ABSTRACT
eIF4E plays a conserved role in initiating protein synthesis, but with multiple eIF4E isoforms present in many organisms, these proteins also adopt specialized functions. Previous RNAi studies showed that ife-3, encoding the sole canonical eIF4E isoform of Caenorhabditis elegans, is essential for viability. Using ife-3 gene mutations, we show here that it is maternal ife-3 function that is essential for embryogenesis, but ife-3 null progeny of heterozygous animals are viable. We find that zygotic ife-3 function promotes body growth and regulates germline development in hermaphrodite worms. Specifically, the normal transition from spermatogenesis to oogenesis in the hermaphrodite germline fails in ife-3 mutants. This failure to switch is reversed by inhibiting expression of the key masculinizing gene, fem-3, suggesting ife-3 resembles a growing number of genes that promote the sperm/oocyte switch by acting genetically as upstream inhibitors of fem-3.
KEY WORDS: C. elegans, EIF4E, Germline, Sex-determination, Spermatogenesis, Oogenesis
INTRODUCTION
Eukaryotic initiation factor 4-complex (eIF4) recruits mature mRNAs to ribosomes as the first step of translation (Gingras et al., 1999). The factor eIF4E recognizes a methylated guanosine cap at the mRNA 5′ end, aiding recruitment of the complex to the mRNA. Multiple eIF4E homologs are common among organisms (Hernández and Vazquez-Pianzola, 2005), allowing them to adopt specialized functions. The nematode Caenorhabditis elegans encodes five eIF4E proteins (IFE-1 to -5) that reflect a diversity partially conserved across the animal kingdom: IFE-3 resembles the canonical eIF4E-1 isoforms of mammals and insects; IFE-4 is a member of the divergent 4E-HP group of eIF4E proteins; and IFE-1, -2, and -5 are closely related isoforms that make a nematode-specific sub-group (Hernández and Vazquez-Pianzola, 2005; Jankowska-Anyszka et al., 1998; Keiper et al., 2000).
Worm eIF4E homologs vary in expression pattern and the effects of their loss. IFE-2 is enriched in the soma, but also functions in the germline. Its loss inhibits general somatic mRNA translation, as well as temperature-dependent translation of germline mRNAs required for meiotic crossover repair (Hansen et al., 2007; Song et al., 2010; Syntichaki et al., 2007). IFE-4 is expressed somatically, and its absence reduces neuronal and egg-laying gene expression, resulting in impaired egg laying (Dinkova et al., 2005). IFE-1, -3, and -5 are germline-enriched (Amiri et al., 2001). No function is known for IFE-5, but IFE-1 loss partially impairs oogenesis, and disrupts spermatogenesis at high temperatures (Amiri et al., 2001; Henderson et al., 2009; Kawasaki et al., 2011). RNA-mediated inhibition (RNAi) studies show IFE-3 is essential for embryogenesis (Keiper et al., 2000). Using ife-3 gene mutations, we report here additional novel roles for IFE-3 in postembryonic development, particularly in promoting the transition of the hermaphrodite germline from a spermatogenic to an oogenic tissue.
RESULTS
Zygotic ife-3 is not essential for viability, but is important for normal body size
The wild-type C. elegans hermaphrodite, being able to produce both sperm and oocytes, is self-fertile. In an analysis of worms mutated for formin family genes, we had reported that a deletion allele tm2133 of the formin gene daam-1 is linked to recessive hermaphrodite sterility (Mi-Mi et al., 2012). However, daam-1(+) transgenes do not restore fertility to homozygous daam-1(tm2133) hermaphrodites, and RNAi against daam-1 does not induce sterility in wild-type hermaphrodites, suggesting an unidentified linked mutation as the cause (R.S.M., unpublished observations; King et al., 2009). To identify such a mutation, we stably balanced daam-1(tm2133) against the genomic transposition nT1[qIs51] in the heterozygous strain XA8002, and sequenced the genome of this strain. No identified point mutations or small deletions in XA8002 are likely to cause sterility (supplementary material Table S1), but over several regions near daam-1, the sequence coverage was decreased approximately 50%, suggesting these regions are deleted from one copy of Chromosome V (ChrV) in XA8002 (Fig. 1A, supplementary material Table S2). The largest putative deletion spans eleven genes (Table 1). Using single-worm PCR, we confirmed that this large deletion, designated here as upsDf41, is present on the daam-1(tm2133)-containing ChrV homolog in XA8002 (Fig. 1B).
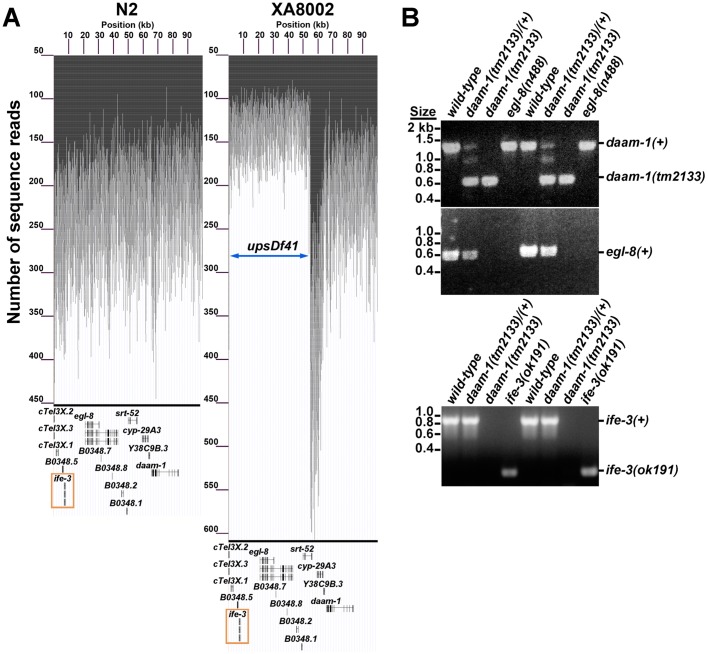
Fig. 1.
A large genomic deletion, upsDf41, is linked to daam-1(tm2133) and eliminates ife-3. (A) The locations of sequence reads obtained by whole genome sequence analysis were plotted against their position on the first 100,000 bp of ChrV for the wild-type strain N2, and the heterozygous daam-1(tm2133)/nT1 strain XA8002. The number of sequence reads for each position over the first 100 kb of ChrV for N2 (left) vary around a relatively constant average of ∼200–250 reads per position, excluding the telomeric region. The number of sequence reads per position for XA8002 also vary around ∼200–250 reads per position for ChrV positions greater than +70,000, but over the first ∼50 kb of ChrV, the average number of reads per position is ∼100–150, marking the genomic deficiency upsDf41. The average number of reads for the neighboring 8 kb is at least 50% greater, indicating the presence of additional copies of that sequence in XA8002. The exon positions of known genes are displayed beneath, including ife-3 (boxed). (B) PCR of single worms using primers for daam-1, ife-3, and egl-8 shows that the 700 bp deletion tm2133 of daam-1 is linked to an absence of ife-3 and egl-8. PCR of control animals bearing the 686 bp deletion (ok191) of ife-3, or the allele (n488) lacking one priming site for egl-8, demonstrate the specificity of primers used.
Table 1.
