Abstract
Patient-specific 3D models obtained by the segmentation of volumetric diagnostic images play an increasingly important role in surgical planning. Surgeons use the virtual models reconstructed through segmentation to plan challenging surgeries. Many solutions exist for the different anatomical districts and surgical interventions. The possibility to bring the 3D virtual reconstructions with native radiological images in the operating room is essential for fostering the use of intraoperative planning. To the best of our knowledge, current DICOM viewers are not able to simultaneously connect to the picture archiving and communication system (PACS) and import 3D models generated by external platforms to allow a straight integration in the operating room. A total of 26 DICOM viewers were evaluated: 22 open source and four commercial. Two DICOM viewers can connect to PACS and import segmentations achieved by other applications: Synapse 3D® by Fujifilm and OsiriX by University of Geneva. We developed a software network that converts diffuse visual tool kit (VTK) format 3D model segmentations, obtained by any software platform, to a DICOM format that can be displayed using OsiriX or Synapse 3D. Both OsiriX and Synapse 3D were suitable for our purposes and had comparable performance. Although Synapse 3D loads native images and segmentations faster, the main benefits of OsiriX are its user-friendly loading of elaborated images and it being both free of charge and open source.
Keywords: Digital Imaging and Communications in Medicine (DICOM), PACS, 3D imaging (imaging three-dimensional), 3D segmentation
Introduction
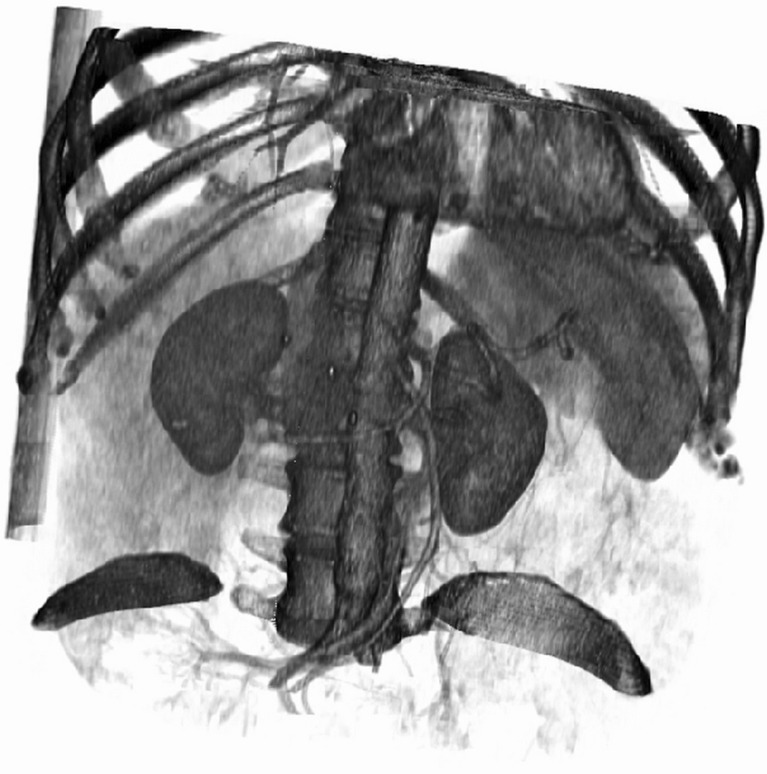
Segmentation of anatomical structures in medical images is crucial in clinical research. Segmentation provides measurements of the size and shape of the anatomy and enables easy three-dimensional (3D) visualization of anatomical variations and the localization of pathological evidence (e.g., lesions or aneurysms). Many software applications and radiological suites for visualization are able to supply a direct surface or volume rendering. Despite the ability to obtain a virtual representation quickly, the utility of direct volume rendering 3D visualization is limited to some surgical fields (e.g., endovascular surgery and orthopedic surgery) where the main anatomical structures to be visualized are well contrasted. The physicians of other specialties do not use these direct 3D surgical planning facilities because of the lack of quantitative information, the inadequate efficacy in rendering organ parenchyma, and limitations in discerning the structures [1]. Figure 1 shows an example of direct surface rendering of an abdominal CT dataset.
Fig. 1.
The surface rendering of an abdominal CT dataset; to distinguish renal arteries and veins in the renal osteons, it is not possible
Instead, patient-specific 3D models obtained by the segmentation of volumetric diagnostic images play an increasingly important role in surgical planning [2]. Users can construct 3D models using manual, semiautomatic, and completely automatic approaches [3, 4]. The semiautomatic methods combine the efficiency and repeatability of automatic systems with the tuning provided by human experts, who need to specify some parameters and monitor the results of each step while maintaining control of the final result [5–8].
