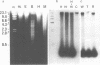
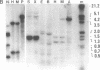
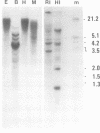
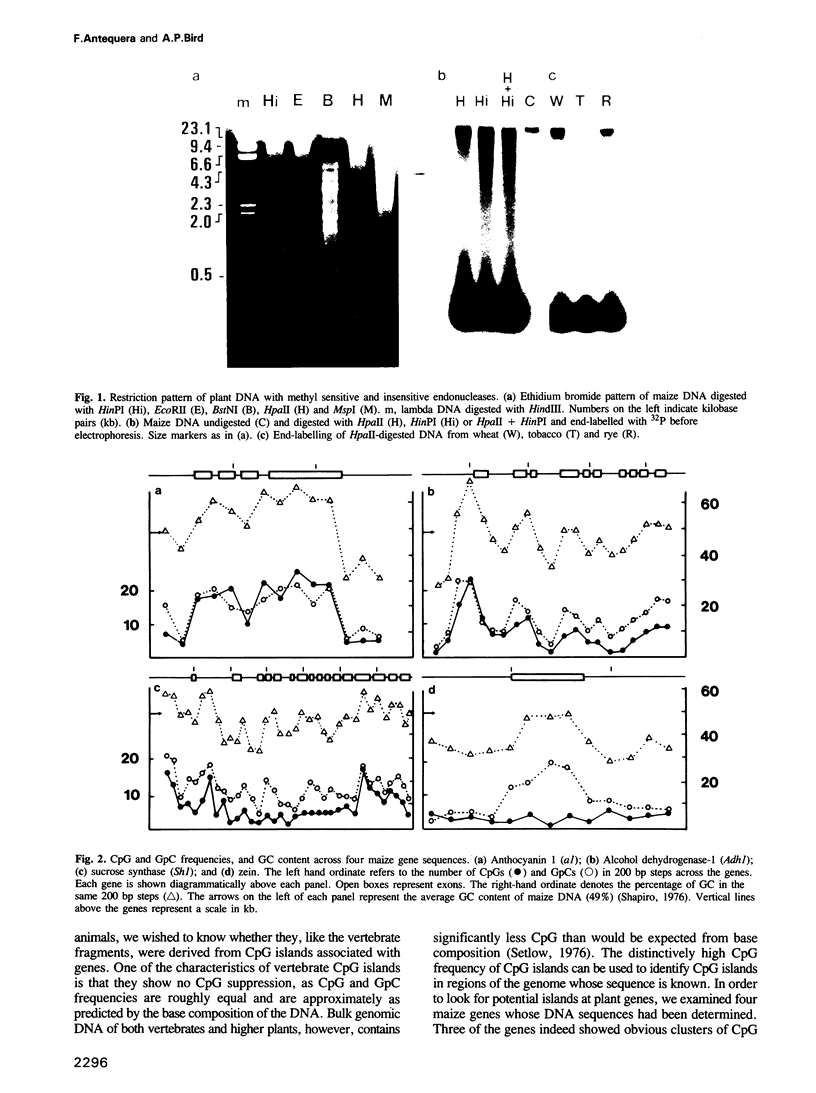
Abstract
The genomes of many higher plant species are the most highly methylated among eukaryotes. We report here that in spite of their heavy methylation, genomic DNAs from four plant species contain a fraction that is very rich in non-methylated sites. The fraction was characterized in maize where it represents about 2.5% of the total nuclear genome. In order to establish the genomic origin of the fraction, three maize genes containing clustered CpG were tested for methylation and were found to be non-methylated in the CpG-rich regions. By contrast, tested CpGs were methylated in a gene whose sequence showed no clustering of CpG. These observations suggest that the CpG-rich fraction of plants is at least partially derived from non-methylated regions that are associated with genes. A similar phenomenon has been described in vertebrate genomes. We discuss the evolution of CpG islands in both groups of organisms, and their possible uses in mapping and gene isolation in plants.
