Abstract
Copy-number variants (CNVs) have been the predominant focus of genetic studies of structural variation, and chromosomal microarray (CMA) for genome-wide CNV detection is the recommended first-tier genetic diagnostic screen in neurodevelopmental disorders. We compared CNVs observed by CMA to the structural variation detected by whole-genome large-insert sequencing in 259 individuals diagnosed with autism spectrum disorder (ASD) from the Simons Simplex Collection. These analyses revealed a diverse landscape of complex duplications in the human genome. One remarkably common class of complex rearrangement, which we term dupINVdup, involves two closely located duplications (“paired duplications”) that flank the breakpoints of an inversion. This complex variant class is cryptic to CMA, but we observed it in 8.1% of all subjects. We also detected other paired-duplication signatures and duplication-mediated complex rearrangements in 15.8% of all ASD subjects. Breakpoint analysis showed that the predominant mechanism of formation of these complex duplication-associated variants was microhomology-mediated repair. On the basis of the striking prevalence of dupINVdups in this cohort, we explored the landscape of all inversion variation among the 235 highest-quality libraries and found abundant complexity among these variants: only 39.3% of inversions were canonical, or simple, inversions without additional rearrangement. Collectively, these findings indicate that dupINVdups, as well as other complex duplication-associated rearrangements, represent relatively common sources of genomic variation that is cryptic to population-based microarray and low-depth whole-genome sequencing. They also suggest that paired-duplication signatures detected by CMA warrant further scrutiny in genetic diagnostic testing given that they might mark complex rearrangements of potential clinical relevance.
Main Text
Structural variation (SV) is a major source of genomic diversity and a common cause of human disease. Most human-disease studies and population-based characterization of SVs to date have focused on copy-number variants (CNVs). This emphasis has been dictated both by technical limitations on delineating copy-neutral classes of SV and by the considerable and well-established risk conferred by large-dosage imbalances in many developmental and neuropsychiatric disorders.1–5 Microarray-based technologies are the conventional method for CNV detection, and chromosomal microarray (CMA) is currently the recommended first-tier diagnostic screen for developmental abnormalities of unknown etiology, as well as for genome-wide prenatal genetic diagnostic testing. These methods are capable of detecting DNA copy gains and losses but are blind to balanced genomic alterations.6,7
In a recent whole-genome sequencing (WGS) study of youth with early-onset neuropsychiatric disorders, we detected a spectrum of complex chromosomal rearrangements that involved an apparent duplication-associated mechanism.8 Recent cytogenetic studies have also described complex duplication-associated rearrangements and their plausible mechanisms.9,10 Although the impact of deletion on gene function is generally predictable, the consequence of duplication is less certain. Duplications can occur in tandem or can involve insertion of the duplicated copy into a distant locus, potentially with deleterious effects through altered dosage or gene disruption at the insertion site. These secondary consequences beyond a mere increase in DNA dosage are invisible to CMA. The above studies suggest that these complex rearrangements associated with duplications are more common than has been appreciated by population sequencing or clinical diagnostic evaluation.