Various fully automatic approaches have been proposed, but the principal limitation of these approaches is the inadequate robustness in cases where the target anatomy widely differs from the standard [2]. Furthermore, automatic software applications are designed to work with a single anatomic district at a time and are not designed to be general-purpose applications [9–12].
The semiautomatic approach requires user interaction. Indeed, the user sets different parameters, depending on the approach chosen, to steer the final result. Although semiautomatic applications are often used for segmenting medical images, simpler readily available user interfaces would facilitate more widespread use of semiautomatic methods in daily clinical practice. The usability and the quickness of the tool are two crucial features in the selection of one application over another.
There are many systems for constructing patient-specific 3D models by segmentation of volumetric images. However, all software applications remain very specific and addressed to a single or only a few anatomical districts. It was beyond the aim of this study to rank the segmentation tools, given the vast amount of methods and parameters, the different anatomy targets of the software applications, and the lack of uniformity in their evaluation.
The final aim of segmentation in surgical use is to plan challenging surgical interventions [2, 13]. Patient-specific 3D planning is even more useful when used in the operating room and offered to the surgeon during the intervention as a sort of map. A limit preventing this application from becoming a routine practice is that even if some operating rooms are equipped with computers connected to the picture archiving and communication system (PACS), no software currently allows connecting with PACS and loading the patient images, importing a patient segmentation obtained by different software, rendering the segmentation in 3D, and fusing the information with a direct volume rendering, which is a very useful step at times.
In this context, our goals were (1) to investigate the software applications used most often for surgical planning and (2) to evaluate the selected platforms to find a single platformable to load a patient 3D reconstruction obtained by various segmentation applications and to connect with PACS in order to exploit 3D patient-specific models in the operating room.
Methods
We analyzed a wide array of applications equipped for viewing and segmenting Digital Imaging Communication in Medicine (DICOM) volumetric data images. The selected software applications were running on a Windows Operating System (OS; Intel Core i7-3770, CPU at 3.40 GHz, 8 GB RAM, 64 bit OS), with the exception of OsiriX DICOM Viewer. Among these, we separated the open-source solutions from the commercial solutions.
We evaluated the performance of the following commercial applications: Amira®, Ginko-CADx-pro®, Mimics®, and the Synapse 3D® platform by Fujifilm in use at the University Hospital Cisanello of Pisa. Finally, the OsiriX DICOM Viewer was studied; it was running on a Macintosh OS (MacbookPro, CPU at 2.6 GHz, 8 GB RAM, 32 bit OS).
The evaluation took into account eight parameters:
Data import
Data export
Metadata
2D viewer
3D viewer
Support
OS
Usability
In the first group, we classified the applications capable of importing directories, complete series, sets of images, and the applications that can import only single images. The second group was for software that can export series from images and that can anonymize the patient information. The anonymize tool is very useful for research purposes. The metadata group refers to applications that can visualize the entire study header and that are good for identifying the header of a single image. The 2D viewer group refers to the visualization features, the applications that permit the tuning and the amplitude of the windowing, the definition of the Color LookUp Table (CLUT) in 2D, the designation and the tuning of histograms, the overlay information, the possibility of measuring distances and diameters, and the ability to add notes in the images. The 3D viewer group comprises different features: the slice scrolling option, the multi-planar reconstruction (MPR) or the curve multi-planar reconstruction (CPR), the maximum intensity projection option (MIP), the possibility of rendering the volume (VR), the possibility of varying the CLUT in VR modality, and the opportunity to create a model by shaded surface display. The applications have also been classified based on the support that the producers offer, in the form of manuals, guides, forums, and the available source code. The operating system in which the software applications run determines the OS group. The last group concerns the graphical user interface (GUI), the speed, and the usability of the system. Table 1 shows the features of the software studied.
Table 1.
