Abstract
Acetaldehyde is an ethanol-derived definite carcinogen that causes oesophageal squamous cell carcinoma (ESCC). Aldehyde dehydrogenase 2 (ALDH2) is a key enzyme that eliminates acetaldehyde, and impairment of ALDH2 increases the risk of ESCC. ALDH2 is produced in various tissues including the liver, heart, and kidney, but the generation and functional roles of ALDH2 in the oesophagus remain elusive. Here, we report that ethanol drinking increased ALDH2 production in the oesophagus of wild-type mice. Notably, levels of acetaldehyde-derived DNA damage represented by N2-ethylidene-2′-deoxyguanosine were higher in the oesophagus of Aldh2-knockout mice than in wild-type mice upon ethanol consumption. In vitro experiments revealed that acetaldehyde induced ALDH2 production in both mouse and human oesophageal keratinocytes. Furthermore, the N2-ethylidene-2′-deoxyguanosine levels increased in both Aldh2-knockout mouse keratinocytes and ALDH2-knockdown human keratinocytes treated with acetaldehyde. Conversely, forced production of ALDH2 sharply diminished the N2-ethylidene-2′-deoxyguanosine levels. Our findings provide new insight into the preventive role of oesophageal ALDH2 against acetaldehyde-derived DNA damage.
Oesophageal cancer is the eighth most common cancer worldwide and the sixth leading cause of cancer-related death1. The overall 5-year survival of patients with oesophageal cancer is poor, at 15–25%2. Squamous cell carcinoma is the most frequent histologic type of oesophageal cancer, particularly in East Asian countries2. Chronic alcohol consumption and gene variants encoding enzymes involved in alcohol metabolism are closely associated with the risk of oesophageal squamous cell carcinoma (ESCC)3.
Alcoholic beverages contain varying ethanol concentrations. Ingested ethanol is absorbed mainly from the duodenum and jejunum, and then metabolized to acetaldehyde by alcohol dehydrogenase in the liver4. Acetaldehyde is a highly reactive compound that causes DNA damage5,6. It reacts with deoxyguanosine, a substrate of DNA, and forms DNA adducts such as N2-ethylidene-2′-deoxyguanosine (N2-ethylidene-dG), N2-ethyl-2′-deoxyguanosine (N2-Et-dG) and 1,N2-propano-2′-deoxyguanosine7, which are involved in mutagenesis8,9. Among them, N2-ethylidene-dG is the most sensitive marker for acetaldehyde exposure10,11, and is commonly analysed to detect acetaldehyde-derived DNA damage11,12,13.
Aldehyde dehydrogenase 2 (ALDH2) is a mitochondrial enzyme that detoxifies acetaldehyde to acetic acid14. Approximately 35–45% of East Asian individuals have a single nucleotide polymorphism (G1510A) in the ALDH2 gene15,16, which results in a Glu504Lys replacement with reduced ability to oxidize acetaldehyde. Therefore, their blood, salivary, and expiratory acetaldehyde levels after alcohol drinking are highly elevated compared with wild-type ALDH2 homozygotes17,18,19. Accordingly, heavy alcohol consumers with this mutant ALDH2 allele are at risk of ESCC because of the potential exposure of their oesophageal tissues to high amounts of acetaldehyde20,21,22,23.
Based on extensive epidemiological data, acetaldehyde associated with consumption of alcoholic beverages is defined as a ‘group 1 carcinogen’ for the oesophagus by the International Agency for Research on Cancer24. However, little is known about how oesophageal epithelial cells are affected by acetaldehyde. In fact, ALDH2 is produced in various tissues, including the liver, heart, and kidney25, but its production and functional roles in oesophageal epithelium remain elusive. In this study, we addressed the production and role of ALDH2 in oesophageal epithelium. We found that ALDH2 production was increased by acetaldehyde in oesophageal squamous epithelium and suppressed acetaldehyde-derived DNA damage.
Results
Effects of ethanol drinking on ALDH2 production and DNA damage in the oesophagus of Aldh2+/+ and Aldh2–/– mice
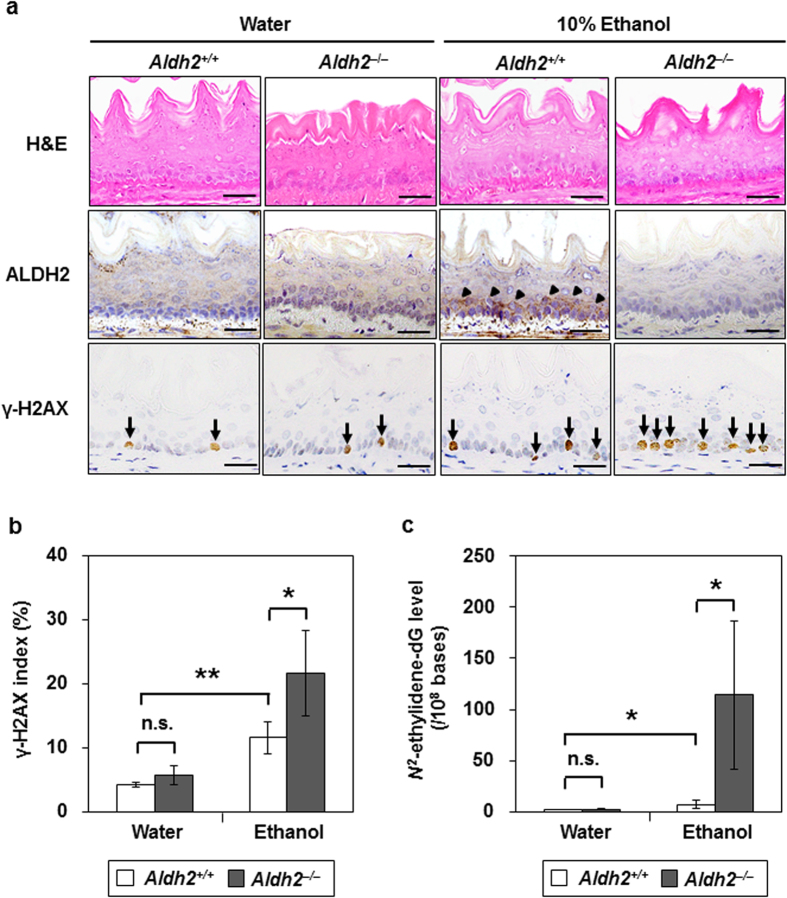
To examine whether ALDH2 was induced in the oesophagus by alcohol drinking and how it influenced alcohol-induced acetaldehyde-derived DNA damage in vivo, Aldh2 wild-type (Aldh2+/+) and Aldh2-knockout (Aldh2–/–) mice were given 10% ethanol to drink for 8 weeks. Neither strain showed obvious histological changes in the oesophagus following ethanol drinking (Fig. 1a, upper panels). Of note, ALDH2 levels increased in the basal and parabasal cells of the oesophageal epithelium in 6 out of 10 Aldh2+/+ mice subjected to ethanol drinking, whereas they did not increase in Aldh2+/+ mice without ethanol drinking. Aldh2–/– mice did not exhibit ALDH2 production in the presence or absence of ethanol drinking (0 of 10 mice) (Fig. 1a, middle panels; Table 1).