Genes affected by upsDf41
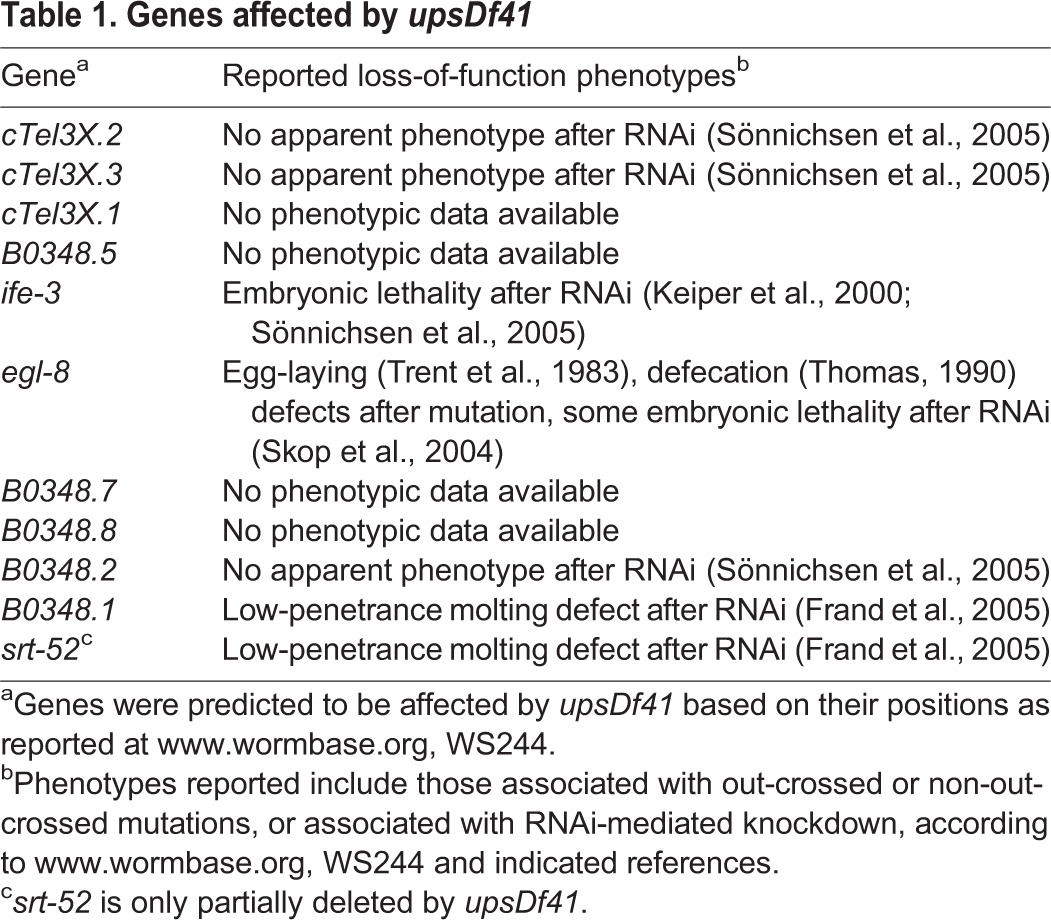
Among the genes disrupted by upsDf41, only ife-3 had been reported to be essential, with RNAi against ife-3 resulting in 100% embryonic lethality (Keiper et al., 2000). However, we were able to isolate upsDf41 homozygous worms that completely lacked ife-3 (Fig. 1B). We also quantitatively tested for association between absence of ife-3 and embryonic lethality. To avoid the embryonic lethality associated with the genomic transposition nT1[qIs51] in XA8002, we first crossed upsDf41 into a wild-type background. We then isolated individual upsDf41/+ heterozygous hermaphrodites and wild-type positive control hermaphrodites, and allowed them to lay eggs, and tracked the fate of their progeny. For worms of both genotypes, nearly 100% of their eggs hatched, and nearly 100% of the resultant larvae grew to adulthood (Table 2). Thus, absence of ife-3 from the zygotic genome does not result in lethality under standard growth conditions.
Table 2.
Zygotic ife-3 is not required for viability
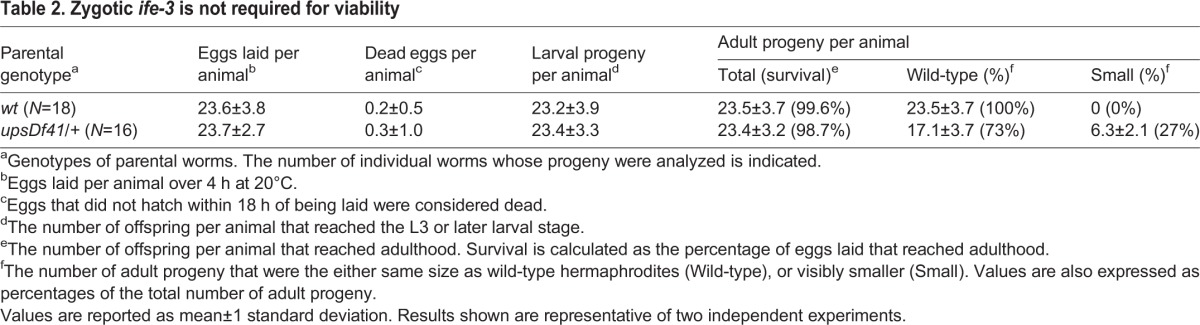
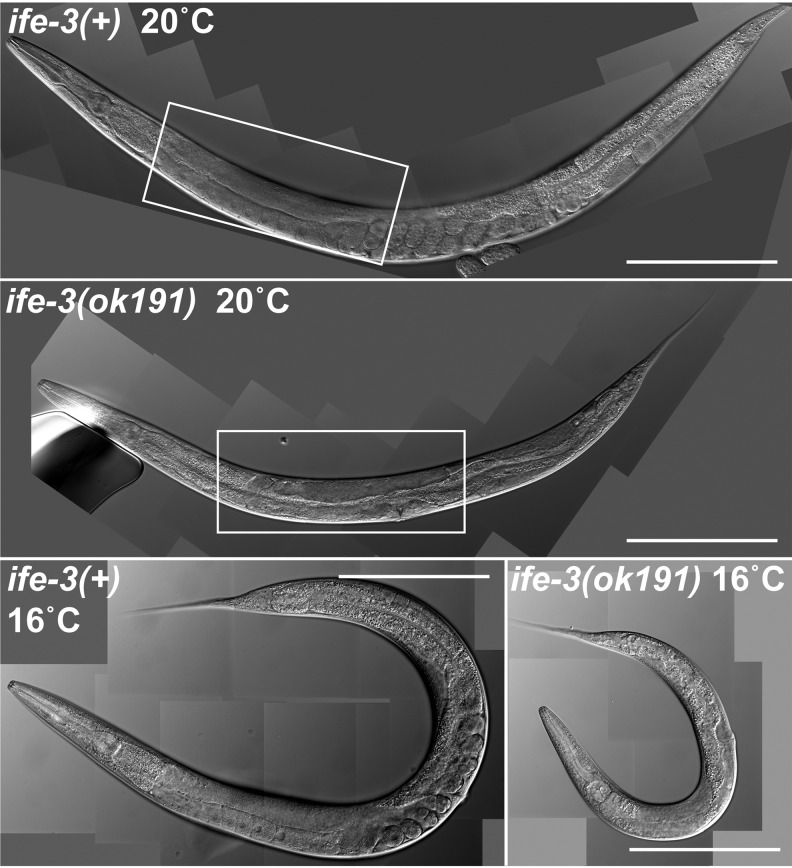
However, while the adult progeny of wild-type animals appeared wild-type, approximately 27% of the adult progeny of the upsDf41/+ worms were small, suggesting homozygosity of upsDf41 or daam-1(tm2133) results in poor growth (Table 2). Confirming this, heterozygous XA8002 worms have a normal body size but their upsDf41 daam-1(tm2133) homozygous progeny are small (supplementary material Fig. S1A). To test whether absence of ife-3 contributes to the small size of upsDf41 homozygotes, we obtained from the Caenorhabditis Genetics Center (University of Minnesota) the worm strain KX10, which is heterozygous for the smaller deletion ife-3(ok191) affecting only the immediate upstream sequence and exon 1 of ife-3 (Wormbase). For ease of analysis, we stably balanced ife-3(ok191) with nT1[qIs51] in the strain DWP70. As nT1[qIs51] encodes a recessive lethal allele and a pharyngeal-expressed GFP, we could unambiguously distinguish GFP-expressing ife-3(ok191)/+ heterozygous progeny from GFP-lacking ife-3(ok191) homozygous progeny. Similar to upsDf41, homozygotes for ife-3(ok191) are smaller than wild-type or heterozygous animals (Fig. 2). This effect is exaggerated at a cooler growth temperature (Fig. 2). Thus, absence of zygotic ife-3 is not lethal, but results in poor growth exacerbated by cold.
Fig. 2.