Assessment of a wide array of software applications for viewing and segmenting DICOM images
| Software | Aeskulaps | Amide | BiolmageXD | Dicompyler | Endrov | Eview Box | Fusion Viewer | Gimias | ImageJ | image Vis3D | In-Vesalius | ITK-Snap 1.5* | ITK-Snap 2.2 | Mayam | Mango | Micro View | MITK 3 m3 | Mito | Osirix | 3D Slicer | Seg 3D | Synedra View P | Amira® | Ginko-CADx-pro® | Mimics® | Synapse 3D® | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Features | |||||||||||||||||||||||||||
| Data import | Images | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||
| Set | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||||
| Series | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||
| Directory | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||
| Data export | Images | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||
| Series | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
| Anonimyze | X | X | X | X | X | X | |||||||||||||||||||||
| Support | doc | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||
| wiki | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
| forum | X | X | X | X | X | X | X | X | X | ||||||||||||||||||
| Source Code | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||
| OS | Windows | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |
| Linux | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||
| Macintosh | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||
| 2D viewer | Windowing | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||
| CLUT | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||||||
| Histogram | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| Info | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||
| Measure | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||
| Notes | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||
| 3D viewer | Slice scroll | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||
| MPR | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||
| CPR | X | ||||||||||||||||||||||||||
| MIP | X | X | X | X | X | X | X | X | X | X | |||||||||||||||||
| VR | X | X | X | X | X | X | X | X | X | X | X | X | X | X | X | ||||||||||||
| CLUT | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| SSD | X | X | X | X | X | X | X | X | X | X | X | X | |||||||||||||||
| Usability | GUI | 6 | 7 | 7 | 6 | 4 | 4 | 3 | 7 | 4 | 5 | 8 | 7 | 7 | 7 | 5 | 6 | 8 | 7 | 8 | 8 | 7 | 7 | 8 | 8 | 9 | 9 |
| Clarity | 5 | 7 | 6 | 6 | 5 | 5 | 3 | 6 | 5 | 5 | 7 | 7 | 7 | 6 | 5 | 6 | 8 | 8 | 8 | 7 | 7 | 6 | 7 | 7 | 9 | 8 | |
| Speed | 7 | 7 | 6 | 7 | 4 | 5 | 5 | 5 | 5 | 7 | 6 | 7 | 7 | 6 | 5 | 6 | 7 | 7 | 9 | 6 | 6 | 5 | 8 | 8 | 8 | 9 |
X features of the software applications
aOpen-source application ITK-Snap provided with the EndoCAS pipeline [14]
Limiting the evaluation to the features of a wide array of software is not exhaustive; therefore, we selected a restricted amount of applications based on the aforementioned characteristics, taking into account interviews with medical personnel, radiologists, and surgeons, and evaluated those applications capable of loading segmentations achieved by external platforms and linking to PACS. We interviewed eight radiologists and eight surgeons. The goal of the interviews was to know which applications are used most often in clinical practice in order to focus the study to their performances. The interviews took into account the usability of the tools, the preferred GUI, and the time that physicists needed to solve the models.
As expected, in many cases, surgeons prefer quantitative images and information about volumes offered by the segmentation. Although some authors evaluated the goodness of segmentation using qualitative and/or quantitative methods, we did not assess this because we focused our survey on pinpointing the software applications used daily by clinical experts.
For the purpose of using a single platform in the operating room to offer 3D segmentation to the surgeon, we tested the ability of the software to import segmentations made by other applications. Considering that there are many strategies to save a 3D model, we reduced the problem to the most used 3D file format, the visual toolkit (VTK) format, given the fact that the visualization toolkit libraries are nowadays a de facto standard for 3D medical elaboration (www.vtk.org). Most of the studied software applications save their 3D segmentation in VTK format.
Synedra View P, Aeskulaps, Fusion-viewer, Dicompyler, Endrow, and Santec are simple DICOM viewers and are not equipped with segmentation tools. Clinicians can exploit these open-source applications to visualize the DICOM sets, but these viewers are unable to segment the surfaces, to give quantitative information, and to render the volumes; furthermore, they are not capable of importing segmentations from other software, so they were not suitable for our integration purposes.
Among the analyzed platforms, no software offered a straightforward solution to import VTK 3D segmentation, and only two solutions emerged from the first selection that were able to connect with PACS and to somehow import third-party segmentation. Thus, we subjected the following applications to further evaluation:
Synapse 3D® by Fujifilm (http://3dimaging.fujimed.com/) and OsiriX by the University of Geneva (http://osirix-viewer.com/).
As said, the import step is not straightforward. Both OsiriX and Synapse 3D cannot directly load an external segmentation, but they are faster and more adept in segmenting a dataset.
The performance of the selected software was tested using the segmentation of the same CT data volume composed of 109 CT images, 2.50 mm thick. The DICOM series refers to a patient with infiltrating hepatic lesions in segments S5-6-8 and with a portal vein thrombosis of the right main branch.
To evaluate the applications, we measured the following parameters: tload and tVR. We averaged 10 measures of the time required to load the series (tload). To calculate tload, we used the harmonic mean mH, as defined in (1), where n is the number of the measures and xi indicates the value recorded. We used the same procedure to estimate the fastest time to render the volume (tVR) and the average time to switch from one kind of rendering to another.
| 1 |
The estimation of tload and tVR was repeated for the two applications examined.
To reach our first aim, we established a procedure to supply the patient-specific 3D model to surgeons in the operating room through PACS.
