Abstract
De novo mutations are recognized both as an important source of genetic variation and as a prominent cause of sporadic disease in humans. Mutations identified as de novo are generally assumed to have occurred during gametogenesis and, consequently, to be present as germline events in an individual. Because Sanger sequencing does not provide the sensitivity to reliably distinguish somatic from germline mutations, the proportion of de novo mutations that occur somatically rather than in the germline remains largely unknown. To determine the contribution of post-zygotic events to de novo mutations, we analyzed a set of 107 de novo mutations in 50 parent-offspring trios. Using four different sequencing techniques, we found that 7 (6.5%) of these presumed germline de novo mutations were in fact present as mosaic mutations in the blood of the offspring and were therefore likely to have occurred post-zygotically. Furthermore, genome-wide analysis of “de novo” variants in the proband led to the identification of 4/4,081 variants that were also detectable in the blood of one of the parents, implying parental mosaicism as the origin of these variants. Thus, our results show that an important fraction of de novo mutations presumed to be germline in fact occurred either post-zygotically in the offspring or were inherited as a consequence of low-level mosaicism in one of the parents.
Introduction
In humans, DNA replication is estimated to entail one error in every 108 base pairs, giving rise to 30–100 genome-wide de novo mutations in each new generation.1–3 Whereas neutral or benign de novo point mutations contribute to normal genetic variation, single detrimental de novo mutations have been established to cause a number of rare developmental disorders4–6 and are increasingly recognized as a major contributor to common sporadic disorders, such as intellectual disability (ID) and autism.7,8 De novo mutations are thought to occur predominantly in the egg or sperm cell and thus result in an embryo with a constitutive mutation. However, de novo mutations can also appear post-zygotically, leading to embryonic mosaicism, a state in which two or more genetically distinct cell populations in an individual develop from a single fertilized egg.
Several reports have shown a high frequency of mosaicism for copy-number variations (CNVs) from cleavage-stage embryos9 to fully differentiated tissues.10–12 Similarly, there is increasing evidence of a high prevalence of mosaicism for single-nucleotide variants (SNVs) as a result of mutations appearing from early embryogenesis onward13,14 and throughout adult life.15,16 Currently, post-zygotic de novo mutations receive growing attention in developmental diseases.17–19 The timing of the event plays a key role in the clinical phenotype by determining not only the proportion of affected cells in the organism but also the type of tissues involved.18 Despite its pervasiveness, however, the true extent of mosaicism for SNVs remains unclear. This is largely a consequence of the technological limitations to accurately detecting these mutations; on one side, mutations with low levels of mosaicism are often below the threshold of sensitivity and specificity for automated and systematic detection of traditional sequencing methods,20 and on the other hand, mutations with a higher percentage of affected cells are easily detected by traditional sequencing methods, but it remains technically challenging to differentiate them from germline de novo mutations. Indeed, to discriminate post-zygotic from germline de novo mutations by sequencing DNA, it is crucial to distinguish biologically relevant allele imbalances from technical artifacts.
To gain insight into the frequency of post-zygotic events among de novo mutations, we performed a systematic evaluation of de novo mutations identified by trio-based whole-genome sequencing (WGS) of 50 individuals with severe ID and their parents. Previous analysis of WGS data from this cohort recently pointed to germline de novo mutations as the major cause of ID in the affected individuals.21 Additionally, these data indicated the presence of de novo mutations of somatic origin.21 By systematically assessing allelic ratios by various sequencing techniques, we show here that a proportion of previously reported de novo mutations did not occur during gametogenesis but, in fact, arose as post-zygotic events in the proband or were present as low-level somatic mutations in one of the parents.
Material and Methods
Defining a Set of De Novo Mutations from WGS of Parent-Proband Trios
This study was performed in accordance with the ethical standards of the Medical Ethics Committee of the Radboud University Medical Center. All participants or their legal representatives gave informed consent. WGS of 50 parent-proband trios and subsequent de novo mutation detection were performed as described previously.21 In brief, trio-based WGS was performed by Complete Genomics (CG) at 80-fold coverage. Sequence reads were mapped to the reference genome (UCSC Genome Browser hg19), and variants were called with CG software v.2.4. De novo mutations were called with CG’s cgatools calldiff program, which detects the differences between the genotypes of two samples and assigns a somatic score on the basis of sequencing quality and comparison of paired samples. Mutations whose scores comparing offspring to each parent were ≥5 were called as high-confidence de novo mutations (a total of 4,081 were detected in the 50 trios). The original report identified a set of 127 de novo mutations affecting either genome-wide coding sequence or specifically the non-coding sequence of known ID-associated genes.21 This set served as the starting point for the current study.