Growth defect of ife-3(ok191) mutants. Wild-type and ife-3(ok191) hermaphrodites were grown to young adulthood at the indicated temperatures. Viewed through DIC microscopy, mutant hermaphrodites are smaller than wild-type animals, particularly at a cooler growth temperature. The boxed regions are reproduced at higher magnification in Fig. 3A. Bars=200 µm.
Maternal effect lethality of ife-3 mutants
We observed that hermaphrodites homozygous for ife-3(ok191) or for upsDf41 (isolated as GFP-negative progeny of DWP70 or XA8002, respectively) produce eggs only under particular conditions (see below), but these eggs always terminate development as a mass of cells with no obvious morphogenesis (Table 3 and data not shown). These results indicate that while zygotically-encoded ife-3 is not essential for embryogenesis, a maternal supply of ife-3 is required for this process. Notably, this embryonic lethality phenotype matches the effects of RNAi against ife-3 (Keiper et al., 2000; Sönnichsen et al., 2005).
Table 3.
Zygotic ife-3 is required for egg formation at 20°C, and maternal ife-3 is required for embryogenesis
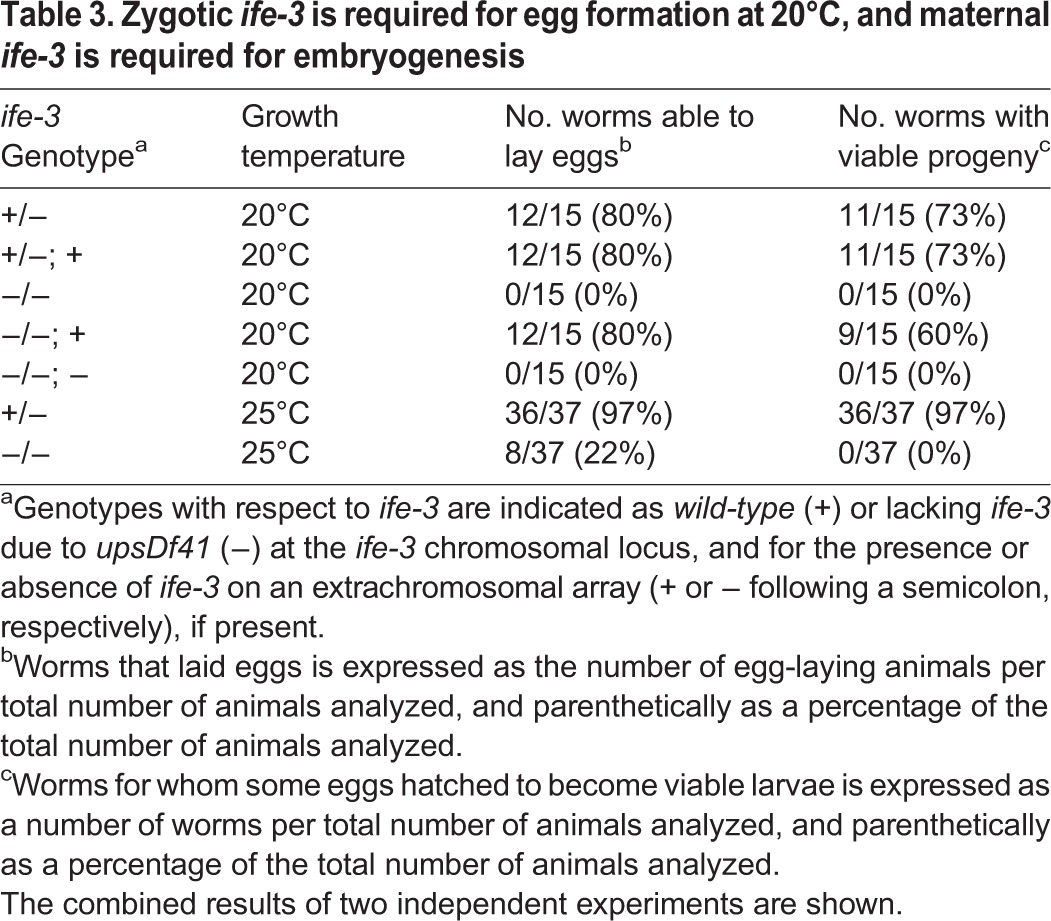
To test whether the maternal effect lethality associated with upsDf41 depends on ife-3, we microinjected XA8002 worms with wild-type ife-3 sequence to create the complex extrachromosomal array upsEx40[ife-3(+)]. Using co-injected mCherry-expressing markers to follow the inheritance of upsEx40[ife-3(+)] among the progeny of injected animals, we found that upsDf41 homozygous progeny that inherit upsEx40[ife-3(+)] are fertile, laying eggs that hatch as larvae and grow to adulthood (Table 3). Conversely, upsDf41 hermaphrodites that inherit a control array lacking ife-3 remain sterile. Thus, maternal ife-3, but not daam-1 or any other gene affected by upsDf41 (Table 1), is required for embryogenesis.
Masculinization of the ife-3 mutant hermaphrodite germline
While ife-3(ok191) or upsDf41 homozygous hermaphrodites produce inviable eggs under certain circumstances (described below), under normal growth conditions these hermaphrodites do not produce eggs (Table 3). Egg production in the C. elegans hermaphrodite begins with the proliferation of germ stem cells in the distal tips of two gonad arms. As these germ cells move in the proximal direction through the arms, they progressively differentiate. During late larval development, germ cells differentiate into spermatids that accumulate in the proximal gonad arms. In early adulthood, spermatogenesis stops and maturing germ cells instead differentiate into oocytes. These much larger cells accumulate in the proximal gonad arms, and push the smaller spermatids into the adjoining spermathecae, where they fully differentiate into sperm (Ward and Carrel, 1979). Throughout the fertile adulthood of hermaphrodites, oocytes are engulfed by the spermatheca during ovulation, where they are fertilized by resident sperm and then pushed into the uterus to commence development as embryos. Fertilized embryos rapidly synthesize eggshells, and after a brief period are laid through the vulva to continue development ex utero.
In young adult wild-type hermaphrodites, developing embryos are easily viewed in the uterus using DIC microscopy, and oocytes are visible as large cells lined up in the proximal gonad (Fig. 3A). In contrast, young adult ife-3(ok191) or upsDf41 homozygous hermaphrodites grown at 16°C or 20°C lack visible oocytes or embryos (Figs 2 and 3A, supplementary material Fig. S1B). Sperm and spermatids can be viewed using DAPI stain of DNA, which reveals their punctate nuclei, or immunofluorescence stain of major sperm protein (MSP), a major cytosolic component of sperm (Kosinski et al., 2005). In young adult wild-type hermaphrodites, sperm are present in the spermatheca (Figs 3B and 4A). In ife-3(ok191) or upsDf41 homozygous hermaphrodites grown at 20°C or lower, sperm/spermatids are also present, but in the proximal gonad rather than the spermatheca, likely as a secondary consequence of the absence of oocytes to push them to their proper place (Figs 3B and 4A, supplementary material Fig. S1B).
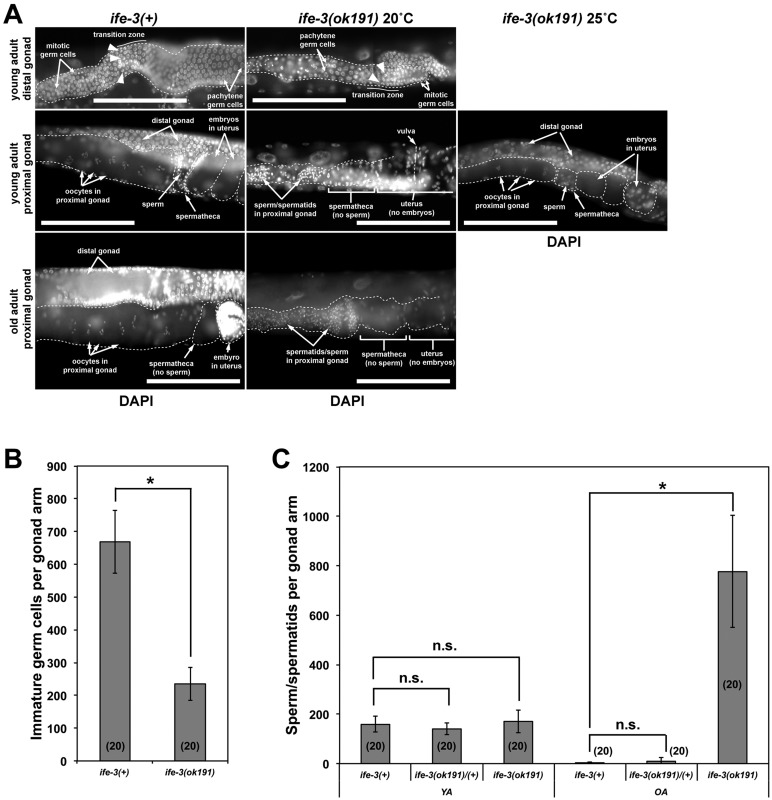
Fig. 3.