The procedure consists of three phases:
Preparing the 3D segmentation model
Converting the segmentation
Importing and processing the segmentation in the PACS-connected operating room software
Preparing the 3D segmentation model
This step involves preparing the 3D model for the surgeons. The radiologists, technicians, or surgeons themselves could use their preferred segmentation software; the only crucial phase is saving the 3D model in VTK format.
Converting the segmentation
This second step is aimed at translating the 3D model obtained by segmentation in a DICOM format that is readable by both selected platforms, OsiriX and Synapse 3D.
To this aim, we developed a simple software network in the MevisLab environment (www.mevislab.net). The solution comprises two crucial modules: Dicom import and Dicom tool. The Dicom import module allows users to import a segmentation realized by external software applications. The Dicom tool can be used to translate the segmentation and export it in the DICOM format.
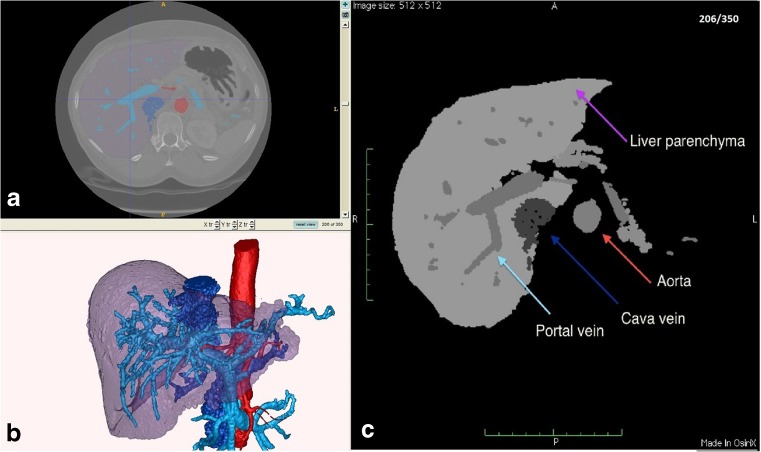
This step entails creating a discretized DICOM dataset out of the VTK segmentation. The exported DICOM dataset contains as many grey levels as the segmented labels contained in the source model (Fig. 2). Furthermore, the exported DICOM file is tagged in the metadata as an elaboration of the selected exams of that patient in order to be loaded and recognized in the right patient directory by PACS.
Fig. 2.
Processing the segmentation in order to import the DICOM format in OsiriX platform. a Original CT dataset and segmentation, b 3D model by segmentation, and c dataset elaborated and saved as DICOM: the arrows indicate the labels previously segmented
The discretized dataset can then be imported in the OsiriX or Synapse 3D environment.
Importing and processing the segmentation in the PACS-connected operating room software
We imported the segmentations both in Synapse 3D and OsiriX. Our rationale originated from the fact that a discretized dataset can be easily segmented and the original 3D model can be easily rebuilt inside both platforms. In this step, the OsiriX platform performed better than the Synapse 3D. In effect, Synapse 3D is endowed with a compositor tool that allows importing and fusion between the segmented label and the patient directory. The last solution is slow and not intuitive. Indeed, the compositor tool requires the user to import label by label to load the entire 3D model, tuning each label when linked to the others. This entails many user adjustments and is a slow and cumbersome process.
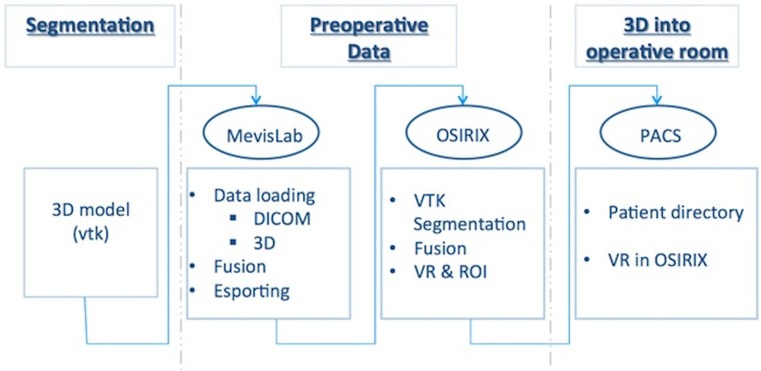
The process is faster in the OsiriX environment. OsiriX is sufficient for loading the exported DICOM data, using the fusion-image plug-in included in the platform, and then for segmenting the discretized 3D dataset and saving everything. Our final aim was to integrate the original segmentation in the CT exam of the patient directly in the OsiriX platform. The entire procedure is an “offline” task that can be done before the intervention. In the operating room, the surgeon can quickly recall the patient directory perioperatively and see the 3D planning using PACS. Figure 3 shows the flow diagram of the defined process.
Fig. 3.
