Abstract
Background
Flavobacterium psychrophilum is the etiologic agent of bacterial coldwater disease in salmonids. Earlier research showed that a rifampicin-passaged strain of F. psychrophilum (CSF 259-93B.17) caused no disease in rainbow trout (Oncorhynchus mykiss, Walbaum) while inducing a protective immune response against challenge with the virulent CSF 259–93 strain. We hypothesized that rifampicin passage leads to an accumulation of genomic mutations that, by chance, reduce virulence. To assess the pattern of phenotypic and genotypic changes associated with passage, we examined proteomic, LPS and single-nucleotide polymorphism (SNP) differences for two F. psychrophilum strains (CSF 259–93 and THC 02–90) that were passaged with and without rifampicin selection.
Results
Rifampicin resistance was conveyed by expected mutations in rpoB, although affecting different DNA bases depending on the strain. One rifampicin-passaged CSF 259–93 strain (CR) was attenuated (4 % mortality) in challenged fish, but only accumulated eight nonsynonymous SNPs compared to the parent strain. A CSF 259–93 strain passaged without rifampicin (CN) accumulated five nonsynonymous SNPs and was partially attenuated (28 % mortality) compared to the parent strain (54.5 % mortality). In contrast, there were no significant change in fish mortalities among THC 02–90 wild-type and passaged strains, despite numerous SNPs accumulated during passage with (n = 174) and without rifampicin (n = 126). While only three missense SNPs were associated with attenuation, a Ser492Phe rpoB mutation in the CR strain may contribute to further attenuation. All strains except CR retained a gliding motility phenotype. Few proteomic differences were observed by 2D SDS-PAGE and there were no apparent changes in LPS between strains. Comparative methylome analysis of two strains (CR and TR) identified no shared methylation motifs for these two strains.
Conclusion
Multiple genomic changes arose during passage experiments with rifampicin selection pressure. Consistent with our hypothesis, unique strain-specific mutations were detected for the fully attenuated (CR), partially attenuated (CN) and another fully attenuated strain (B17).
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-015-0518-1) contains supplementary material, which is available to authorized users.
Keywords: Rifampicin, Attenuation, Flavobacterium psychrophilum, SNP, Methylome
Background
Serial passage of bacteria with exposure to rifampicin may result in rifampicin-resistant microorganisms that may be useful as live-attenuated vaccines [1]. Rifampicin (Rif) is a potent, broad-spectrum antibiotic from the rifamycin group that inhibits the β-subunit of prokaryotic DNA-dependent RNA polymerase (RNAP). The antibiotic acts by directly blocking elongation of mRNA transcripts and drug resistance is normally conferred by point mutations in rpoB gene that encodes the β-subunit of the RNAP [2–5], although alternative mechanisms of rifampicin resistance have been described [6]. Recently, rifampicin passage was used to generate live-attenuated vaccines against a number of bacterial diseases of fish including columnaris disease, edwardsiellosis, enteric septicemia of catfish and motile aeromonad septicemia [7–12] or brucellosis in cattle [1]. Similarly, a live-attenuated strain CSF259-93B.17 of F. psychrophilum was developed by passage with rifampicin and infection with this strain induces a protective immune response in rainbow trout (Oncorhynchus mykiss, Walbaum) against challenge with the virulent parent CSF 259–93 strain [13]. Further analysis of the CSF259-93B.17 (B17) strain revealed a point mutation in the rpoB gene and numerous proteomic changes as compared to the parent strain [14].
Although the method of passaging pathogens with rifampicin has been successfully used to generate live vaccines for more than two decades, the mechanism of attenuation from this procedure remains unknown. That is, it is not clear if loss of virulence is directly associated with point mutations within the rpoB gene that confer resistance to rifampicin, or if accumulation of random mutations resulting from repeated passages with antibiotic selection pressure lead to attenuation, or perhaps a combination of both [9, 15–18]. This is an important question because knowing the mechanisms of attenuation provides information to better assess the likelihood that an attenuated strain might revert to a virulent phenotype in the future. Furthermore, knowing the mechanisms involved could lead to more efficient strategies to develop live-attenuated strains that do not rely on random chance.
In this study we passaged two pathogenic strains of F. psychrophilum, CSF 259–93 and THC 02–90, on media with and without rifampicin, and applied next-generation genome sequencing techniques and other methods to analyze changes associated with these culture conditions. The choice of these two strains was based on the fact that they belong to two distinct genetic lineages of F. psychrophilum [19], and that both are highly virulent to salmonids. These characteristics make these strains suitable for bacterial challenge in our rainbow trout model for assessment of attenuation of passaged strains [13, 19].
Results
Growth comparison
Growth kinetics of the parent F. psychrophilum strains (CSF 259–93 and THC 02–90) and strains passaged with and without rifampicin were determined in TYES broth at 16 °C and assessed with endpoint optical density measurements and the area-under-the-curve (AUC) comparisons (Fig. 1). In general, the parental strains (CW and TW) grew slightly faster than their passaged counterparts, but only the CR strain grew significantly slower when compared to its parental CW strain (P < 0.005). For THC 02–90 strains, even though the wild-type TW strain exhibited slightly better growth, there was no statistical difference between growth rates of TW, TN and TR strains for both endpoint OD and AUC measurements. All measurements of growth included three independent biological replicates per strain.
Fig. 1.
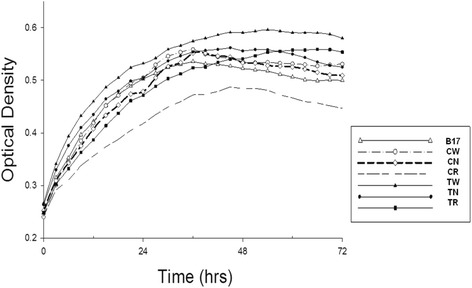
Growth curves of F. psychrophilum CSF 259–93 and THC 02–90 parent and passaged strains. Independent triplicates of each strain were grown statically in TYES broth at 16 °C with optical density measurements at 450–580 nm preceded by brief shacking. Abbreviations used: CW - F. psychrophilum CSF 259–93 parent strain, CN and CR – CSF 259–93 passaged 17 times with no and with rifampicin, respectively; B17 – rifampicin attenuated F. psychrophilum CSF 259-93B.17 strain [13]; TW - THC 02–90 parent strain and TN and TR – THC 02–90 passaged for 17 times with and without rifampicin, respectively
Cell and colony morphology
Gram staining and microscopy showed no apparent differences in size or shape of cells from the six F. psychrophilum strains (data not shown). When grown on TYES agar the CR strain exhibited reduced yellow pigmentation compared with the CW and CN strains (data not shown). Additionally, when cultured on gliding motility agar the CR colonies showed decreased spreading indicative of impaired gliding motility (Fig. 2). There was no obvious motility impairment for the remaining strains and the TR strain appeared to be the most motile. From a qualitative perspective, all three THC 02–90 strains appeared to be more motile than the CSF 259–93 strains.
Fig. 2.
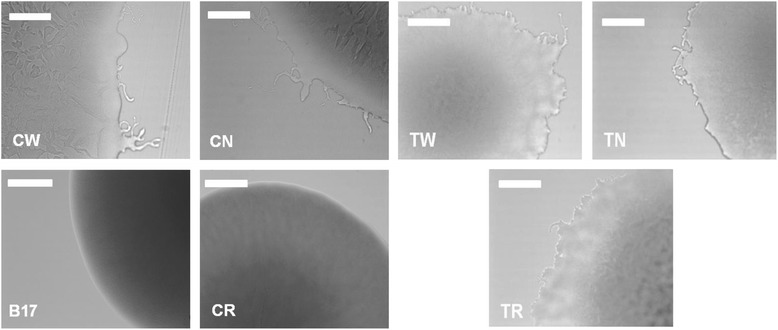
Light micrographs showing representative colony edges after 3-day incubation at 16 °C on gliding motility agar. CW – wild-type CSF 259.93; CN – CSF 259–93 passaged without rifampicin; CR– CSF 259–93 passaged with rifampicin; B17 – attenuated CSF 259.93B.17; TW – wild-type THC 02–90; TN – THC 02–90 passaged without rifampicin; TR – THC 02–90 passaged with rifampicin; white bars indicate 200 μm scale
rpoB mutations
Analysis of rpoB mutations from rifampicin resistant CR, TR and B17 strains obtained through genome sequencing, validated later using sequencing of PCR amplified rpoB (data not shown), revealed the presence of point mutations that are distinctive for these three strains. While the completely attenuated B17 strain harbors a Gln474Arg mutation, the rpoB of the CR strain is changed at Ser492Phe. The rpoB gene of the TR strain has a double mutation with Asp477Tyr and Pro496Ser substitutions (numbering based on NCBI reference sequence NC_009613.3).
Carbohydrate and protein characterization
Bacterial proteinase-K digested carbohydrate extractions and whole-cell lysates were prepared from each strain and analyzed by SDS-PAGE and 2D PAGE, respectively. There were no visible differences in LPS banding patterns among the six strains, although there were visual differences in band intensities (Additional file 1: Figure S1). Proteomic analysis of the CN strain showed increased synthesis of a protein with a molecular mass of approximately 25 kDa as compared to its parent strain. Additionally, whole-cell lysates the CR strain revealed that synthesis of three proteins of 20, 35 and 200 kDa was qualitatively increased. Both TN and TR appeared to have increased expression of a similar protein with approximate molecular mass of 35 kDa when compared to their parent TW strain (Fig. 3).