We hypothesized that a fraction of duplications delineated by microarray are misclassified and mark complex SVs that are not detectable by analyses restricted to dosage imbalance. We performed long-insert, or “jumping library,”11 WGS for 259 subjects who had been diagnosed with autism spectrum disorder (ASD [MIM: 209850]) and had been previously screened for CNVs by CMA as part of the Simons Simplex Collection (SSC).12,13 All subjects provided informed consent to participate in the SSC, and this study was approved by the institutional review board of Partners HealthCare. Karyotype analysis for cytogenetically visible chromosome rearrangements has never been performed on the SSC. Jumping-library WGS enables high physical coverage of rearrangement breakpoints via mapped inserts between paired reads at a resolution proportional to the size of the insert.14 We have previously demonstrated that the method can be optimized to yield sufficient physical coverage of the genome to capture both balanced and unbalanced SV in basic research and in clinical diagnostic practice.8,14–17 We generated jumping libraries according to published protocols with a median insert size of 3.75 kb, and subjects were sequenced to an average of 94.1× haploid physical coverage.18 We performed analyses by integrating three algorithms that we customized for long-insert libraries: LUMPY,19 cn.MOPS,20 and our SV classifier.8 These computational methods detect both anomalous read pairs that cluster in proximity to SV breakpoints and genomic intervals with aberrant sequencing depth indicating copy gain or loss. Sanders and colleagues have previously described methods for all CMA analyses used here.13
WGS revealed that a surprisingly high proportion of individuals in this cohort harbored a complex genomic rearrangement associated with duplications. The most abundant class of complex variation was marked by a dosage signature of two closely located duplications (“paired duplications”) flanking a cryptic inversion (termed dupINVdup here; Figure 1). We observed at least one dupINVdup in 8.1% (21/259) of all sequenced ASD probands (Table 1). We recently described one example of a dupINVdup in a subject with an early-onset neuropsychiatric disorder featuring an inversion of 5.25 Mb, and similar variants have been confirmed from cytogenetic studies.8,9 The dupINVdup inversions in this cohort ranged in size from 39.6 kb to 4.3 Mb, whereas their corresponding flanking duplications ranged from 611 bp to 587 kb. Some dupINVdup variants, such as a dupINVdup with overlapping duplication breakpoints to form a triplication, contained additional complexity (Table S1).
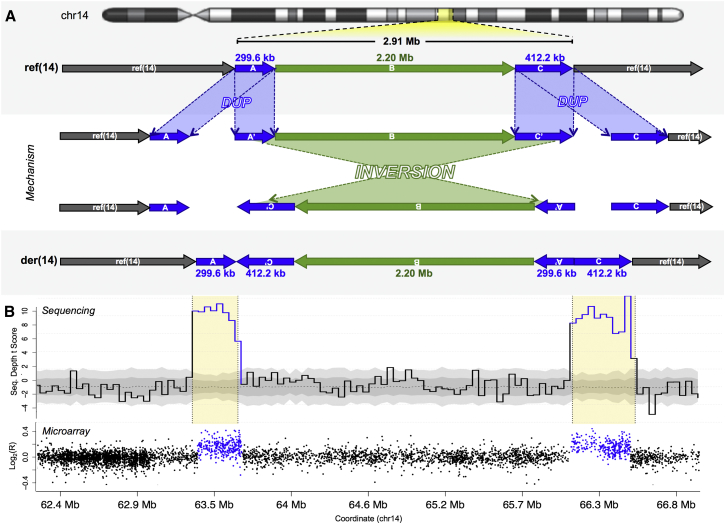
Figure 1.
Paired Duplications Mark Cryptic Inversions
(A) Duplication of two loci in proximity (segments A and C, in blue) flanks an inversion (segment B, in green) of the interval between the paired-duplication breakpoints. This example involves the rearrangement of 2.91 Mb of chromosome 14 (gray) in an ASD proband.
(B) WGS clearly delineated the two flanking duplications (top), which were confirmed by microarray (bottom). Sequencing depth (top) is represented by the binwise t-score of the scaled physical sequence depth from mapped inserts in this ASD proband when normalized against all other probands in the cohort (n = 259). Blue bins indicate a statistically significant sequencing-depth alteration that exceeds a Bonferroni-corrected threshold. Gray shading represents either one (dark gray) or two (light gray) binwise median absolute deviations across all probands. Flanking duplications as delineated by clustered read pairs are highlighted in yellow. Microarray intensities (bottom) are plotted as log2 marker-intensity ratios; all markers corresponding to the microarray duplication calls are shaded blue. All coordinates listed are based upon the GRCh37 reference genome build version 71 (UCSC Genome Browser).
Table 1.