Flow diagram of the global procedure tailored for the OsiriX platform: The first phase is the “segmentation” block, the second phase is the “preoperative data” block, and the third phase is the “3D into operative room” block
Based on our studies, the OsiriX platform allows time-effective importing of data and fast rendering to take advantage of 3D models. Additionally, the platform links easily to PACS allowing for rapid visualization and elaboration of clinical cases studied.
Finally, three cases of pancreatic tumor were studied, and the 3D models of related patients were provided by means of ITK-SNAP and Seg3D open-source software. To segment the aforementioned clinical cases, we applied a previously investigated procedure [15–18]. Finally, a 3D model of a liver tumor case was constructed using the commercial solution Synapse 3D, as in Lo Presti et al., Takahashi, Akinari Miyazaki et al., and Yukio Oshiro et al. [19–22]. The volumetric models were imported into the OsiriX platform and fused with the matching radiological images. In such a retrospective study, Ethics Committee approval was not required because all patients provided informed consent for any research involving clinical information.
Results
We compared the features of the DICOM viewers used most often in clinical practice with equivalent applications. We first compared the characteristics regarding the data and the operating systems: we required a platform capable of importing and exporting native data and 3D models constructed using external applications. Secondly, the platforms must be linked to PACS to facilitate the use of the 3D reconstructions in the operating room. A single PACS-connected platform that allows users to load both a native DICOM dataset of a patient and the pertaining 3D model entails that surgeons can take advantage of the virtual anatomy replicas before and during the surgery. Surgeons can exploit the 3D models in order to facilitate the planning and as an aid during the operation. Starting from the applications listed in Table 1, only the following applications were able to segment a DICOM volume dataset and to visualize a 3D model obtained by segmentation: In-Vesalius, ITK-SNAP, Mito, 3D Slicer, Seg3D, OsiriX, Amira®, Ginkgo-CADx-pro®, Mimics®, and Synapse 3D®. Among these applications, we distinguished those capable of connecting to PACS and loading 3D models obtained by external platforms in the standard de facto VTK format. We implemented a software network to export the VTK segmentation in DICOM format. Both Synapse 3D and OsiriX proved capable of importing the elaborated dataset. Particularly, the OsiriX platform was very simple and fast. In addition, the OsiriX producers released the source code, enabling many users to implement tailored plug-ins, such as the ones written by Vides et al. [23]. Table 2 shows a detailed description of the import and export of data, the kind of support provided by the producers and by the community and the machines in which the two chosen solutions run.
Table 2.
Description of basic features of viewing and segmenting software applications
| Software | Osirix | Synapse 3D® | |
|---|---|---|---|
| Data import | Images | x | x |
| Set | x | x | |
| Series | x | x | |
| Directory | x | x | |
| Data export | Images | x | x |
| Series | x | ||
| Anonimyze | x | x | |
| Support | doc | x | x |
| wiki | |||
| forum | |||
| Source Code | x | ||
| OS | Windows | x | |
| Linux | x | ||
| macintosh | x | x | |
Table 2 shows the features of the aforementioned applications but does not give information about the performance of those applications. To rate them, we executed the segmentation of a CT dataset. The dataset comprised 109 CT images; the voxel spacing was 0.703125 × 0.703125 × 2.50 mm and the data dimensions were 512 × 512 × 109 voxels. We accomplished the specific segmentation using each software application. Table 3 reports the maximum time requested to perform the loading of the dataset mentioned and the time required to render the volume.
Table 3.
Performance of the software applications
| Software | t load (s) | t VR (s) |
|---|---|---|
| Osirix | 12 | 9 (2***) |
| Synapse 3D® | 5 | 2 |
aTwo seconds were needed to switch between CPU mode and GPU mode
We also recorded answers from interviewees regarding the following parameters nonobjectively and dependently by the questioned users: the graphic user interface, the clarity of the functions, and the general speed to obtain a virtual model. Table 4 shows the evaluation of these parameters. Particularly for the speed, we distinguished the time spent segmenting the native dataset and the time spent importing and loading the 3D model. The related maximum segmentation rate was attributed to Synapse 3D, which allows expert users to complete a virtual reconstruction of the liver, the hepatic tumors, the cava vein, the portal vein, the aorta, and the biliary ducts in less than 10 min [15]. To rate the applications, we normalized the time spent to achieve a virtual model in comparison with the time taken up using Synapse 3D. Table 4 also shows a total evaluation of the presented software taking into account all the parameters studied and the performance–cost ratio for the commercial solutions.
Table 4.
Total evaluation of Osirix and Synapse 3D® platforms
| Software | Usability | Total evaluation | |||
|---|---|---|---|---|---|
| GUI | Clarity | Segmentation | Loading rate | ||
| Osirix | 8 | 8 | 8 | 9 | 9 |
| Synapse 3D® | 9 | 8 | 9 | 7 | 8.5 |
Although Synapse 3D performs faster than OsiriX, the times are comparable. We used the free version of OsiriX, and after testing the software, we purchased and adopted the FDA-cleared version, also for clinical use.