Figure 1. Effects of ethanol drinking in Aldh2+/+ and Aldh2–/– mice oesophagus.
The mice were allowed to drink 10% ethanol or water alone (controls) for 8 weeks. Data are presented as the mean ± SD. (a) Hematoxylin and eosin (H&E) and immunohistochemical staining for ALDH2 (arrowheads) and γ-H2AX (arrows). Scale bar = 50 μm. (b) The index of γ-H2AX staining is defined as the proportion of cells staining positive for γ-H2AX in the oesophageal basal cell layer (**p < 0.01, *p < 0.05 between the indicated groups; n = 5 in each group; n.s., not significant). (c) N2-ethylidene-dG level in the oesophagus of Aldh2+/+ and Aldh2–/– mice with or without ethanol consumption (*p < 0.05 between the indicated groups; n = 5 in each group).
Table 1. ALDH2 protein production in the oesophagus of Aldh2 +/+ and Aldh2 –/– mice treated with or without ethanol drinking.
| Groups | ALDH2 level | |
|---|---|---|
| Negative | Positive | |
| Aldh2+/+ mice | ||
| Water | 10 | 0 |
| Ethanol | 4 | 6 |
| Aldh2–/– mice | ||
| Water | 10 | 0 |
| Ethanol | 10 | 0 |
We defined positive of ALDH2 protein levels when more than 50% of the cells were stained with anti-ALDH2 antibody in the basal and parabasal layers of the oesophageal epithelium. Ethanol drinking induced ALDH2 in the oesophageal epithelium in 6 out of 10 Aldh2+/+ mice, whereas there was no induction in the water group of Aldh2+/+ mice (p = 0.011 by Fisher’s exact test).
Next, we examined the levels of phosphorylated histone H2AX (γ-H2AX), a well-established marker of DNA damage26, and N2-ethylidene-dG in the oesophagus. γ-H2AX indexes as well as N2-ethylidene-dG levels in Aldh2+/+ mice with ethanol drinking were significantly higher than those without ethanol drinking (p < 0.001; Fig. 1b), (p = 0.02; Fig. 1c), respectively. Moreover, both levels were significantly elevated in Aldh2–/– mice with ethanol drinking compared to Aldh2+/+ mice with ethanol drinking (p = 0.02; Fig. 1b), (p = 0.03; Fig. 1c), respectively, although there was no significant difference between Aldh2–/– and Aldh2+/+ mice without ethanol drinking (Fig. 1b,c). Thus, ethanol drinking induced the production of ALDH2 in the oesophagus, and acetaldehyde-derived oesophageal DNA damage was enhanced in the absence of Aldh2 gene expression.
Effects of acetaldehyde on DNA damage and ALDH2 production in human oesophageal keratinocytes
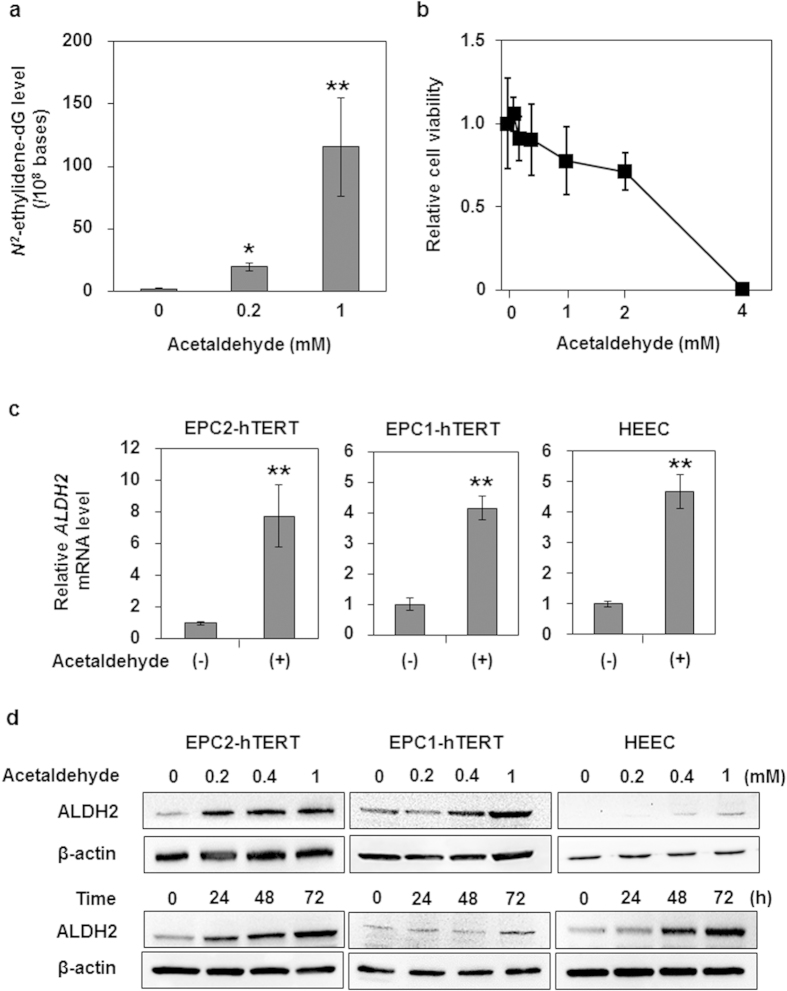
To examine how acetaldehyde affects oesophageal keratinocytes, we treated human oesophageal keratinocytes immortalized with human telomerase reverse transcriptase (hTERT; EPC2-hTERT cells) with acetaldehyde and assessed DNA damage and cell viability. As shown in Fig. 2a, acetaldehyde induced DNA adduct formation in a dose-dependent manner at doses of less than or equal 1 mM that did not induce substantive cell death (Fig. 2b).