Sequencing Methods Used for Assessing the Post-zygotic State of De Novo Mutations
PCR amplicons for amplicon-based deep sequencing (ADS) and Sanger sequencing were generated according to standard PCR protocols. ADS was performed on an Ion Torrent Personal Genome Machine (Life Technologies) as described previously.21 In brief, raw sequencing reads were mapped to the reference genome with the Burrows-Wheeler Aligner (BWA), and the alignment files were then analyzed in the Integrative Genomics Viewer (IGV).22 For Sanger sequencing, PCR products were sequenced after enzymatic clean up.
Sequencing using single-molecule molecular inversion probes (smMIPs) was performed according to previously published protocols.23 In brief, smMIPs targeting the selected de novo mutation and a total of 112 bp of surrounding sequence were designed in house and ordered from Integrated DNA Technologies. The smMIPs were pooled and phosphorylated, after which the genomic regions of interest were captured with the probes and amplified. Sequencing was performed on the NextSeq 500 Desktop Sequencer (Illumina), and the reads were aligned with our in-house bioinformatics pipeline for analysis of molecular inversion probes. Through the use of molecular barcodes, we were able to remove PCR duplicates. Read counts for the positions of interest were extracted from the alignment files through IGV.
Assessment of the Allelic-Ratio Distribution of True Heterozygous Variants
To define the parameters of technical variation in WGS, ADS, and Sanger sequencing, we determined for each technology the allelic ratio of inherited SNVs as a proxy for true heterozygous mutations. The allelic ratio was defined as the proportion of variant reads from the total number of sequencing reads covering a given base pair and is expressed here as a percentage. We established the distribution of the allelic ratio for true heterozygous variants in WGS data by determining the allelic ratio of 115 inherited SNVs (coding, synonymous variants absent from dbSNP138 or present at a frequency below 1.5%) from WGS data of a single individual. To minimize the risk of false-positive variant calls, we used a second independent set of 109 inherited SNVs to determine the distribution of the allelic ratio in ADS and Sanger sequencing. This set was randomly selected from a larger set of 442 rare, coding variants inherited from either parent in ten probands, and variants were selected to have a coverage ≥ 20-fold in WGS and an allelic ratio between 40% and 60%. Variants on the X chromosome and/or located in established disease-associated genes were excluded. For ADS experiments, after mapping with the BWA, variants were visualized with the IGV, and allelic ratios were determined by assessment of the number of total reads and each respective base at this position. For Sanger sequencing, the chromatogram trace files were visualized with Vector NTI (Life Technologies), and intensities per dye per variant base were used for calculating the allelic ratio.
Identification of Post-zygotic Events in Probands
A set of 127 de novo mutations identified by WGS were re-sequenced by ADS and Sanger sequencing. For 107 (84%) of these variants, allelic ratios could be determined for all three sequencing techniques. We calculated the individual Z score per method for each mutation by using the values from sequencing heterozygous variants with each sequencing method as a reference. To calculate Z scores, we first obtained the difference between the allelic ratio and the mean allelic ratio and then divided that by the SD for heterozygous variants on that sequencing technique. Subsequently, we combined these scores into a single Z score for each de novo mutation by summing the individual Z scores and dividing this total by the square root of the number of scores. The critical value for statistical significance was established as 0.05 after Benjamini-Hochberg correction for multiple testing. To exclude amplification bias as the cause of a deviation in the allelic ratio, we re-sequenced de novo SNVs with a statistically significant combined Z score by ADS with a second independent primer pair. Finally, we used smMIPs as an independent technique to validate the presence of these variants as mosaic mutations (a set of seven heterozygous mutations served as a reference).
Identification of Parental Mosaicism in WGS Data
To detect low-level parental mosaicism for SNVs mimicking germline de novo mutations in the child, we re-analyzed the WGS data of the 50 parent-offspring trios. To this end, we used all 4,081 high-confidence candidate de novo mutations identified in the probands, because these have previously been shown to have a de novo validation rate of 78%.21 We then filtered for de novo variants for which at least two reads carrying the same mutation in the raw sequencing data were found in either one of the parents. We sequenced the position of interest by ADS in the DNA of the transmitting parent to validate parental mosaicism for the remaining 11 mutations. We established the position-specific sequencing error rate by sequencing the same position by ADS in the DNA of the non-transmitting parent in an independent sequencing run to avoid any contamination or barcode bleed-through. Then, the fraction of reads showing a non-reference allele at the corresponding base pair was calculated. The presence of the variant as a mosaic mutation in the transmitting parent was confirmed if the proportion of variant reads for the position and nucleotide of interest was significantly higher than the sequencing error established for that base-pair position from the non-transmitting parent.
