Abstract
The tenacious association between strains of the heterotrophic alphaproteobacterial genus Acidiphilium and chemolithotrophic iron oxidizing bacteria has long been known. In this context the genome of the heterotroph Acidiphilium sp. JA12-A1, an isolate from an iron oxidizing mixed culture derived from a pilot plant for bioremediation of acid mine drainage, was determined with the aim to reveal metabolic properties that are fundamental for the syntrophic interaction between Acidiphilium sp. JA12-A1 and the co-occurring chemolithoautotrophic iron oxidizer. The genome sequence consists of 4.18 Mbp on 297 contigs and harbors 4015 protein-coding genes and 50 RNA genes. Additionally, the molecular and functional organization of the Acidiphilium sp. JA12-A1 draft genome was compared to those of the close relatives Acidiphilium cryptum JF-5, Acidiphilium multivorum AIU301 and Acidiphilium sp. PM DSM 24941. The comparative genome analysis underlines the close relationship between these strains and the highly similar metabolic potential supports the idea that other Acidiphilium strains play a similar role in various acid mine drainage communities. Nevertheless, in contrast to other closely related strains Acidiphilium sp. JA12-A1 may be able to take up phosphonates as an additional source of phosphor.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-015-0040-y) contains supplementary material, which is available to authorized users.
Keywords: Acidiphilium sp. JA12-A1, acid mine drainage, AMD, microbial community, acidophilic bacteria
Introduction
Strains of the alphaproteobacterial genus Acidiphilium have first been isolated from supposed pure cultures of iron oxidizing bacteria such as Acidithiobacillus ferrooxidans [1]. Later on, Acidiphilium spp. have also been identified as characteristic members of the microbial communities in acid mine drainage and mining associated water bodies [2–5]. Although the physiological role of these heterotrophic acidophiles within the microbial community has not yet been completely elucidated, the tenacious association between them and the chemolithoautotrophic iron oxidizers has often been reported to be problematic for the isolation of the iron oxidizing bacteria [1, 6, 7]. Several attempts have been undertaken to investigate the interaction between the iron oxidizing bacterium Acidithiobacillus ferrooxidans and Acidiphilium spp. In a co-culture with Acidiphilium acidophilum the increased growth rate and ferrous iron oxidation rate of Acidithiobacillus ferrooxidans have indicated a stimulating influence of Acidiphiliumacidophilus on Acidithiobacillus ferrooxidans [8]. A stable isotope probe based proteome analysis of an Acidithiobacillus ferrooxidans/Acidiphilium cryptum mixed culture has revealed carbon dioxide transfer from the heterotroph to the iron oxidizing bacterium [9]. Based on the absence of organic carbon and energy sources in the cultivation media of iron oxidizing bacteria it has been suggested that Acidiphilium spp. benefit in turn from secreted metabolites and remnants of the biomass from the iron oxidizers by utilizing them as carbon and energy sources [10–12].
Since such an interaction is not only relevant for the isolation and cultivation of iron oxidizing bacteria but also for the general understanding of the ecology of microbial communities in AMD, we were interested in elucidating the potential of Acidiphilium for such a syntrophic interaction. Therefore we sequenced and analyzed the genome of Acidiphilium sp. JA12-A1 with special focus on transport systems for the uptake of nutrients, the pathways of nutrient assimilation and the general energy metabolism. The resulting permanent draft genome was also compared to the genomes of the close relatives Acidiphilium cryptum JF-5, Acidiphilium multivorum AUI301 and Acidiphilium sp. PM DSM 24941 regarding the genome structure and the functional organization.
Organism Information
Classification and features
Strain Acidiphilium sp. JA12-A1 was detected as the heterotrophic contamination in the mixed culture JA12 of a novel chemolithoautotrophic iron oxidizing bacterium [13], which is related to “Ferrovum myxofaciens” P3G [7, 14]. The iron oxidizing mixed culture originated from a pilot plant for the biological remediation of AMD close to a lignite mining site in Lusatia, Germany [5, 13, 15]. Acidiphilium sp. JA12-A1 was isolated from the mixed culture by cultivation in SJH medium [16, 17] (Table 1, Additional file 1).
Table 1.
Classification and general features of Acidiphilium sp. JA12-A1 [32]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [32] | |
| Phylum Proteobacteria | TAS [33–35] | ||
| Class Alphaproteobacteria | TAS [34, 36] | ||
| Order Rhodospirillales | TAS [37, 38] | ||
| Family Acetobacteraceae | TAS [39, 40] | ||
| Genus Acidiphilium | TAS [2, 41, 42] | ||
| Species Acidiphilium sp. | TAS [2] | ||
| Strain: JA12-A1 | TAS [2] | ||
| Gram stain | Negative | NAS | |
| Cell shape | Rod | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Not reported | ||
| Temperature range | Mesophile | NAS | |
| Optimum temperature | 30 °C | NAS | |
| pH range; Optimum | Not reported | ||
| Carbon source | Heterotroph (galactose, glucose, tryptic soy broth, fructose, yeast extract) | NAS | |
| MIGS-6 | Habitat | Acid mine drainage | NAS |
| MIGS-6.3 | Salinity | Not reported | |
| MIGS-22 | Oxygen requirement | Aerobic, anaerobic | NAS |
| MIGS-15 | Biotic relationship | Free-living | NAS |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | Lignite mining site, Lusatia, Germany | NAS |
| MIGS-5 | Sample collection | 2011 | NAS |
| MIGS-4.1 | Latitude | 51° 28' 10.38'' N | NAS |
| MIGS-4.2 | Longitude | 14° 28' 22.19'' E | NAS |
| MIGS-4.4 | Altitude | 125.45 m | NAS |
aEvidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [43]
The complete 16S rRNA gene sequence of Acidiphilium sp. JA12-A1 was compared to the non-redundant nucleotide collection of the NCBI using NCBI MegaBLAST [18, 19]. The analysis of the 100 best hits revealed a sequence similarity of 99 % to 16S rRNA gene fragments of Acidiphilium multivorum AUI301, Acidiphilium cryptum JF-5, Acidiphilium organovorum TFC, Acidiphilium sp. SJH, and “Acidiphiliumsymbioticum” and others, and a sequence similarity of 95 % to Acidiphilium acidophilum MS Silver, Acidiphilium angustum ATCC 35903 and Acidiphilium rubrum. These gene fragments also formed the basis for the calculation of a dendrogram illustrating the phylogenetic neighborhood of Acidiphilium sp. JA12-A1 (Fig. 1).