Furthermore, we analyzed the import and export of 3D models obtained by segmentation. Three cases of pancreatic tumor were studied, and the 3D reconstructions of related patients were provided by means of ITK-SNAP and Seg3D open-source software. To segment the aforementioned clinical cases, we applied a procedure consolidated and admitted in the clinical practice. We sent the models to the MevisLab tool, where we defined a network capable of loading the volumetric model and converting the 3D data to a format compatible both with the OsiriX software and PACS.
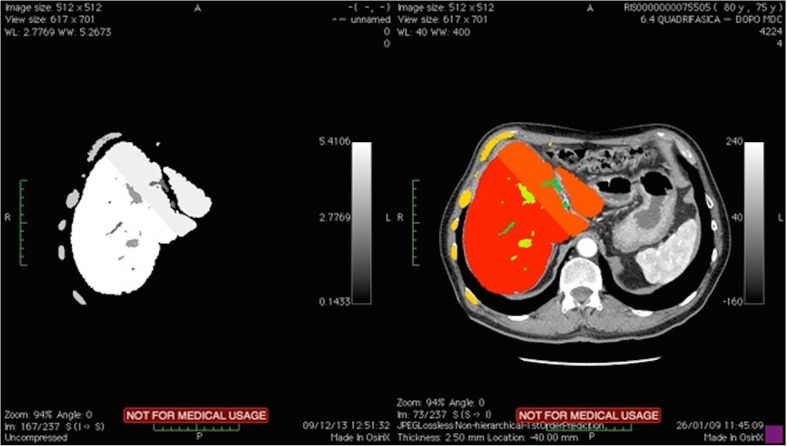
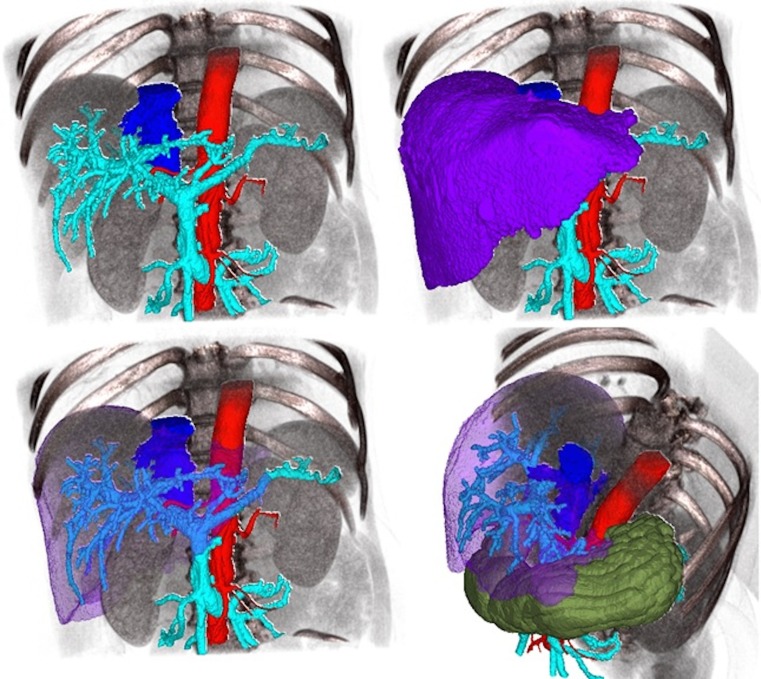
Finally, the 3D model of a liver tumor case was constructed using Synapse 3D. Also in this case, we exported the model to OsiriX by means of the MevisLab tool. Figure 4 shows the degree of fusion between the imported data and the original dataset of the patient.
Fig. 4.
Fusion image between a 3D model imported into OsiriX platform and the matching volumetric dataset. The imported model (visualized in 2D) (left side). The labels segmented and fused with the CT dataset (right side)
At the end of the procedure, in the OsiriX environment, we merged the models imported to the radiological data of the corresponding patient. Thanks to our procedure, we were able to bring the patient-specific 3D model into the operating room to support the information currently available to the specialist. Figure 5 shows the results and the capabilities of the 3D model. Particularly, Fig. 5 shows the 3D model of the liver case analyzed, exported from MevisLab to OsiriX, segmented, and rendered using the latter platform.
Fig. 5.
