive controls, and signal/background (S/B) ratio were calculated from three representative plates (Fig. 3B) (Z′ > 0.7, % CV << 10 %, and S/B 2.45). From the HTS results, we selected 206 compounds for further analysis based on potency of PERK inhibition >80% at 10 µM and on structure tractability from a medicinal chemistry perspective (e.g., lack of reactive functionality, predicted to have reasonable physicochemical properties, such as solubility, and opportunities for introducing structural modifications and diversity through readily available synthetic means). The screening results are presented in Figure 3A.
Figure 3.
High-throughput screen of a small-molecule compounds library. (A) Graph representing % of PERK inhibition in the fluorescence resonance energy transfer (FRET)–based kinase assay and high-throughput screening results. (B) Statistics for the high-throughput screen were determined (Z′; % coefficient of variation [CV] of positive and negative controls and signal-to-background [S/B] ratio). The background was run in the absence of PERK.
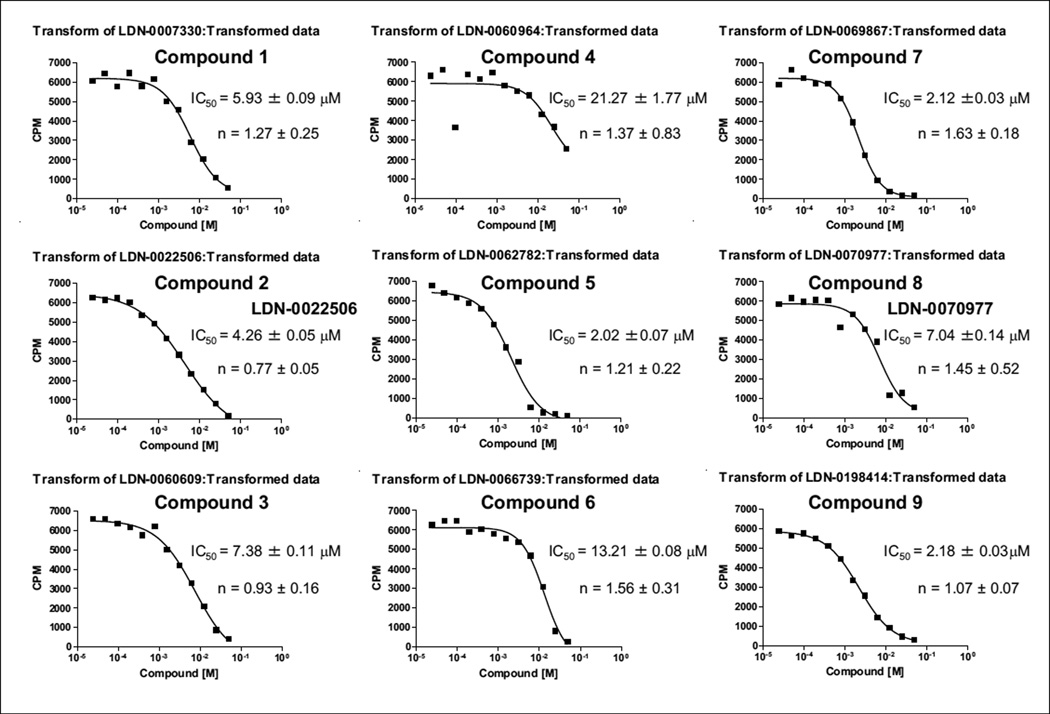
Initial hits were confirmed in a 5-point dose response in the range of 0.3 to 30 µM in the HTS assay (data not shown). Nine compounds were then selected based on IC50 and confirmed in a 12-point dose response (0.024–50 µM). From the curves generated from these experiments, we were able to determine the IC50 of each compound, which ranged from 2.2 to 22 µM, and the activity of the nine hit compounds was confirmed in the radiometric assay in at least three independent experiments (Fig. 4).
Figure 4.