Figure 2. Effects of acetaldehyde treatment on human oesophageal keratinocytes.
Data are presented as the mean ± SD. (a) N2-ethylidene-dG levels in DNA isolated from EPC2-hTERT cells treated with the indicated concentrations of acetaldehyde for 72 h (**p < 0.01, *p < 0.05 vs. the cells treated with 0 mM acetaldehyde; n = 3). (b) Cell viability of EPC2-hTERT cells treated with various concentrations of acetaldehyde for 72 h. The ratio of viable cells is expressed relative to the cells treated with 0 mM acetaldehyde (n = 6 for each acetaldehyde concentration). (c) ALDH2 mRNA expression levels in EPC2-hTERT, EPC1-hTERT, and HEEC cells after treatment with acetaldehyde (1 mM) for 72 h. The mRNA levels for the ALDH2 gene relative to the cells treated with 0 mM acetaldehyde were determined by quantitative real-time reverse transcription PCR; the gene for β-actin served as an internal control (**p < 0.01 vs. the cells treated with 0 mM acetaldehyde; n = 3). (d) ALDH2 protein production levels in EPC2-hTERT, EPC1-hTERT, and HEEC cells after treatment with the indicated concentrations of acetaldehyde for 72 h (upper panels) or with acetaldehyde (0.2 mM) for the indicated time points (lower panels). ALDH2 protein production levels were determined by western blotting, and β-actin served as a loading control.
Next, we hypothesized that ALDH2 levels would be increased in oesophageal keratinocytes by acetaldehyde administration, as an autonomous cytoprotective mechanism. To test this hypothesis, we treated three independent human oesophageal keratinocyte cell lines (EPC2-hTERT, EPC1-hTERT, and HEEC) carrying wild-type ALDH2 with acetaldehyde. We found that each of these cell lines displayed enhanced expression of ALDH2 mRNA and protein levels upon stimulation with acetaldehyde in times and dose-dependent manners (Fig. 2c,d). These data indicate that acetaldehyde directly increased ALDH2 production in oesophageal epithelial cells.
Effects of depletion of ALDH2 on acetaldehyde-induced DNA damage
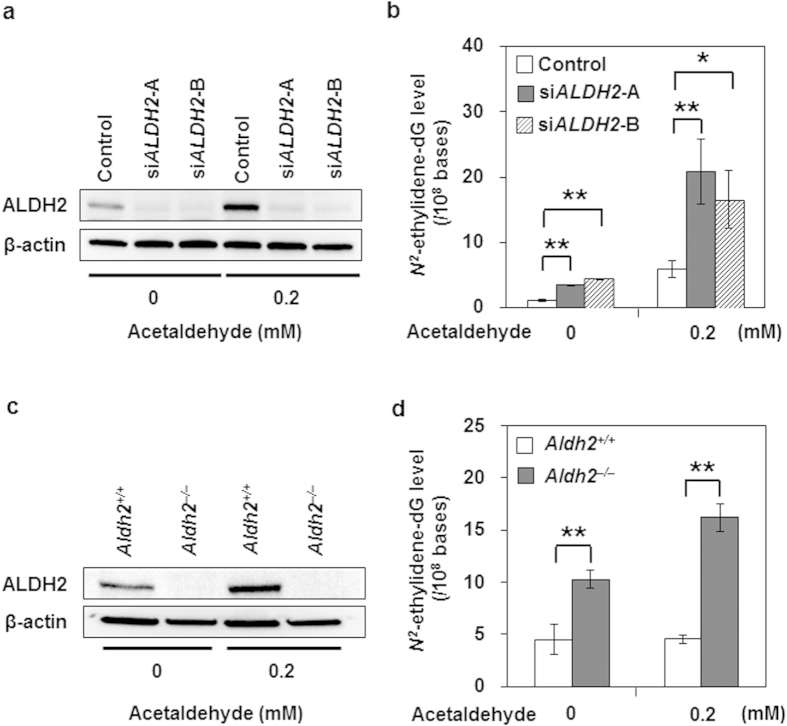
To determine the functional role of ALDH2 in human oesophageal keratinocytes, we knocked down ALDH2 expression by small interfering RNA (siRNA) in EPC2-hTERT cells. ALDH2-knockdown by siRNA (siALDH2-A and siALDH2-B) reduced ALDH2 protein production, whereas a nonsilencing siRNA sequence did not (Fig. 3a). The basal N2-ethylidene-dG level was significantly higher in both ALDH2-knockdown cells than in the control cells (p < 0.001 and p < 0.001, respectively) (Fig. 3b). Furthermore, acetaldehyde treatment markedly increased the N2-ethylidene-dG level in both ALDH2-knockdown cells compared with control cells (p = 0.007 and p = 0.016, respectively) (Fig. 3b).
Figure 3. Facilitation of acetaldehyde-derived DNA damage caused by ALDH2 depletion in human and mouse oesophageal keratinocytes.
EPC2-hTERT cells were treated with 0 or 0.2 mM acetaldehyde for 72 h following transfection with siRNA targeting ALDH2 mRNA translation (siALDH2). Oesophageal keratinocytes isolated from Aldh2+/+ or Aldh2–/– mice were treated with 0 or 0.2 mM acetaldehyde for 72 h. Data are presented as the mean ± SD. (a) Western blotting showing the effect of RNA interference for ALDH2 on EPC2-hTERT cells; β-actin served as a loading control for whole cell lysates. (b) N2-ethylidene-dG levels in EPC2-hTERT cells treated with or without siALDH2-A or siALDH2-B in 0 or 0.2 mM acetaldehyde (**p < 0.01, *p < 0.05 between pairs of indicated groups; n = 3 in each group). (c) Western blotting showing ALDH2 levels in mouse oesophageal keratinocytes in 0 or 0.2 mM acetaldehyde; β-actin served as a loading control. (d) N2-ethylidene-dG level in oesophageal keratinocytes isolated from Aldh2+/+ and Aldh2–/– mice in 0 or 0.2 mM acetaldehyde (**p < 0.01 between pairs of indicated groups; n = 5 in each group).