Characteristics of Duplication-Associated Complex Rearrangements
| Class | Additional Complexity |
Variants |
Cohort Frequency (n = 259) |
Median Size (kb) |
|||
|---|---|---|---|---|---|---|---|
| Total | Private | Polymorphic | Duplication | Inversion | |||
| dupINVdup | no | 16 | 15 | 1 | 18 (6.9%) | 48.0 | 272.3 |
| yes | 3 | 3 | 0 | 3 (1.2%) | 36.5 | 169.5 | |
| dupDELdup | no | 7 | 7 | 0 | 7 (2.7%) | 58.1 | – |
| yes | 2 | 2 | 0 | 2 (0.8%) | 26.1 | 18.0 | |
| Other complex duplication inversions | – | 8 | 4 | 4 | 15 (5.7%) | 63.0 | 76.4 |
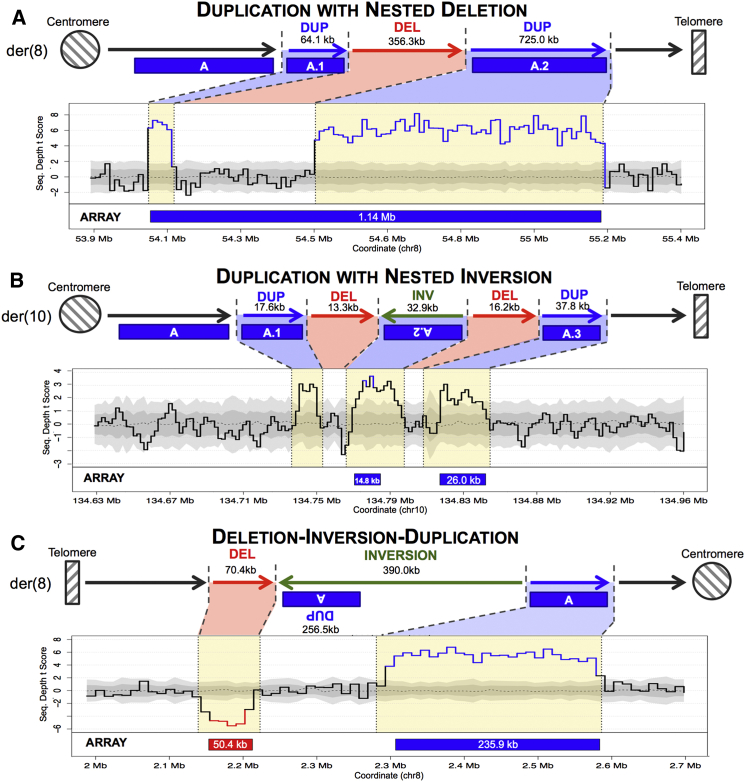
We discovered additional paired-duplication sequence signatures that marked other duplication-associated complex rearrangements in addition to dupINVdup variants. One such signature was a duplication with a nested deletion (dupDELdup; Figure 2A), which appeared as a paired duplication because an internal deletion in the duplicated region caused the deleted segment to remain at a normal diploid copy number. In two complex variants, an interstitial inversion occurred within a larger duplication, and this inversion was flanked by a deletion, thus presenting with a three-duplication signature (Figure 2B). Other complex duplication-associated variants included an inversion flanked by a 3′ duplication and a 5′ deletion (Figure 2C), three inverted tandem repeats,21 three inversions flanked by a single duplication, and a duplicated inversion with a nested deletion. Six subjects each harbored two complex duplication events, but all observed variants were rare in this population such that no variant exceeded a cohort frequency of 1.5% (4/259 individuals). Collectively, 15.8% (41/259) of all subjects in this cohort harbored at least one duplication-associated rearrangement with substantial complexity that was cryptic to CMA and other analyses restricted to dosage imbalance (Table 1).
Figure 2.
Sequencing Identifies a Spectrum of Complex Rearrangements Associated with Duplications Detected by Microarray
Sequencing revealed that 7.6% of rare (≤1% population frequency) duplications detected by microarray at 40-kb resolution or greater were associated with cryptic complex rearrangements. The majority of these complex duplications were paired-duplication inversions (dupINVdup) as described in Figure 1; however, we also observed a spectrum of complex duplication-mediated rearrangements, such as duplications with nested deletions misclassified by microarray as single large duplications (A), complex duplication inversions with internal deletions (B), or rare duplications flanking inversions with distal deletions (C). See Figure 1 for a description of sequencing-depth plots.