Fig. 3.
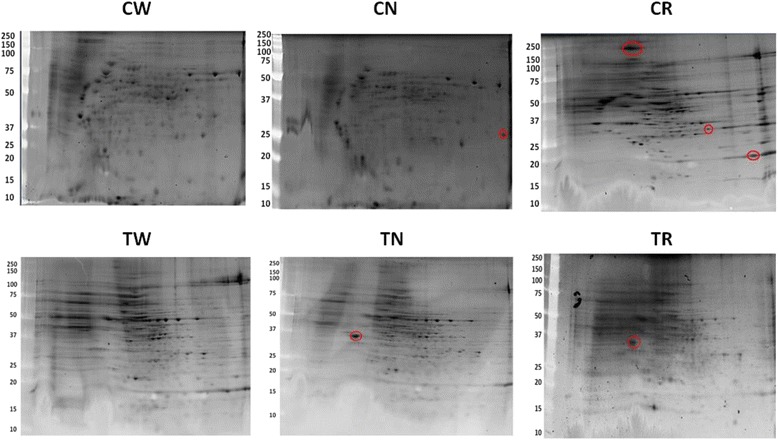
2D PAGE analysis of whole-cell lysates from the F. psychrophilum strains. Proteins were stained with SyPro Ruby gel stain and molecular weight markers (kDa) are indicated on the left of each gel. Red ovals designate proteins reproducibly greater abundance as compared to the respective (CW or TW) parent strain. CW – wild-type CSF 259.93; CN – CSF 259–93 passaged without rifampicin; CR– CSF 259–93 passaged with rifampicin; B17 – attenuated CSF 259.93B.17; TW – wild-type THC 02–90; TN – THC 02–90 passaged without rifampicin; TR – THC 02–90 passaged with rifampicin
Analysis of single-nucleotide polymorphisms
Initial 454 sequencing of a CW and B17 strain yielded 179,155 reads totaling 63.1 Mb for the CW strain. When compared to the CSF 259–93 reference genome, 99.3 % of the CW reads assembled into 109 contigs covering 2,769,031 bp with mean length of contig size of 69.8 kb. Most (99.8 %) of bases were Q40+ quality and an additional 17,852 bp were assembled into 66 shorter contigs. Sequencing of the B17 strain yielded 449,264 reads totaling 167 Mb, and assembly to reference sequence resulted in 98.9 % bases assembled into 107 contigs covering 2,779,773 bp. Most (99.6 %) of the base pairs were Q40+ and the mean contig size was 70.7 kb. An additional 25,684 bp were included in 60 shorter contigs. Results of IonTorrent sequencing of CW, CN, CR, TW, TN and TR strains are shown in Table 1.
Table 1.
Summary of output from IonTorrent and 454 sequencing for seven F. psychrophilum strains
| Sample | Mb totala | Number of reads (in millions) | Mean read length (in bp) |
|---|---|---|---|
| CSF 259–93 wild-type (CW)b | 603 | 2.46 | 245 |
| CSF 259–93 passaged w/o Rif (CN)b | 346 | 1.37 | 254 |
| CSF 259–93 passaged with Rif (CR)b | 772 | 2.97 | 261 |
| THC 02–90 wild-type (TW)b | 620 | 2.38 | 260 |
| THC 02–90 passaged w/o Rif (TN)b | 676 | 2.78 | 244 |
| THC 02–90 passaged with Rif (TR)b | 666 | 2.78 | 240 |
| CSF 259–93 wild-type (CW)c | 63.1 | 0.18 | N/A |
| CSF 259-93B.17 (B17)c | 167 | 0.45 | N/A |
aMb total = total number of mega bases of DNA sequences
bResults from Ion Torrent sequencing, 316 chip
cResults from 454 sequencing
We based our SNP analysis on polymorphisms with read frequency ≥80 % and we focused on mutations that led to predicted nonsynonymous amino acid changes (Table 2). Our wild-type CSF 259–93 strain exhibited 19 SNPs leading to nonsynonymous amino acid changes when compared to the reference genome of reported for CSF 259–93 (GenBank Accession number: CP007627.1). These 19 SNPs included codon changes in 14 genes leading to nonsynonymous amino acid changes (Table 3) and may reflect mutations that have accumulated over years of passage in different labs although these two sequenced strains retain their virulence against rainbow trout. Analysis of the B17 strain’s genome after 454 sequencing revealed SNPs resulting in 14 nonsynonymous amino acid changes in codons of 14 genes when compared to its parental CW strain (Table 4). Genomes from CN and CR strains accumulated 5 and 8 SNPs, respectively, when compared to their parental CSF 259–93 strain (CW). These SNPs led to missense mutations in codons of 4 genes in the CN strain (Table 5), and 7 in case of CR (Table 6). After comparison with the genome of their parental THC 02–90 strain (TW), TN and TR strains displayed numerous differences. The TN strain accumulated 126 SNPs resulting in missense mutations in codons of 48 genes (Additional file 1: Table S1), and in the TR strain 174 SNPs led to 64 genes with codons leading to nonsynonymous amino acid substitutions (Additional file 1: Table S2). Interestingly, despite the large number of missense mutations in the THC 02–90 passaged strains there was no evidence for commensurate changes in the proteome for these strains relative to the wild-type proteome (Fig. 3).
Table 2.
Single nucleotide polymorphisms (SNPs) in F. psychrophilum strains used in our experiments
| NONSYNONYMOUS SNPs | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| SYNONYMOUS SNPs | CW | CN | CR | TW | TN | TR | ERGO | B17 | |
| CW | n.a. | 5 | 8 | 85 | 113 | 123 | 19 | 14 | |
| CN | 32 | n.a. | 3 | 105 | 141 | 117 | 25 | N.D. | |
| CR | 34 | 33 | n.a. | 113 | 158 | 108 | 28 | N.D. | |
| TW | 161 | 170 | 174 | n.a. | 126 | 174 | 100 | N.D. | |
| TN | 233 | 142 | 146 | 177 | n.a. | 48 | 102 | N.D. | |
| TR | 278 | 287 | 291 | 257 | 180 | n.a. | 132 | N.D. | |
| ERGO | 37 | 75 | 78 | 206 | 208 | 238 | n.a. | 33 | |
| B17 | 45 | N.D. | N.D. | N.D. | N.D. | N.D. | 50 | n.a. | |
CW wild type CSF 259–93, CN CSF 259–93 passaged without Rif, CR CSF 259–93 passaged with Rif, TW wild type THC 02–90, TN THC 02–90 passaged without Rif, THC 02–90 passaged with Rif, B17 – CSF 259-93B.17
N.D. not determined due to these genomes being sequenced at different times and by different methods
n.a. = not applicable
Table 3.
Analysis of single nucleotide polymorphisms (SNPs) from F. psychrophilum CSF 259–93 used in our laboratory
| Reference positiona | Allele frequency (in %)b | SNPc | Coveraged | Annotatione | Amino acid changee | Gene/Proteinf |
|---|---|---|---|---|---|---|
| 721449 | 100 | C > A | 72 | FPSM_00632 | Gly111Trp | Hypothetical protein |
| 742016 | 100 | A > G | 70 | FPSM_00646 | Ser615Gly | Type II restriction-modification system methylation subunit |
| 899151 | 100 | C > A | 84 | FPSM_00800 | Phe215Leu | RmuC family protein |
| 899152 | 100 | G > A | 92 | FPSM_00800 | Gly216Glu | |
| 921140 | 96.7 | C > A | 90 | FPSM_00823 | Pro254Thr | Hypothetical protein (similar to FP1973-GldK gliding motility protein) |
| 1257208 | 100 | C > G | 98 | FPSM_01126 | Trp148Ser | Putative membrane spanning protein |
| 1333946 | 97.7 | A > T | 43 | FPSM_01204 | Phe507Ile | Glucose/galactose transporter |
| 1334321 | 98.7 | A > C | 78 | FPSM_01204 | Tyr382Asp | |
| 1387849 | 95.1 | T > G | 61 | FPSM_01258 | Leu133Arg | Phage related protein |
| 1387851 | 100 | T > G | 99 | FPSM_01258 | Tyr134Asn | |
| 1537329 | 98.7 | C > G | 75 | FPSM_01388 | Arg547Thr | TonB-dependent outer membrane receptor |
| 1630279 | 92 | A > C | 25 | FPSM_01473 | Thr160Pro | Transposase |
| 1664395 | 100 | C > G | 81 | FPSM_01493 | Val213Leu | Cation efflux system plasma membrane protein (similar to FP0880 – probable multidrug resistance protein precursor AcrB/D/F family) |
| 1664410 | 99 | G > C | 96 | FPSM_01493 | Pro208Ala | |
| 1970401 | 97.4 | T > C | 116 | FPSM_01788 | Ser383Gly | Von Willebrand factor type A domain protein (FP0550 – yfbK probable outer membrane protein precursor) |
| 1970443 | 100 | C > T | 122 | FPSM_01788 | Asp369Asn | |
| 2066236 | 100 | C > A | 76 | FPSM_01870 | His300Asn | Hypothetical protein |
| 2514049 | 98.5 | C > G | 130 | FPSM_02290 | Leu293Val | 3-oxoacyl-(acyl carrier-protein) synthase III (FP1374 fabH1) |
| 2737510 | 100 | C > G | 119 | FPSM_02485 | Leu163Phe | tRNA pseudouridine synthase B (FP2357 truB) |
aReference position (in bp) for reference sequence (listed under annotation field) are based on CSF 259–93 sequence from ERGO-Integrated genomics
bAllele frequency refers to the proportion of sequences showing a given single-nucleotide polymorphism (SNP) at the started reference position
cShows the SNP change represents the change from nucleotide X to Y (X > Y) at the reference position
dCoverage refers to the total number of sequencing reads that align to each base within the sample DNA
ePredicted amino acid change for the identified SNP
fAnnotation shows the name and putative function for the identified gene
Table 4.