Fig. 1.
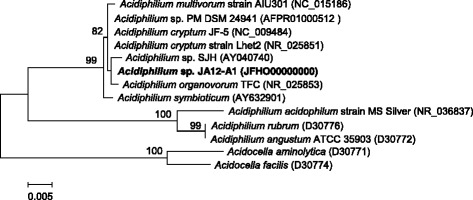
Dendrogram of strains of the genus Acidiphilium - based on partial 16S rRNA gene sequences. The dendrogram was calculated with MEGA5 [20] using the Maximum Likelihood method based on the Jukes-Cantor model [21]. The analyzed sequences were aligned by CLUSTALW [22]. The clustering of the sequences was tested by the bootstrap approach with 1000 repeats. The length of the tree branches was scaled according to the number of substitutions per site (see size bar). All strains used in the analysis, except Acidiphilium cryptum JF-5 and Acidiphilium sp. SJH, are type strains of their respective species [23–30] with Acidiphilium cryptum representing the genus Acidiphilium as the designated type species [2]. Acidocella aminolytica (D300771) and Acidocella facilis (D30774) were used as outgroup. The 16S rRNA gene sequence for Acidiphilium sp. PM DSM 24941 can be found under the locus tag APM_R0045 on contig Ctg_00688 (AFPR01000512) of the whole genome shotgun sequence. Whole genome sequences are only available for Acidiphilium cryptum JF-5, Acidiphilium multivorum AIU301, Acidiphilium sp. PM DSM 24941 and Acidiphilium angustum ATCC 35903 (GOLD project IDs: Gc00559, Gc01862, Gi09776, Gi0051610; accession numbers: NC_009484, NC_015186; AFPR00000000, JNJH00000000)
The 16S rRNA gene sequences cluster into two distinct subgroups within the genus Acidiphilium. The novel strain Acidiphilium sp. JA12-A1 belongs to the same subgroup as Acidiphilium cryptumJF-5,Acidiphilium multivorum AIU301 and Acidiphilium sp. PM DSM 24941.
In terms of physiological features Acidiphilium sp. JA12-A1 appears to be closely related to the type strain Acidiphilium cryptum Lhet2 [2]: Acidiphilium sp. JA12-A1 is a Gram-negative, rod-shaped (ca. 1.9 μm × 0.7 μm), motile alphaproteobacterium, which lives under acidophilic conditions. It has a chemoorganotrophic lifestyle growing with galactose, fructose, yeast extract and soy broth as growth substrates. In the mixed culture with the iron oxidizer “Ferrovum” sp. JA12 [31] the proportion of Acidiphilium sp. JA12-A1 was estimated by terminal restriction fragment length polymorphism (T-RFLP) analysis to vary between 1 % and 50 % depending on the ferrous iron concentration and growth phase (unpublished results). An electron micrograph of Acidiphilium sp. JA12 is provided in Fig. 2.
Fig. 2.
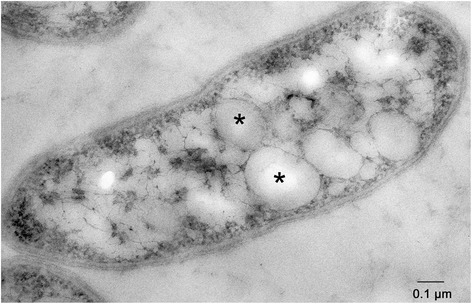
Transmission electron micrograph of Acidiphilium sp. JA12-A1 (ultrathin section, post-staining with 4 % uranyl acetate). PHB granula are marked by asterisks. The cells were harvested at the beginning of the fast growth phase
Genome sequencing information
Genome project history
The genome of Acidiphilium sp. JA12-A1 was sequenced to obtain genetic information on physiological properties that may play a fundamental role in its tenacious association with the co-occurring iron oxidizing bacterium in the mixed culture JA12. The permanent draft genome sequence is available at the NCBI with the accession number JFHO00000000 (genome project number 238988). The cultivation and genome sequence analysis was undertaken at the TU Bergakademie Freiberg while the genome sequencing and annotation was performed at Göttingen Genomics Laboratory (G2L). Table 2 provides a summary of the project information according to MIGS compliance [32].