Computational Modeling of Sequencing Coverage for the Identification of Mosaicism
To assess the ability of identifying mosaic variants from sequencing data, we simulated the effect of sequencing coverage on variant identification for different levels of mosaicism. To distinguish low-level mosaicism from sequencing artifacts, we assumed that automated variant-calling algorithms require the variant to be present in ≥5 sequencing reads and constitute ≥5% of the total number of reads at the position of interest. We used a binomial distribution to calculate the probability of reaching both these requirements for different depths of coverage and various levels of mosaicism. Assuming that a mosaic variant is identified, we also modeled the deviation of the allelic ratio from 50% (representing true heterozygosity), which is necessary for distinguishing a mosaic from a germline variant. Reads for heterozygous variants at different sequencing depths were simulated (n = 10,000) on the basis of a binomial distribution. We calculated the SD of this distribution and the level of mosaicism at which a mosaic variant could be reliably distinguished from a heterozygous variant for different thresholds of significance. Lastly, we determined the sequence coverage required for identifying low-level parental mosaicism. In this case, the position of interest was readily identified because the offspring presented with an apparently de novo mutation at this position. For this, we considered that at least two variant reads were sufficient for distinguishing the variant from a background sequencing error. Finally, we applied a binomial model for different sequencing depths and levels of mosaicism to calculate the probability of obtaining two variant reads in the sequencing data.
Results
Determining the Technical Variation for WGS, ADS, and Sanger Sequencing
In this study, we set out to distinguish mosaic mutations from true germline de novo mutations (Figure S1) by sequencing. To gain insight into the sensitivity of WGS, ADS, and Sanger sequencing, we re-sequenced two different sets of inherited germline mutations as a proxy for true heterozygosity (Figure 1A and Figure S2). We subsequently determined the distribution of the allelic ratios per technology (Table S1 and Figure S3). With an allelic ratio of 48.2 ± 4.4% (average ± SD), ADS showed to be the most precise technique for identifying true heterozygosity. In comparison, WGS showed an allelic ratio of 50.5 ± 8.9%, and Sanger sequencing had a ratio of 51.4 ± 8.7% (Table S2). On the basis of the obtained distributions for the allelic ratio, we determined that de novo mutations with an allelic ratio below 32.8% for WGS, 39.3% for ADS, and 33.9% for Sanger sequencing had a statistically significant deviation from the expected ratio for true heterozygous mutations and might, as such, reflect mosaic mutations.
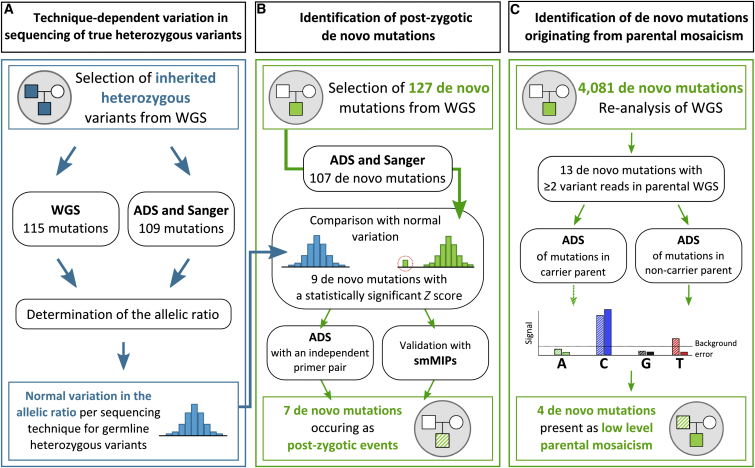
Figure 1.
Workflow for the Detection of Mosaic Mutations among a Subset of Apparently De Novo Mutations
(A) Assessment of technique-dependent variation in sequencing of two groups of heterozygous germline variants (in blue) for determining the distribution of allelic ratios for three different techniques (WGS, ADS, and Sanger sequencing).
(B) Previously identified de novo mutations were re-sequenced by ADS and Sanger sequencing for determining the variant ratio. With the use of the combined Z score, nine putative somatic variations were identified. They were then validated by ADS with a second independent primer pair and smMIPs. Seven of nine were confirmed to deviate in allelic ratio, suggesting a non-germline event.