Single nucleotide polymorphisms (SNPs) from F. psychrophilum CSF 259-93B.17 strain
| Reference positiona | Allele frequency (in %)b | SNPc | Coveraged | Annotatione | Amino acid changee | Gene/Proteinf |
|---|---|---|---|---|---|---|
| 43576 | 100 | A > T | 5 | FPSM_00026 | Met1Leu | Multimodular transpeptidase-transglycosylase PBP 1A |
| 155877 | 100 | A > T | 43 | FPSM_00129 | Leu71Phe | Putative exported protein |
| 283611 | 80 | A > T | 10 | FPSM_00250 | Asn522Ile | Amino acid permease |
| 325469 | 100 | T > A | 4 | FPSM_00287 | Met1Leu | Thioredoxin |
| 488182 | 100 | A > C | 4 | FPSM_00425 | Gln466Pro | DNA primase |
| 514297 | 86.4 | C > T | 162 | FPSM_00445 | Ala11Thr | Malonyl-CoA-[acyl-carrier-protein] transacylase |
| 548737 | 100 | A > T | 5 | FPSM_00479 | Met1Leu | Ribosome recycling factor (RRF) |
| 846079 | 100 | T > A | 4 | FPSM_00743 | Phe98Tyr | Putative membrane spanning protein |
| 856484 | 85.7 | T > A | 7 | FPSM_00754 | Met13Leu | Hypothetical protein |
| 1334056 | 100 | A > C | 39 | FPSM_01204 | Phe470Cys | Glucose/galactose transporter |
| 1480580 | 80 | A > T | 5 | FPSM_01337 | Asn41Ile | Hypothetical protein |
| 1498680 | 100 | T > A | 4 | FPSM_01353 | Asn123Ile | Phosphoribosyl-ATP cyclohydrolase |
| 1522077 | 80 | T > A | 5 | FPSM_01375 | Met1Leu | Protein translation elongation factor P (EF-P) |
| 1537329 | 100 | C > G | 51 | FPSM_01388 | Arg547Thr | TonB-dependent outer membrane receptor |
aReference position (in bp) for reference sequence (listed under annotation field) are based on CSF 259–93 sequence from ERGO-Integrated genomics
bAllele frequency refers to the proportion of sequences showing a given single-nucleotide polymorphism (SNP) at the started reference position
cShows the SNP change represents the change from nucleotide X to Y (X > Y) at the reference position
dCoverage refers to the total number of sequencing reads that align to each base within the sample DNA
ePredicted amino acid change for the identified SNP
fAnnotation shows the name and putative function for the identified gene. Annotation is presented from the CSF 259–93 sequence from ERGO-Integrated genomics
Table 5.
Single nucleotide polymorphisms (SNPs) from F. psychrophilum CSF 259–93 strain passaged with no rifampicin (CN)
| Reference positiona | Allele frequency (in %)b | SNPc | Coveraged | Annotatione | Amino acid changee | Gene/Proteinf |
|---|---|---|---|---|---|---|
| 575838 | 100 | C > G | 71 | FPSM_00502 | Val638Leu | Ribonucleoside-diphosphate reductase large chain (nrdA) |
| 824811 | 94 | A > G | 50 | FPSM_00723 | Ile62Thr | GTP-binding protein YihA |
| 2606362 | 100 | G > T | 25 | FPSM_02361 | Trp4Leu | Putative membrane spanning protein |
| 2606399 | 95 | A > T | 20 | FPSM_02361 | Leu16Phe | |
| 2718910 | 100 | T > C | 83 | FPSM_02464 | Tyr418Cys | Di-tripeptide transporter |
aReference position (in bp) for reference sequence (listed under annotation field) are based on CSF 259–93 sequence from ERGO-Integrated genomics
bAllele frequency refers to the proportion of sequences showing a given single-nucleotide polymorphism (SNP) at the started reference position
cShows the SNP change represents the change from nucleotide X to Y (X > Y) at the reference position
dCoverage refers to the total number of sequencing reads that align to each base within the sample DNA
ePredicted amino acid change for the identified SNP
fAnnotation shows the name and putative function for the identified gene. Annotation is presented from the CSF 259–93 sequence from ERGO-Integrated genomics
Table 6.
Single nucleotide polymorphisms (SNPs) from F. psychrophilum CSF 259–93 strain passaged with rifampicin (CR)
| Reference positiona | Allele frequency (in %)b | SNPc | Coveraged | Annotatione | Amino acid changee | Gene/Proteinf |
|---|---|---|---|---|---|---|
| 575838 | 100 | C > G | 71 | FPSM_00502 | Val638Leu | Ribonucleoside-diphosphate reductase large chain (nrdA) |
| 824811 | 94 | A > G | 50 | FPSM_00723 | Ile62Thr | GTP-binding protein YihA |
| 1327825 | 100 | G > A | 138 | FPSM_01200 | Asp275Val | Hypothetical protein |
| 2111614 | 81.9 | C > A | 144 | FPSM_01917 | Thr365Lys | Phytoene desaturase (crtI) |
| 2319546 | 98.8 | G > A | 167 | FPSM_02089 | Ser492Phe | DNA-directed RNA polymerase beta chain RpoB |
| 2606236 | 100 | G > T | 73 | FPSM_02361 | Trp4Leu | Putative membrane spanning protein |
| 2606399 | 100 | A > T | 65 | FPSM_02361 | Leu16Phe | |
| 2788973 | 97.1 | T > A | 105 | FPSM_02533 | Ile35Phe | Transcriptional regulator, TetR family |
aReference position (in bp) for reference sequence (listed under annotation field) are based on CSF 259–93 sequence from ERGO-Integrated genomics
bAllele frequency refers to the proportion of sequences showing a given single-nucleotide polymorphism (SNP) at the started reference position
cShows the SNP change represents the change from nucleotide X to Y (X > Y) at the reference position
dCoverage refers to the total number of sequencing reads that align to each base within the sample DNA
ePredicted amino acid change for the identified SNP
fAnnotation shows the name and putative function for the identified gene. Annotation is presented from the CSF 259–93 sequence from ERGO-Integrated genomics
DNA methylation analysis
We used Allora EC assemblies from the Pacific Biosciences sequencer to evaluate different DNA methylation patterns for CR and TR strain. RS ALLORA assembly EC.1 protocol of SMRT portal version 1.3.3 was used. Reads shorter than 100 bp and with quality below 0.85 were discarded. Postfiltered reads for the CR strain consisted of 385 Mb in 107,567 reads. The reads had an average length of 3581 bp and an average quality of 0.873. 100 % of the reads were assembled into 7 contigs with a sum of 2.27 Mb, a max contig size of 908,349 and an N50 of 511,868. Postfiltered reads for the TR strain had an average quality of 0.877 and totaled 449 Mb in 124,950 reads averaging 3590 bp. One hundred percent of the reads assembled into 7 contigs with a sum of 2.26 Mb and a max contig size of 710,139 bp and an N50 of 371,120 bp. DNA methylation was detected on both 7 contig assemblies and the corresponding motifs were identified with RS Modification and Motif Analysis.1 protocol of SMRT portal version 1.3.3.
Upon release of SMRT portal version 2.0.1 the existing PacBio data was reassembled using the RS HGAP Assembly 1 protocol. For the strain CR, less strict filtering allowed 830 Mb of data to enter the assembly with an average read length of 3658 bp and average quality of 0.847. Preassembly yield was 0.73 on a self-calculated minimum seed read size of 8038 bp, and generated 54 Mb in 7385 pre-assembled reads with an N50 of 8649 bp. The Celera assembler component of the HGAP algorithm assembled the pre-assembled reads into a single contig. Quiver was used to polish the assembly and found consensus concordance of 99.987 % with uniform 210× coverage and a polished contig size of 2,905,139 bp. For the strain TR, 927 Mb of data entered the assembly with an average read length of 3720 bp and average quality of 0.849. Preassembly yield was 0.73 on a self-calculated minimum seed read size of 8480 bp, and generated 54.8 Mb in 7137 pre-assembled reads with an N50 of 8987 bp. HGAP assembled the pre-assembled reads into two contigs. Quiver was used to polish the assembly and found consensus concordance of 99.991 % with uniform 232× coverage and polished contig sizes of 2,810,854 bp and 18,318 bp.
Genome wide methylation was detected by mapping the Pacific Biosciences reads onto the HGAP contigs and the corresponding modification motifs were identified using the RS Modification and Motif Analysis.1 protocol of SMRT portal version 2.0.1. Comparison of the CR and TR modifications resulted in identification of 8 DNA motifs in the CR and 16 in the TR that exhibited different methylation (Additional file 1: Table S3 and S4). No motifs were shared between these two strains.