Evaluation of compounds by radiometric assay. Compounds were evaluated at 1 µM eIF2α, 1 µM adenosine triphosphate (ATP), and 8 nM PERK. The IC50 for each compound was averaged from the three independent experiments, and one representative experiment is shown. The sigmoidal dose-response (variable slop) equation was used to obtain curve fits.
Mechanism-of-Action Studies
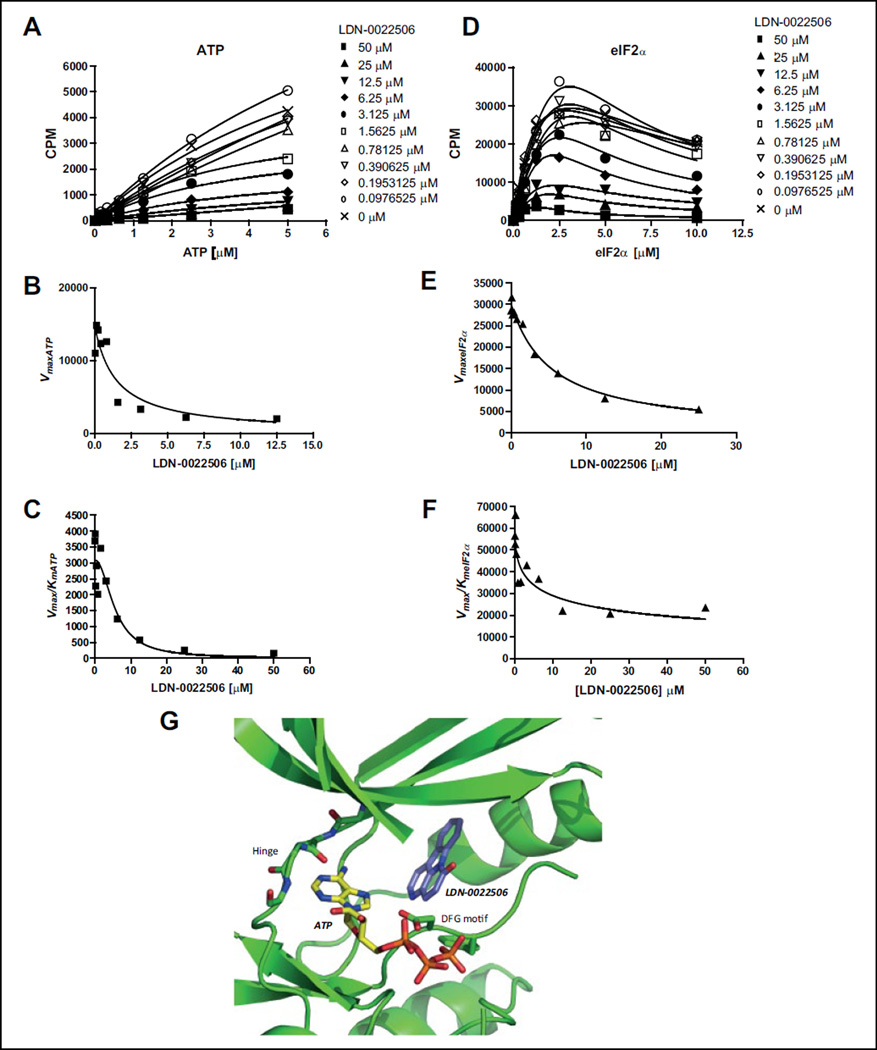
The mechanism of inhibition of one of the compounds, LDN-0022506, was assessed in a radiometric assay and data were analyzed using the method of replots.19 First we determined, at a single eIF2α (10 µM) concentration, the dependence of v0 on ATP concentration at several concentrations of LDN-0022506 (Fig. 5A) and, at a single ATP (10 µM) concentration, the dependence of v0 on eIF2α concentrations at several concentrations of LDN-0022506 (Fig. 5D). Next (Fig. 5A), we analyzed the dependence of v0 on ATP concentration at each concentration of LDN-0022506 to calculate apparent values (Vmax)ATP and (Vmax/ Km)ATP. For Figure 5D, we analyzed the dependence of v0 on eIF2α concentration at each concentration of LDN-0022506 to calculate apparent (Vmax)eIF2α and (Vmax/Km)eIF2α. Replots of apparent values of (Vmax)X (X = ATP or eIF2α) vs [LDN-0022506] and apparent values of (Vmax/Km)X vs [LDN-0022506] were then constructed, which shows a specific, mechanism-based pattern. When ATP was the variable substrate, we found that apparent values of (Vmax)ATP and (Vmax/Km)ATP both depended on LDN-0022506 concentration (Fig. 5B,C). These patterns indicate that the compound LDN-0022506 is noncompetitive with ATP. As a comparison, (Vmax)ATP should be independent of a competitive inhibitor. When eIF2α was the variable substrate, both apparent values of (Vmax)eIF2α and (Vmax/Km)eIF2α titrated with the compound LDN-0022506 (Fig. 5E,F), suggesting that the compound LDN-0022506 is also a noncom-petitive inhibitor of eIF2α and can bind to all the enzyme forms accumulated in the steady-state, E, E:ATP, E:eIF2α, and E:eIF2α:ATP (Suppl. Fig. S6).14,15,19
Figure 5.