To confirm the enhancement of acetaldehyde-derived DNA damage by ALDH2 impairment, we also conducted ex vivo experiments using mouse oesophageal keratinocytes isolated from Aldh2+/+ and Aldh2–/– mice and treated with acetaldehyde. ALDH2 production was absent in Aldh2–/– oesophageal keratinocytes irrespective of acetaldehyde treatment, whereas it was increased by this treatment in keratinocytes of Aldh2+/+ mice (Fig. 3c). Consistent with the results of the siRNA experiments (Fig. 3b), the basal N2-ethylidene-dG level was significantly higher in Aldh2–/– cells than in Aldh2+/+ cells (p < 0.001) (Fig. 3d). Moreover, after acetaldehyde treatment, the N2-ethylidene-dG level was significantly elevated in Aldh2–/– cells, whereas it was not increased in Aldh2+/+ cells (p < 0.001) (Fig. 3d). These results collectively suggest that endogenous ALDH2 plays a role in preventing acetaldehyde-derived DNA damage in oesophageal keratinocytes.
Effects of ALDH2 overexpression on acetaldehyde-derived DNA damage
Finally, we examined whether ALDH2 overexpression would decrease acetaldehyde-derived DNA damage. The control EPC2-hTERT cells, transduced with a lentiviral control vector bearing a FLAG-tag but lacking the ALDH2 coding site, showed production of endogenous ALDH2 protein (52.6 kDa). EPC2-hTERT cells stably overexpressing wild-type ALDH2 or mutant ALDH2, designated as EPC2-ALDH2 or EPC2-ALDH2MT cells respectively, showed substantial production of both exogenous FLAG-tagged ALDH2 protein (53.6 kDa) and endogenous ALDH2 protein (52.6 kDa) (Fig. 4a). Basal N2-ethylidene-dG levels were significantly lower in EPC2-ALDH2 cells than in either control cells (p = 0.032) or EPC2-ALDH2MT cells (p = 0.02) (Fig. 4b). Most importantly, the N2-ethylidene-dG production induced by acetaldehyde treatment was also significantly smaller in EPC2-ALDH2 cells than in control (p < 0.001) or EPC2-ALDH2MT cells (p = 0.001) (Fig. 4b).
Figure 4. Reduction of acetaldehyde-derived DNA damage by overexpression of ALDH2 in EPC2-hTERT cells.
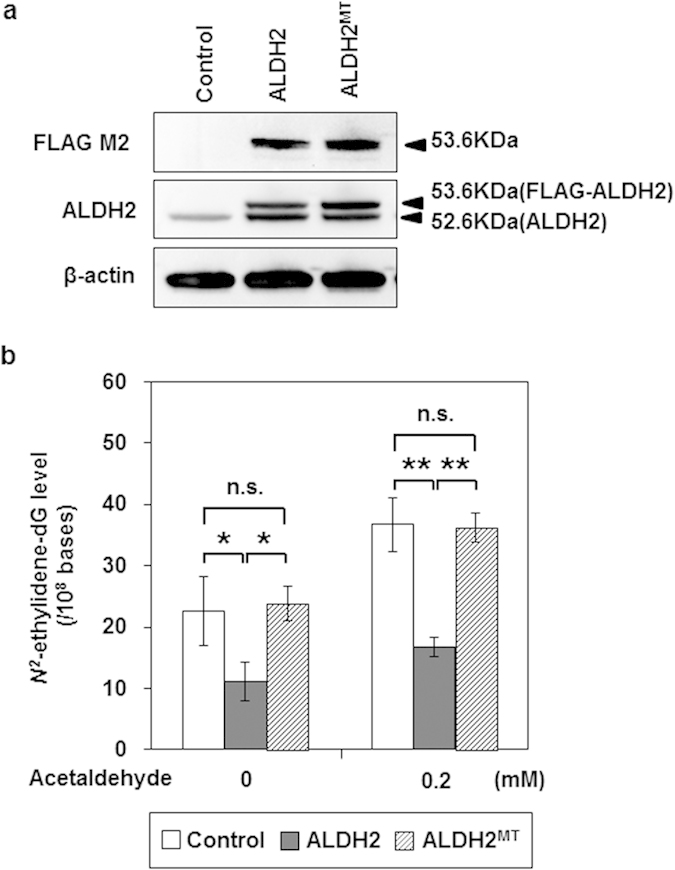
EPC2-hTERT cells were transduced with lentiviral pLEX-ALDH2 or pLEX-ALDH2MT vectors to establish EPC2-ALDH2 or EPC2-ALDH2MT cells, respectively, which stably overexpressed wild-type or mutant ALDH2. As control cells, we transduced EPC2-hTERT cells with a lentiviral control vector bearing a FLAG-tag without an ALDH2 coding site. Data are presented as the mean ± SD. (a) Western blotting showing overproduction of FLAG-tagged ALDH2 protein (53.6 kDa) and non-FLAG-tagged ALDH2 (52.6 kDa) in EPC2-ALDH2 and EPC2-ALDH2MT cells using antibodies to FLAG M2 and ALDH2; β-actin served as a loading control. (b) N2-ethylidene-dG levels in EPC2-ALDH2, EPC2-ALDH2MT, and control cells treated with 0 or 0.2 mM acetaldehyde for 72 h (**p < 0.01, *p < 0.05 between the indicated groups; n = 3 in each group; n.s., not significant).
Discussion
Here, we found that ethanol drinking increased ALDH2 production in the oesophagus of Aldh2+/+ mice. Compared with Aldh2+/+ mice, the Aldh2–/– mice displayed greatly enhanced acetaldehyde-derived DNA damage in their oesophagus. We also found that acetaldehyde directly enhanced ALDH2 production in both mouse and human oesophageal keratinocytes in vitro, and that acetaldehyde-derived DNA damage was augmented in both human ALDH2-knockdown keratinocytes and Aldh2–/– mouse keratinocytes. Furthermore, overexpression of wild-type ALDH2, but not mutant ALDH2, reduced acetaldehyde-derived DNA damage in human keratinocytes. These data indicate that oesophageal ALDH2 plays a protective role in reducing acetaldehyde-derived DNA damage in keratinocytes.