Assessment of attenuation
Challenge experiments with the CN strain demonstrated partial loss of virulence with cumulative percent mortality (CPM) of 28.1 % (standard error of the mean (SEM) ± 2.0 %) (Fig. 4). The CR strain appears to be almost completely attenuated inflicting only 4 % mortality (SEM ± 2.3 %). CPM of fish challenged with the wild-type CSF 259–93 strain (CW) reached 54.5 % (SEM ± 6.9 %). Log-rank analysis of mortality patterns (Fig. 4a) demonstrated that observed changes in virulence were statistically significant for survival in CN treatment compared to CW (P = 0.002) and for survival of CR group compared to CW (P < 0.0001). Changes in virulence between CN and CR were also statistically significant (P = 0.0003). Cumulative percent mortalities in case of TW, TN and TR were 74.3 % (SEM ± 3.5 %), 69.7 % (SEM ± 1.2 %) and 69.4 % (SEM ± 2.9 %), respectively (Fig. 4b). Log-rank analysis among the three THC 02–90 groups indicated no statistically significant differences in CPM values (TW vs. TR P = 0.27, TN vs. TR P = 0.27 and TN vs. TR P = 0.97). There was no mortality in PBS mock infected group. For all mortalities that we tested, culture of the kidney, liver and spleen tissue confirmed the presence of yellow-pigmented bacterial colonies that resembled typical morphology of F. psychrophilum.
Fig. 4.
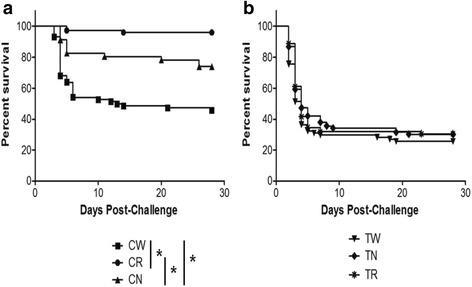
Percent survival curves of rainbow trout (Oncorhynchus mykiss, Walbaum) fry challenged with different F. psychrophilum strains. a Challenges with different CSF 259–93 strains with (panel b). showing comparisons between different THC 02–90 strains. Statistically significant values (P < 0.005) are indicated by asterisks. CW – wild-type CSF 259.93; CN – CSF 259–93 passaged without rifampicin; CR– CSF 259–93 passaged with rifampicin; B17 – attenuated CSF 259.93B.17; TW – wild-type THC 02–90; TN – THC 02–90 passaged without rifampicin; TR – THC 02–90 passaged with rifampicin
Discussion
Repeated laboratory passage of pathogens in the presence of rifampicin has been used to generate live-attenuated vaccines [1, 7–12], but the mechanism of rifampicin-induced attenuation remains unknown. There are a variety of potential confounding factors with this type of passage experiment that make cause-and-effect interpretations challenging. Passage can produce changes in global protein expression profiles [12, 13] or altered lipopolysaccharide (LPS) biosynthesis/colony roughness [7]. These changes could be attributed to altered activity of RpoB in the Rif resistant DNA-dependent RNA polymerase (RNAP) present in rifampicin resistant bacteria [14]. That is, the presence and activity of the mutated rpoB is one hypothesis for the mechanism of attenuation. Alternatively, rifampicin-associated attenuation may result from accumulation of spontaneous mutations acquired in the course of in vitro passaging. Serial passage of a pathogenic bacterium without any antibiotics can also induce changes in LPS profile and colony roughness, which have been correlated with attenuation [18]. This finding is consistent with the role of random mutations in the attenuation process, although the rate of mutation may differ depending on the stress imposed on the passaged microorganism [20, 21]. For example, for the current study the two strains that were passaged with rifampicin, CR and TR, accumulated more SNPs compared to their matched strains that were passaged without the antibiotic (CN and TN, 33.3 and 29.4 % respectively).
In the present study, passage did not affect in vitro growth characteristics except for the CR strain, which was compromised to some extent (Fig. 1). Others have reported that acquisition of rifampicin resistance can negatively impact growth rates although this probably depends on the exact mutation that is acquired in rpoB [22–24]. When grown as a colony on agar plates, the CR colonies had smooth edges and the CR strain lacked gliding motility, both changes that could be explained by either altered function of a mutated rpoB or other SNPs in relevant genes. For example, colonies of the CR grown on TYES and GMA media have reduced yellow pigmentation compared to the CW and CN strains. This phenotype may be attributed to the Thr365Lys mutation in the phytoene desaturase (crtI) gene, which encodes an enzyme involved in carotenoid biosynthetic pathway in Flavobacterium sp. [25–27]. Microscopic analysis of gliding motility and colony morphologies of TW, TN and TR strains did not reveal any phenotypic changes, despite the large number of SNPs that were accumulated by the TN and TR strains.
Polyacrylamide gel electrophoresis of LPS fractions revealed no obvious differences in LPS profiles, including no apparent changes from an Ile376Leu substitution in O-antigen acetylase (FPSM_01556) in the TR strain. 2D-PAGE analyses of proteins revealed probable differences in protein synthesis among rifampicin resistant F. psychrophilum strains (B17, CR and TR), which is consistent with our previous studies [14]. It is important to emphasize that while rpoB mutations might alter transcriptional regulation, other mutations in these strains could have contributed to this effect. Specifically, SNPs in proteins directly involved in gene/protein expression such as DNA primase, ribosome recycling factor, elongation factor P (EF-P), ATP-dependent RNA helicase (FPSM_01657) in the B17 or transcriptional regulator FPSM_00453 and ATP-dependent RNA helicase DbpA (FPSM_01345) in the TR strain.
Remarkably, considerable reduction in virulence of the CN (28.1 % mortality) and CR (4 % mortality) strains occurred despite relatively few SNPs leading to nonsynonymous amino acid changes (5 and 8, respectively). There were overlapping synonymous mutations among these strains and synonymous SNPs have been reported to potentially affect protein function [28]. Aside from SNPs occurring in the rpoB sequence, there was no overlap in SNPs for the attenuated B17 strain and the CR and CN strains developed in the current project. Noteworthy, 3 genes (nrdA – encoding a large chain of ribonucleotide-diphosphate reductase, yihA – encoding a GTP-binding protein and a putative membrane spanning protein FPSM_02361) out of 4 that harbor SNPs in the CN strain are identical with the CR strain. We surmise that the probability of these identical SNPs arising independently is unlikely. Instead, they most likely arose from the original culture used to initialize the passage experiments (i.e., a founder effect), or a cross-over contamination event occurred sometime early in the course of the experiment; if this was a contamination event, it must have occurred early in the experiment because these strains have distinct rpoB mutations making it easy to differentiate the final cultures. One of these shared SNPs is in nrdA, which has been implicated in pathogenesis of Pseudomonas aeruginosa [29]. Additionally, in Escherichia coli yihA is a GTPase that is essential for normal cell morphology and coordination of division; thus SNPs in this gene may contribute to attenuation [30, 31]. Because adhesion is an essential process in host-pathogen interaction and pathogenesis, mutation in the putative membrane spanning protein FPSM_02361 may have a direct effect on reduced virulence of CN and CR strains. Collectively, these findings are consistent with involvement of mutational events in the process of attenuation.
The CR strain was mostly attenuated (4 % mortality) and was characterized by slower growth in culture, lost gliding motility, three observable changes in protein synthesis, and altered carotenoid metabolism that is probably due to mutated phytoene desaturase (ctrI). As part of a gene cluster involved in carotenoid biosynthesis, mutations in ctrI were shown to alter colony pigmentation of other pathogens [32]. Importantly, mutations of ctr genes are able to decrease resistance to oxygen radicals and reduce growth in macrophages of the fish pathogen, Mycobacterium marinum [33]. Additionally, golden pigment synthesized through carotenoid pathway in Staphylococcus aureus enhances virulence by promoting resistance to respiratory burst of neutrophils, and the ∆crtM strain of S. aureus was also attenuated in mice model [34].
Additional SNPs unique to the attenuated CR strain included those found in a hypothetical protein (FPSM_01200), a transcriptional regulator (TetR family protein, FPSM_02533) and the Ser492Phe substitution in rpoB. The FPSM_01200 hypothetical protein has a predicted function of an N-acetylglucosamine (NAG) kinase, an enzyme important for bacterial cell wall and LPS metabolism. Moreover, importance of NAG metabolism was implied in initiation of murine intestine colonization by E. coli [35] and in bacterial signaling and growth in mucus of P. aeruginosa [36]. Together, these missense mutations may affect the virulence of the CR strain and its ability to colonize host and survive in challenged fish. In our analysis, however, we cannot discount the possible effects of silent SNPs on phenotype changes as described by Kimchi-Safraty and co-workers [28].