Inhibition mechanism study of LDN-0022506. (A) Plot of initial velocities vs [ATP] at different concentrations of LDN-0022506, all at a fixed eIF2α concentration of 1 µM. (B, C) LDN-0022506 concentration dependencies of (Vmax)ATP and (Vmax/Km)ATP apparent values derived from analysis of the data of panel A. (D) Plot of initial velocities vs [eIF2α] at different concentrations of LDN-0022506, all at a fixed ATP concentration of 1 µM. (E, F) LDN-0022506 concentration dependencies of (Vmax)eIF2α and (Vmax/Km)eIF2α apparent values derived from analysis of the data of panel D. Experiments were repeated a minimum of three times. One representative experiment is shown. (G) ATP molecule and LDN-0020506 were docked into the crystal structure of PERK kinase domain (4G31). Docking calculations were carried out using induced fit docking with Glide and Prime as part of the Schrodinger First Discovery Suite.
To independently assess mechanism of inhibition, ATP and LDN-0020506 were docked into the crystal structure of PERK kinase domain (4G31) in a completely unbiased fashion. Induced fit docking (using Schrodinger First Discovery Suite (Portland, OR) was carried out to allow subtle but necessary receptor flexibility to accommodate both ligands. In the first step, LDN-0020506 was docked into the crystal structure of PERK kinase domain using an unbiased search grid. The best docked pose for the inhibitor was in the allosteric pocket or the type III pocket (Fig. 5G).20 The inhibitor appeared to favor this hydrophobic region of the kinase, comprising residues I623, L648, Y653, F955, and L957. The inhibitor also did not bind to the hinge region of the kinase, which is the typical ATP binding site. In the next step, ATP was docked in an unbiased fashion into the PERK kinase domain. These docking calculations produced a strong low-energy pose, where the adenine group of the ATP molecule made two strong hydrogen bonds with the hinge region of the kinase and the phosphate groups made several hydrogen bonds with the glycine-rich loop and the catalytic loop. Alignment of the docked ATP conformation with several x-ray structures of kinase bound to ATP molecules showed that our docked pose and the x-ray structures had RMSD <1.0Å. The inhibitor and ATP molecule appeared to bind at two distinct sites on the kinase, supporting the experimental observation where the inhibitor was determined to be noncompetitive. Further studies on the mechanism of action of LDN-0070977 are needed for a more in-depth understanding of its activity.