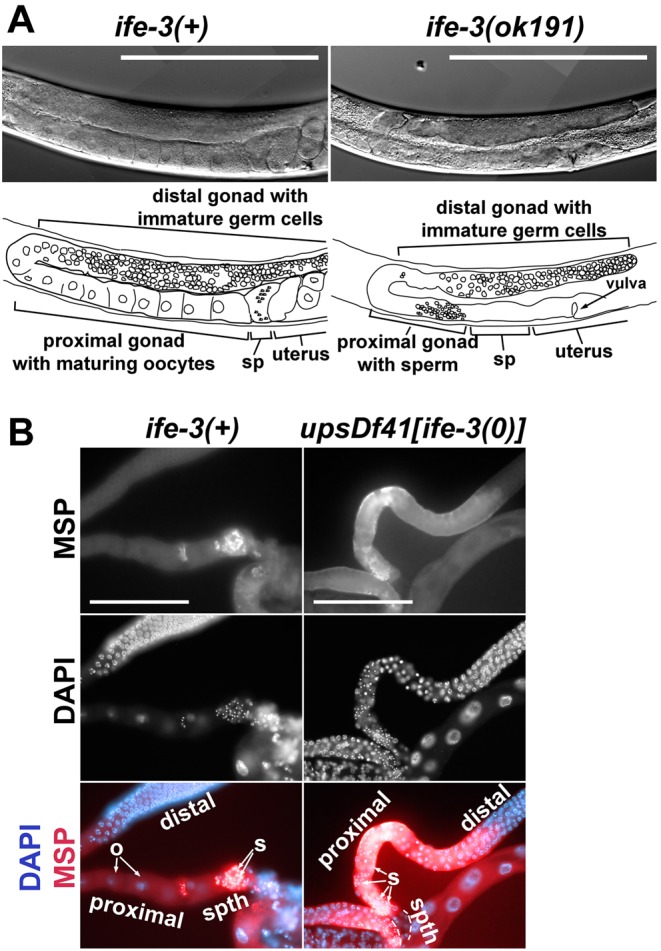
The gonads of ife-3 mutant hermaphrodites grown at 20°C lack oocytes and embryos, but contain sperm. (A) Gonad arms of wild-type and ife-3(ok191) hermaphrodites grown to young adulthood at 20°C were viewed through DIC microscopy. In the wild-type gonad, immature germ cells occupy the distal region of a gonad arm, while large oocytes are lined up in the proximal arm, tiny indistinct-appearing sperm occupy the spermatheca (sp), and embryos occupy the uterus. In the ife-3(ok191) gonad, immature germ cells occupy the distal arm, but oocytes are absent from the proximal gonad, and the spermatheca and uterus appear empty. Bars=200 µm. (B) Dissected gonad arms of young adult wild-type or upsDf41 homozygous hermaphrodites (which lack ife-3) were stained with DAPI to reveal germline nuclei, and with antibodies specific to MSP, a cytosolic component of sperm and spermatids. In the wild-type gonad arm, large oocytes (o) lacking MSP occupy the proximal region, while small MSP-rich sperm (s) with punctate nuclei are visible in the spermatheca (spth). In the upsDf41 gonad arm, MSP is enriched in small spermatids (s) with punctate nuclei in the proximal region, as well as more diffusely in the gonad, consistent with ongoing sperm production, while the spermatheca is empty of germ cells. In both strains, nuclei of immature germ cells are visible in the distal portion of the gonad arms. Bars=100 µm.
Fig. 4.
Spermatogenesis continues through adulthood in ife-3(ok191) hermaphrodites. (A) Wild-type and ife-3(ok191) hermaphrodites were stained with DAPI. (Top) Distal gonad arms. In both trains, spherical mitotic germ cell nuclei are near the distal tip, crescent shaped nuclei (arrowheads) of germ cells entering meiosis are present in a neighboring transition zone, and germ cells arrested in pachytene with nuclei containing thread-like chromatin strands occupy the remainder. (Middle) Proximal gonad region of young adults. In the wild-type hermaphrodite, large oocytes with nuclei containing six condensed chromatin bodies are lined up in the proximal arm, while punctate sperm nuclei occupy the spermatheca. In an ife-3(ok191) hermaphrodite grown at 20°C, the proximal gonad lacks oocytes but contains sperm/spermatid with punctate nuclei, while the spermatheca and uterus are empty of germ cells. In an ife-3(ok191) hermaphrodite grown at 25°C, the proximal gonad resembles that of a wild-type hermaphrodite, with oocytes lined up in the proximal arm, sperm present in the spermatheca, and developing embryos in the uterus. (Bottom) Proximal gonad region of old adults. In the wild-type hermaphrodite, oocytes are lined up in the proximal arm, while sperm have been depleted from the spermatheca. In an ife-3(ok191) hermaphrodite grown at 20°C, the proximal gonad contains many spermatids, while the spermatheca and uterus remain empty. Bars=100 µm. (B) The number of immature germ cell nuclei, which includes stem cells, meiotic cells in the transition zone, and pachytene-arrested cells, were counted in the gonad arms of DAPI-stained young adult wild-type and ife-3(ok191) homozygous hermaphrodites. The mutant gonad arms contain fewer immature germ cells. Results shown are typical of two independent experiments. (C) The number of sperm/spermatids, identified as punctate DAPI-stained nuclei, declines from young adulthood (YA) to old adulthood (OA) in wild-type and heterozygous hermaphrodites, but increases in ife-3(ok191) homozygotes. Results shown are typical of three independent experiments. For B and C, the number of gonad arms analyzed per experiment is indicated parenthetically, and values are expressed as mean±one standard deviation. * indicates P<0.0001. n.s. indicates no significant difference.
DAPI stain also shows that the distal portions of ife-3(ok191) and upsDf41 hermaphrodite gonad arms are small and contain fewer germ cells than in wild-type animals (Fig. 4A,B). However, they contain recognizable germ stem cells in the distal tips, immature germ cells entering meiosis in the transition zone, and germ cells arrested in pachytene, as typical for wild-type animals (Fig. 4A, supplementary material Fig. S1B). Moreover, young adult ife-3(ok191) or upsDf41 homozygous hermaphrodite gonads house a similar number of sperm as young adult wild-type hermaphrodites (Fig. 4C, supplementary material Fig. S1C). However, where wild-type hermaphrodites deplete their sperm over subsequent days through self-fertilization of oocytes, the homozygous mutant hermaphrodites continue to accumulate sperm through adulthood (Fig. 4C, supplementary material Fig. S1C). The number of sperm accumulated in these mutants is much higher than the maximal ∼160 sperm per arm created in wild-type hermaphrodites (Hodgkin, 1988), indicating that germ cells that would normally become oocytes instead differentiate into sperm.
The persistence of spermatogenesis and absence of oogenesis in the ife-3 mutant germline does not reflect a global larval developmental arrest of the mutants, as ife-3(ok191) or upsDf41 homozygous hermaphrodites complete the final cuticle molt that marks adulthood, and their vulvas open normally at the same time as their heterozygous siblings (data not shown). Also, while their germline is masculinized, the soma of ife-3 mutants remains that of hermaphrodites, with whip-shaped tails rather than fan-shaped tails like male worms, and with two-armed gonads rather than one-armed gonads like male worms (Fig. 2). Thus, ife-3 mutants grown at 16°C or 20°C have a masculinization of germline (mog) phenotype.