Table 2.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Improved high-quality draft |
| MIGS-28 | Libraries used | Two genomic libraries: 454 pyrosequencing shotgun library, Illumina paired-end library (1 kb insert size) |
| MIGS 29 | Sequencing platforms | 454 GS FLX Titanium, Illumina GAII |
| MIGS 31.2 | Fold coverage | 18.7 × 454, 54.8 × Illumina |
| MIGS 30 | Assemblers | Newbler 2.8, MIRA 3.4 |
| MIGS 32 | Gene calling method | YACOP, Glimmer |
| Locus Tag | ACIDI | |
| Genbank ID | JFHO01000000 | |
| GenBank Date of Release | 2014-05-20 | |
| GOLD ID | Gi0008223 | |
| BIOPROJECT | PRJNA238988 | |
| MIGS 13 | Source Material Identifier | TU BAF Acidi |
| Project relevance | Environmental and biotechnological |
Growth conditions and genomic DNA preparation
Acidiphilium sp. JA12-A1 was cultivated in liquid SJH medium [16, 17] at 30 °C. It was continuously shaken on a rotary shaker at 120 rpm. The cells were harvested by centrifugation at 10,000 × g. The DNA was isolated using the Ultra Clean™ Microbial DNA Isolation Kit (MoBio, Carlsbad, CA) according to the manufacturer’s instructions.
Genome sequencing and assembly
Genome sequencing of Acidiphilium sp. JA12-A1 was performed via a hybrid approach using the 454 GS-FLX TitaniumXL system (Titanium GS70 chemistry, Roche Life Science, Mannheim, Germany) and the Genome Analyzer II (Illumina, San Diego, CA). Shotgun libraries were prepared according to the manufacturer's protocols, resulting in 126,343 reads for 454 shotgun and 10,136,209 112-bp paired-end Illumina reads. We used all 126.343 454 shotgun reads and 3,000,000 of the 112-bp paired-end Illumina reads for the initial hybrid de-novo assembly, which was calculated using the MIRA 3.4 [44] and Newbler 2.8 (Roche Life Science, Mannheim, Germany) software. The final assembly contained 297 contigs with a 73.5-times coverage on average.
Genome annotation
The software tools YACOP and Glimmer [45] were used for automatic gene prediction, while identification of rRNA and tRNA genes was performed using RNAmmer and tRNAscan, respectively [46, 47]. An automatic annotation was performed within the integrated microbial genomes-expert review (IMG-ER) system [48, 49] and subsequently curated manually by using the Swiss-Prot, TrEMBL, and InterPro databases [50].
Genome Properties
The draft genome of Acidiphilium sp. JA12-A1 consists of 4.18 Mbp on 298 contigs, of which 99 have a length of at least 10 kbp. Genome features are summarized in Table 3. The average G + C content is 66.9 %. The draft genome encodes 4065 genes in total, of which 4015 (98.8 %) are predicted protein coding genes and 50 (1.2 %) are RNA genes. 2663 (65.5 %) genes are assigned to COG groups (Table 4), 1238 (30.5 %) are connected to KEGG pathways and 520 (12.8 %) are assigned to the transporter classification. A comparison of genome features of Acidiphilium sp. JA12-A1 to the genomes of Acidiphilium cryptum JF-5, Acidiphilium multivorum AUI301 and Acidiphilium sp. PM, DSM 24941 is provided in Table 5.
Table 3.
Genome statistics Acidiphilium sp. JA12-A1
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 4,184,331 | 100.0 |
| DNA coding (bp) | 3,699,946 | 88.4 |
| DNA G + C (bp) | 2,801,106 | 66.9 |
| DNA scaffolds | 298 | |
| Total genes | 4,065 | 100.0 |
| Protein coding genes | 4,015 | 98.8 |
| RNA genes | 50 | 1.2 |
| Pseudo genes | 293 | 7.2 |
| Genes in internal clusters | 3,092 | 76.1 |
| Genes with function prediction | 3,193 | 78.6 |
| Genes assigned to COGs | 2,663 | 65.5 |
| Genes with Pfam domains | 3,191 | 78.5 |
| Genes with signal peptides | 268 | 6.6 |
| Genes with transmembrane helices | 857 | 21.1 |
| CRISPR repeats | Not reported |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 147 | 5.0 | Translation, ribosomal structure and biogenesis |
| A | 0 | 0.0 | RNA processing and modification |
| K | 180 | 6.1 | Transcription |
| L | 157 | 5.3 | Replication, recombination and repair |
| B | 2 | 0.1 | Chromatin structure and dynamics |
| D | 167 | 5.7 | Cell cycle control, Cell division, chromosome partitioning |
| V | 35 | 1.2 | Defense mechanisms |
| T | 77 | 2.6 | Signal transduction mechanisms |
| M | 167 | 5.7 | Cell wall/membrane biogenesis |
| N | 44 | 1.5 | Cell motility |
| U | 77 | 2.6 | Intracellular trafficking and secretion |
| O | 107 | 3.6 | Posttranslational modification, protein turnover, chaperones |
| C | 260 | 8.8 | Energy production and conversion |
| G | 247 | 8.3 | Carbohydrate transport and metabolism |
| E | 294 | 10.0 | Amino acid transport and metabolism |
| F | 66 | 2.2 | Nucleotide transport and metabolism |
| H | 125 | 4.2 | Coenzyme transport and metabolism |
| I | 164 | 5.6 | Lipid transport and metabolism |
| P | 124 | 4.2 | Inorganic ion transport and metabolism |
| Q | 89 | 3.0 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 320 | 10.8 | General function prediction only |
| S | 241 | 8.2 | Function unknown |
| - | 1,400 | 34.4 | Not in COGs |
The total is based on the total number of protein coding genes in the genome
Table 5.