Inhibition of PERK Kinase Activity in Cultured Cells
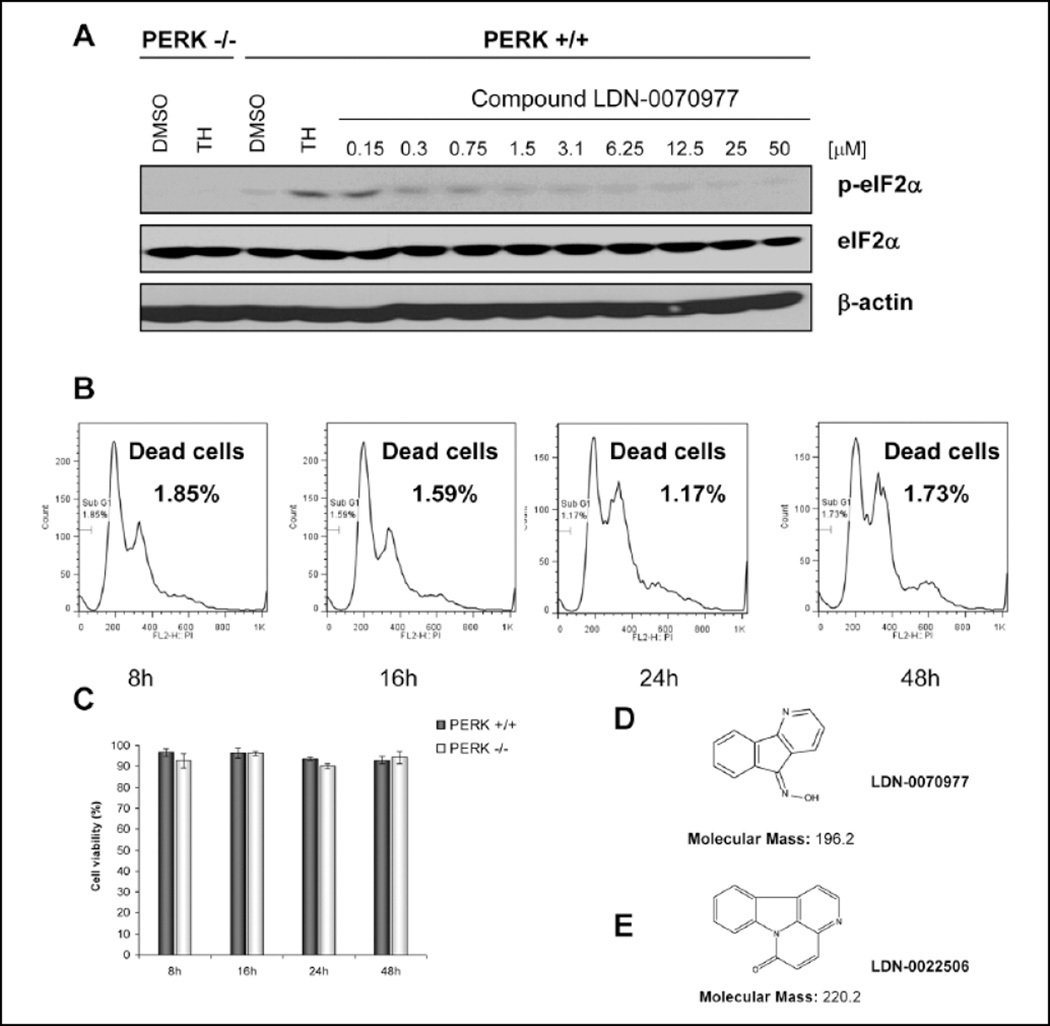
To begin to assess biological activity of the PERK inhibitors, we used wild-type MEFs and a cellular model of ER-dependent stress, thapsigargin. Thapsigargin is a non-competitive inhibitor of a family of enzymes known as SERCA (sarcoplasmic/endoplasmic reticulum Ca2+ ATPase). Thapsigargin causes a depletion of calcium stores within the sarcoplasmic and endoplasmic reticula, thereby impeding folding of proteins that are processed in the ER. We pretreated cells with LDN-0070977 at a concentration of 50 µM for either 1 or 24 h and next treated cells with thapsigargin (500 nM final concentration) for 1 h. PERK activity was assessed by evaluating eIF2α phosphorylation by Western blot using the anti–phospho-eIF2α(S51)– specific antibody. Thapsigargin treatment resulted in increased eIF2α phosphorylation (Fig. 6A, lane 4), as predicted. In contrast, treatment of cells with either LDN-0070977 or LDN-0022506 inhibited stress-dependent eIF2α phosphorylation. Because LDN-0070977 exhibited more profound activity in the cells, it was used in additional experiments. LDN-0070977 was titrated over a range of 0.15 to 50 µM. Significant inhibition of PERK action was noted at 0.3 µM and higher concentrations (Fig. 6A). Treatment of cells for 1 h with LDN-0070977 was sufficient to achieve PERK-dependent inhibition of eIF2α phosphorylation. Of note, compounds LDN-0070977 and LDN-0022506 displayed simple chemical structures with molecular weights of 196.2 and 220.2, respectively (Fig. 6D,E).