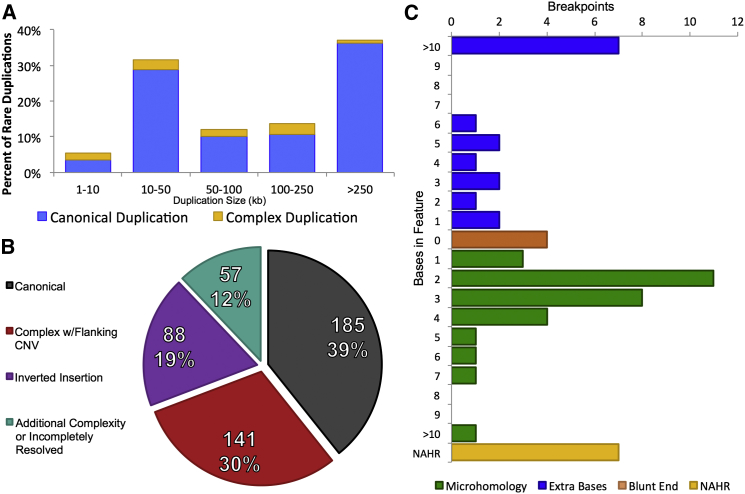
We evaluated the breakpoint sequences of these duplication-associated complex variants to provide insight into potential mechanisms of formation. Analysis of all breakpoints suggested predominantly microhomology-mediated mechanisms (Table 1; Figure 3). Among all resolved dupINVdup breakpoints (Table S1), 55.6% (20/36) exhibited 2–29 bp of microhomology at one or more breakpoints, suggesting microhomology-mediated mechanisms, such as break-induced replication, in the formation of the rearrangement.22,23 The breakpoints of five dupINVdup variants lay within pairs of repetitive genomic features with high sequence homology (>100 bp of >90% sequence homology, on average)—two pairs of Alu repeats, two pairs of LINE repeats, and one pair of tandem repeats—indicating non-allelic homologous recombination as a possible mechanism of formation for these rearrangements.24 Only three dupINVdup variants were more consistent with non-homologous end joining, although two of these breakpoints displayed a single base of microhomology. Analysis of a subset of 20 breakpoints from other complex duplication events demonstrated microhomology patterns similar to those observed for dupINVdup variants alone. In sum, we predicted microhomology-mediated repair to occur for 51.8% (29/56) of all breakpoints. We also observed that 28.6% (16/56) of all breakpoints included non-templated inserted sequences; this proportion is comparable to the previous finding that 24.8% of breakpoints were from large, cytogenetically detectable balanced chromosomal abnormalities.17
Figure 3.
Characteristics of Complex Duplication and Inversion Rearrangements
(A) Sizes of complex duplications (gold bars) are compared to those of all rare duplications identified in four or fewer individuals in this cohort (allele frequency ≤ 1.5%; blue bars).
(B) Characterization of all 471 inversion variants detected among the 235 highest-quality proband WGS libraries (>60× haploid physical coverage) revealed that only 185/471 (39.3%) of inversion variants were simple, or canonical, inversions.
(C) Microhomology-mediated breakpoint formation (green) was the predominant feature among all breakpoints of complex duplication-mediated rearrangements identified in this cohort. Notably, seven breakpoints also featured the insertion of non-templated sequence (blue) in excess of 10 bp.
Given the surprisingly high frequency of dupINVdup variants and other CNV-associated inversions in our analyses, we scrutinized all inversion breakpoints detected in the genomes of 235 probands with the highest-quality sequencing metrics (>60× physical coverage) to determine the fraction of genomic inversion variants that comprised simple, or canonical, inverted sequence without concomitant CNV or additional complexity. Remarkably, of all 471 independent inversion variants identified, only 39.3% (185/471) represented canonical inversions. The remainder of inverted segments could be classified into three broader categories: inversion with one or more flanking CNVs (141/471 [29.9%]), inverted insertion (88/471 [18.7%]), and more-complex rearrangements involving inverted sequence (57/471 [12.1%]) (Figure 3C). Notably, 88.1% (415/471) of all inversion variants were fully resolved at jumping-library resolution. The remaining variants had one incompletely resolved breakpoint, and these “single-end mapped” inversions might harbor additional complexity. We also observed an inverse correlation between inversion complexity and population frequency. Complex inversion with concomitant duplication occurred in just 3.8% (12/317) of polymorphic inversion variants (observed in more than one proband), whereas 20.1% (31/154) of private inversion variants (observed in only one individual) harbored one or more flanking duplications (chi-square test, p = 2.1 × 10−8). These findings indicate possible selective pressure against complex duplication-associated inversion rearrangements and warrant further study. Notably, inversion breakpoint complexity might be yet more prevalent for smaller genomic variants than detected here via large-insert libraries.