Our alternative hypothesis that rpoB can be directly involved in attenuation is potentially supported by the fact that RpoB is crucial in bacterial transcription and allosteric changes of the mutant protein may produce global alterations such as phenotype changes. The potential involvement of rpoB Ser492Phe mutation in attenuation of the CR strain is supported by existence of phenotypic (e.g. loss of gliding motility) and proteomic changes, and by a Gln474Arg rpoB mutation in the completely attenuated B17 strain [13, 14]. Furthermore, in other bacteria different rpoB mutations lead to different phenotypic changes [22–24, 37–39]. In Brucella spp. virulence and colony roughness of RifR phenotypes also varied depending on the position and character of single amino acid substitutions of RpoB [17]. Mutations providing resistance to rifampicin are clearly not universally responsible for attenuation because presumptively virulent RifR strains of Mycobacterium tuberculosis have been described [40–42].
Unlike attenuated strains derived from CSF 259–93, the TN and TR strains from wild-type THC 02–90 accumulated a large number of SNPs from serial passage regardless of the presence of rifampicin, and yet these changes had no apparent effect on virulence. The large numbers of SNPs present in the TN and TR strains as compared to the CN and CR strains may be attributed to the fact that the CSF 259–93 and THC 02–90 belong to different genetic lineages of F. psychrophilum. These two lineages are generally associated with different fish hosts, and the lineage to which THC 02–90 belongs is amenable to some genetic manipulations, which have not been successful with the lineage of strains to which CSF 259–93 belongs [19, 43–46]. It is notable that 54 and 46 % of the nonsynonymous mutations found in the TN and TR strains, respectively, occurred in subtilisin-like proteases that are putative cell surface proteins with predicted leucine-rich repeats. Repeat regions in DNA sequences are notoriously difficult to assemble correctly; particularly when using short read technologies such as employed herein [47, 48]. Notably, however, these same regions are also found in the CSF 259–93 genome and yet there were no mutations found in these gene sequences for the CN and CR strains. Given that all of these passaged strains introduced in this study were developed and sequenced at the same time using the same chemistries, we submit that this is not a sequencing artifact but that there is probably a distinctly different mutation process underway between the CSF 259–93 strains and the THC 02–90 strains. More strains from the two lineages need to be tested to determine if this is a lineage-level difference or if THC 02–90 is unique in this regard.
Additional changes at the SNP level, such as a SNP in nrdA in CR, and mutation in the FPSM_00026 gene encoding multimodular transpeptidase-transglycosylase PBP 1A and mreC in the B17 also support a mutation-dependent attenuation hypothesis because these genes have been associated with virulence (e.g. nrdA in P. aeruginosa [29], PBP 1A in group B streptococci [49], and MreC in Salmonella [50]). Alternatively, if rpoB mutation is more important, then the exact position of the mutations may be important. For example, TR was not attenuated, but it also had distinctly different mutations in the rpoB (Asp477Tyr and Pro496Ser) as compared to single amino acid substitutions of the mostly attenuated CR (Ser492Phe) and the fully attenuated B17 (Gln474Arg) strains. The large differential in SNP accumulation between passaged strains of CSF 259–93 and THC 02–90 are consistent with differential mutation process and coincidently may reflect differences at the lineage level [19].
Unfortunately, there are no molecular tools allowing for universal allelic exchange of genes in F. psychrophilum [44, 51]. Therefore, there is no direct way of assessing rpoB involvement in the process of rifampicin-induced attenuation. Introduction of the mutant rpoB allele from the B17, CR and TR into the wild-type F. psychrophilum would be the most direct method to investigate the role of rifampicin-resistant RNA polymerase in attenuation of these bacterial strains. Other options to characterize of attenuation in the CN, CR and B17 strains would be comparison of transcriptomes or more in-depth analysis of DNA methylation patterns between these and the virulent CSF 259–93 strain to examine possible global transcriptional changes from rpoB mutations or altered DNA methylation that might contribute to differential transcriptional regulation.
Differential DNA methylation may be associated with loss of virulence as altered DNA methylation affects these traits in other bacterial species [52, 53]. Others have found evidence for different methylation patterns among F. psychrophilum strains using restriction enzyme analysis [44, 54, 55]. Consequently, it was not entirely unexpected to find differences in methylation motifs based on the PacBio analysis (Additional file 1: Tables S3 and S4). It is remarkable, however, that there were no shared motifs between these two strains. Unfortunately, this also means that the comparison of the CR and TR strains provides no insight into the potential contribution of methylation to attenuation. Nevertheless, given that methylation and restriction enzymes can function to defend bacteria from foreign DNA, it is tempting to speculate that the two lineages of F. psychrophilum have diverged due to the influence of different phage communities. CSF 259–93 is most closely associated with freshwater fisheries whereas the THC 02–90 lineage is most closely associated with anadromous fish. It is likely that phage communities differ substantially between these ecosystems.
Conclusions
Our findings demonstrate that the two F. psychrophilum strains passaged with rifampicin harbored from 27.6 % (for THC 02–90) to 33.33 % (for CSF 259–93) more SNPs than the paired strains that were passaged without the antibiotic. Importantly, passaged THC 02–90 strains, regardless of rifampicin presence in media, revealed considerably more SNPs (roughly 20-fold) than the CSF 259–93 strains, and that difference is correlated with the fact that these two strains belong to two genetically divergent lineages [19]. We also present data consistent with distinctly different methylation motifs between these two lineages. We observed almost complete attenuation of CSF 259–93 passaged with rifampicin (CR) and significant loss of virulence of the CSF 259–93 strain passaged without antibiotic (CN). These reductions of virulence of the CN (28.1 % mortality) and CR (4 % mortality) strains occurred despite a limited number of SNPs, 6 and 9, respectively, as compared to 31 SNPs of the fully attenuated B17 strain. The B17 strain shares no SNPs with the CN or CR strains, although both B17 and CR share a loss of gliding motility. There are several additional SNP’s that could collectively contribute to reduced virulence.
Methods
Generation of rifampicin resistant F. psychrophilum strains
F. psychrophilum CSF-259-93 [19] and THC 02–90 [56] were used as parent strains to generate rifampicin resistant strains. Previously frozen glycerol stocks of F. psychrophilum CSF-259-93 (hereafter referred to as CW) and THC 02–90 (hereafter referred to as TW) were plated for isolation on tryptone yeast extract salts (TYES; 0.4 % tryptone, 0.04 % yeast extract, 0.05 % MgSO4 × 7 H2O, 0.05 % CaCl2 × 2 H2O, pH 7.2) agar and incubated at 16 °C for 5 days [57]. Subsequently, single colonies were passed to TYES agar containing 10 μg/ml of rifampicin (Sigma, St. Louis, MO, USA) and incubated at 15 °C for 6 days. Two of the resulting colonies were then selected, designated CSF 259–93. AR and THC 02–90. AR, and independently passed to TYES agar containing increasing concentrations of rifampicin (initially every 10 μg/ml per passage and 25 μg/ml after 100 μg/ml). This process was repeated until the CSF 259–93. AR and THC 02–90. AR strains achieved growth at Rif concentrations of 250 μg/ml. This required 17 passages with 259-93AR strain designated as 259–93. AR17 (hereafter designated as CR) and 17 passages for the THC 02–90. AR17 (hereafter designated as TR) strain. Following each passage, a portion of the recovered cells was harvested, resuspended in sterile 100 % glycerol and frozen at −80 °C. The rifampicin resistant CSF 259-03B.17 (hereafter designated as B17 strain) used in our experiments was independently isolated by LaFrentz and coworkers [13].
Passaging of F. psychrophilum strains without rifampicin
Previously frozen virulent CSF 259–93 and THC 02–90 strains were plated for isolation on TYES agar and incubated at 16 °C for 5 days. Two of the resultant colonies were selected for subsequent passages on TYES agar (total of 17 passages) leading to generation of CSF 259–93. N17 (hereafter referred to as CN) and THC 02–90. N17 strains (hereafter referred to as TN). Following each passage, a portion of the recovered cells was harvested, resuspended in sterile 100 % glycerol and frozen at −80 °C.
Growth comparison and bacterial cultures
A Bioscreen C system and EZ Experiment software (Growth Curves USA) were used to measure optical density of cultures to examine possible differences for in vitro growth kinetics. Briefly, bacterial strains were cultured statically in 5 ml of TYES broth from previously frozen stocks. After 3 days of incubation at 16 °C the cultures were diluted with TYES broth to OD 0.3 (at 600 nm) and loaded onto the Bioscreen C system. We used 10 × 10 honeycomb microplates, with triplicates of every strain (in 200 μl TYES broth per well) being statically incubated for 5 days at 16 °C with 10 s shaking before each measurement. The optical density measurements were collected using a 450–580 nm bandwidth filter every 3 h.
F. psychrophilum strains (CW, CN, CR, TW, TN, TR and B17) used later for genomic DNA or protein extractions, and for fish challenge were grown statically in 25 ml of TYES broth at 16 °C for 3 days.