Figure 6.
Compound LDN-0070977 inhibits PERK phosphorylation with no toxicity to the cells. (A) Mouse embryonic fibroblast (MEF) cells were pretreated with LDN-0070977 inhibitor in the range of 0.15 to 50 µM for 1 h; next cells were treated with 500 nM thapsigargin (TH) to activate PERK. PERK−/− MEFs were used as a control. Cells were lysed and analyzed by Western blot. (B, C) Compound toxicity was measured in 3T3 cells by propidium iodide (B) and trypan blue staining (C). (D) Structure of LDN-0070977 compound. (E) Structure of LDN-0022506 compound.
Finally, we assessed whether LDN-0070977 exhibited any general cellular toxicity. MEFs were treated with the inhibitor for 8, 16, 24, or 48 h at a concentration of 50 µM. After treatment with inhibitor, the cells were stained with propidium iodide (PI) for quantitative determination of DNA content, and the percentage of dead cells was quantified by flow cytometry. The cells were viable when the highest concentration (50 µM) of inhibitor was used and we did not observe a significant number of dead cells (Fig. 6B). As an independent measure for toxicity, we stained cells with trypan blue. The same inhibitor concentration and incubation times were used as in the previously described experiment (Fig. 6C). No toxicity was observed by either method.
Discussion
We have interrogated the steady-state kinetics of PERK kinase toward two of its primary substrates, eIF2α and ATP. The data demonstrate that PERK follows a random/ steady-state ordered mechanism for eIF2α phosphorylation. We also established the optimal conditions for utilization of eIF2α phosphorylation as the basis of an HTS to identify small-molecule inhibitors of PERK. We developed optimal buffer conditions for PERK-eIF2α kinase reaction, which contain 20 mM HEPES (pH 7.2), 10 mM MgCl2, 150 mM NaCl, and 0.01% Tween 20. Also, we found the Km values for PERK substrates (KeIF2α = 1 µM and KATP = 1 µM).
With regard to assay development, we optimized the kinase buffer composition, enzyme and its substrate concentration, kinetics reaction time determination, and assay automation. Following assay optimization, 79,552 compounds were evaluated for their ability to inhibit PERK-dependent phosphorylation of eIF2α. From this initial screen, two compounds with low µM IC50 were selected for additional analysis, including mechanism-of-inhibitor activity (whether it is a competitive, uncompetitive, or non-competitive mechanism) and cellular activity. Compound LDN-0022506 is noncompetitive for both ATP and eIF2α, and compound LDN-0070977 demonstrates better activity in the cells, is not toxic, and inhibits PERK with better activity compared with the other eIF2α-related kianses such as PKR (IC50 23.3 µM) and HRI (IC50 59.6 µM) (data not shown). Compound LDN-0022506 is a naturally occurring β-carboline (canthin-6-one), initially isolated in 1952 from Pentaceras australis.21 Since then, more than 40 naturally occurring analogues have been reported that exhibit various biological activities,22 including anticancer23,24 and antiparasitic.25 Canthin-6-one also has a range of biological activities; however, it is difficult to attribute its effects to a specific molecular target. The 1-methoxy analogue of LDN-0022506 (1-methoxy-canthin-6-one) has been reported to target c-Jun NH2-terminal kinase (JNK).26 Of the limited number of published PERK kinase inhibitors, none are reported to exhibit a noncompetitive mechanism of action, including GlaxoSmithKline’s potent ATP competitive inhibitor GSK2606414.27–29
Future efforts are needed to optimize these hit compounds by conducting structure-activity relationship studies that will be guided by improving PERK kinase inhibitory potency and selectivity as assessed by kinome profiling as well as cell-based functional activity. In addition, derivatives that lack PERK inhibitory activity will serve as valuable controls for determining PERK-dependent cellular effects. Given the noncompetitive mechanism of inhibition exhibited by LDN-0022506, we anticipate that imparting increased PERK specificity to this compound series may be more feasible compared with inhibitors that bind to the highly conserved ATP-binding site. Ultimately, these studies could identify derivatives with suitable biological and chemical properties to justify testing in animal disease models.