Our in vivo study revealed that ethanol drinking induced ALDH2 production in the basal and parabasal layers of the mouse oesophagus. It is controversial whether ALDH2 protein is produced in the oesophagus. Yin et al. reported that agarose isoelectric focusing did not show ALDH2 expression in human oesophageal mucosa27. By contrast, in an immunohistochemistry study, Morita et al. found that ALDH2 was produced in the oesophageal epithelium and that the expression levels were closely associated with the patients’ drinking habits28. Our data are in agreement with those reported by Morita et al. Moreover, we demonstrated that oesophageal ALDH2 production was induced by acetaldehyde exposure in human and mouse oesophageal keratinocytes. These results suggest that the increased oesophageal ALDH2 levels induced by ethanol drinking are triggered by the direct exposure of oesophageal mucosal cells to acetaldehyde rather than to ethanol per se.
To determine the role of oesophageal ALDH2 in acetaldehyde-derived DNA damage, we conducted in vitro and ex vivo experiments in which the same amount of acetaldehyde was administered to both human and mouse oesophageal keratinocytes with genetic modifications to ALDH2 production levels, and found a strong negative association between the extent of acetaldehyde-derived DNA damage and ALDH2 levels. These results indicate that oesophageal ALDH2 might act genoprotectively for acetaldehyde as an autonomous defence response to acetaldehyde exposure. Thus, DNA damage might be caused by acetaldehyde exposure that exceeds the innate defence capacity of oesophageal keratinocytes.
Immunohistochemical analysis of γ-H2AX in our in vivo study showed that DNA damage was accumulated at the basal layer of the oesophageal epithelium in Aldh2–/– mice following ethanol drinking. Acetaldehyde-derived DNA adducts are known to induce DNA–DNA crosslinking, which leads to gene mutations and double-strand DNA breaks, and ultimately to the induction of γ-H2AX expression6,29. This result indicates that the basal cells in the oesophageal epithelium might be susceptible to DNA damage induced by acetaldehyde.
In our in vivo study, obvious histologic abnormalities were not found in Aldh2–/– mice with ethanol drinking for 8 weeks, although DNA adducts levels highly increased in those mice. Similarly, N2-ethylidene-dG levels increased in acetaldehyde-treated ALDH2-knockdown cells; however, those cells were not transformed in the present study. Moreover, proliferative and migrative activities of those cells were not influenced by acetaldehyde treatment (supplementary Figure S1). Accordingly, it is still unclear whether DNA adducts are the direct initiators of alcohol-related carcinogenesis in the oesophagus.
In our study, overexpression of wild-type ALDH2 in the oesophageal mucosal cells reduced acetaldehyde-derived DNA damage. Recently, ALDH2 is reported as an important mediator of endogenous cytoprotection in the heart when it is subjected to ischemia30,31. In addition, overexpression of the ALDH2 transgene ameliorates acute cardiac toxicity of ethanol32 as well as acetaldehyde-induced cell injury in human cardiac myocytes or umbilical vein endothelial cells33,34. These findings support the notion that ALDH2 induction or activation might have a preventive potential to alcohol- or acetaldehyde-induced cytotoxicity in the oesophagus.
A limitation of this study is that we could not demonstrate ALDH2 enzymatic activity in oesophageal tissues and keratinocytes. To show such activity, we conducted experiments to quantify the reduction of nicotinamide adenine dinucleotide production by ALDH2 as described previously35. We could measure the ALDH activity in mouse liver (0.9 ± 0.22 nmol/min/mg protein); however, its activity in the mouse oesophagus and human oesophageal keratinocytes were undetectable. Because the ALDH2 expression level determined by western blotting was very low in the mouse oesophagus compared with the liver (Supplement Figure S2), the above experiments might not be able to determine ALDH activity in the mouse oesophagus. Additional research is warranted to elucidate the relationship between ALDH2 expression level and its activity.
In conclusion, we have demonstrated here that ALDH2 production in oesophageal keratinocytes is induced by acetaldehyde and that oesophageal ALDH2 acts cytoprotectively as a local defence mechanism against acetaldehyde-derived DNA damage.
Methods
Mouse preparation and alcohol drinking
Six-week-old male Aldh2–/– mice36, backcrossed with C57BL/6 mice, were obtained from the Department of Environmental Health, University of Occupational and Environmental Health (Kitakyushu, Japan). Age-matched control C57BL/6 male mice, which carry the wild-type Aldh2, were purchased from Charles River Laboratories Japan Inc. (Yokohama, Japan). All experiments conformed to the relevant regulatory standards and were approved by the Institutional Animal Care and Use Committee of Kyoto University (Med Kyo 14522). The genotype of Aldh2 was determined by polymerase chain reaction (PCR) amplification as described36. Aldh2+/+ and Aldh2–/– mice were allowed to drink water with or without 10% ethanol supplementation freely for 8 weeks as described37. Mice were euthanized painlessly under anesthesia with diethyl ether inhalation followed by cervical dislocation. Oesophageal tissues were collected in 4% Paraformaldehyde Phosphate Buffer Solution (Wako Pure Chemical Industries, Ltd., Osaka, Japan) for immunohistochemistry (n = 10 each group) or frozen in liquid nitrogen and stored at –80 °C for analysis of DNA adducts (n = 5 each group).
Oesophageal keratinocyte culture and treatments
Primary mouse oesophageal epithelial cells (keratinocytes) were isolated from C57BL/6 mice and grown as described38. Two independent nontransformed human oesophageal keratinocyte cell lines, EPC1-hTERT and EPC2-hTERT, were obtained from the Cell Culture Core of the Center for Molecular Studies in Digestive and Liver Diseases at the University of Pennsylvania39,40. They were cultured in Keratinocyte Serum-Free Media (Life Technologies Corp., Carlsbad, CA, USA), supplemented with 40 μg/mL of Bovine Pituitary Extract, 1 ng/mL Epidermal Growth Factor, and 1% penicillin/streptomycin. A primary human oesophageal epithelial cell line, HEEC41, was purchased from ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in Epithelial Cell Medium-2 (EpiCM-2). The ALDH2 genotype of human oesophageal keratinocytes was assessed using SmartAmp ALDH2 Typing Kits (DNAFORM, Yokohama, Japan). Cells were treated with acetaldehyde (0–4 mM) (Sigma-Aldrich, St. Louis, MO, USA) for 0–72 h. Cell viability was determined by sulfonated tetrazolium salt WST-1 assays according to the manufacturer’s instructions (Roche Applied Science, Penzberg, Germany).