Microscopy
Bacterial cell morphology was examined with Gram staining of heat-fixed bacteria obtained from TYES broth cultures that were incubated statically for 3 days at 16 °C. Colony morphology was examined with isolated colonies growing on TYES agar and gliding motility was observed by stab inoculation of gliding motility agar (GMA; 0.8 % nutrient broth and 0.75 % agar), similar to procedures described by Perez-Pascual and co-workers [58]. Both TYES and GMA plates were incubated for 3 days at 16 °C. Bacterial colonies were observed under 200× magnification with a Leica microscope equipped with an EC-3 digital camera. Leica Application Suite EZ (LAS EZ) software was used for image acquisition and analysis (Leica Microsystems). At least three independent biological and technical replicates per media type per strain were used.
rpoB and 16S rRNA analysis
To ensure culture purity before whole-genome sequencing, rpoB sequencing and 16S rRNA analysis were performed. Briefly, genomic DNA was isolated from 5 ml TYES cultures of the parent and passaged strains of F. psychrophilum with QIAamp DNA Mini Kit (QIAgen) according to the manufacturer’s instructions. External primers for rpoB amplification were 5′-AAAATCGGAACGGATTACGG-3′ and 5′-TTTTGAATTGTTTTTAAAGAGGTATTG-3′, and PCR involved 35 cycles of 94 °C for 30 s, 45 °C for 30 s and 68 °C for 4 min. Primers used to amplify internal rpoB segments for sequencing were described previously by Gliniewicz and coworkers [14]. All reactions included 2 mM MgSO4, 1 × PCR buffer, 0.2 mM dNTP mixture, 1 U of HiFi Taq polymerase (Invitrogen) with 5 ng DNA template and 0.1 μM of each primer in 50 μl reaction volume. 16S rRNA analysis was performed according to method described by Soule and coworkers with 16S_336fwd and 16S_517rvs primers used for target amplification and MaeIII digestion of amplified 16S rRNA to distinguish CSF 259–93 and THC 02–90 strains [19].
One-dimensional SDS-PAGE
Protein electrophoresis followed Laemmli (1970) with some modifications. Prior to electrophoresis 2 ml cultures of parent and passaged strains of F. psychrophilum (OD595 0.6) were centrifuged, cells were resuspended in 0.5 ml PBS and diluted 1:2 in sample buffer containing a reducing agent (100 mM β-mercaptoethanol) and boiled for 5 min. Proteins from bacterial whole-cell lysate were separated using pre-cast Any-kD polyacrylamide gels (Bio-Rad). Gels were used in a Mini-PROTEAN 3 electrophoresis cell (Bio-Rad) at 120 V for 70 min. Proteins were stained with Coomassie Blue and Precision Plus protein standards (Bio-Rad) were used to estimate the molecular mass of proteins. Analysis of carbohydrate extractions was conducted according to the method of LaFrentz and co-workers [13] and LPS fractions were silver stained with Pierce Silver Stain Kit according to manufacturer’s instruction (Thermo Scientific, USA). ChemiDoc XRS system and Image Lab 4.1 software were used for visualization (Bio-Rad).
Two-dimensional PAGE
Two-dimensional PAGE (2D PAGE) of F. psychrophilum proteins was performed as described earlier by Gliniewicz and co-workers [14]. Briefly, proteins from whole-cell lysates of F. psychrophilum strains were subjected to alkylation reduction and cleaned with 2D clean-up kits according to the manufacturer’s instructions (BioRad). Protein concentrations were estimated by using a QuickStart Bradford kit (BioRad). Samples from each strain with ~250 μg/μl protein per strip, were applied to immobilized pH gradient (IPG) strips (11 cm, pH 3–10) by passive rehydration for 1 h. Rehydrated IPG strips were covered with mineral oil and incubated overnight at room temperature. First dimension isoelectric focusing (IEF) was performed using a Protean IEF Cell (BioRad) with 0–250 V for 15 min, 0–8000 V rapid ramp for 40,000 Vh with 50 μA per strip. IPG strips were subsequently treated with DTT (2 % w/v) and iodoacetamide (2.5 % w/v) dissolved in equilibration buffer (6 M urea, 0.357 M Tris–HCl, pH 8.8, 2 % SDS, 20 % glycerol) with gentle rocking for 10 min. Second dimension separation was accomplished by application of IPG strips onto polyacrylamide gels with a linear 10–20 % Tris–HCl gradient and electrophoresis with Criterion Gel System (BioRad) and resolved at 200 V for 55 min. Gels were stained with SyPro Ruby Protein Stain according to the manufacturer’s directions. Digital images were collected using a Fluor-S Multi Imager (BioRad) and Quantity One software was used for comparison of protein profiles between bacterial strains (BioRad). For our discussion we only noted differences in protein profiles that were consistent across three biological replicates each having three technical replicates.
Genomic DNA extraction
Genomic DNA of the parent and passaged strains of F. psychrophilum were isolated with QIAamp DNA Mini Kit according to the manufacturer’s instructions (QIAgen) from 20 ml TYES broth cultures incubated at 16 °C for 5 days. DNA quality was analyzed with NanoDrop ND-1000 spectrophotometer (Thermo Scientific) and by agarose gel electrophoresis and visualization with ethidium bromide.
High throughput DNA sequencing
Genomic DNA libraries for CW and CSF 259-93B.17 strains were bar-coded by ligating adapters incorporating molecular identifiers during robust long-library construction. The libraries were quantified by fluorescence on a VictorX 384 well plate fluorometer (Parkin-Elmer). Quantified libraries were titrated to yield 8 % labeling of DNA capture beads using small volume emulsion PCR (emPCR) reactions and enough beads for sequencing were obtained by pooling the titration beads with a single medium volume emPCR reaction. Beads (1 × 106) for each strain were combined and used to load half of one 70 × 75 titanium picotiter plate. Sequencing was performed on a Roche 454 FLX Titanium instrument. Later, genomic DNA of six strains of F. psychrophilum (CW, CN, CR, TW, TN and TR) was subject to whole-genome sequencing with IonTorrent technology. Briefly, genomic DNA samples were fragmented using a Bioruptor 300 (Diagenode) for 37 min using a 30 s on-off cycling and temperature controlled sonication bath and had a peak apparent molecular weight of 300 nucleotide base pairs. The fragmented DNA was size selected and purified using a Pippin Prep elecro-elution device (Sage Science) set for a tight range harvest at 315 bp. Library construction was performed using the Ion Fragment Plus library kit (Life technologies). Library quantification and amplification efficiency was determined by real-time PCR. An appropriate number of library molecules were used to achieve 20 % bead labeling. Emulsion PCR and bead harvesting was performed on Ion One-Touch/One Touch ES instruments (Life technologies). Percent DNA bead labeling and number of beads harvested was determined using Sybr Gold stained beads and a guava easyCyte flow cytometer (Millipore). Sequencing was performed on an Ion Torrent PGM running Torrent Suite software version 2.0.1 (Life technologies). All strains were sequenced separately using a single 316 chip for each strain. Sequencing experiments were conducted in the Molecular Biology and Genomics Core at Washington State University. This whole genome shotgun project is available at DDBJ/EMBL/GenBank under the accession JRWA00000000, JRWB00000000, JRWC00000000, JRWD00000000, JRWE00000000, JRWF00000000. The version described in this paper is version JRWX01000000 where X is A, B, C, D, E, or F.
Single-nucleotide polymorphism analysis
CLC Genomic Workbench 5.0 (CLC bio) with default parameters was used for contig assembly, SNP detection and analysis of amino acid substitutions. For SNP detection, The Quality Based Variant Detection toolbox in CLC was used, with the following minor modifications to default settings. A minimum coverage of 4 was required with a variant frequency of 80 %. Only variants that resulted in an amino acid substitution in a coding sequence are reported. F. psychrophilum CSF 259–93 genome of 2,900,735 bp was used as reference (GenBank Accession number: CP007627) [59]. For the analysis of TN and TR strains the SNPs were compared with the sequenced parent TW and CSF 259–93 parent strains, and the F. psychrophilum JIP 02/86 genome (NCBI Reference Sequence: NC_009613.3).
DNA methylation analysis
Two F. psychrophilum strains (CR and TR) were selected for analysis of possible changes of DNA methylation patterns. Large-fragment libraries were created from 5 μg of genomic DNA from each strain. DNA was sheared at 20× speed code 15 through a large aperture ruby of a DigiLab Hydroshear Plus device (DigiLab, USA) and a larger-fragment library was constructed using the PacBio DNA Template Kit for 3–10 kb fragments (Pacific Biosciences, USA). Short-fragment libraries were constructed from 5 μg of genomic DNA from each strain with the DNA sheared at 20× speed code 6 through a small aperture ruby of a DigiLab Hydroshear Plus device (DigiLab, USA) device and prepared using the PacBio DNA Template Kit for 250 bp – 3 kb fragments (Pacific Biosciences, USA). The resulting libraries were quantified using a Qbit fluorometer (Life Technologies, USA) and size range determined on an Agilent 2100 Bioanlayzer using the DNA12000 kit (Agilent Technologies, USA). Peak size distributions of libraries were approximately 10 kb and 2 kb for the large and short library, respectively. The large libraries were annealed to primer, bound to C2 polymerase, loaded with magnetic beads and observed on 5 SMRT cells per library using single 120 min movies (Pacific Biosciences, USA). The short libraries were annealed to primer, bound to C2 polymerase, diffusion loaded into 4 SMRT cells per library, and observed using two 45 min movies per cell. Cumulatively, 13 movies from 9 SMRT cells were collected for each strain. Allora and HGAP software were used for assembly and analysis of the data (Pacific Biosciences, USA).
Fish rearing conditions
Rainbow trout (mean weight 1 g) were used to assess virulence of six F. psychrophilum strains. Fish were stocked in 19-liter flow-through tanks (25 fish per tank) supplied with dechlorinated municipal and fed 1.4 mm pelleted trout food (Rangen, Inc.) at 1 % body weight per day. Water temperature was maintained at 14 °C throughout the challenge experiments. The fish had no previous history of F. psychrophilum infection.