Implication of PERK Activation in Disease Development
Data from many laboratories suggest that PERK activation can play important roles in tumorigenesis. Ex vivo deletion of PERK kinase in mouse mammary tumor cell lines or PERK knockdown in human cancer cell lines suppresses growth of secondary tumors in an orthotropic mouse model.30 Conversely, PERK may also promote tumor growth in vivo. When PERK wild-type transformed cells with oncogenic Ki-RasV12 were injected into nude mice, faster and larger tumor growth was observed compared with PERK−/− control.31 More recent published data support the finding that PERK-eIF2α phosphorylation plays a significant role in chronic myeloid leukemia progression and STI571 (imatinib mesylate) treatment resistance.32
Increased phosphorylation of eIF2α has been also observed in the hippocampus of patients with Alzheimer disease (AD), indicating that PERK is activated in this disease.33,34 A link between the UPR activation and β-amyloid plaque production is further suggested by the observation that UPR activation induces the amyloid precursor protein (APP) expression in the central nervous system of guinea pigs.35 Proteolytic processing of APP largely occurs in the ER, coincident with the localization of PERK. The discovery of small-molecule inhibitors should be beneficial for the treatment of a number of diseases (including cancer, diabetes, heart disease, and AD) wherein PERK signaling may be implicated. Such small molecules may also prove useful in the interrogation of UPR signaling in an experimental setting. It is hoped that the hit compounds identified will prove useful in the development of highly potent, specific small molecular regulators of the PERK enzyme.
Supplementary Material
Acknowledgment
The authors thank Ken Auerbach for outstanding technical assistance.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Institutes of Health (NIH) grant P01 CA104838 (JAD); Polish National Science Centre (NCN) “HARMONIA 5” grant; Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grants (T32) T32 2T32CA009140; and National Center for Drug Discovery in Neurodegeneration, NIH National Institute of Neurological Disorders and Stroke (NINDS) Cooperative Agreement Award (U24) NS04933901 and the Harvard NeuroDiscovery Center.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Shi Y, Vattem KM, Sood R, et al. Identification and Characterization of Pancreatic Eukaryotic Initiation Factor 2 Alpha-Subunit Kinase, PEK, Involved in Translational Control. Mol. Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang XZ, Harding HP, Zhang Y, et al. Cloning of Mammalian Ire1 Reveals Diversity in the ER Stress Responses. Embo J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haze K, Yoshida H, Yanagi H, et al. Mammalian Transcription Factor ATF6 Is Synthesized as a Transmembrane Protein and Activated by Proteolysis in Response to Endoplasmic Reticulum Stress. Mol. Biol. Cell. 1999;10:3787–3799. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto K, Sato T, Matsui T, et al. Transcriptional Induction of Mammalian ER Quality Control Proteins Is Mediated by Single or Combined Action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Rutkowski DT, Dubois M, et al. ATF6alpha Optimizes Long-Term Endoplasmic Reticulum Function to Protect Cells from Chronic Stress. Dev. Cell. 2007;13:351–364. doi: 10.1016/j.devcel.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Hamanaka RB, Bobrovnikova-Marjon E, Ji X, et al. PERK-Dependent Regulation of IAP Translation during ER Stress. Oncogene. 2009;28:910–920. doi: 10.1038/onc.2008.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fels DR, Koumenis C. The PERK/eIF2alpha/ATF4 Module of the UPR in Hypoxia Resistance and Tumor Growth. Cancer Biol. Ther. 2006;5:723–728. doi: 10.4161/cbt.5.7.2967. [DOI] [PubMed] [Google Scholar]