At the relatively high temperature of 25°C, the mog phenotype is only partially penetrant, with approximately one-fifth of ife-3(ok191) or upsDf41 homozygous hermaphrodites producing oocytes that become fertilized and commence development (Table 3, Fig. 4A). However, as described above, the resultant eggs do not hatch due to maternal effect lethality during embryonic morphogenesis (Table 3). We also rarely observed homozygous mutant hermaphrodites producing oocytes and laying inviable eggs at 20°C, but only when all food had been consumed, suggesting starvation might also bypass the requirement for ife-3 in the spermatogenesis-to-oogenesis switch. For either permissive condition (25°C or starvation), we only observed egg production when the condition was introduced before the final L4 larval stage, when the spermatogenesis-to-oogenesis transition normally occurs, suggesting the mog phenotype is not reversible once established.
ife-3 functions upstream of the masculinizing gene fem-3
A key point of control of the spermatogenesis-to-oogenesis transition is regulation of the pro-spermatogenic gene fem-3 (Ellis and Schedl, 2007). Excess fem-3 activity results in a mog phenotype similar to ife-3 mutants, while absence of fem-3 produces “female” animals that fail to produce sperm, but accumulate large numbers of unfertilized oocytes in their proximal gonad (Hodgkin, 1986; Ahringer and Kimble, 1991; Barton et al., 1987). Many genes that promote the spermatogenesis-to-oogenesis transition act as upstream inhibitors of fem-3 activity.
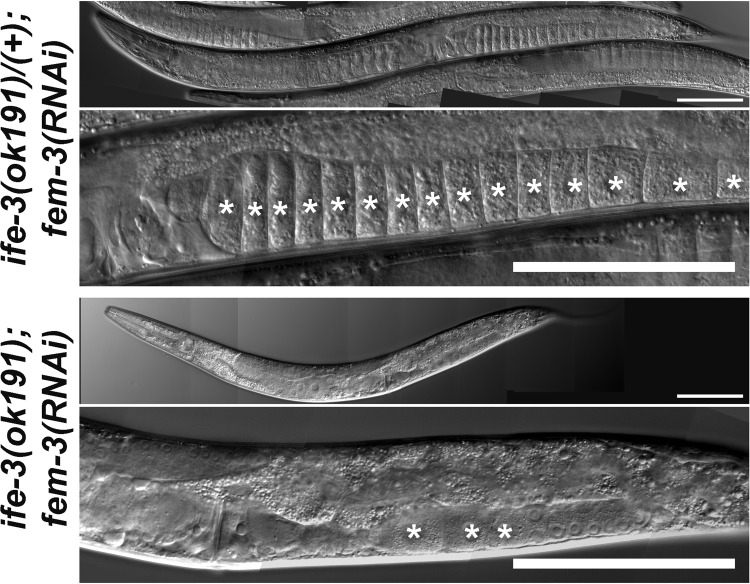
To determine whether ife-3 might also act as an upstream inhibitor of fem-3, we tested whether inhibition of fem-3 expression bypasses the need for ife-3 in promoting oogenesis. We induced RNAi against fem-3 by microinjecting fem-3-encoding dsRNA into the gonads of wild-type hermaphrodites and DWP70 hermaphrodites with ife-3(ok191) balanced by nT1[qIs51]. To ensure the ife-3(ok191) mog phenotype was fully penetrant, worms were maintained at 20°C. Demonstrating the efficacy of fem-3(RNAi), we found that 10 to 47% of the progeny of four treated wild-type animals developed as females that lacked fertilized embryos and contained an excessive number of oocytes in their proximal gonad arms, both of which were apparent through a dissecting microscope. Similarly, for five fem-3(RNAi)-treated DWP70 animals, 50 to 80% of their GFP-positive heterozygous progeny appeared female when viewed using a dissecting microscope. Absence of embryos and excess of oocytes in the proximal gonad arms of these females was confirmed using DIC microscopy (compare Figs 2 and 3A to Fig. 5). We were unable to unambiguously identify GFP-negative ife-3(ok191) homozygous progeny that were female using a dissecting microscope due to the fact that these animals normally lack embryos and have small gonads. However, we examined nine randomly selected GFP-negative progeny using DIC microscopy, and found that seven contained oocytes and smaller oocyte-like cells in their proximal gonad arms, and six of these appeared to be true females that lacked fertilized embryos (Fig. 5). Thus, inhibition of fem-3 bypasses the requirement for ife-3 in promoting oogenesis, indicating fem-3 is epistatic to ife-3.
Fig. 5.
fem-3 is epistatic to ife-3. Progeny of DWP70 hermaphrodites treated for RNAi against fem-3 were examined by DIC microscopy. Shown are whole worms and magnified views of one gonad arm. Young adult heterozygous progeny were often female, characterized by a lack of fertilized embryos in their uterus and an excessive number of oocytes (marked by *) in their proximal gonad arms. Young adult ife-3(ok191) progeny were also apparently female in that they also lacked embryos and contained oocytes. However, they had only a few small oocytes (*), suggesting a reversal of the mog phenotype by fem-3(RNAi), but not reversal of the poor body or gonad growth phenotypes. Bars=100 µm.
DISCUSSION
We demonstrate that the canonical eIF4E isoform of C. elegans encoded by ife-3 is required for proper body growth, and is a novel regulator of sex-determination in the hermaphrodite germline. This differs from previous studies showing that RNAi-mediated knockdown of ife-3 prevents embryonic morphogenesis (Keiper et al., 2000; Sönnichsen et al., 2005). This discrepancy is likely due to RNAi eliminating an essential maternal gene function and masking zygotic gene functions (Ahringer, 2006). Confirming this phenomenon for ife-3, we found that animals completely lacking ife-3 due to a large genomic deletion are still able grow to adulthood so long as they are the progeny of heterozygous parents (Table 2; Fig. 1B). On rare occasions when ife-3 homozygous mutants produce embryos, which receive no maternal ife-3 activity, these embryos arrest before completion of morphogenesis, matching the effects of RNAi (Table 3). Thus, maternal but not zygotic ife-3 is essential for embryogenesis.
Mutants for ife-3 have stunted growth (Fig. 2, supplementary material Fig. S1A), a phenotype reasonably expected from loss of an eIF4E homolog that promotes protein synthesis. Also, ife-3 mutant gonads are smaller and contain fewer germ cells than those of wild-type animals (Fig. 4A,B). A somewhat less intuitive effect of absence of ife-3 is a disruption of hermaphrodite germline sex-determination. Germline sex-determination in C. elegans is regulated by successive layers of genes that alternatively promote spermatogenesis or oogenesis, with each layer inhibiting the activity of the layer immediately downstream (for review, see Ellis and Schedl, 2007). The final output of spermatogenesis or oogenesis depends on the balance of these successively opposing influences. In male C. elegans worms, defined genetically by a single X chromosome, her-1 gene activity pushes this path toward spermatogenesis throughout their fertile life. In XX hermaphrodites, sex-determination is also initially tipped toward spermatogenesis during larval development, but somehow switches toward oogenesis in adulthood. We find that this switch normally requires ife-3, as XX ife-3 mutants grown under standard conditions have a normal hermaphrodite soma but a masculinized germline (mog phenotype) that produces sperm and fails to produce oocytes (Figs 3 and 4A,C, supplementary material Fig. S1B,C).
Another key gene involved in the spermatogenesis-to-oogenesis transition is fem-3. Loss-of-function alleles of fem-3 result in female XX worms that produce oocytes but never produce sperm, while gain-of-function alleles result in the XX worms with the mog phenotype (Hodgkin, 1986; Barton et al., 1987). Thus, fem-3 promotes spermatogenesis during larval development, but its activity must be restrained to permit the transition to oogenesis in adulthood. Many genes that promote the spermatogenesis-to-oogenesis transition function genetically as upstream inhibitors of fem-3. Thus, while mutations in these genes cause a mog phenotype in XX animals, the additional loss of fem-3 reverses this effect to produce XX females. Similarly, the mog phenotype of ife-3 mutants is reversed by RNAi-mediated inhibition of fem-3. This suppression of the ife-3 phenotype is specific to sex-determination, as fem-3(RNAi) does not restore normal body growth or gonad size to ife-3 mutants, leaving small female worms with few, small oocytes (Fig. 5).