(C) Identification of de novo mutations originating from parental mosaicism. Of 4,081 high-confidence de novo mutations identified by WGS, 13 were identified to have two or more variant reads in parental DNA. With the use of ADS data from the non-carrier parent for correcting for the background sequencing error, four mutations appearing as de novo in the child were identified as low-level mosaicism in one of the parents.
Identification of Post-zygotic De Novo Mutations in Probands
Our next objective was to determine the proportion of post-zygotic events among a subset of de novo mutations in our cohort. For this, we studied a pre-defined set of 107 de novo mutations by using WGS, ADS, and Sanger sequencing (Figure 1B).21 As we did for the inherited variants, we determined each mutation’s allelic ratio for each sequencing technique. After calculation of the mean allelic ratio across the three sequencing techniques, nine de novo mutations showed a statistically significant deviation from the expected ratio for true germline heterozygosity (Figures S4 and S5). To exclude technical artifacts resulting from biased allele amplification during PCR, which would thereby falsely suggest the presence of mosaicism, we generated a second independent amplicon with different PCR primers to re-sequence all nine mutations by ADS (Tables 1, S1, and S3). This analysis confirmed a statistically significant deviation in the allelic ratio for eight out of nine de novo mutations. Of note, three of these mutations had been previously reported as possible mosaic mutations.21
Table 1.
De Novo Mutations Occurring as Post-zygotic Events in Offspring
| Gene | OMIM Accession Number | Mutation at gDNA Level (hg19) | Location | Predicted Mutation at cDNA Level (GenBank Accession Number) | Predicted Protein Substitution | p Valuea | Average Allelic Ratio |
|---|---|---|---|---|---|---|---|
| KANSL2 | 615488 | chr12:49072911C>A | exon 4 | c.453G>T (NM_017822.3) | p.( = ) | 6.94E−21 | 20.8% |
| CREBL2 | 603476 | chr12:12788868G>C | exon 2 | c.173G>C (NM_001310.2) | p.Arg58Pro | 6.40E−19 | 21.0% |
| PIAS1 | 603566 | chr15:68468014T>A | exon 10 | c.1209T>A (NM_016166.1) | p.Asp403Glu | 1.84E−18 | 22.9% |
| PNKP | 605610 | chr19:50367525C>T | intron 5 | c.579−32G>A (NM_007254.3) | NA | 7.05E−17 | 22.7% |
| HIVEP2 | 143054 | chr6:143092683C>T | exon 5 | c.3193G>A (NM_006734.3) | p.Ala1065Thr | 2.20E−14 | 25.2% |
| DPYD | 274270 | chr1:97588236C>T | intron 21 | c.2623−24048G>A (NM_000110.3) | NA | 3.17E−10 | 29.7% |
| NEK1 | 604588 | chr4:170359295T>G | exon 27 | c.2703A>C (NM_001199397.1) | p.Lys901Asn | 3.67E−08 | 29.4% |
The following abbreviation is used: NA, not applicable.
p values were corrected by Benjamini-Hochberg for multiple testing. The level of the mutation was calculated as the average variant ratio for each mutant from all sequencing methods.
To validate these findings with an independent test, we set out to sequence the eight candidate mosaic mutations by using smMIPs for increased depth and accuracy. By sequencing germline mutations within the same assay, we first established for this technique the average and SD of the allelic ratio for true heterozygosity—this was shown to be 47.1 ± 3.3%. Unique smMIPs could be designed for all but one candidate mosaic event, located in an intron of SETBP1 (OMIM: 611060). The remaining seven mutations were tested and confirmed to be present as mosaic events with allelic ratios between 20.8% and 29.7%. Translating these allelic ratios into percentages of cells carrying the mutation predicted that the mutations must be present in 41.6%–59.4% of the cells in blood. Thus, our results indicate that at least 7/107 (6.5%) de novo mutations detected in our cohort did not occur in the germline of the parent but instead arose post-zygotically in the offspring.