Keywords: CpG islands, DNA methylation, Zea mays DNA
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H., Nicholls R. D., Higgs D. R. Non-methylated CpG-rich islands at the human alpha-globin locus: implications for evolution of the alpha-globin pseudogene. EMBO J. 1987 Apr;6(4):999–1004. doi: 10.1002/j.1460-2075.1987.tb04851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Brown W. R., Bird A. P. Long-range restriction site mapping of mammalian genomic DNA. 1986 Jul 31-Aug 6Nature. 322(6078):477–481. doi: 10.1038/322477a0. [DOI] [PubMed] [Google Scholar]
- Chomet P. S., Wessler S., Dellaporta S. L. Inactivation of the maize transposable element Activator (Ac) is associated with its DNA modification. EMBO J. 1987 Feb;6(2):295–302. doi: 10.1002/j.1460-2075.1987.tb04753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D. N., Taggart M. H., Bird A. P. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983 Feb 11;11(3):647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. S., Gerlach W. L., Pryor A. J., Bennetzen J. L., Inglis A., Llewellyn D., Sachs M. M., Ferl R. J., Peacock W. J. Molecular analysis of the alcohol dehydrogenase (Adh1) gene of maize. Nucleic Acids Res. 1984 May 11;12(9):3983–4000. doi: 10.1093/nar/12.9.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Freeling M., Bennett D. C. Maize Adh1. Annu Rev Genet. 1985;19:297–323. doi: 10.1146/annurev.ge.19.120185.001501. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Graham D. E. The isolation of high molecular weight DNA from whole organisms or large tissue masses. Anal Biochem. 1978 Apr;85(2):609–613. doi: 10.1016/0003-2697(78)90262-2. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Naveh-Many T., Cedar H., Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981 Aug 27;292(5826):860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- Luthe D. S., Quatrano R. S. Transcription in Isolated Wheat Nuclei: I. ISOLATION OF NUCLEI AND ELIMINATION OF ENDOGENOUS RIBONUCLEASE ACTIVITY. Plant Physiol. 1980 Feb;65(2):305–308. doi: 10.1104/pp.65.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Devereux J., Wilson D. R., Sheldon E., Larkins B. A. Cloning and sequence analysis reveal structural variation among related zein genes in maize. Cell. 1982 Jul;29(3):1015–1026. doi: 10.1016/0092-8674(82)90465-2. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Gierl A., Klösgen R. B., Wienand U., Peterson P. A., Saedler H. The Spm (En) transposable element controls the excision of a 2-kb DNA insert at the wx allele of Zea mays. EMBO J. 1984 May;3(5):1021–1028. doi: 10.1002/j.1460-2075.1984.tb01922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Shepherd N., Tacke E., Gierl A., Rohde W., Leclercq L., Mattes M., Berndtgen R., Peterson P. A., Saedler H. Influence of transposable elements on the structure and function of the A1 gene of Zea mays. EMBO J. 1987 Feb;6(2):287–294. doi: 10.1002/j.1460-2075.1987.tb04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B., Werr W., Starlinger P., Bennett D. C., Zokolica M., Freeling M. The Shrunken gene on chromosome 9 of Zea mays L is expressed in various plant tissues and encodes an anaerobic protein. Mol Gen Genet. 1986 Dec;205(3):461–468. doi: 10.1007/BF00338083. [DOI] [PubMed] [Google Scholar]
- Vodkin L. O., Rhodes P. R., Goldberg R. B. cA lectin gene insertion has the structural features of a transposable element. Cell. 1983 Oct;34(3):1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]
- Werr W., Frommer W. B., Maas C., Starlinger P. Structure of the sucrose synthase gene on chromosome 9 of Zea mays L. EMBO J. 1985 Jun;4(6):1373–1380. doi: 10.1002/j.1460-2075.1985.tb03789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler S. R., Baran G., Varagona M., Dellaporta S. L. Excision of Ds produces waxy proteins with a range of enzymatic activities. EMBO J. 1986 Oct;5(10):2427–2432. doi: 10.1002/j.1460-2075.1986.tb04517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]