Generation of the ALDH2 transgene construct
To establish a stably transfected cell line expressing ALDH2, we produced a lentiviral plasmid vector. The ALDH2 open reading frame was PCR-amplified using Mammalian Gene Collection Human ALDH2 Sequence-Verified cDNA (Clone ID: 5477768) (Thermo Fisher Scientific Inc., Waltham, MA, USA) as a template with PrimeSTAR Max DNA Polymerase (Takara Bio Inc., Otsu, Japan) and ALDH2-Forward (5′–AAAAAGAATTCATGTTGCGCGCTGCCGCC–3′) and ALDH2-Reverse (5′–AAAAACTCGAGTTATGAGTTCTTCTGAGG–3′) primers. The amplified ALDH2 fragment was ligated into the EcoRI and XhoI sites of a pCMV-Tag2B vector (Stratagene Corp., La Jolla, CA, USA) by Ligation Mix (Takara Bio Inc.), resulting in the creation of pCMV-ALDH2. pCMV-ALDH2 was cut by NheI and XhoI to include the coding site for FLAG-tagged ALDH2. The lentiviral plasmid pLEX-MCS vector42 (Thermo Fisher Scientific Inc.) was cut by SpeI and XhoI. The site of FLAG-ALDH2 was cloned into a pLEX-MCS vector. The construct sequence was confirmed by sequencing, and designated as the pLEX-ALDH2 plasmid vector. To produce recombinant lentiviruses, pMD2.G and psPAX2 (Addgene, Cambridge, MA, USA) were cotransfected with the pLEX-ALDH2 vector into HEK293T cells using Lipofectamine LTX (Life Technologies Corp.). Finally, a recombinant ALDH2-coding lentivirus was transduced into EPC2-hTERT cells, and the cells were selected by puromycin (1 μg/ml) (Sigma-Aldrich). We constructed a control vector bearing a FLAG-tag and lacking the ALDH2 coding site. We also constructed a lentiviral plasmid vector including a mutant ALDH2 with a nucleotide substitution (G1510A; Glu504Lys) in exon 12 of the ALDH2 locus—this point mutation causes catalytic deficiency of human ALDH215—using the PrimeSTAR Mutagenesis Basal Kit (Takara Bio Inc.) according to the manufacturer’s protocol, and designated this as the pLEX-ALDH2MT vector. The mutated primers were as follows: forward (5′–ATACACTAAAGTGAAAACTGTCACAG–3′) and reverse (5′–TTTCACTTTAGTGTATGCCTGCAGCC–3′).
Immunohistochemistry
Immunohistochemistry was performed as described43. Primary antibodies and the titers used were as follows: rabbit polyclonal anti-ALDH2 (Ag7452; 15310-1-AP; Proteintech Group, Inc., Chicago, IL, USA; 1:500); rabbit monoclonal anti-phospho-histone H2AX (Ser139; 20E3; Cell Signaling Technology, Inc., Danvers, MA, USA; 1:480). Immunostained tissues were assessed using a Keyence BIOREVO BZ-9000 microscope (Keyence Corp., Osaka, Japan). For the staining of ALDH2, we defined the sample as positive when more than 50% of the cells were stained with anti-ALDH2 in the basal and parabasal layer of the oesophageal epithelium together. We present the ratio of cells positive for phosphorylated histone H2AX (γ-H2AX) to the total number of cells, determined using a Hybrid Cell Count BZ-H2C system (Keyence Corp.), as a staining index.
DNA isolation, digestion, and quantification of N2-ethylidene-dG
DNA was isolated from tissue specimens or cell pellets as described11. To quantify acetaldehyde-derived DNA damage, we measured the N2-ethylidene-dG level in the oesophagus of mice and cultured cells as described37. Briefly, NaBH3CN (100 mM) (Sigma-Aldrich), a reducing reagent, was added to DNA samples. This converts N2-ethylidene-dG to stable N2-Et-dG. As the endogenous N2-Et-dG level in tissues is extremely low, the N2-Et-dG level that is converted from N2-ethylidene-dG indicates the endogenous N2-ethylidene-dG level10. The DNA adduct standard, N2-Et-dG, and its stable isotope, [U-15N5]-labeled N2-Et-dG, were synthesized as described11. DNA samples were digested as described11, and subjected to liquid chromatography tandem mass spectrometry (LC/MS/MS). LC/MS/MS analyses were performed using a Shimadzu LC system (Shimadzu Corp., Kyoto, Japan) interfaced with a Quattro Ultimo triple-stage quadrupole mass spectrometer or an ACQUITY UPLC H-Class system interfaced with a XEVO-TSQ triple-stage quadrupole mass spectrometer (Waters Corp., Milford, MA, USA) as reported11. Shim-pack XR-ODS columns (3.0 × 75 mm, 2.2 μm; Shimadzu Corp.) or ACQUITY UPLC BEH C18 columns (2.1 × 100 mm, 1.7 μm; Waters Corp.) were used to separate the samples.
TaqMan gene expression assays
RNA was isolated using RNeasy Plus Mini Kits (QIAGEN Inc., Hilden, Germany) and cDNA was synthesized using PrimeScript RT reagent kits (Takara Bio Inc.) according to the manufacturer’s instructions. Real-time reverse-transcription PCR was done with TaqMan Gene Expression Assays (Life Technologies Corp.) for ALDH2 (Assay ID; Hs01007998_m1) and for β-actin (Assay ID; Hs99999903_m1) using a LightCycler 480 Instrument II (Hoffmann-La Roche Ltd, Basel, Switzerland) as described44. All PCR reactions were performed in triplicate. The relative mRNA expression level was normalized to that of β-actin as an internal control.
Western blot analysis
Whole cell lysates were prepared as described45. The denatured protein samples (20 μg) were fractionated on Any kD Mini-PROTEAN TGX Precast Gels (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Primary antibodies and the titers used for western blotting were as follows: goat polyclonal anti-ALDH2 (N-14) (sc-48838; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA; 1:1000); mouse monoclonal anti-FLAG M2 (F1805; Sigma-Aldrich; 1:1000); rabbit monoclonal anti-β-actin (13E5; Cell Signaling Technology, Inc.; 1:2000). These were then reacted with the appropriate horseradish peroxidase-conjugated secondary antibody (GE Healthcare, Little Chalfont, UK). The signal was visualized using an Immobilon Western Chemiluminescent Horseradish Peroxidase Substrate (Merck Millipore, Darmstadt, Germany) and was exposed using a ChemiDoc XRS system equipped with Image Lab software (Bio-Rad Laboratories, Inc.).