Parental Mosaicism as a Source of Seemingly De Novo Mutations
Gonadal mosaicism in a healthy parent can lead to the transmission of disease-causing mutations and recurrence of disorders with seemingly de novo ocurrence.24 In some cases, mosaicism might not be restricted to the germ cells; it was recently shown that healthy individuals with gonadal mosaicism for disease-causing CNVs, revealed by recurrence of the disease in the offspring, carried low levels of mosaicism for this CNV in blood.25 Following this idea, we aimed to determine whether any of the seemingly germline de novo events in our cohort of 50 probands had actually occurred as somatic mutations in one of the parents (Figure 1C). For this, we re-analyzed all 4,081 high-confidence de novo mutations previously detected by WGS in the probands and selected those de novo mutations in which two or more variant reads could be detected in the raw sequence data in one of the respective parents. Thirteen such mutations were identified, but two could not be amplified by PCR and were excluded from further analysis. We performed ADS on the remaining 11 mutations to determine whether we could detect the variant in DNA from the carrier parent. After stringent correction for the background sequencing error, four of these mutations were confirmed to be present in the blood of one of the parents. These low-level parental mosaic mutations showed an average allelic ratio of 3.54% (range 0.22%—6.15%; Tables 2 and S4). Of note, these low-level parental mosaic mutations, of which three were transmitted by the father and one by the mother, were not detected in the parental DNA by Sanger sequencing (Figure S6).
Table 2.
De Novo Mutations Originating from Parental Mosaicism
| Genomic Location | Gene | OMIM Accession Number | Gene Location | Origin | Total Reads (ADS) | Variant Reads | p Valuea |
|---|---|---|---|---|---|---|---|
| chr13:78303535A>T | SLAIN1 | 610491 | intron | father | 31,470 | 6.15% | <0.001 |
| chr18:25210178C>T | – | – | intergenic | father | 34,149 | 2.56% | <0.001 |
| chr5:11327458C>T | CTNND2 | 604275 | intron | mother | 12,754 | 5.25% | <0.001 |
| chr5:147855052G>A | HTR4 | 602164 | intron | father | 20,927 | 0.22% | <0.05 |
p values were corrected for multiple testing by Bonferroni correction.
Modeling the Effect of Sequence Coverage on the Detection of Mosaic Mutations
Evidently, sufficient sequencing coverage is required for reliably identifying mosaic mutations. To investigate the impact of coverage on the detection of mosaic mutations, we modeled the probability of detecting both post-zygotic mutations in a proband and low-level parental mosaicism given different sequencing coverage.
The detection of post-zygotic de novo mutations requires two essential steps: calling the variant in the proband and identifying a significant deviation of the allelic distribution. Modeling under the assumption that ≥5 variant reads are required for variant calling and that these constitute ≥5% of the total number of sequence reads indicates that at least 100-fold coverage is required for calling 90% of mosaic variants with an allelic ratio equal to 10% or higher (Figure S7). Increased sequencing coverage decreases the SD in the allelic ratio, which reduces technical variation (Figure S8) and allows for better discrimination between true heterozygosity and mosaicism. Provided that a post-zygotic mutation is called, we also modeled the required deviation in the allelic ratio of a mosaic variant for it to be reliably distinguished from a heterozygous variant (Figure S9). Our model indicated that at least 100-fold coverage is required for distinguishing mosaic mutations with allelic ratios < 40% from germline mutations with 95% probability.
The analysis for parental mosaicism for de novo mutations identified in a proband requires a different approach; the identification of parental mosaicism for a seemingly de novo mutation in the offspring is guided by the presence of the variant in the proband. As a consequence, the only requirement for the identification of parental mosaicism is to distinguish the variant reads in the parent from the background sequencing error at the respective genomic location. Under the assumption that two variant reads in the parent are sufficient for this, we modeled the coverage required for identifying low-level parental mosaicism (Figure S10), which showed that at least 140-fold coverage is needed for detecting low-level mosaicism of ≥5% with ≥95% probability.
Discussion
The aim of our study was to investigate the presence of non-germline events among de novo mutations. Our results show that 6.5% (7/107) of a subset of de novo mutations were present as mosaic mutations in the blood of the proband, strongly suggestive of a post-zygotic origin. Extrapolating our results to published genome-wide de novo mutation rates3,21 suggests that each individual carries at least two to seven de novo mutations of post-zygotic origin. Additionally, from a group of 4,081 mutations presumed to be de novo in the offspring, we detected four mutations that were in fact inherited from one of the parents in whom the mutation was present as a low-level mosaic mutation. Although this represents only 0.1% of all high-quality de novo mutations, parental mosaicism for a seemingly de novo mutation in the offspring was observed in 4 out of 50 trios. On the basis of the stringent criteria that we used to validate variants as mosaics and our modeling data, we anticipate that our results are most likely an underestimation of the true number of mosaic mutations present in blood.