Inhibition of fem-3 is at least in part through suppression of its translation. The fem-3 3′UTR bears a cis-acting point mutation element (PME) that when mutated, results in masculinizing gain-of-function alleles (Ahringer and Kimble, 1991; Barton et al., 1987). Attachment of the fem-3 3′UTR is sufficient to inhibit expression of exogenous reporter genes such as lacZ, while a fem-3(gf) 3′UTR with a mutated PME has no such effect (Gallegos et al., 1998). Fem-3 binding factor (FBF), encoded by fbf-1 and fbf-2, promotes the spermatogenesis-to-oogenesis transition by binding to the PME in vivo. Consequently, fbf-1/fbf-2 mutants have a mog phenotype, and wild-type FBF cannot bind the PME of mog-causing fem-3(gf) alleles (Zhang et al., 1997). Six additional genes, mog-1, -2, -3, -4, -5, and -6, are also important for PME-dependent inhibition of fem-3, as loss-of-function alleles result in a mog phenotype and also prevent PME-containing 3′UTRs from inhibiting expression of lacZ (Graham and Kimble, 1993; Graham et al., 1993; Gallegos et al., 1998). However, it is not clear how the products of the mog genes contribute to this inhibition.
Interestingly, the mog genes and ife-3 share mutant phenotypes beyond the mog. mog mutants also have poor body growth and small gonads (Graham and Kimble, 1993; Graham et al., 1993). Also, when oogenesis is induced in mog mutants (e.g. through inhibition of fem-3) and resultant mog oocytes are fertilized, the progeny die as embryos or larvae, demonstrating maternal effect lethality. The MOG gene products share no similarity with IFE-3, but they are also involved in RNA metabolism, with mog-1, -4, and -5 encoding homologs of mRNA splicing factors, mog-2 encoding a U2 snRNP protein, mog-3 encoding a small nucleolar preRNA-splicing protein, and mog-6 encoding a divergent cyclophilin that associates with MEP-1, an RNA-binding protein also required for PME-mediated inhibition of lacZ (Belfiore et al., 2002, 2004; Kasturi et al., 2010; Puoti and Kimble, 1999, 2000; Zanetti et al., 2011). The mog genes and ife-3 appear to be part of an even wider set of genes that are involved in RNA functions, and whose mutation results in poor growth, maternal effect lethality, and germline masculinization, including ddx-23 encoding an mRNA splicing factor, mag-1 encoding a mago nashi homolog, and R07E14 encoding a homolog of Y-14 (Kawano et al., 2004; Konishi et al., 2008; Li et al., 2000). These similarities hint at the possibility of some commonality in the function of these genes.
As eIF4E proteins generally act to promote mRNA translation, it seems unlikely that ife-3 participates directly in PME-mediated inhibition of fem-3 translation, but may promote translation of some other factor that is more directly involved. Alternatively, eIF4E-dependent inhibition of translation can arise in response to eIF4E-binding proteins (4E-BPs), as demonstrated by the Drosophila 4E-BP, Bicoid. Association of Bicoid with the caudal mRNA 3′UTR inhibits translation when the mRNA is associated with the eIF4E isoform encoded by d4EHP, but not when it is associated with the isoform encoded by deIF4E (Cho et al., 2005). In light of this, it might be significant that mutation of 4E-BP-encoding ifet-1 mildly masculinizes the C. elegans hermaphrodite germline (Sengupta et al., 2013). Unfortunately, we were unable to determine directly whether ife-3 is required for PME-dependent inhibition using the lacZ reporter developed by Gallegos and colleagues (1998), as loss of ife-3 inhibited expression of the reporter in a PME-independent manner (data not shown).
An intriguing alternative model for how ife-3 might promote the spermatogenesis-to-oogenesis transition is provided by a suggestion by Goodwin and Ellis (2002). They suggest the tipping in the balance between spermatogenesis to oogenesis in the germline might be tied to growth of the germline in terms of cell size or number. Based on such a model, ife-3 mutants might be defective in the spermatogenesis-to-oogenesis transition as a simple consequence of their poor body growth and small gonad size. Such a model might also help account for the recurrent pairing of poor body/gonad growth and mog phenotype among many sex-determination mutants.
An interesting question is why the other eIF4E isoforms cannot replace ife-3 function. Two possible contributions are differential expression patterns, and differential specificity for mRNA 5′ caps. In terms of expression, IFE-3 is enriched in the germline over the soma (Amiri et al., 2001). Unfortunately, our transgene rescue experiments shed little light on where ife-3 function is required, as we used the endogenous ife-3 promoter to drive expression ife-3 from a complex extrachromosomal array, which can support expression in the germline or the soma. However, maternal effect lethality and the mog phenotype often correlate with germline expression of the affected gene. We therefore might have expected suppression of these phenotypes by ife-1, -2, or -5, all of which have some function and/or enriched expression in the germline, but not ife-4, which is soma-enriched (Amiri et al., 2001; Dinkova et al., 2005; Song et al., 2010). However, the eIF4E isoforms encoded by ife-1, -2, and -5 all differ from IFE-3 in their recognition of mRNA caps. In C. elegans, mRNAs acquire a 5′-5′ linked monomethylated guanosine (MMG) cap, but this is often replaced with a trimethylated guanosine (TMG) cap during the trans-splicing (Blumenthal, 2012). While IFE-3 and -4 bind MMG caps exclusively, IFE-1, -2, and -5 bind both cap structures (Jankowska-Anyszka et al., 1998; Keiper et al., 2000). Thus, TMG-capped mRNAs in the germline might prevent these isoforms from efficiently binding MMG-capped IFE-3 targets. Whether expression pattern and cap-specificity are sufficient to explain the unique roles of ife-3, or whether factors such as 4E-BPs contribute, remains to be determined.
MATERIALS AND METHODS
Plasmids, primers and molecular techniques
pCR2.1-ife-3 was constructed by PCR amplification of genomic ife-3, including 2026 bp upstream and 386 bp downstream sequence, followed by TOPO-TA cloning (Thermo Fisher Scientific, Life Technologies, Grand Island, NY, USA). For production of double-stranded RNA (dsRNA) for fem-3(RNAi), the fem-3 coding region was PCR amplified from wild-type genomic DNA and TOPO-TA cloned. The fem-3 sequence was then subcloned between two opposing T7 promoters in plasmid L4440 (Timmons and Fire, 1998), and dsRNA was synthesized in vitro with T7 polymerase using standard protocols. Supplementary material Table S3 lists all primers used in this study.
Strains, growth conditions, and genetics
Worms were maintained under standard conditions at 20°C (Brenner, 1974), except where indicated. Strains N2 [wild-type] (Brenner, 1974), MT1083 [egl-8(n488) V] (Trent et al., 1983), and KX10 [ife-3(ok191)/unc-34(e566) V] were supplied by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA). FX02133 [upsDf41 daam-1(tm2133)/+ V], the original source of upsDf41 and daam-1(tm2133), was provided by Shohei Mitani (National BioResource Project for the Experimental Animal Nematode C. elegans, Tokyo Women's Medical University School of Medicine, Tokyo, Japan).
The genomic rearrangement nT1 behaves as a reciprocal translocation between ChrIV and ChrV (Edgeley et al., 2006). The variant nT1[qIs51], encoding pharyngeal GFP and a recessive lethal mutation, balances ife-3(ok191) in DWP70 [+/nT1[qIs51] IV; ife-3(ok191)/nT1[aIs51]V] and upsDf41 daam-1(tm2133) in XA8002 [+/nT1[qIs51] IV; upsDf41 daam-1(tm2133)/nT1[qIs51] V]. DWP72 [+/nT1[qIs51] IV; upsDf41 daam-1(tm2133)/nT1[qIs51] V; upsEx40[ife-3]] carrying the ife-3-containing extrachromosomal array upsEx40, was constructed through microinjection of XA8002 with pCR2.1-ife-3 (25 ng/µl), mCherry marker plasmids (2.5 ng/µl pCJF90 [Pmyo-2::mCherry], 10 ng/µl pGH8 [Prab-3::mCherry], and 5 ng/µl pCJF104 [Pmyo-3::mCherry]) (Frøkjær-Jensen et al., 2008), and carrier DNA (75 ng/µl pRS315) (Sikorski and Hieter, 1989). RNAi was induced against fem-3 by microinjection of fem-3-encoding dsRNA into the germline of young adult hermaphrodites (Wang and Barr, 2005). The F1 progeny resulting from eggs laid 24 h after injection were microscopically examined after 4 days growth.