RNA interference
To suppress endogenous ALDH2 induction, FlexiTube siRNA (QIAGEN Inc.) for ALDH2 (two independent sequences: ALDH2-A, SI03059973; ALDH2-B, SI03087070) were employed. AllStars Negative Control (1027280) (QIAGEN Inc.) was used as a nonsilencing control. The siRNAs (10 nM) were transfected transiently using HiPerFect transfection reagent (QIAGEN Inc.), according to the manufacturer’s instructions.
Statistical analysis
Data are presented as the mean ± standard deviation. The data were first tested for normality of distribution. The differences between two groups were analysed using two-tailed paired Student’s t tests for equal variance data or using Welch’s t test for the unequal variance data presented in Figs 1b,c and 2a,c and 3d. Student’s t test followed by the Bonferroni post hoc test was used to analyse the data presented in Fig. 3b. One-way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis was used to analyse the data shown in Fig. 4b. Fisher’s exact test was used to assess the relationship between ALDH2 expression and drinking ethanol in Aldh2+/+ mice (data presented in Table 1). Statistical analyses were performed using SPSS software (version 17; IBM Corp., Armonk, NY, USA). P-values < 0.05 were considered significant.
Additional Information
How to cite this article: Amanuma, Y. et al. Protective role of ALDH2 against acetaldehyde-derived DNA damage in oesophageal squamous epithelium. Sci. Rep. 5, 14142; doi: 10.1038/srep14142 (2015).
Supplementary Material
Acknowledgments
This work was supported by the National Cancer Center and Development Fund (22-36 to M.M.); Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (24590917 to M.M., 25460926 to S.O., 23221006 to T.M., 25130706 to T.C. and 24229005 to T.C.); Raptor Pharmaceuticals (to H.N.); National Institutes of Health grants (P01CA098101 to K.W., H.N., K26-RR032714 to H.N., K01DK103953 to K.W. and F32-CA174176 to K.W.). We are grateful to the NIH/NIDDK Center for Molecular Studies in Digestive and Liver Diseases (P30-DK050306). We thank the Center for Anatomical, Pathological and Forensic Medical Researches, Kyoto University Graduate School of Medicine, for technical assistance in preparing microscope slides for immunohistochemistry.
Footnotes
Author Contributions Y.A., S.O. and M.M. were responsible for study concept and design. Y.A., M.T. and Y.N. were responsible for acquisition of data. Y.A., S.O., H.N., T.M. and M.M. were responsible for analysis and interpretation of data. Y.A., S.O., Y.I., K.W., H.N., T.C. and M.M. were responsible for writing or revision of the manuscript. Y.I., M.T., S.M., O.K., Y.N., S.M., T.O., T.K., H.N. and T.M. were responsible for technical or material support.
References
- Ferlay J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–2917 (2010). [DOI] [PubMed] [Google Scholar]
- Pennathur A., Gibson M. K., Jobe B. A. & Luketich J. D. Oesophageal carcinoma. The Lancet 381, 400–412 (2013). [DOI] [PubMed] [Google Scholar]
- Cui R. et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 137, 1768–1775 (2009). [DOI] [PubMed] [Google Scholar]
- Crabb D. W., Matsumoto M., Chang D. & You M. Overview of the role of alcohol dehydrogenase and aldehyde dehydrogenase and their variants in the genesis of alcohol-related pathology. Proc. Nutr. Soc. 63, 49–63 (2004). [DOI] [PubMed] [Google Scholar]
- Seitz H. K. & Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer 7, 599–612 (2007). [DOI] [PubMed] [Google Scholar]
- Brooks P. J. & Zakhari S. Acetaldehyde and the genome: beyond nuclear DNA adducts and carcinogenesis. Environ. Mol. Mutagen. 55, 77–91 (2014). [DOI] [PubMed] [Google Scholar]
- Yu H. S. et al. Formation of acetaldehyde-derived DNA adducts due to alcohol exposure. Chem. Biol. Interact. 188, 367–375 (2010). [DOI] [PubMed] [Google Scholar]
- Upton D. C. et al. Replication of N2-ethyldeoxyguanosine DNA adducts in the human embryonic kidney cell line 293. Chem. Res. Toxicol. 19, 960–967 (2006). [DOI] [PubMed] [Google Scholar]
- Stein S., Lao Y., Yang I. Y., Hecht S. S. & Moriya M. Genotoxicity of acetaldehyde- and crotonaldehyde-induced 1,N2-propanodeoxyguanosine DNA adducts in human cells. Mutat. Res. 608, 1–7 (2006). [DOI] [PubMed] [Google Scholar]
- Wang M. et al. Identification of an acetaldehyde adduct in human liver DNA and quantitation as N2-ethyldeoxyguanosine. Chem. Res. Toxicol. 19, 319–324 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T. et al. Increased formation of hepatic N2-ethylidene-2′-deoxyguanosine DNA adducts in aldehyde dehydrogenase 2-knockout mice treated with ethanol. Carcinogenesis 28, 2363–2366 (2007). [DOI] [PubMed] [Google Scholar]
- Nagayoshi H. et al. Increased formation of gastric N(2)-ethylidene-2′-deoxyguanosine DNA adducts in aldehyde dehydrogenase-2 knockout mice treated with ethanol. Mutat. Res. 673, 74–77 (2009). [DOI] [PubMed] [Google Scholar]
- Yukawa Y. et al. Combination of ADH1B*2/ALDH2*2 polymorphisms alters acetaldehyde-derived DNA damage in the blood of Japanese alcoholics. Cancer Sci. 103, 1651–1655 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyosov A. A., Rashkovetsky L. G., Tahir M. K. & Keung W. M. Possible role of liver cytosolic and mitochondrial aldehyde dehydrogenases in acetaldehyde metabolism. Biochemistry 35, 4445–4456 (1996). [DOI] [PubMed] [Google Scholar]
- Yoshida A., Huang I. Y. & Ikawa M. Molecular abnormality of an inactive aldehyde dehydrogenase variant commonly found in Orientals. Proc. Natl. Acad. Sci. USA 81, 258–261 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi S., Matsushita S., Murayama M., Takagi S. & Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am. J. Psychiatry 152, 1219–1221 (1995). [DOI] [PubMed] [Google Scholar]