- 8.Cullinan SB, Zhang D, Hannink M, et al. Nrf2 Is a Direct PERK Substrate and Effector of PERK-Dependent Cell Survival. Mol. Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schroder M. The Unfolded Protein Response. Mol. Biotechnol. 2006;34:279–290. doi: 10.1385/MB:34:2:279. [DOI] [PubMed] [Google Scholar]
- 10.Kim I, Xu W, Reed JC. Cell Death and Endoplasmic Reticulum Stress: Disease Relevance and Therapeutic Opportunities. Nat. Rev. Drug Discov. 2008;7:1013–1030. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 11.Bobrovnikova-Marjon E, Hatzivassiliou G, Grigoriadou C, et al. PERK-Dependent Regulation of Lipogenesis during Mouse Mammary Gland Development and Adipocyte Differentiation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16314–16319. doi: 10.1073/pnas.0808517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui W, Li J, Ron D, et al. The Structure of the PERK Kinase Domain Suggests the Mechanism for Its Activation. Acta Crystallogr. D Biol. Crystallogr. 2011;67:423–428. doi: 10.1107/S0907444911006445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segal IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. New York: Wiley-Interscience; 1975. [Google Scholar]
- 14.Stein RL. Kinetics of Enzyme Action: Essential Principles for Drug Hunters. New York: John Wiley; 2011. [Google Scholar]
- 15.Liu M, Dobson B, Glicksman MA, et al. Kinetic Mechanistic Studies of Wild-Type Leucine-Rich Repeat Kinase 2: Characterization of the Kinase and GTPase Activities. Biochemistry. 2010;49:2008–2017. doi: 10.1021/bi901851y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Poulose S, Schuman E, et al. Development of a Mechanism-Based High-Throughput Screen Assay for Leucine-Rich Repeat Kinase 2—Discovery of LRRK2 Inhibitors. Anal. Biochem. 2010;404:186–192. doi: 10.1016/j.ab.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M, Kang S, Ray S, et al. Kinetic, Mechanistic, and Structural Modeling Studies of Truncated Wild-Type Leucine-Rich Repeat Kinase 2 and the G2019S Mutant. Biochemistry. 2011;50:9399–9408. doi: 10.1021/bi201173d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park YW, Cummings RT, Wu L, et al. Homogeneous Proximity Tyrosine Kinase Assays: Scintillation Proximity Assay versus Homogeneous Time-Resolved Fluorescence. Anal. Biochem. 1999;10:94–104. doi: 10.1006/abio.1999.4029. [DOI] [PubMed] [Google Scholar]
- 19.Liu M, Choi S, Cuny GD, et al. Kinetic Studies of Cdk5/p25 Kinase: Phosphorylation of Tau and Complex Inhibition by Two Prototype Inhibitors. Biochemistry. 2008;47:8367–8377. doi: 10.1021/bi800732v. [DOI] [PubMed] [Google Scholar]
- 20.Gavrin LK, Saiah E. Approaches to Discover Non-ATP Site Kinase Inhibitors. Med. Chem. Commun. 2013;4:41–51. [Google Scholar]
- 21.Haynes HF, Nelson ER, Price JR. Alkaloids of the Australian Rutaceae: Pentaceras australis Hook. F. I Isolation of the Alkaloids and Identification of Canthin-6-One. Aust. J. Sci. Res. A Phys. Sci. 1952;5:387–400. [Google Scholar]