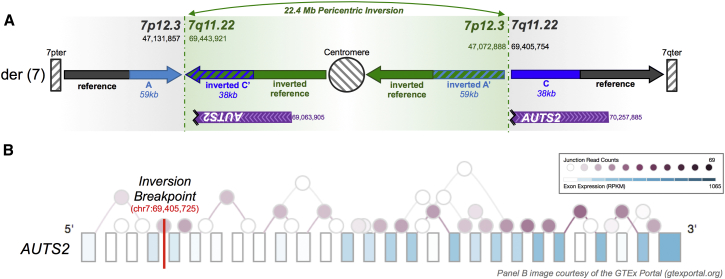
The functional impact and pathogenic potential of duplication-associated complex variants are likely to be variant specific and locus specific. A dupINVdup can alter gene dosage by producing extra copies of genes on the inverted segment or represent loss-of-function (LoF) variants in which the inversion directly disrupts a gene. We predict that the dupINVdup variants identified in this cohort duplicate 27 genes, disrupt the coding sequence of three genes, and produce one possible fusion gene (KCNH5 [MIM: 605716] and FUT8 [MIM: 602589]) (Table S1). We assessed inheritance for each dupINVdup event and found that only one (a dupINVdup on chr13) arose de novo, suggesting that most events were stably inherited from a parent (Table S1). In this cohort, we did not observe a paternal bias in the transmission of dupINVdup variants (63.6% maternal, 31.8% paternal, and 4.5% de novo). Unfortunately, WGS of all parents and unaffected siblings was outside of the scope of these analyses, and a large-scale healthy control population with deep coverage from large-insert WGS is not available to permit further interpretation of pathogenicity. Nonetheless, we predict that one dupINVdup duplicates AMBP (MIM: 178760), a gene recently implicated in ASD risk by analysis of de novo LoF variants from exome sequencing studies.25,26 Further, in an independent analysis of a proband presenting with a neuropsychiatric phenotype and dysmorphic features, we detected a dupINVdup variant that involved a 22.4-Mb pericentric inversion that directly disrupted AUTS2 (MIM: 607270), a known pathogenic locus in ASD (Figure 4).16,28–30 The 22.4-Mb inversion in this rearrangement was confirmed by karyotype. Collectively, these data suggest that a subset of dupINVdup and other duplication-associated complex rearrangements are likely to contribute to ASD etiology. However, most variants were inherited from an unaffected parent. Given the lack of a comparison population-based control cohort, we conservatively interpret the majority of these complex duplication-associated rearrangements as representing standing classes of genomic variation that has not been systematically captured by population-based CMA and short-read sequencing. Determining the overall contribution of these complex variants to human disease will require large-scale WGS studies.
Figure 4.
A Large Pericentric dupINVdup Directly Disrupts AUTS2 in a Proband with a Neuropsychiatric Phenotype
(A) A de novo dupINVdup involves a 22.4-Mb pericentric inversion that is flanked by 59- and 38-kb duplications (blue) and directly disrupts AUTS2, a known pathogenic locus in ASD. Karyotype analysis confirmed the presence of the pericentric inversion, corroborating the proposed structure of the dupINVdup variants detected herein. The karyotype interpretation was inv(7)(p13q11.23).
(B) Cortex expression data from the GTEx consortium portal27 (see Web Resources) are shown for the most common AUTS2 transcript. The breakpoint of this de novo disruptive dupINVdup is shown in red.