Our initial selection of potential mosaic variants was based on results obtained with relatively high-coverage (80-fold) WGS. We have shown that, for trio-based WGS, 80-fold sequencing coverage is sufficient for identifying post-zygotic events among de novo mutations. However, statistical modeling of the probability of detecting mosaicism given various sequencing depths showed that, with this coverage, there is only an 80% probability of obtaining sufficient reads for identifying mosaicism present in ≥10% of the alleles (corresponding to ≥20% of the cells studied; Figure S7). Similarly, with this coverage, we were only able to reliably distinguish somatic events with allelic ratios below 39% from germline mutations (Figure S9). This suggests that post-zygotic variants with allelic ratios at either extreme in the proband could have gone unidentified in our study. On the other hand, the probability of obtaining at least two sequence reads for identifying ≥5% parental mosaicism is only 78% with 80-fold sequencing coverage, suggesting that the identification of these mutations can also be optimized by higher-sequencing coverage (Figure S10). Indeed, the low-level parental mosaic variants identified in our study had a significantly higher sequencing depth in the carrier parent than did the other de novo or post-zygotic mutations studied (Figure S11). Our results and statistical modeling highlight the importance of high sequencing coverage in the design of trio-based WGS studies. Currently, most WGS studies are performed at 30-fold coverage.13,26,27 If we assume that sequence quality is comparable to that of our study, this entails that fewer than 20% of mosaic variants with an allelic ratio between 10% and 33% can be identified with 30-fold sequencing coverage. Additionally, at this sequencing coverage, only mosaic mutations with an allelic ratio below 35% can be reliably distinguished from true heterozygous variants. Furthermore, our modeling suggests that there is less than a 20% probability of identifying parental mosaicism with an allelic ratio of less than 5% with WGS at 30-fold coverage. Given these results, our findings underline the need for increased sequencing coverage in WGS for the accurate identification of mosaicism.
Despite the aforementioned limitations, we have shown that WGS is a powerful method for genome-wide discovery of mosaic events. In this study, we used three additional techniques to confirm mosaicism of SNVs. After identifying de novo mutations by WGS, we first evaluated their status as post-zygotic events by ADS and Sanger sequencing. A limitation of both of these techniques is that they might show an allelic imbalance as a result of biased amplification of one allele over the other.28 For the most part, significant deviations in the allelic ratio secondary to technical artifacts observed in Sanger sequencing and ADS were method-specific rather than reproducible PCR artifacts (Figure S12). We have attempted to remedy this problem by using smMIPs, which provide targeted high sequence coverage and the ability to identify individual captured molecules23 and thus prevent any allelic-ratio deviations resulting from PCR amplification bias.
The presence of parental gonosomal mosaicism as the cause of a sporadic disorder in a family places the subsequent offspring at higher risk for recurrence of the disease than when the mutation is caused by a germline de novo mutation.29 Considering this, the presence of parental mosaicism in 4 out of 50 individuals of our cohort stresses the importance of a thorough follow-up in families affected by a disorder due to a de novo mutation.30 Notably, the lower limit of detection by Sanger sequencing has been reported to be close to only 10%,25 whereas the highest level of parental mosaicism here detected was only 6.15% and could not be identified by Sanger sequencing (Figure S6). Because Sanger sequencing is commonly used in diagnostics, parental mosaicism below the threshold of detection of this method could account for recurrence of de novo disorders within families24,31 and explain unsolved pedigrees with an apparently recessive inheritance of disorders otherwise known to be dominant.32 Under these circumstances, high-coverage next-generation sequencing should be favored over Sanger sequencing for the detection of low-level parental mosaicism and might even be warranted as a standard follow-up test for each pathogenic de novo mutation. Related to this, the frequent detection of mosaic events might partially explain the occurrence of known dominant pathogenic mutations within large-scale variant databases of healthy individuals, such as the NHLBI Exome Sequencing Project Exome Variant Server. This point needs to be taken into account when these databases are used for clinical interpretation of possible pathogenic mutations. Also, previous studies have shown that certain mutations found as true heterozygous events in one tissue could be detected at low levels or be completely absent in another.33 Clearly, further studies of mosaic mutations and their impact on phenotypic variation require an in-depth analysis of different tissues.
In summary, our results show that a proportion of de novo mutations presumed to be germline actually either occurred post-zygotically in the offspring or were inherited from low-level mosaicism in one of the parents. This indicates that de novo mutations do not arise solely during gametogenesis but also as post-zygotic mutations, suggesting that our genomes might be much more dynamic than previously considered. As the contribution of de novo mutations to human disease becomes increasingly apparent, this conclusion might very well have clinical implications. Pathogenic variants in the mosaic state require particular attention as to their detection via sequencing methods. Furthermore, their influence on the risk of recurrence of a disease underlines the importance of identifying mosaicism to offer accurate genetic counseling in sporadic disorders caused by de novo mutations.