- Vakevainen S., Tillonen J., Agarwal D. P., Srivastava N. & Salaspuro M. High salivary acetaldehyde after a moderate dose of alcohol in ALDH2-deficient subjects: strong evidence for the local carcinogenic action of acetaldehyde. Alcohol. Clin. Exp. Res. 24, 873–877 (2000). [PubMed] [Google Scholar]
- Yokoyama A. et al. Salivary acetaldehyde concentration according to alcoholic beverage consumed and aldehyde dehydrogenase-2 genotype. Alcohol. Clin. Exp. Res. 32, 1607–1614 (2008). [DOI] [PubMed] [Google Scholar]
- Muto M. et al. Association between aldehyde dehydrogenase gene polymorphisms and the phenomenon of field cancerization in patients with head and neck cancer. Carcinogenesis 23, 1759–1765 (2002). [DOI] [PubMed] [Google Scholar]
- Yokoyama A. et al. Cancer screening of upper aerodigestive tract in Japanese alcoholics with reference to drinking and smoking habits and aldehyde dehydrogenase-2 genotype. Int. J. Cancer 68, 313–316 (1996). [DOI] [PubMed] [Google Scholar]
- Matsuo K. et al. Gene-environment interaction between an aldehyde dehydrogenase-2 (ALDH2) polymorphism and alcohol consumption for the risk of esophageal cancer. Carcinogenesis 22, 913–916 (2001). [DOI] [PubMed] [Google Scholar]
- Muto M. et al. Risk of multiple squamous cell carcinomas both in the esophagus and the head and neck region. Carcinogenesis 26, 1008–1012 (2005). [DOI] [PubMed] [Google Scholar]
- Brooks P. J., Enoch M. A., Goldman D., Li T. K. & Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 6, e50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secretan B. et al. A review of human carcinogen—Part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 10, 1033–1034 (2009). [DOI] [PubMed] [Google Scholar]
- Stewart M. J., Malek K. & Crabb D. W. Distribution of messenger RNAs for aldehyde dehydrogenase 1, aldehyde dehydrogenase 2, and aldehyde dehydrogenase 5 in human tissues. J. Investig. Med. 44, 42–46 (1996). [PubMed] [Google Scholar]
- Mah L. J., El-Osta A. & Karagiannis T. C. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia 24, 679–686 (2010). [DOI] [PubMed] [Google Scholar]
- Yin S. J. et al. Alcohol and aldehyde dehydrogenases in human esophagus: comparison with the stomach enzyme activities. Alcohol. Clin. Exp. Res. 17, 376–381 (1993). [DOI] [PubMed] [Google Scholar]
- Morita M. et al. Expression of aldehyde dehydrogenase 2 in the normal esophageal epithelium and alcohol consumption in patients with esophageal cancer. Front. Biosci. 10, 2319–2324 (2005). [DOI] [PubMed] [Google Scholar]
- Clingen P. H. et al. Histone H2AX phosphorylation as a molecular pharmacological marker for DNA interstrand crosslink cancer chemotherapy. Biochem. Pharmacol. 76, 19–27 (2008). [DOI] [PubMed] [Google Scholar]
- Chen C. H. et al. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 321, 1493–1495 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Ferreira J. C. & Mochly-Rosen D. ALDH2 activator inhibits increased myocardial infarction injury by nitroglycerin tolerance. Sci. Transl. Med. 3, 107ra111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Li J., Gao F. & Ren J. Aldehyde dehydrogenase 2 ameliorates acute cardiac toxicity of ethanol: role of protein phosphatase and forkhead transcription factor. J. Am. Coll. Cardiol. 54, 2187–2196 (2009). [DOI] [PubMed] [Google Scholar]
- Li S. Y. et al. Attenuation of acetaldehyde-induced cell injury by overexpression of aldehyde dehydrogenase-2 (ALDH2) transgene in human cardiac myocytes: role of MAP kinase signaling. J. Mol. Cell. Cardiol. 40, 283–294 (2006). [DOI] [PubMed] [Google Scholar]
- Li S. Y. et al. Overexpression of aldehyde dehydrogenase-2 (ALDH2) transgene prevents acetaldehyde-induced cell injury in human umbilical vein endothelial cells: role of ERK and p38 mitogen-activated protein kinase. J. Biol. Chem. 279, 11244–11252 (2004). [DOI] [PubMed] [Google Scholar]
- Ushida Y. & Talalay P. Sulforaphane accelerates acetaldehyde metabolism by inducing aldehyde dehydrogenases: relevance to ethanol intolerance. Alcohol Alcohol. 48, 526–534 (2013). [DOI] [PubMed] [Google Scholar]
- Kitagawa K. et al. Aldehyde dehydrogenase (ALDH) 2 associates with oxidation of methoxyacetaldehyde; in vitro analysis with liver subcellular fraction derived from human and Aldh2 gene targeting mouse. FEBS Lett. 476, 306–311 (2000). [DOI] [PubMed] [Google Scholar]
- Yukawa Y. et al. Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am. J. Cancer Res. 4, 279–284 (2014). [PMC free article] [PubMed] [Google Scholar]
- Kalabis J. et al. A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. J. Clin. Invest. 118, 3860–3869 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H. et al. Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation. Mol. Cancer Res. 1, 729–738 (2003). [PubMed] [Google Scholar]
- Ohashi S. et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology 139, 2113–2123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyros O. et al. Dickkopf-1, the Wnt antagonist, is induced by acidic pH and mediates epithelial cellular senescence in human reflux esophagitis. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G557–574 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itatani Y. et al. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology 145, 1064–1075.e1011 (2013). [DOI] [PubMed] [Google Scholar]
- Andl C. D. et al. Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. J. Biol. Chem. 278, 1824–1830 (2003). [DOI] [PubMed] [Google Scholar]
- Takaoka M. et al. Epidermal growth factor receptor regulates aberrant expression of insulin-like growth factor-binding protein 3. Cancer Res. 64, 7711–7723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka M. et al. Ha-Ras(G12V) induces senescence in primary and immortalized human esophageal keratinocytes with p53 dysfunction. Oncogene 23, 6760–6768 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.