Comparison of genome features of Acidiphilium sp. JA12-A1 to close relatives
| Genome features | Genome name | |||
|---|---|---|---|---|
| A. cryptum JF-5a | A. multivorum AIU301b | Acidiphilium sp. PM DSM 24941c | Acidiphilium sp. JA12-A1d | |
| Sequencing status | Finished | Finished | Draft | Permanent draft |
| Genome size (Mbp) | 4.0 | 4.2 | 3.9 | 4.2 |
| Number of plasmids | 8 | 8 | 9 | Not reported |
| GC (percentage) | 67.1 % | 67.0 % | 66.4 % | 66.9 % |
| Total gene count | 3,701 | 4,004 | 3,908 | 4,065 |
| Number of CDS genes (percentage) | 3,637 (98.3 %) | 3,948 (98. 6 %) | 3,859 (98.8 %) | 4,015 (98.8 %) |
| Number of RNA genes | 64 (1.7 %) | 56 (1.4 %) | 49 (1.3 %) | 50 (1.2 %) |
| Number of genes assigned to COGs (percentage) | 2,830 (79.1 %) | 3,188 (76.5 %) | 3,116 (79.7 %) | 2,663 (65.5 %) |
| Number of genes connected to KEGG pathways (percentage) | 1,197 (32.3 %) | 1,283 (32.0 %) | 1,133 (29.0 %) | 1,238 (30.5 %) |
| Number of genes assigned to enzymes (percentage) | 1,055 (28.5 %) | 1,107 (27.7 %) | 965 (24.7 %) | 1,076 (26.5 %) |
| Number of genes assigned to transporter classification (percentage) | 524 (14.1 %) | 562 (14.0 %) | 573 (14.7 %) | 520 (12.8 %) |
| Number of genes coding transmembrane proteins (percentage) | 817 (22.1 %) | 880 (22.0 %) | 839 (21.5 %) | 857 (21.1 %) |
| Number of genes with signal peptides (percentage) | 240 (6.5 %) | 266 (6.6 %) | 232 (5.9 %) | 268 (6.6 %) |
aaccession number: NC_009484; bNC_015186; cAFPR00000000; dJFHO00000000
Insights from the genome sequence
In order to understand the potential interaction between Acidiphilium sp. JA12-A1 and the iron oxidizer “Ferrovum” sp. JA12 in the mixed culture we analyzed the genome of Acidiphilium sp. JA12-A1 with special focus on genes that may be involved in the utilization of “Ferrovum” derived organic substances as an energy source and as growth substrates.
The genome analysis revealed six genes that encode for putative oligo- and polysaccharide hydrolyzing enzymes, among which we identified α-amylases or amylase-related enzymes, β-glucosidase, endoglucanase, a trehalase and a glycogen-debranching enzyme. Acidiphilium sp. JA12-A1 may use these enzymes to break down polysaccharides that are part of the cell envelope of the iron oxidizer “Ferrovum” or that are excreted as slimes. Applying the EBI InterProScan to the sequences of these enzymes resulted in predicted N-terminal signal peptides in the β-glucosidase and endoglucanase which indicates a potential excretion of these enzymes.
The genome of Acidiphilium sp. JA12-A1 encodes a variety of transport systems to take up secreted organic compounds or the products of the hydrolysis of polysaccharides. These transport systems comprise annotated sugar transporters or sugar phosphate permeases of the major facilitator family, 15 ABC-transport systems for mono- and disaccharides and a phosphotransferase system (PTS) of the fructose type. The ABC-transporters are predicted to take up ribose, xylose, galactose or similar monosaccharides. The PTS in Acidiphilium sp. JA12-A1 consists, similar to the PTS of other Acidiphilium strains, of two fusion proteins (HPr/EI/EIIA and EIIB/EIIC).
Based on the genome sequence we reconstructed the metabolic pathways that may enable Acidiphilium sp. JA12-A1 to gain energy by the complete aerobic oxidation of organic compounds, preferably of monosaccharides. Although we did not identify the fructose-6-phosphate kinase, one of the key enzymes of the glycolysis, Acidiphilium sp. JA12-A1 may bypass the reaction via the activity of enzymes of the pentosephosphate pathway, thus still being able to convert glucose to acetyl-CoA. Acetyl-CoA is further oxidized to carbon dioxide by the citrate cycle and the electrons are transferred to oxygen by the protein complexes of the aerobic respiratory chain. We also identified gene clusters encoding the subunits of a photosynthetic reaction center, associated cytochromes and proteins involved in the biogenesis of the reaction center proteins that may enable Acidiphilium sp. JA12-A1 to use light as additional energy source.
In addition to the aerobic respiration Acidiphilium sp. JA12-A1 may also be able to reduce ferric iron under microaerobic or anaerobic conditions as it has been described for other Acidiphilium strains [51, 52]. Despite of the experimental evidence for the ferric iron reduction, the proteins that are involved in the direct reduction of ferric iron in acidophiles have still not been identified [53]. The genome analysis of Acidiphilium sp. JA12-A1 also failed to reveal any further details of the electron transfer processes to ferric iron.
Apart from providing the source of energy the sugar compounds also appear to be the preferred carbon source for the biomass production in Acidiphilium sp. JA12-A1. We inferred the pathways that are necessary for the conversion of the monosaccharides to the precursors of the biomass production, such as the amino sugar and nucleotide sugar metabolism, the citrate cycle, the fatty acid synthesis and the purine and pyrimidine metabolism. Besides the synthesis of biomass there is genetic evidence for the storage of carbon compounds as polyhydroxybutyrate (PHB) which is further supported by transmission electron microscopic analysis of representative cells showing PHB granula (Fig. 2). Acidiphilium sp. JA12-A1 also appears to be able to fix carbon dioxide heterotrophically, since its genome encodes a pyruvate carboxylase and a pyruvate carboxykinase.