To determine the viability of the progeny of upsDf41 daam-1(tm2133)/+ heterozygous animals (Table 2), young adults were placed individually on plates with fresh bacterial lawns, and allowed to lay eggs for 4 h at 20°C. The adults were then removed, and the number of eggs counted on each plate. The following day (day 2), unhatched (dead) eggs were counted, larvae were counted on day 3, and adults were counted on day 4.
To assay for the ability of hermaphrodites to lay eggs (Table 3), L1 larvae were placed individually on plates with fresh bacterial lawns, and allowed to grow at the indicated temperatures. Each animal was examined for the presence of eggs or larvae each day of their fertile adulthood.
Whole genome sequence analysis
Purified genomic DNA samples from N2 and XA8002 were submitted to the Cornell University Biotechnology Resource Center (Cornell University, Ithaca, NY, USA) for DNA library preparation and single-end 50 nucleotide sequencing (50× coverage) using Illumina HiSeq instrumentation. Illumina 1.8 datasets were uploaded to the Galaxy website (https://main.g2.bx.psu.edu/root), FASTQ groomed (Blankenberg et al., 2010), trimmed to remove 1 nucleotide from each sequence read end, and mapped as single-end reads against the C. elegans reference genome WS220 (http://www.wormbase.org) using Bowtie for Illumina (Langmead et al., 2009). To identify small nucleotide polymorphisms (SNPs), the resultant SAM format files were filtered to remove unmapped reads, and converted to a BAM format, from which a pileup was generated (Li et al., 2009). The pileup data were filtered to remove reads with quality lower than 20, and reads for genomic positions with sequence coverage lower than 3 (Li et al., 2009). SNPs present in ≥15% of reads at a given position were considered to represent potentially heterozygous or homozygous SNPs in XA8002 rather than sequencing errors. SNPs between ChrV coordinates 1 to 1,540,027, encompassing the sequence to the left of daam-1 plus approximately 1 cM to its right, were considered daam-1-linked (supplementary material Table S1). To identify small deletions or insertions, FASTQ groomed files were mapped with BWA for Illumina (Li and Durbin, 2009), and searched using the Indel Analysis tool (supplementary material Table S1). To identify large genomic deletions or duplications, SAM datasets were converted to an interval format. Intervals that mapped to ChrV 1 to 154,000 were visualized against the C. elegans genome using the USCS genome browser (Genome Bioinformatics Group of University of California at Santa Cruz, Santa Cruz, CA, USA). Visually identified stretches over which the average number of sequence reads per position was approximately 50% lower than surrounding areas were considered possible heterozygous deletions. One region over which reads per position was increased >50% was considered to possibly indicate an amplified sequence.
Staining and microscopic analysis
Live animals were visualized by differential interference contrast (DIC) microscopy as previously described (Mi-Mi et al., 2012). To visualize DNA, worms were fixed 4 min in fresh Carnoy's fixative (60% ethanol, 10% chloroform, 30% glacial acetic acid), rehydrated 3 min each in a 90%, 70%, 50%, and 25% ethanol series, washed in PBS, and stained 10 min in PBS with 1 µg/ml 4′6-diamidino-2-phenylindole (DAPI) before two final PBS washes. To stain for MSP, dissected worms were fixed 1 h in 100 mM potassium phosphate, pH 7.2 with 1.8% formaldehyde, washed with PBS/0.1% Tween-20 (PBS/T), post-fixed 5 min in −20°C methanol, and blocked 1 h in PBST with 1 mg/ml bovine serum albumin (PBS/TB) at room temperature. Incubations with MSP-specific antibody 4A5 (Kosinski et al., 2005) (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA, USA) at 1:200 dilution in PBS/TB, and Texas Red-labeled goat anti-mouse antibody (Rockland Immunochemicals, Limerick, PA, USA) at 1:8000 dilution in same, were each overnight at 4°C, with PBS/T washes between. DAPI (100 ng/ml) was included in the final wash.
Fluorescence and DIC images were acquired using a Cool-SNAP HQ2 digital monochrome charge-coupled device camera (Photometrics, Tuscon, AZ, USA) mounted on an Eclipse 90i upright research microscope (Nikon, Tokyo, Japan), and driven by NIS-Elements AR acquisition and analysis software (version 3.1; Nikon, Tokyo, Japan). Low magnification images (supplementary material Fig. S1A) were acquired using a DP-20 digital camera on an SZ61TR stereoimaging microscope, and driven by DP2-BSW software (Olympus, Center Valley, PA, USA). Images were processed linearly to enhance contrast using Photoshop CS4 (Adobe, San Jose, CA, USA).
Statistical analysis
Quantitative data are presented as mean±one standard deviation. Results from experiments involving two data sets were subjected to unpaired, two-tailed Student's t-tests, with P≤0.05 considered statistically significant. Results from experiments involving three or more groups were subjected to one-factor analysis of variance followed by Fisher's least significant difference post hoc testing, with differences between groups exceeding the 95% confidence interval considered statistically significant.
Supplementary Material
Acknowledgements
We thank the Caenorhabditis Genetics Center (funded by the NIH Office of Research Infrastructure Programs P40 OD010440) and Shohei Mitani (the National BioResource Project for the Experimental Animal Nematode C. elegans, Tokyo Women's Medical University, Tokyo, Japan) for providing worm strains. Monoclonal antibody 4A5, developed by David Greenstein (University of Minnesota, Saint Paul, MN, USA), was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242 (USA). We also thank Wormbase.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
This work was conceived and designed by R.S.M. and D.P., and executed by R.S.M., S.V., and D.P. Data were interpreted by R.S.M. and D.P. The article was prepared by R.S.M., S.V., and D.P.
Funding
Research reported in this publication was supported by the National Institute of Arthritis and Musculoskeletal And Skin Diseases of the National Institutes of Health [grant number R01AR064760], and by the Hendricks Fund [grant number 56283].