Assessment of attenuation
Fish were challenged by intraperitoneal injection with a 30-gauge needle with 25 μl of F. psychrophilum culture (OD595 = 0.2) resuspended in 25 μl of PBS. Triplicate groups of 25 fish were challenged with each F. psychrophilum strain or with PBS as a mock infected control group of 25 fish. An OD of 0.2 correlated to ~1.5 × 108 colony forming units (cfu)/ml and was estimated using the 6 × 6 drop plate method [60]. Mortalities were recorded daily for 28 days and kidney, liver and spleen tissues were streaked onto TYES agar to confirm the presence of yellow-pigmented bacteria that were presumptive F. psychrophilum. The University of Idaho Institute for Animal Care and Use Committee approved all animal husbandry and experimental challenge procedures.
Statistical analysis
SigmaPlot version 12 (Systat Software, Inc.) was used to calculate the areas-under-the-curve (AUC) for in vitro growth curves. NCSS ver. 7.1.19 software (NCSS, LCC) was used for one-way ANOVA with Tukey’s post hoc tests of endpoint and AUC data. Log-rank Mantel-Cox test was used to compare fish survival rates among different treatments. GraphPad Prism software (GraphPad Software, Inc.) was used for statistical analyses and for graph preparation.
Acknowledgements
Dr. S. Aguilar from Washington State University, and Dr. A. Long and T. Fehringer from University Idaho provided technical assistance. Dr. C. Moffitt and Dr. Z. Penney from University of Idaho provided help with micrographs. This project was supported in part by the University of Idaho/Washington State University Aquaculture Initiative, the Western Regional Aquaculture Center, and Washington State Agricultural Research Center.
Additional file
SDS-PAGE analysis of carbohydrate extractions from different F. psychrophilum CSF 259-93 and THC 02-90 strains. Table S1: Single nucleotide polymorphisms (SNPs) from F. psychrophilum THC 02-90 strain passaged without rifampicin (TN). Table S2: Single nucleotide polymorphisms (SNPs) from F. psychrophilum THC 02-90 strain passaged with rifampicin (TR). Table S3: DNA methylation motifs unique for the rifampicin passaged CSF 259-93 strain (CR). Table S4: DNA methylation motifs unique for the rifampicin passaged THC 02-90 strain (TR). (DOCX 637 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
KG, KC, KRS and DRC conceived the study. KG, LHO, MW, GDW and KKL were involved in DNA sequencing, sequence alignments and analysis of DNA methylation patterns. KG carried out proteomic and LPS characterization, growth kinetics and microscopy analysis of the strains used in the study. KC provided the B17 strain and supervised the disease challenged experiments. KG, LHO, KRS and DRC drafted the manuscript. KG and DRC performed the statistical analysis. All authors read and approved the final version of the manuscript.
Contributor Information
Karol Gliniewicz, Email: karol@uidaho.edu.
Mark Wildung, Email: wildung@wsu.edu.
Lisa H. Orfe, Email: lorfe@vetmed.wsu.edu
Gregory D. Wiens, Email: greg.wiens@ars.usda.gov
Kenneth D. Cain, Email: kcain@uidaho.edu
Kevin K. Lahmers, Email: klahmers@vt.edu
Kevin R. Snekvik, Email: ksnek@wsu.edu
Douglas R. Call, Email: drcall@wsu.edu
References
- 1.Schurig GG, Roop RM, 2nd, Bagchi T, Boyle S, Buhrman D, Sriranganathan N. Biological properties of RB51; a stable rough strain of Brucella abortus. Vet Microbiol. 1991;28(2):171–88. doi: 10.1016/0378-1135(91)90091-S. [DOI] [PubMed] [Google Scholar]
- 2.Wehrli W, Knusel F, Schmid K, Staehelin M. Interaction of rifamycin with bacterial RNA polymerase. Proc Natl Acad Sci U S A. 1968;61(2):667–73. doi: 10.1073/pnas.61.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manten A, Van Wijngaarden LJ. Development of drug resistance to rifampicin. Chemotherapy. 1969;14(2):93–100. doi: 10.1159/000220615. [DOI] [PubMed] [Google Scholar]
- 4.Jin DJ, Gross CA. Mapping and sequencing of mutations in the Escherichia coli rpoB gene that lead to rifampicin resistance. J Mol Biol. 1988;202(1):45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 5.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, et al. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104(6):901–12. doi: 10.1016/S0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 6.Tupin A, Gualtieri M, Roquet-Baneres F, Morichaud Z, Brodolin K, Leonetti JP. Resistance to rifampicin: at the crossroads between ecological, genomic and medical concerns. Int J Antimicrob Agents. 2010;35(6):519–23. doi: 10.1016/j.ijantimicag.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Klesius PH, Shoemaker CA. Development and use of modified live Edwardsiella ictaluri vaccine against enteric septicemia of catfish. Adv Vet Med. 1999;41:523–37. doi: 10.1016/S0065-3519(99)80039-1. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence M, Banes M. Tissue persistence and vaccine efficacy of an O-polysaccaride mutant strain of Edwarsiella ictaluri. J Aquat Anim Health. 2005;17:228–32. doi: 10.1577/H04-049.1. [DOI] [Google Scholar]
- 9.Olivares-Fuster O, Arias CR. Development and characterization of rifampicin-resistant mutants from high virulent strains of Flavobacterium columnare. J Fish Dis. 2011;34(5):385–94. doi: 10.1111/j.1365-2761.2011.01253.x. [DOI] [PubMed] [Google Scholar]
- 10.Pridgeon JW, Klesius PH. Development and efficacy of novobiocin and rifampicin-resistant Aeromonas hydrophila as novel vaccines in channel catfish and Nile tilapia. Vaccine. 2011;29(45):7896–904. doi: 10.1016/j.vaccine.2011.08.082. [DOI] [PubMed] [Google Scholar]
- 11.Shoemaker CA, Klesius PH, Evans J. In ovo methods for utilizing the modified live Edwardsiella ictaluri vaccine against enteric septicemia in channel catfish. Aquaculture. 2002;203:221–7. doi: 10.1016/S0044-8486(01)00631-7. [DOI] [Google Scholar]
- 12.Sun Y, Liu CS, Sun L. Isolation and analysis of the vaccine potential of an attenuated Edwardsiella tarda strain. Vaccine. 2010;28(38):6344–50. doi: 10.1016/j.vaccine.2010.06.101. [DOI] [PubMed] [Google Scholar]
- 13.LaFrentz BR, LaPatra SE, Call DR, Cain KD. Isolation of rifampicin resistant Flavobacterium psychrophilum strains and their potential as live attenuated vaccine candidates. Vaccine. 2008;26(44):5582–9. doi: 10.1016/j.vaccine.2008.07.083. [DOI] [PubMed] [Google Scholar]
- 14.Gliniewicz K, Plant KP, LaPatra SE, LaFrentz BR, Cain K, Snekvik KR, et al. Comparative proteomic analysis of virulent and rifampicin-attenuated Flavobacterium psychrophilum. J Fish Dis. 2012;35(7):529–39. doi: 10.1111/j.1365-2761.2012.01378.x. [DOI] [PubMed] [Google Scholar]
- 15.Adone R, Ciuchini F, Marianelli C, Tarantino M, Pistoia C, Marcon G, et al. Protective properties of rifampin-resistant rough mutants of Brucella melitensis. Infect Immun. 2005;73(7):4198–204. doi: 10.1128/IAI.73.7.4198-4204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu YH, Deng T, Sun BG, Sun L. Development and efficacy of an attenuated Vibrio harveyi vaccine candidate with cross protectivity against Vibrio alginolyticus. Fish Shellfish Immunol. 2012;32(6):1155–61. doi: 10.1016/j.fsi.2012.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Marianelli C, Ciuchini F, Tarantino M, Pasquali P, Adone R. Genetic bases of the rifampin resistance phenotype in Brucella spp. J Clin Microbiol. 2004;42(12):5439–43. doi: 10.1128/JCM.42.12.5439-5443.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swain P, Behera T, Mohapatra D, Nanda PK, Nayak SK, Meher PK, et al. Derivation of rough attenuated variants from smooth virulent Aeromonas hydrophila and their immunogenicity in fish. Vaccine. 2010;28(29):4626–31. doi: 10.1016/j.vaccine.2010.04.078. [DOI] [PubMed] [Google Scholar]
- 19.Soule M, Cain K, LaFrentz S, Call DR. Combining suppression subtractive hybridization and microarrays to map the intraspecies phylogeny of Flavobacterium psychrophilum. Infect Immun. 2005;73(6):3799–802. doi: 10.1128/IAI.73.6.3799-3802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42(5):373–97. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Alberola N, Campoy S, Barbe J, Erill I. Analysis of the SOS response of Vibrio and other bacteria with multiple chromosomes. BMC Genomics. 2012;13:58. doi: 10.1186/1471-2164-13-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall AR, Iles JC, MacLean RC. The fitness cost of rifampicin resistance in Pseudomonas aeruginosa depends on demand for RNA polymerase. Genetics. 2011;187(3):817–22. doi: 10.1534/genetics.110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mariam DH, Mengistu Y, Hoffner SE, Andersson DI. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2004;48(4):1289–94. doi: 10.1128/AAC.48.4.1289-1294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Neill AJ, Huovinen T, Fishwick CW, Chopra I. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob Agents Chemother. 2006;50(1):298–309. doi: 10.1128/AAC.50.1.298-309.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermott JC, Britton G, Goodwin TW. Carotenoid biosynthesis in a Flavobacterium sp.: stereochemistry of hydrogen elimination in the desaturation of phytoene to lycopene, rubixanthin and zeaxanthin. Biochem J. 1973;134(4):1115–7. doi: 10.1042/bj1341115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDermott JC, Brown DJ, Britton G, Goodwin TW. Alternative pathways of zeaxanthin biosynthesis in a Flavobacterium species. Biochem J. 1974;144(2):231–43. doi: 10.1042/bj1440231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasamontes L, Hug D, Tessier M, Hohmann HP, Schierle J, van Loon AP. Isolation and characterization of the carotenoid biosynthesis genes of Flavobacterium sp. strain R1534. Gene. 1997;185(1):35–41. doi: 10.1016/S0378-1119(96)00624-5. [DOI] [PubMed] [Google Scholar]