Extended insights
Although there are four genome sequences of species belonging to the genus Acidiphilium to compare the genome of strain JA12-A1 with, we focused our comparative genomics approach on Acidiphilium cryptum JF-5, Acidiphilium multivorum AUI301, Acidiphilium sp. PM DSM 24941 and Acidiphilium sp. JA12-A1. A comparison of the genomes of Acidiphilium sp. JA12-A1 and Acidiphilium angustum ATCC 35903 confirmed the phylogenetic distance and revealed that these genomes cannot be meaningfully aligned (results not shown). Therefore, the circular representation of the genome comparisons (Fig. 3) and the Venn diagram summarizing orthologous genes between the genomes are limited to strains belonging to the same phylogenetic cluster as Acidiphilium sp. JA12-A1 (Fig. 4).
Fig. 3.
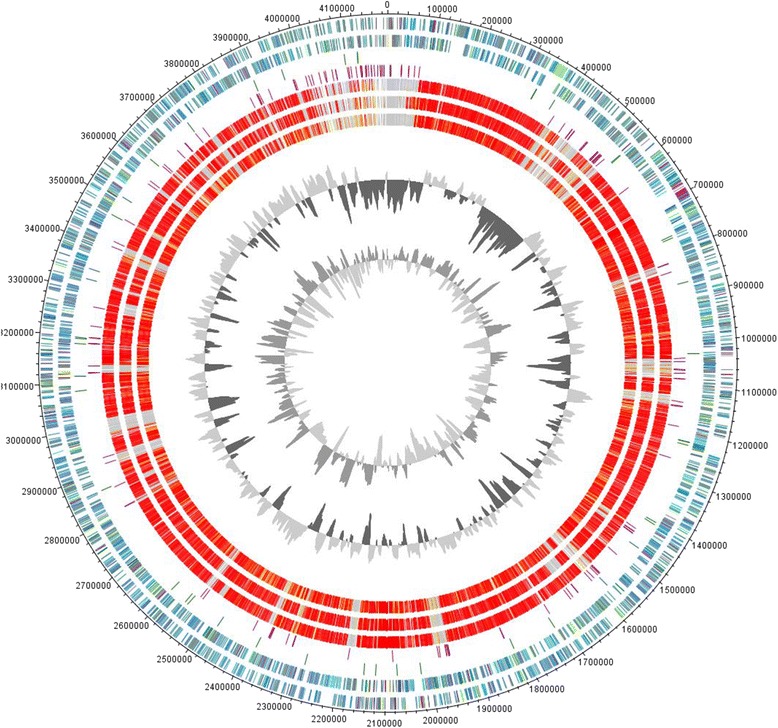
Circular representation of the genome comparison of Acidiphilium sp. JA12-A1 with other Acidiphilium strains. A: The genes encoded by the leading and the lagging strand (outer circles 1 and 2) of Acidiphilium sp. JA12-A1 are marked in COG colors in the artificial chromosome map. The genes for tRNAs and transposases in Acidiphilium sp. JA12-A1 are shown in circles 3 and 4, respectively. The presence of orthologous genes is indicated for the genomes of Acidiphilium cryptum JF-5 (CP000689-CP000697), Acidiphilium multivorum AIU301 (AP012035-AP012043) and Acidiphilium sp. PM DSM 24941 (circle 5 to 7). The two innermost plots represent the GC-content and the GC-skew
Fig. 4.
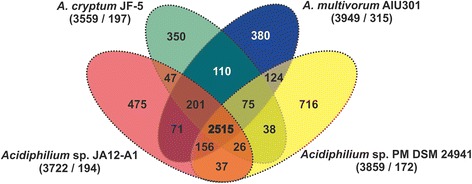
Venn diagramm of the genome comparison of Acidiphilium sp. JA12-A1 with other Acidiphilium strains. Venn diagram showing the orthologous genes between Acidiphilium sp. JA12-A1, Acidiphilium cryptum JF-5 (CP000689-CP000697), Acidiphilium multivorum AIU301 (AP012035-AP012043) and Acidiphilium sp. PM DSM 24941 (AFPR00000000). Ortholog detection was done with the Proteinortho software (blastp) with an similarity cutoff of 50 % and an E-value of 1e-10. The total number of genes and paralogs, respectively, are depicted under the corresponding species name. Open reading frames (ORFs) that were classified as pseudogenes, were not included in this analysis
The circular representation of genome sequences of four Acidiphilium strains revealed a high structural similarity of the genomes (Fig. 3). To identify orthologous genes between all four organisms, we performed a whole genome comparison. To prepare the data for analysis we used the scripts ncbi_ftp_download v0.2, cat_seq v0.1 and cds_extractor v0.6 [54] and Proteinortho v5.04 [55] with a similarity cutoff of 50 % and an E-value of 1e-10. Paralogous genes detected for all genomes were not included into this approach. All four strains have a core genome comprising 2515 genes, which is up to 70 % of the genes present in a single genome (Fig. 4). Acidiphilium JA12-A1 has 2943 orthologous genes in common with Acidiphilium multivorum AIU301, 2789 with Acidiphilium cryptum JF-5 and 2734 with Acidiphilium sp. PM DSM 24941. We detected the highest number of orthologous genes (2901) between Acidiphilium cryptum JF-5 and Acidiphilium multivorum AIU301. Acidiphilium sp. PM DSM 24941 and Acidiphilium multivorum AIU301 have 2870 in common, while Acidiphilium cryptum JF-5 and Acidiphilium sp. PM DSM 24941 share 2654 genes. Acidiphilium sp. PM DSM 24941 harbors the highest number of singletons (716) followed by Acidiphilium JA12-A1 with 475, Acidiphilium multivorum AIU301 with 381 and Acidiphilium cryptum JF-5 with 350, respectively. This, therefore, confirms the high degree of similarity among the various Acidiphilium strains as already concluded from the 16S rRNA gene based phylogeny (Fig. 1). Moreover, the high degree of congruence of the selected genome features provided in Table 5 demonstrates the high similarity among the four genomes with respect to the functional organization, (e.g. number of genes assigned to various COG functional categories (not shown), and pathways of the central metabolism).