Acknowledgments
We thank Drs. Bregje van Bon, Marjolein Willemsen, Bert de Vries, Tjitske Kleefstra, and Han Brunner from the Department of Human Genetics of the Radboud University Medical Center for the inclusion of affected individuals. We also thank Richard Leach, Robert Klein, and Rick Tearle from Complete Genomics for whole-genome sequencing. This work was in part financially supported by grants from the Netherlands Organization for Scientific Research (916-14-043 to C.G., 916-12-095 to A.H., and SH-271-13 to C.G. and J.A.V.) and the European Research Council (ERC Starting Grant DENOVO 281964 to J.A.V.). R.A.-H. was supported by a Radboud University Medical Center grant.
Published: June 5, 2015
Footnotes
Supplemental Data include 12 figures and 4 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.05.008.
Web Resources
The URLs for data presented herein are as follows:
Integrated Genomics Viewer, http://www.broadinstitute.org/igv/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Nachman M.W.M., Crowell S.L. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conrad D.F., Keebler J.E.M., DePristo M.A., Lindsay S.J., Zhang Y., Casals F., Idaghdour Y., Hartl C.L., Torroja C., Garimella K.V., 1000 Genomes Project Variation in genome-wide mutation rates within and between human families. Nat. Genet. 2011;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoischen A., van Bon B.W.M., Gilissen C., Arts P., van Lier B., Steehouwer M., de Vries P., de Reuver R., Wieskamp N., Mortier G. De novo mutations of SETBP1 cause Schinzel-Giedion syndrome. Nat. Genet. 2010;42:483–485. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 5.Rivière J.-B., van Bon B.W.M., Hoischen A., Kholmanskikh S.S., O’Roak B.J., Gilissen C., Gijsen S., Sullivan C.T., Christian S.L., Abdul-Rahman O.A. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012;44:440–444. doi: 10.1038/ng.1091. S1–S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng S.B., Bigham A.W., Buckingham K.J., Hannibal M.C., McMillin M.J., Gildersleeve H.I., Beck A.E., Tabor H.K., Cooper G.M., Mefford H.C. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vissers L.E.L.M., de Ligt J., Gilissen C., Janssen I., Steehouwer M., de Vries P., van Lier B., Arts P., Wieskamp N., del Rosario M. A de novo paradigm for mental retardation. Nat. Genet. 2010;42:1109–1112. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 8.O’Roak B.J., Deriziotis P., Lee C., Vives L., Schwartz J.J., Girirajan S., Karakoc E., Mackenzie A.P., Ng S.B., Baker C. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 2011;43:585–589. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanneste E., Voet T., Le Caignec C., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 2009;15:577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 10.O’Huallachain M., Karczewski K.J., Weissman S.M., Urban A.E., Snyder M.P. Extensive genetic variation in somatic human tissues. Proc. Natl. Acad. Sci. USA. 2012;109:18018–18023. doi: 10.1073/pnas.1213736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McConnell M.J., Lindberg M.R., Brennand K.J., Piper J.C., Voet T., Cowing-Zitron C., Shumilina S., Lasken R.S., Vermeesch J.R., Hall I.M., Gage F.H. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abyzov A., Mariani J., Palejev D., Zhang Y., Haney M.S., Tomasini L., Ferrandino A.F., Rosenberg Belmaker L.A., Szekely A., Wilson M. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dal G.M., Ergüner B., Sağıroğlu M.S., Yüksel B., Onat O.E., Alkan C., Özçelik T. Early postzygotic mutations contribute to de novo variation in a healthy monozygotic twin pair. J. Med. Genet. 2014;51:455–459. doi: 10.1136/jmedgenet-2013-102197. [DOI] [PubMed] [Google Scholar]