- 28.Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315(5811):525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 29.Sjoberg BM, Torrents E. Shift in ribonucleotide reductase gene expression in Pseudomonas aeruginosa during infection. Infect Immun. 2011;79(7):2663–9. doi: 10.1128/IAI.01212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arigoni F, Talabot F, Peitsch M, Edgerton MD, Meldrum E, Allet E, et al. A genome-based approach for the identification of essential bacterial genes. Nat Biotechnol. 1998;16(9):851–6. doi: 10.1038/nbt0998-851. [DOI] [PubMed] [Google Scholar]
- 31.Dassain M, Leroy A, Colosetti L, Carole S, Bouche JP. A new essential gene of the ‘minimal genome’ affecting cell division. Biochimie. 1999;81(8–9):889–95. doi: 10.1016/S0300-9084(99)00207-2. [DOI] [PubMed] [Google Scholar]
- 32.Ashour J, Hondalus MK. Phenotypic mutants of the intracellular actinomycete Rhodococcus equi created by in vivo Himar1 transposon mutagenesis. J Bacteriol. 2003;185(8):2644–52. doi: 10.1128/JB.185.8.2644-2652.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao LY, Groger R, Cox JS, Beverley SM, Lawson EH, Brown EJ. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect Immun. 2003;71(2):922–9. doi: 10.1128/IAI.71.2.922-929.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, Bastian JF, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202(2):209–15. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A. 2004;101(19):7427–32. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol. 2011;193(4):909–17. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin DJ, Gross CA. Characterization of the pleiotropic phenotypes of rifampin-resistant rpoB mutants of Escherichia coli. J Bacteriol. 1989;171(9):5229–31. doi: 10.1128/jb.171.9.5229-5231.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moeller R, Vlasic I, Reitz G, Nicholson WL. Role of altered rpoB alleles in Bacillus subtilis sporulation and spore resistance to heat, hydrogen peroxide, formaldehyde, and glutaraldehyde. Arch Microbiol. 2012;194(9):759–67. doi: 10.1007/s00203-012-0811-4. [DOI] [PubMed] [Google Scholar]
- 39.Moorman DR, Mandell GL. Characteristics of rifampin-resistant variants obtained from clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1981;20(6):709–13. doi: 10.1128/AAC.20.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dinic L, Akande P, Idigbe EO, Ani A, Onwujekwe D, Agbaji O, et al. Genetic determinants of drug-resistant tuberculosis among HIV-infected patients in Nigeria. J Clin Microbiol. 2012;50(9):2905–9. doi: 10.1128/JCM.00982-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahmo A, Hamdar Z, Kasaa I, Dabboussi F, Hamze M. Genotypic detection of rifampicin-resistant M. tuberculosis strains in Syrian and Lebanese patients. J Infect Public Health. 2012;5(6):381–7. doi: 10.1016/j.jiph.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 42.Tang K, Sun H, Zhao Y, Guo J, Zhang C, Feng Q, et al. Characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Sichuan in China. Tuberculosis (Edinb) 2013;93(1):89–95. doi: 10.1016/j.tube.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez B, Alvarez J, Menendez A, Guijarro JA. A mutant in one of two exbD loci of a TonB system in Flavobacterium psychrophilum shows attenuated virulence and confers protection against cold water disease. Microbiology. 2008;154(Pt 4):1144–51. doi: 10.1099/mic.0.2007/010900-0. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez B, Secades P, McBride MJ, Guijarro JA. Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum. Appl Environ Microbiol. 2004;70(1):581–7. doi: 10.1128/AEM.70.1.581-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsrud AL, LaFrentz SA, LaFrentz BR, Cain KD, Call DR. Differentiating 16S rRNA alleles of Flavobacterium psychrophilum using a simple PCR assay. J Fish Dis. 2007;30(3):175–80. doi: 10.1111/j.1365-2761.2007.00795.x. [DOI] [PubMed] [Google Scholar]
- 46.Gomez E, Perez-Pascual D, Fernandez L, Mendez J, Reimundo P, Navais R, et al. Construction and validation of a GFP-based vector for promoter expression analysis in the fish pathogen Flavobacterium psychrophilum. Gene. 2012;497(2):263–8. doi: 10.1016/j.gene.2012.01.069. [DOI] [PubMed] [Google Scholar]
- 47.Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–72. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundquist A, Ronaghi M, Tang H, Pevzner P, Batzoglou S. Whole-genome sequencing and assembly with high-throughput, short-read technologies. PLoS One. 2007;2(5) doi: 10.1371/journal.pone.0000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones AL, Mertz RH, Carl DJ, Rubens CE. A streptococcal penicillin-binding protein is critical for resisting innate airway defenses in the neonatal lung. J Immunol. 2007;179(5):3196–202. doi: 10.4049/jimmunol.179.5.3196. [DOI] [PubMed] [Google Scholar]
- 50.Bulmer DM, Kharraz L, Grant AJ, Dean P, Morgan FJ, Karavolos MH, et al. The bacterial cytoskeleton modulates motility, type 3 secretion, and colonization in Salmonella. PLoS Pathog. 2012;8(1) doi: 10.1371/journal.ppat.1002500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rhodes RG, Pucker HG, McBride MJ. Development and use of a gene deletion strategy for Flavobacterium johnsoniae to identify the redundant gliding motility genes remF, remG, remH, and remI. J Bacteriol. 2011;193(10):2418–28. doi: 10.1128/JB.00117-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, et al. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol. 2012;30(12):1232–9. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar R, Mukhopadhyay AK, Ghosh P, Rao DN. Comparative transcriptomics of H. pylori strains AM5, SS1 and their hpyAVIBM deletion mutants: possible roles of cytosine methylation. PLoS One. 2012;7(8):e42303. doi: 10.1371/journal.pone.0042303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen YC, Davis MA, Lapatra SE, Cain KD, Snekvik KR, Call DR. Genetic diversity of Flavobacterium psychrophilum recovered from commercially raised rainbow trout, Oncorhynchus mykiss (Walbaum), and spawning coho salmon, O. kisutch (Walbaum) J Fish Dis. 2008;31(10):765–73. doi: 10.1111/j.1365-2761.2008.00950.x. [DOI] [PubMed] [Google Scholar]
- 55.Soule M, LaFrentz S, Cain K, LaPatra S, Call DR. Polymorphisms in 16S rRNA genes of Flavobacterium psychrophilum correlate with elastin hydrolysis and tetracycline resistance. Dis Aquat Organ. 2005;65(3):209–16. doi: 10.3354/dao065209. [DOI] [PubMed] [Google Scholar]
- 56.Chakroun C, Grimont F, Urdaci MC, Bernardet JF. Fingerprinting of Flavobacterium psychrophilum isolates by ribotyping and plasmid profiling. Dis Aquat Organ. 1998;33(3):167–77. doi: 10.3354/dao033167. [DOI] [PubMed] [Google Scholar]
- 57.Holt R, Rohovec J, Fryer J. Bacterial cold-water disease. In: Inglis V, Roberts R, Bromage N, editors. Bacterial disease of fish. Oxford: Blackwell Scientific Publications; 1993. pp. 3–22. [Google Scholar]
- 58.Perez-Pascual D, Menendez A, Fernandez L, Mendez J, Reimundo P, Navais R, et al. Spreading versus biomass production by colonies of the fish pathogen Flavobacterium psychrophilum: role of the nutrient concentration. Int Microbiol. 2009;12(4):207–14. [PubMed] [Google Scholar]
- 59.Wiens GD, LaPatra S, Welch TJ, Rexroad CE, Call DR, Cain K, et al. Complete genome sequence of Flavobacteirum psychrophilum strain CSF259-93 used to select rainbow trout for increased genetic resistance against bacterial coldwater disease. Genome Announcements--Prokaryotes, In Press. [DOI] [PMC free article] [PubMed]
- 60.Chen CY, Nace GW, Irwin PL. A 6 × 6 drop plate method for simultaneous colony counting and MPN enumeration of Campylobacter jejuni, Listeria monocytogenes, and Escherichia coli. J Microbiol Methods. 2003;55(2):475–9. doi: 10.1016/S0167-7012(03)00194-5. [DOI] [PubMed] [Google Scholar]