Despite the high similarity in genome organization and content there are also unique genes in each of the Acidiphilium species that were included in this comparative genome analysis. For instance, Acidiphilium sp. JA12-A1, Acidiphilium cryptum JF-5 and Acidiphilium multivorum AUI301 contain a cluster of homologous genes encoding phosphonate C-P-lyases which are required for utilization of organic phosphate compounds. However, of those only Acidiphilium sp. JA12-A1 encodes a putative phosphonate specific ABC transporter. ABC transporter encoding genes are usually clustered. In the case of Acidiphilium sp. JA12 the genes are spread within the genome indicating that these have possibly been acquired via horizontal gene transfer.
Conclusions
The microbial communities of AMD and mining associated water bodies have been investigated in some detail over the last decades [3, 5, 10–12, 14, 56–58]. All of these reports agree on the supposed role of heterotrophic microorganisms, including members of the genus Acidiphilium, regarding their utilization of organic substances secreted by other community members or derived from microbial cell decay.
Analyzing the genome sequence of the novel strain Acidiphilium sp. JA12-A1 we inferred such an interspecies carbon transfer in an iron oxidizing mixed culture derived from a pilot plant for the biological remediation of AMD. The potential carbon transfer involves Acidiphilium sp. JA12-A1 excreting polysaccharide hydrolyzing enzymes, such as β-glucosidases or endoglucanases, to break down cell envelope polysaccharides from decaying cells and from the co-occurring iron oxidizer that is related to F. myxofaciens P3G [7]. Monosaccharides originating from polysaccharide hydrolysis or from lysed cells are taken up by Acidiphilium sp. JA12-A1 via specific uptake systems to produce bacterial biomass. Alternatively, the monosaccharides or parts thereof are oxidized to gain energy for the cellular metabolism. Under aerobic conditions the electron donor is completely oxidized to carbon dioxide which is the preferred carbon source for the autotrophic iron oxidizer. However, the iron oxidizer may not only profit from the local increase of the carbon dioxide availability but also from the removal of organic compounds by Acidiphilium sp. JA12-A1, since chemolithoautotrophic iron oxidizers have long been known to be sensitive to organic compounds [59]. The sum of these potential interactions may account for the tenacious association of both organisms in the mixed culture and provide an explanation for the difficulties encountered when attempting to obtain pure cultures of the iron oxidizing bacteria.
In order to experimentally substantiate such an interspecies carbon transfer we suggest to analyze, similar to the study of Kermer et al. [9], secreted metabolites in combination with a stable isotope approach (13C-labelled carbon dioxide) since this may reveal the actual metabolites that are utilized by Acidiphilium sp. JA12-A1 in the mixed culture. This approach may not only extend our knowledge of the proposed interspecies carbon transfer [9], but also elucidate whether Acidiphilium sp. JA12-A1 incorporates carbon dioxide heterotrophically by carboxylation reactions under the conditions provided within the mixed culture. In Acidiphilium rubrum the incorporation of carbon dioxide was described to be enhanced under aerobic-light conditions with the required energy provided by light utilization via a photosynthetic reaction center and phototrophic pigments [60]. We identified gene clusters homologous to those described for Acidiphilium rubrum and other Acidiphilium strains in the genome of Acidiphilium sp. JA12-A1 hinting at a potential photosynthetic activity. However, since none of the described Acidiphilium strains seems to be capable of using light as sole source of energy [61], it has been proposed that the photosynthetic activity is used to pump protons across the cytoplasmic membrane in order to stabilize the proton balance between the acidic environment and the neutral cytoplasm [60].
Acidiphilium strains are also thought to play a direct role in the iron cycle by regenerating dissolved ferrous iron through the reduction of ferric iron under microaerobic and anoxic conditions [11, 62]. Other studies have shown that ferrous iron is regenerated from the reduction of ferric iron minerals by Acidiphilium spp. and other acidophilic ferric iron reducers [52]. The ferrous iron is then available as an energy source for the iron oxidizers again. Details of the pathway of ferric iron reduction could, however, not be deduced from the genome of Acidiphilium sp. JA12-A1.
The Acidiphilium strains Acidiphilium cryptum JF-5, Acidiphilium multivorum AUI301, Acidiphilium sp. PM DSM 24941 and Acidiphilium sp. JA12-A1, which all belong to the same phylogenetic subgroup within the genus Acidiphilium, show high similarities regarding their structural and functional genome organization. Since they also share important metabolic traits with respect to growth conditions and nutrient requirements the proposed interaction between Acidiphilium sp. JA12-A1 and the iron oxidizer Ferrovum spp. may also be true for other members of the genus Acidiphilium in their natural habitats.