Results of segmentation by using OsiriX platform available for using into the operative room
Discussion
We evaluated several open-source and commercial DICOM viewers and applications for the segmentation of volumetric diagnostic images. An initial analysis was based on the technical features of 22 open-source and four commercial software applications. Interviews with medical personnel showed that physicians need quantitative information about volumes and anatomical districts to perform several surgeries. Based on the interviews, we selected the applications that were used most often in clinical practice, able to import 3D models obtained by segmenting DICOM volumetric images and able to connect with PACS. We then assessed the performance of these applications, taking into account the time spent to load, open and segment a CT volume comprising 109 CT images, 2.50 mm thick. A limit of this dataset is the relatively low resolution in respect to typical data used for segmentation, but we focused on the timing of the software applications in this study. Consequently, using the same dataset was fundamental. To assess only time performance is limiting and does not allow for evaluation of the goodness of the segmentation, as, for instance, by means of quantitative parameters such as accuracy. An exhaustive evaluation of the goodness of the segmentation is outside the scope of the present study, which refers to applications that we consider reliable, since are used daily in an operating suite.
There are software applications suitable for segmenting anatomical structures. Particularly, the commercial solution Synapse 3D is the fastest at segmenting the liver reliably, even for cases in which a priori registration among different phases is mandatory. The aforementioned solution has some trouble completing a virtual model of a pancreatic case; a way to plan a pancreatectomy has been offered using ITK-SNAP.
We created patient-specific 3D models of four clinical cases by segmenting the CT datasets. An expert radiologist validated the virtual replicas to offer an effective reconstruction for planning the surgical procedures.
We developed a procedure in the MevisLab environment to translate the 3D model labels obtained by external segmentation applications in the standard DICOM format. We found two solutions to bring the 3D model into the operating room by means of a single platform: Synapse 3D and OsiriX.
The first solution requires users to merge the labels into the Synapse 3D platform by means of its own “compositor tool” and to tune the thresholds to obtain a 3D model where each label is well contrasted from the others. The latter step requires many user adjustments, which makes the procedure slow and cumbersome.
The second solution entails importing the 3D model and fusing it to the patient directory in the OsiriX platform, which is able to render the 3D model.
Conclusion
We investigated a wide array of open-source DICOM viewers and applications for the segmentation of volumetric diagnostic images. We also reviewed a small number of commercial solutions. We focused on the software applications actually used by surgeons to plan surgeries. To the best of our knowledge, there is no software application usable for general purpose surgical planning; each platform is focused on limited anatomical districts. Furthermore, bringing patient-specific 3D models into the operating room will make surgeries easier, thanks to its intrinsic quantitative information. In addition, it will allow users to hide or highlight target anatomical structures.
We proposed an approach to use a single platform for constructing or loading patient-specific 3D models obtained by other applications, integrated with PACS, and usable in the operating room. We analyzed two solutions that matched our requirements: the Synapse 3D platform and OsiriX platform. Synapse 3D was able to construct the models more quickly but required longer times to load models from external segmentation applications.
Finally, we selected an application in which the import of volumes segmented by other applications is time-effective. We developed a procedure to import the patient-specific 3D models into the operating room by means of the OsiriX platform. This study demonstrates that the optimal solution is to use OsiriX, which can be linked to PACS and in which the segmentations achieved by other applications can be loaded and incorporated to the patient directory using our procedure. While 3D imaging in the operating suite is not a unique concept and is routinely utilized (e.g., in neurosurgery and plastic surgery), our methodology is the first to be applicable for a wide range of surgical fields, and it may encourage broader utilization of this operative strategy given its use of available software.
Acknowledgments
This work has been financed by “OPERA Project, Funded by Tuscany Region within PAR FAS 2007-2013 Action 1.1 P.I.R. 1.1.B.” and “SILS-Study, Funded by Italian Ministry of Health and Tuscany Region through the call “Ricerca Finalizzata 2009.”