- 14.Huang A.Y., Xu X., Ye A.Y., Wu Q., Yan L., Zhao B., Yang X., He Y., Wang S., Zhang Z. Postzygotic single-nucleotide mosaicisms in whole-genome sequences of clinically unremarkable individuals. Cell Res. 2014;24:1311–1327. doi: 10.1038/cr.2014.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie M., Lu C., Wang J., McLellan M.D., Johnson K.J., Wendl M.C., McMichael J.F., Schmidt H.K., Yellapantula V., Miller C.A. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindhurst M.J., Sapp J.C., Teer J.K., Johnston J.J., Finn E.M., Peters K., Turner J., Cannons J.L., Bick D., Blakemore L. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N. Engl. J. Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirley M.D., Tang H., Gallione C.J., Baugher J.D., Frelin L.P., Cohen B., North P.E., Marchuk D.A., Comi A.M., Pevsner J. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N. Engl. J. Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurek K.C., Luks V.L., Ayturk U.M., Alomari A.I., Fishman S.J., Spencer S.A., Mulliken J.B., Bowen M.E., Yamamoto G.L., Kozakewich H.P., Warman M.L. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am. J. Hum. Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohlin A., Wernersson J., Engwall Y., Wiklund L., Björk J., Nordling M. Parallel sequencing used in detection of mosaic mutations: comparison with four diagnostic DNA screening techniques. Hum. Mutat. 2009;30:1012–1020. doi: 10.1002/humu.20980. [DOI] [PubMed] [Google Scholar]
- 21.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W.M., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 22.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiatt J.B., Pritchard C.C., Salipante S.J., O’Roak B.J., Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Natacci F., Baffico M., Cavallari U., Bedeschi M.F., Mura I., Paffoni A., Setti P.L., Baldi M., Lalatta F. Germline mosaicism in achondroplasia detected in sperm DNA of the father of three affected sibs. Am. J. Med. Genet. A. 2008;146A:784–786. doi: 10.1002/ajmg.a.32228. [DOI] [PubMed] [Google Scholar]
- 25.Campbell I.M., Yuan B., Robberecht C., Pfundt R., Szafranski P., McEntagart M.E., Nagamani S.C.S., Erez A., Bartnik M., Wiśniowiecka-Kowalnik B. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am. J. Hum. Genet. 2014;95:173–182. doi: 10.1016/j.ajhg.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen B.S., Spehlmann M.E., Raedler A., Stade B., Thomsen I., Rabionet R., Rosenstiel P., Schreiber S., Franke A. Whole genome and exome sequencing of monozygotic twins discordant for Crohn’s disease. BMC Genomics. 2014;15:564. doi: 10.1186/1471-2164-15-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemirovsky S.I., Córdoba M., Zaiat J.J., Completa S.P., Vega P.A., González-Morón D., Medina N.M., Fabbro M., Romero S., Brun B. Whole genome sequencing reveals a de novo SHANK3 mutation in familial autism spectrum disorder. PLoS ONE. 2015;10:e0116358. doi: 10.1371/journal.pone.0116358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veal C.D., Freeman P.J., Jacobs K., Lancaster O., Jamain S., Leboyer M., Albanes D., Vaghela R.R., Gut I., Chanock S.J., Brookes A.J. A mechanistic basis for amplification differences between samples and between genome regions. BMC Genomics. 2012;13:455. doi: 10.1186/1471-2164-13-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell I.M., Stewart J.R., James R.A., Lupski J.R., Stankiewicz P., Olofsson P., Shaw C.A. Parent of origin, mosaicism, and recurrence risk: probabilistic modeling explains the broken symmetry of transmission genetics. Am. J. Hum. Genet. 2014;95:345–359. doi: 10.1016/j.ajhg.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faivre L., Williamson K.A., Faber V., Laurent N., Grimaldi M., Thauvin-Robinet C., Durand C., Mugneret F., Gouyon J.-B., Bron A. Recurrence of SOX2 anophthalmia syndrome with gonosomal mosaicism in a phenotypically normal mother. Am. J. Med. Genet. A. 2006;140:636–639. doi: 10.1002/ajmg.a.31114. [DOI] [PubMed] [Google Scholar]
- 31.Elalaoui S.C., Kraoua L., Liger C., Ratbi I., Cavé H., Sefiani A. Germinal mosaicism in Noonan syndrome: A family with two affected siblings of normal parents. Am. J. Med. Genet. A. 2010;152A:2850–2853. doi: 10.1002/ajmg.a.33685. [DOI] [PubMed] [Google Scholar]
- 32.Schinzel A., Giedion A. A syndrome of severe midface retraction, multiple skull anomalies, clubfeet, and cardiac and renal malformations in sibs. Am. J. Med. Genet. 1978;1:361–375. doi: 10.1002/ajmg.1320010402. [DOI] [PubMed] [Google Scholar]
- 33.Huisman S.A., Redeker E.J., Maas S.M., Mannens M.M., Hennekam R.C. High rate of mosaicism in individuals with Cornelia de Lange syndrome. J. Med. Genet. 2013;50:339–344. doi: 10.1136/jmedgenet-2012-101477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.