Acknowledgements
This work was funded by the European Social Fund (ESF) through the project GETGEOWEB and the BMBF through the project SURFTRAPII (03G0821B) within the R&D-program GEOTECHNOLOGIEN.
Abbreviations
- AMD
acid mine drainage
- PHB
polyhydroxybutyrate
Additional file
Associated MIGS record. (DOC 73 kb)
Footnotes
Sophie R. Ullrich and Anja Poehlein contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors‘ contributions
MM and MS designed the study. MM supervised the genome analysis. SRU analysed the annotated sequence information with contributions from AP, in particular regarding comparative genome analyses, and drafted the manuscript with contributions from AP, MS and MM. SV performed the genome sequencing. AP performed the assembly and annotation of the sequence data. MH performed electron microscopy of strain JA12-A1. RD supervised the genome sequencing, assembly and annotation. AL contributed bioinformatic expertise to the analysis of the sequence data. JST isolated strain JA12-A1 and prepared genomic DNA. All authors read and approved the final manuscript.
References
- 1.Harrison AP, Jarvis BW, Johnson JL. Heterotrophic bacteria from cultures of autotrophic Thiobacillus ferrooxidans: relationships as studied by means of deoxyribonucleic acid homology. J Bacteriol. 1980;143:448–454. doi: 10.1128/jb.143.1.448-454.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison AP. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int J Sys Evol Microbiol. 1981;31:327–332. [Google Scholar]
- 3.Johnson DB, Rolfe S, Hallberg KB, Iversen E. Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic mine drainage waters at an abandoned Norwegian copper mine. Environ Microbiol. 2001;3:630–637. doi: 10.1046/j.1462-2920.2001.00234.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DB, Hallberg KB. The microbiology of acidic mine waters. Res Microbiol. 2003;154:466–473. doi: 10.1016/S0923-2508(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 5.Heinzel E, Hedrich S, Jannec KE, Glombitza F, Seifert J, Schlömann M. Bacterial diversity in a mine water treatment plant. Appl Environ Microbiol. 2009;75:858–861. doi: 10.1128/AEM.01045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson DB, Kelso WI. Detection of heterotrophic contaminants in cultures of Thiobacillus ferrooxidans and their elimination by subculturing in media containing copper sulphate. Soc General Microbiol. 1983;129:2969–2972. [Google Scholar]
- 7.Johnson DB, Hallberg KB, Hedrich S. Uncovering a microbial enigma: isolation and characterization of the streamer-generating, iron-oxidizing, acidophilic bacterium "Ferrovum myxofaciens". Appl Environl Microbiol. 2014;80:672–680. doi: 10.1128/AEM.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchand EA, Silverstein J. The role of enhanced heterotrophic bacterial growth on iron oxidation by Acidithiobacillus ferrooxidans. Geomicrobiol J. 2003;20:231–244. doi: 10.1080/01490450303878. [DOI] [Google Scholar]
- 9.Kermer R, Hedrich S, Taubert M, Baumann S, Schlömann M, Johnson DB, Seifert J. Elucidation of carbon transfer in a mixed culture of Acidiphilium cryptum and Acidithiobacillus ferrooxidans using protein-based stable isotope probing. J Integr OMICS. 2012;2:37–45. [Google Scholar]
- 10.Harrison AP. The acidophilic Thiobacilli and other acidophilic bacteria that share their habitat. Annu Rev Microbiol. 1984;38:265–292. doi: 10.1146/annurev.mi.38.100184.001405. [DOI] [PubMed] [Google Scholar]
- 11.Baker BJ, Banfield JF. Microbial communities in acid mine drainage. FEMS Microbiol Ecol. 2003;44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Toril E, Aguilera A, Souza-Egipsy V, Lopez Pamo E, Sanchez Espana J, Amils R. Geomicrobiology of La Zarza-Perrunal acid mine effluent (Iberian Pyritic Belt, Spain) Appl Environ Microbiol. 2011;77:2685–2694. doi: 10.1128/AEM.02459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tischler JS, Jwair RJ, Gelhaar N, Drechsel A, Skirl A, Wiacek C, Janneck E, Schlömann M. New cultivation medium for “Ferrovum” and Gallionella-related strains. J Microbiol Methods. 2013;95:138–144. doi: 10.1016/j.mimet.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 14.Hallberg KB, Coupland K, Kimura S, Johnson DB. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl Environ Microbiol. 2006;72:2022–2030. doi: 10.1128/AEM.72.3.2022-2030.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinzel E, Janneck E, Glombitza F, Schlömann M, Seifert J. Population dynamics of iron-oxidizing communities in pilot plants for the treatment of acid mine waters. Environ Sci Technol. 2009;43:6138–6144. doi: 10.1021/es900067d. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DB, McGinness S. A highly efficient and universal solid medium for growing mesophilic and moderately thermophilic, iron-oxidizing, acidophilic bacteria. J Microbiol Methods. 1991;13:113–122. doi: 10.1016/0167-7012(91)90011-E. [DOI] [Google Scholar]
- 17.Johnson DB, Hallberg KB. Techniques for detecting and identifying acidophilic mineral-oxidizing microorganisms. In: Rawlings DE, Johnson DB, editors. Biomining. 1. Berlin: Springer; 2007. pp. 237–261. [Google Scholar]