References
- 1.Udupa JK, Hung HM, Chuang KS. Surface and volume rendering in three-dimensional imaging: a comparison. J Digit Imaging. 1991;4:159–168. doi: 10.1007/BF03168161. [DOI] [PubMed] [Google Scholar]
- 2.Ferrari V, Carbone M, Cappelli C, Boni L, Melfi F, Ferrari M, et al. Value of multidetector computed tomography image segmentation for preoperative planning in general surgery. Surg Endosc. 2012;26:616–626. doi: 10.1007/s00464-011-1920-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haller JW, Banerjee A, Christensen GE, Gado M, Joshi S, Miller MI, et al. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- 4.Goldszal AF, Davatzikos C, Pham DL, Yan MX, Bryan RN, Resnick SM: An image-processing system for qualitative and quantitative volumetric analysis of brain images. J Comput Assist Tomogr 22(5):827–37, 1998 [DOI] [PubMed]
- 5.Zhu SC, Yuille A. Region competition: unifying snakes, region growing, and Bayes/MDL for multiband image segmentation. IEEE Trans Pattern Anal Mach Intell. 1996;18:884–900. doi: 10.1109/34.537343. [DOI] [Google Scholar]
- 6.Caselles V, Kimmel R, Sapiro G. Geodesic active contours. Int J Comput Vis. 1997;22:61–79. doi: 10.1023/A:1007979827043. [DOI] [Google Scholar]
- 7.Sethian JA, Sethian JA. Level Set Methods and Fast Marching Methods: Evolving Interfaces in Computational Geometry, Fluid Mechanics, Computer Vision, and Materials Science. 2. Cambridge: Cambridge University Press; 1999. [Google Scholar]
- 8.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 9.Lamata P, Lamata F, Sojar V, Makowski P, Massoptier L, Casciaro S, et al. Use of the Resection Map system as guidance during hepatectomy. Surg Endosc Other Interv Techn. 2010;24:2327–2337. doi: 10.1007/s00464-010-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrari V, Cappelli C, Megali G, Pietrabissa A: An anatomy driven approach for generation of 3D models from multi-phase CT images. Int J Comput Assist Radiol Surg, 2008
- 11.Cha K, Hadjiiski L, Chan HP, Caoili EM, Cohan RH, Zhou C. CT urography: segmentation of urinary bladder using CLASS with local contour refinement. Phys Med Biol. 2014;59:2767–2785. doi: 10.1088/0031-9155/59/11/2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Street E, Hadjiiski L, Sahiner B, Gujar S, Ibrahim M, Mukherji SK, et al. Automated volume analysis of head and neck lesions on CT scans using 3D level set segmentation. Med Phys. 2007;34:4399–4408. doi: 10.1118/1.2794174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappelli C, Carbone M, Ferrari V, Signori S, De Lio N, Perrone V, Mosca F, Boggi U: Patient-Specific 3D surgical planning to perform cutting edge robotic surgery. The Hamlyn Symposium of Medical Robotics, 2012
- 14.Lo Presti G, Ferrari V, Ferrari M, Mosca F, Giannessi F, Ruffoli R. Segmentation procedure for the generation of a 3D model and solid replica of a human skull. Ital J Anat Embryol. 2012;17(Supplement 2):99. [Google Scholar]
- 15.Lo Presti G, Carbone M, Cappelli C, Ferrari V, Ferrari M, Caramella D: An integrated platform for an effective liver surgical planning through segmentation of multiphase CT datasets. IJCARS 6(Supplement 1), 2013
- 16.Takahashi R: Abdominal Imaging in “SYNAPSE 3D”. FUJIFILM Medical Co., Ltd
- 17.Akinari Miyazaki AC, Hiroshi Y, Matsuo N, Nobuhiro T, Hideaki S, Hiroaki S, Hisashi G, Atsushi I, Ikuko M, Tomoko T, Munemasa R, Taro H, Kazuyoshi N, Kentaro S, Taketo Y. 3D images of pancreatic diseases with multi-detector row CT. J Biliary Tract Pancreas. 2008;29:1259–1262. [Google Scholar]
- 18.Yukio Oshiro RS, Ryu M, Takeguchi T, Ibukuro K, Ohkohchi N. adiosurgery anatomical investigation of the artery to the caudate lobe of the liver. Japan Res Soc Clin Anat. 2010;11:4. [Google Scholar]
- 19.Vides CS, Azpiroz LJ, Jimenez AJ. Plugin for OsiriX: mean shift segmentation. Conf Proc IEEE Eng Med Biol Soc. 2007;2007:3060–3. doi: 10.1109/IEMBS.2007.4352974. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu A, Kawamura T, Kobatake H: Proposal of computer-aided detection system for three dimensional CT images of liver cancer. Int Congr Ser 1281:1157–1162, 2005. doi:10.1016/j.ics.2005.03.070
- 21.Liangjia Z, et al. A complete system for automatic extraction of left ventricular myocardium from CT images using shape segmentation and contour evolution. IEEE Trans Image Process. 2014;23(3):1340–1351. doi: 10.1109/TIP.2014.2300751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammon M, et al. Automated detection and volumetric segmentation of the spleen in CT scans. Röfo. 2012;184(8):734–739. doi: 10.1055/s-0031-1299495. [DOI] [PubMed] [Google Scholar]
- 23.Sboarina A, Foroni RI, Minicozzi A, Antiga L, Lupidi F, Longhi M, et al. Software for hepatic vessel classification: feasibility study for virtual surgery. Int J Comput Assist Radiol Surg. 2010;5:39–48. doi: 10.1007/s11548-009-0380-4. [DOI] [PubMed] [Google Scholar]