The paired duplications associated with the largest dupINVdup were detectable by CMA in the SSC, and the frequency of these complex SVs suggests that such previously uncharacterized rearrangements could have a meaningful impact on clinical diagnostic testing. Given that CMA is the recommended first-tier screen for many developmental disorders and unexplained congenital anomalies, we re-analyzed clinical diagnostic CMA data from 33,573 subjects, 19,556 (58%) of whom were referred for a neurodevelopmental phenotype.16 We surveyed rare (<1% frequency) paired duplications (i.e., two duplications within 5 Mb of each other on the same chromosome) that were detectable at the resolution of CMA, which averages 240 kb across the genomic backbone and 40 kb in some specifically targeted regions.16 Despite the fact that this resolution is lower than that of SNP microarrays and WGS, we discovered that at least 1.4% of affected subjects harbored a paired-duplication signature in these data. These analyses indicate that a subset of the complex variants associated with relatively large flanking duplications are detectable at CMA resolution and can confound genetic diagnostic evaluation when it is limited to interpretation of dosage imbalance alone.
In this brief report, we demonstrate the surprising abundance and considerable complexity of duplication-associated cryptic SVs. These results highlight the benefits of sequence-based technologies that achieve high physical coverage in capturing the spectrum of SV beyond dosage imbalances. They suggest that classes of complex SV such as those described here should be considered in both basic research and clinical diagnostic practice. Indeed, we found that 7.6% of all rare duplications (≤1% population frequency) detected by SNP microarray at 40-kb resolution are part of more-complex rearrangements that are invisible to all microarray technologies. Because CMA-based CNV evaluation is the currently recommended first-tier clinical screen for many developmental abnormalities and prenatal diagnosis,6,7,31 these data suggest that detecting such duplication signatures by CMA in genetic testing might warrant further scrutiny with higher-resolution approaches. These findings might be particularly relevant for prenatal and pediatric populations with developmental abnormalities of unknown etiology.
Acknowledgments
We are grateful to all of the families at the participating Simons Simplex Collection sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, and E. Wijsman). We also thank Matthew State, Bernie Devlin, and Kathryn Roeder for contributing to the generation and analysis of the microarray copy-number-variant data and Patricia Greipp for coordinating efforts at the Mayo Clinic. We thank Dr. Lisa Shaffer, formerly of Signature Genomics, and Dr. Yiping Shen of Boston Children’s Hospital for making the clinical diagnostic chromosomal-microarray data available to us. This work was supported by funding from the Simons Foundation for Autism Research (SFARI 238504), the Nancy Lurie Marks Family Foundation, the March of Dimes, the Charles Hood Foundation, the Brain & Behavioral Research Foundation, and the National Institute of Mental Health (R00MH095867 and GM061354) to M.E.T. H.B. is supported by a T32 fellowship from the National Institute of Child Health and Human Development (HD007396).
Published: June 18, 2015
Footnotes
Supplemental Data include one table and can be found online with this article at http://dx.doi.org/10.1016/j.ajhg.2015.05.012.
Web Resources
The URLs for data presented herein are as follows:
GTEx Portal, http://www.gtexportal.org/
OMIM, http://www.omim.org
UCSC Genome Browser, http://genome.ucsc.edu/
Supplemental Data
References
- 1.Sebat J., Lakshmi B., Malhotra D., Troge J., Lese-Martin C., Walsh T., Yamrom B., Yoon S., Krasnitz A., Kendall J. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glessner J.T., Wang K., Cai G., Korvatska O., Kim C.E., Wood S., Zhang H., Estes A., Brune C.W., Bradfield J.P. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girirajan S., Johnson R.L., Tassone F., Balciuniene J., Katiyar N., Fox K., Baker C., Srikanth A., Yeoh K.H., Khoo S.J. Global increases in both common and rare copy number load associated with autism. Hum. Mol. Genet. 2013;22:2870–2880. doi: 10.1093/hmg/ddt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinto D., Pagnamenta A.T., Klei L., Anney R., Merico D., Regan R., Conroy J., Magalhaes T.R., Correia C., Abrahams B.S. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills R.E., Walter K., Stewart C., Handsaker R.E., Chen K., Alkan C., Abyzov A., Yoon S.C., Ye K., Cheetham R.K., 1000 Genomes Project Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.American College of Obstetricians and Gynecologists Committee on Genetics Committee Opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet. Gynecol. 2013;122:1374–1377. doi: 10.1097/01.AOG.0000438962.16108.d1. [DOI] [PubMed] [Google Scholar]