- 22.Cao R, Peng W, Wang Z, et al. Beta-Carboline Alkaloids: Biochemical and Pharmacological Functions. Curr. Med. Chem. 2007;14:479–500. doi: 10.2174/092986707779940998. [DOI] [PubMed] [Google Scholar]
- 23.Peduto A, More V, de Caprariis P, et al. Synthesis and Cytotoxic Activity of New Beta-Carboline Derivatives. Mini Rev. Med. Chem. 2011;11:486–491. doi: 10.2174/138955711795843383. [DOI] [PubMed] [Google Scholar]
- 24.Cebrian-Torrejon G, Kahn SA, Ferreira ME, et al. Alkaloids from Rutaceae: Activities of Canthin-6-One Alkaloids and Synthetic Analogues on Glioblastoma Stem Cells. MedChemComm. 2012;3:771–774. [Google Scholar]
- 25.Ferreira ME, Nakayama H, de Arias AR, et al. Effects of Canthin-6-One Alkaloids from Zanthoxylum chiloperone on Trypanosoma cruzi-Infected Mice. J. Ethnopharmacol. 2007;109:258–263. doi: 10.1016/j.jep.2006.07.028. [DOI] [PubMed] [Google Scholar]
- 26.Ammirante M, Di Giacomo R, De Martino L, et al. 1-Methoxy-Canthin-6-One Induces c-Jun NH2-Terminal Kinase-Dependent Apoptosis and Synergizes with Tumor Necrosis Factor-Related Apoptosis-Inducing Ligand Activity in Human Neoplastic Cells of Hematopoietic or Endodermal Origin. Cancer Res. 2006;66:4385–4393. doi: 10.1158/0008-5472.CAN-05-3895. [DOI] [PubMed] [Google Scholar]
- 27.Axten JM, Medina JR, Feng Y, et al. Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-di-hydro-1H–indol-5-yl)-7H–p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-Like Endoplasmic Reticulum Kinase (PERK) J. Med. Chem. 2012;55:7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 28.Axten JM, Romeril SP, Shu A, et al. Discovery of GSK2656157: An Optimized PERK Inhibitor Selected for Preclinical Development. ACS Med. Chem. Lett. 2013;4:964–968. doi: 10.1021/ml400228e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atkins C, Liu Q, Minthorn E, et al. Characterization of a Novel PERK Kinase Inhibitor with Antitumor and Antiangiogenic Activity. Cancer Res. 2013;15:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 30.Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, et al. PERK Promotes Cancer Cell Proliferation and Tumor Growth by Limiting Oxidative DNA Damage. Oncogene. 2010;29:3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi M, Naczki C, Koritzinsky M, et al. ER Stress-Regulated Translation Increases Tolerance to Extreme Hypoxia and Promotes Tumor Growth. Embo J. 2005;24:3470–3481. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusio-Kobialka M, Podszywalow-Bartnicka P, Peidis P, et al. The PERK-eIF2alpha Phosphorylation Arm Is a Pro-Survival Pathway of BCR-ABL Signaling and Confers Resistance to Imatinib Treatment in Chronic Myeloid Leukemia Cells. Cell Cycle. 2012;11:4069–4078. doi: 10.4161/cc.22387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoozemans JJ, Veerhuis R, Van Haastert ES, et al. The Unfolded Protein Response Is Activated in Alzheimer’s Disease. Acta Neuropathol. 2005;110:165–172. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- 34.Chang RC, Wong AK, Ng HK, et al. Phosphorylation of Eukaryotic Initiation Factor-2alpha (eIF2alpha) Is Associated with Neuronal Degeneration in Alzheimer’s Disease. Neuroreport. 2002;13:2429–2432. doi: 10.1097/00001756-200212200-00011. [DOI] [PubMed] [Google Scholar]
- 35.Finnie JW, Manavis J, Blumbergs PC, et al. Axonal and Neuronal Amyloid Precursor Protein Immunoreactivity in the Brains of Guinea Pigs Given Tunicamycin. Vet. Pathol. 2000;37:677–680. doi: 10.1354/vp.37-6-677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.