Supplementary material
Supplementary material available online at http://bio.biologists.org/lookup/suppl/doi:10.1242/bio.011585/-/DC1
References
- Ahringer J. (2006). Reverse genetics. In WormBook (ed. The C. elegans Research Community), 10.1895/wormbook.1.47.1http://www.wormbook.org. [DOI] [Google Scholar]
- Ahringer J. and Kimble J. (1991). Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3′ untranslated region. Nature 349, 346-348. 10.1038/349346a0 [DOI] [PubMed] [Google Scholar]
- Amiri A., Keiper B. D., Kawasaki I., Fan Y., Kohara Y., Rhoads R. E. and Strome S. (2001). An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128, 3899-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton M. K., Schedl T. B. and Kimble J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore M., Mathies L. D., Pugnale P., Moulder G., Barstead R., Kimble J. and Pouti A. (2002). The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3′ untranslated region in Caenorhabditis elegans. RNA 8, 725-739. 10.1017/S1355838202028595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfiore M., Pugnale P., Saudan Z. and Puoti A. (2004). Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development 131, 2935-2945. 10.1242/dev.01154 [DOI] [PubMed] [Google Scholar]
- Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J. and Nekrutenko A.. ; Galaxy Team. (2010). Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783-1785. 10.1093/bioinformatics/btq281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T. (2012). Trans-splicing and operons in C. elegans. In WormBook (ed. The C. elegans Research Community) 10.1895/wormbook.1.5.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans . Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho P. F., Poulin F., Cho-Park Y. A., Cho-Park I. B., Chicoine J. D., Lasko P. and Sonenberg N. (2005). A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121, 411-423. 10.1016/j.cell.2005.02.024 [DOI] [PubMed] [Google Scholar]
- Dinkova T. D., Keiper B. D., Korneeva N. L., Aamodt E. J. and Rhoads R. E. (2005). Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol. Cell. Biol. 25, 100-113. 10.1128/MCB.25.1.100-113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgeley M. L., Baillie D. L., Riddle D. L. and Rose A. M. (2006). Genetic balancers. In WormBook (ed. The C. elegans Research Community) 10.1895/wormbook.1.89.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. and Schedl T. (2007). Sex determination in the germ line. In WormBook (ed. The C. elegans Research Community) 10.1895/wormbook.1.82.2, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frand A. R., Russel S. and Ruvkun G. (2005). Functional genomic analysis of C. elegans molting. PLoS Biol. 3, e312 10.1371/journal.pbio.0030312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C., Davis M. W., Hopkins C. E., Newman B. J., Thummel J. M., Olesen S.-P., Grunnet M. and Jorgensen E. M. (2008). Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 40, 1375-1383. 10.1038/ng.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos M., Ahringer J., Crittenden S. and Kimble J. (1998). Repression by the 3′ UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans. EMBO J. 17, 6337-6347. 10.1093/emboj/17.21.6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Raught B. and Sonenberg N. (1999). eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68, 913-963. 10.1146/annurev.biochem.68.1.913 [DOI] [PubMed] [Google Scholar]
- Goodwin E. B. and Ellis R. E. (2002). Turning clustering loops: sex determination in Caenorhabditis elegans . Curr. Biol. 12, R111-R120. 10.1016/S0960-9822(02)00675-9 [DOI] [PubMed] [Google Scholar]
- Graham P. L. and Kimble J. (1993). The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133, 919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P. L., Schedl T. and Kimble J. (1993). More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14, 471-484. 10.1002/dvg.1020140608 [DOI] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.-J. and Kenyon C. (2007). Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95-110. 10.1111/j.1474-9726.2006.00267.x [DOI] [PubMed] [Google Scholar]
- Henderson M. A., Cronland E., Dunkelbarger S., Contreras V., Strome S. and Keiper B. D. (2009). A germline-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J. Cell Sci. 122, 1529-1539. 10.1242/jcs.046771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández G. and Vazquez-Pianzola P. (2005). Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech. Dev. 122, 865-876. 10.1016/j.mod.2005.04.002 [DOI] [PubMed] [Google Scholar]
- Hodgkin J. (1986). Sex determination in the nematode C. eleagns: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114, 15-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. (1988). Sexual dimorphism and sex determination. In The Nematode Caenorhabditis elegans (ed. Wood W. B.), pp. 243-280. Cold Spring Harbor: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Jankowska-Anyszka M., Lamphear B. J., Aamodt E. J., Harrington T., Darzynkiewicz E., Stolarski R. and Rhoads R. E. (1998). Multiple isoforms of eukaryotic protein synthesis initiation factor 4E in Caenorhabditis elegans can distinguish between mono- and trimethylated mRNA cap structures. J. Biol. Chem. 273, 10538-10542. 10.1074/jbc.273.17.10538 [DOI] [PubMed] [Google Scholar]
- Kasturi P., Zanetti S., Passannante M., Saudan Z., Müller F. and Puoti A. (2010). The C. elegans sex determination protein MOG-3 functions in meiosis and binds to the CSL co-repressor CIR-1. Dev. Biol. 344, 593-602. 10.1016/j.ydbio.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Kawano T., Kataoka N., Dreyfuss G. and Sakamoto H. (2004). Ce-Y14 and MAG-1, components of the exon-exon junction complex, are required for embryogenesis and germline sexual switching in Caenorhabditis elegans. Mech. Dev. 121, 27-35. 10.1016/j.mod.2003.11.003 [DOI] [PubMed] [Google Scholar]
- Kawasaki I., Jeong M.-H. and Shim Y.-H. (2011). Regulation of sperm-specific proteins by IFE-1, a germline-specific homolog of eIF4E, in C. elegans. Mol. Cells 31, 191-197. 10.1007/s10059-011-0021-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper B. D., Lamphear B. J., Deshpande A. M., Jankowska-Anyszka M., Aamodt E. J., Blumenthal T. and Rhoads R. E. (2000). Functional characterization of five eIF4E isoforms in Caenorhabditis elegans . J. Biol. Chem. 275, 10590-10596. 10.1074/jbc.275.14.10590 [DOI] [PubMed] [Google Scholar]
- King R. S., Maiden S. L., Hawkins N. C., Kidd A. R. III, Kimble J., Hardin J. and Walston T. D. (2009). The N- or C-terminal domains of DSH-2 can activate the C. elegans Wnt/β-catenin asymmetry pathway. Dev. Biol. 328, 234-244. 10.1016/j.ydbio.2009.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T., Uodome N. and Sugimoto A. (2008). The Caenorhabditis elegans DDX-23, a homolog of yeast splicing factor PRP28, is required for the sperm-oocyte switch and differentiation of various cell types. Dev. Dyn. 237, 2367-2377. 10.1002/dvdy.21649 [DOI] [PubMed] [Google Scholar]
- Kosinski M., McDonald K., Schwartz J., Yamamoto I. and Greenstein D. (2005). C. elegans sperm bud vesicles to deliver a meiotic maturation signal to distant oocytes. Development 132, 3357-3369. 10.1242/dev.01916 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. and Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. and Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754-1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Boswell R. and Wood W. B. (2000). mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 218, 172-182. 10.1006/dbio.1999.9593 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G. and Durbin R.; 1000 Genome Data Processing Subgroup. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi-Mi L., Votra S., Kemphues K., Bretscher A. and Pruyne D. (2012). Z-line formins promote contractile lattice growth and maintenance in striated muscles of C. elegans. J. Cell Biol. 198, 87-102. 10.1083/jcb.201202053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A. and Kimble J. (1999). The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-box protein family. Mol. Cell. Biol. 19, 2189-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puoti A. and Kimble J. (2000). The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc. Natl. Acad. Sci. USA 97, 3276-3281. 10.1073/pnas.97.7.3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta M. S., Low W. Y., Patterson J. R., Kim H.-M., Traven A., Beilharz T. H., Colaiácovo M. P., Schisa J. A. and Boag P. R. (2013). ifet-1 is a broad-scale translational repressor required for normal P granule formation in C. elegans. J. Cell Sci. 126, 850-859. 10.1242/jcs.119834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S. and Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics 122, 19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop A. R., Liu H., Yates J. III, Meyer B. J. and Heald R. (2004). Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305, 61-66. 10.1126/science.1097931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A., Labella S., Korneeva N. L., Keiper B. D., Aamodt E. J., Zetka M. and Rhoads R. E. (2010). A C. elegans eIF4E-family member upregulates translation at elevated temperatures of mRNAs encoding MSH-5 and other meiotic crossover proteins. J. Cell Sci. 123, 2228-2237. 10.1242/jcs.063107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., Alleaume A.-M., Artlet J., Bettencourt P., Cassin E. et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462-469. 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- Syntichaki P., Troulinaki K. and Tavernarakis N. (2007). eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans . Nature 445, 922-926. 10.1038/nature05603 [DOI] [PubMed] [Google Scholar]
- Thomas J. H. (1990). Genetic analysis of defecation in Caenorhabditis elegans . Genetics 124, 855-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L. and Fire A. (1998). Specific interference by ingested dsRNA. Nature 395, 854 10.1038/27579 [DOI] [PubMed] [Google Scholar]
- Trent C., Tsung N. and Horvitz H. R. (1983). Egg-laying defective mutants of the nematode Caenorhabditis elegans . Genetics 104, 619-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. and Barr M. M. (2005). RNA interference in Caenorhabditis elegans . Methods Enzymol. 392, 36-55. 10.1016/S0076-6879(04)92003-4 [DOI] [PubMed] [Google Scholar]
- Ward S. and Carrel J. S. (1979). Fertilization and sperm competition in the nematode Caenorhabditis elegans . Dev. Biol. 73, 304-321. 10.1016/0012-1606(79)90069-1 [DOI] [PubMed] [Google Scholar]
- Zanetti S., Meola M., Bochud A. and Puoti A. (2011). Role of the C. elegans U2 snRNP protein MOG-2 in sex determination, meiosis, and splice site selection. Dev. Biol. 354, 232-241. 10.1016/j.ydbio.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Zhang B., Gallegos M., Puoti A., Durkin E., Fields S., Kimble J. and Wickens M. P. (1997). A conserved RNAi-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390, 477-484. 10.1038/37297 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.