- 8.Brand H., Pillalamarri V., Collins R.L., Eggert S., O’Dushlaine C., Braaten E.B., Stone M.R., Chambert K., Doty N.D., Hanscom C. Cryptic and complex chromosomal aberrations in early-onset neuropsychiatric disorders. Am. J. Hum. Genet. 2014;95:454–461. doi: 10.1016/j.ajhg.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drabova J., Trkova M., Hancarova M., Novotna D., Hejtmankova M., Havlovicova M., Sedlacek Z. A 15 Mb large paracentric chromosome 21 inversion identified in Czech population through a pair of flanking duplications. Mol. Cytogenet. 2014;7:51. doi: 10.1186/1755-8166-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newman S., Hermetz K.E., Weckselblatt B., Rudd M.K. Next-generation sequencing of duplication CNVs reveals that most are tandem and some create fusion genes at breakpoints. Am. J. Hum. Genet. 2015;96:208–220. doi: 10.1016/j.ajhg.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischbach G.D., Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–195. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talkowski M.E., Ernst C., Heilbut A., Chiang C., Hanscom C., Lindgren A., Kirby A., Liu S., Muddukrishna B., Ohsumi T.K. Next-generation sequencing strategies enable routine detection of balanced chromosome rearrangements for clinical diagnostics and genetic research. Am. J. Hum. Genet. 2011;88:469–481. doi: 10.1016/j.ajhg.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talkowski M.E., Ordulu Z., Pillalamarri V., Benson C.B., Blumenthal I., Connolly S., Hanscom C., Hussain N., Pereira S., Picker J. Clinical diagnosis by whole-genome sequencing of a prenatal sample. N. Engl. J. Med. 2012;367:2226–2232. doi: 10.1056/NEJMoa1208594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Talkowski M.E., Rosenfeld J.A., Blumenthal I., Pillalamarri V., Chiang C., Heilbut A., Ernst C., Hanscom C., Rossin E., Lindgren A.M. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiang C., Jacobsen J.C., Ernst C., Hanscom C., Heilbut A., Blumenthal I., Mills R.E., Kirby A., Lindgren A.M., Rudiger S.R. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 2012;44:390–397. doi: 10.1038/ng.2202. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanscom C., Talkowski M. Design of large-insert jumping libraries for structural variant detection using illumina sequencing. Curr. Protoc. Hum. Genet. 2014;80:1–9. doi: 10.1002/0471142905.hg0722s80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Layer R.M., Chiang C., Quinlan A.R., Hall I.M. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014;15:R84. doi: 10.1186/gb-2014-15-6-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klambauer G., Schwarzbauer K., Mayr A., Clevert D.A., Mitterecker A., Bodenhofer U., Hochreiter S. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroth G.P., Ho P.S. Occurrence of potential cruciform and H-DNA forming sequences in genomic DNA. Nucleic Acids Res. 1995;23:1977–1983. doi: 10.1093/nar/23.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastings P.J., Ira G., Lupski J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu W., Zhang F., Lupski J.R. Mechanisms for human genomic rearrangements. PathoGenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium G.T., GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bakkaloglu B., O’Roak B.J., Louvi A., Gupta A.R., Abelson J.F., Morgan T.M., Chawarska K., Klin A., Ercan-Sencicek A.G., Stillman A.A. Molecular cytogenetic analysis and resequencing of contactin associated protein-like 2 in autism spectrum disorders. Am. J. Hum. Genet. 2008;82:165–173. doi: 10.1016/j.ajhg.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sultana R., Yu C.E., Yu J., Munson J., Chen D., Hua W., Estes A., Cortes F., de la Barra F., Yu D. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics. 2002;80:129–134. doi: 10.1006/geno.2002.6810. [DOI] [PubMed] [Google Scholar]
- 30.Beunders G., Voorhoeve E., Golzio C., Pardo L.M., Rosenfeld J.A., Talkowski M.E., Simonic I., Lionel A.C., Vergult S., Pyatt R.E. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 2013;92:210–220. doi: 10.1016/j.ajhg.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wapner R.J., Martin C.L., Levy B., Ballif B.C., Eng C.M., Zachary J.M., Savage M., Platt L.D., Saltzman D., Grobman W.A. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012;367:2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.