- 18.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 19.Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA. Database indexing for production MegaBLAST searches. Bioinformatics. 2008;24:1757–1764. doi: 10.1093/bioinformatics/btn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jukes TH, Cantor CR. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. 1. New York: Academic Press; 1969. pp. 21–132. [Google Scholar]
- 22.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 23.Wakao N, Nagasawa N, Matsuura T, Matsukura H, Matsumoto T, Hiraishi A, Hiraishi A, Sakurai Y, Shiota H. Acidiphilium multivorum sp. nov., an acidophilic chemoorganotrophic bacterium from pyritic acid mine drainage. J Gen Appl Microbiol. 1994;40:143–159. doi: 10.2323/jgam.40.143. [DOI] [Google Scholar]
- 24.Validation List no. 52 Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Sys Bacteriol. 1995;45:197–198. doi: 10.1099/00207713-45-1-197. [DOI] [Google Scholar]
- 25.Lobos JH, Chisolm TE, Bopp LH, Holmes DS. Acidiphilium organovorum sp. nov., an acidophilic heterotroph isolated from a Thiobacillus ferrooxidans culture. Int J Sys Bacteriol. 1986;36:139–144. doi: 10.1099/00207713-36-2-139. [DOI] [Google Scholar]
- 26.Hiraishi A, Nagashima KVP, Matsuura K, Shimada K, Takaichi S, Wakao N, Katayama Y. Phylogeny and photosynthetic features of Thiobacillus acidophilus and related acidophilic bacteria: its transfer to the genus Acidiphilium as Acidiphilium acidophilum comb. nov. Int J Sys Bacteriol. 1998;48:1389–1398. doi: 10.1099/00207713-48-4-1389. [DOI] [PubMed] [Google Scholar]
- 27.Wichlacz PL, Unz RF, Langworthy TA. Acidiphilum angustum sp. nov., Acidiphilum facilis sp. nov., and Acidiphilum rubrum sp. nov.: acidophilic heterophilic bacteria isolated from acidic coal mine drainage. Int J Sys Bacteriol. 1986;36:197–201. doi: 10.1099/00207713-36-2-197. [DOI] [Google Scholar]
- 28.Validation List no. 48 Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Evol Microbiol. 1994;44:182–183. [Google Scholar]
- 29.Kishimoto N, Kosako Y, Tano T. Acidiphilium aminolytica sp. nov.: an acidophilic chemoorganotrophic bacterium isolated from acidic mineral environment. Curr Microbiol. 1993;27:131–136. doi: 10.1007/BF01576009. [DOI] [PubMed] [Google Scholar]
- 30.Validation List no. 56 Validation of the publication of new names and new combinations previously effectively published outside the IJSB. Int J Syst Evol Microbiol. 1996;46:362–363. [Google Scholar]
- 31.Mosler S, Poehlein A, Voget S, Daniel R, Kipry J, Schlömann M, Mühling M. Predicting the metabolic potential of the novel iron oxidising bacterium "Ferrovum" sp. JA12 using comparative genomics. AMR. 2013;825:153–156. doi: 10.4028/www.scientific.net/AMR.825.153. [DOI] [Google Scholar]
- 32.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW, editors. Bergey's Manual of Systematic Bacteriology. Volume 1. 2. New York: Springer; 2001. pp. 119–169. [Google Scholar]
- 35.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey's Manual of Systematic Bacteriology. Volume 2 (Part B) 2. New York: Springer; 2005. p. 1. [Google Scholar]
- 36.Validation List no. 106 Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Int J Syst Evol Microbiol. 2005;55:2235–2238. doi: 10.1099/ijs.0.64108-0. [DOI] [Google Scholar]
- 37.Validation List No. 107 List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol. 2006;56:1–6. doi: 10.1099/ijs.0.64188-0. [DOI] [PubMed] [Google Scholar]
- 38.Pfennig N, Trüper HG. Higher taxa of the phototrophic bacteria. Int J Syst Evol Microbiol. 1971;21:17–18. [Google Scholar]
- 39.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Evol Microbiol. 1980;30:225–420. [Google Scholar]
- 40.Henrici AT. The Biology of Bacteria. In: Henrici AT, editor. The Biology of Bacteria. Second. Chicago: Heath and Co; 1939. pp. 1–494. [Google Scholar]
- 41.Gillis M, De Ley J. Intra- and intergeneric similarities of the ribosomal ribonucleic acid cistrons of Acetobacter and Gluconobacter. Int J Syst Evol Microbiol. 1980;30:7–27. [Google Scholar]
- 42.Kishimoto N, Kosako Y, Wakao N, Tano T, Hiraishi A. Transfer of Acidiphilium facilis and Acidiphilium aminolytica to the genus Acidocella gen. nov., and emendation of the genus Acidiphilium. Syst Appl Microbiol. 1995;18:85–91. doi: 10.1016/S0723-2020(11)80453-4. [DOI] [Google Scholar]
- 43.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology, The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. In: Wingender E, editor. Proceedings of the German Conference on Bioinformatics: 4-6 October 1999; Hannover. Braunschweig: GBF-Braunschweig; 1999. pp. 45–56. [Google Scholar]
- 45.Tech M, Merkl R. YACOP: enhanced gene prediction obtained by a combination of existing methods. In Silico Biol. 2003;3:441–451. [PubMed] [Google Scholar]
- 46.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markowitz VM, Mavromatis K, Ivanova NN, Chen IM, Chu K, Kyrpides NC. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 49.Markowitz VM, Chen IM, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zdobnov EM, Apweiler R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 51.Johnson DB, Bridge TAM. Reduction of ferric iron by acidophilic heterotrophic bacteria: evidence for constitutive and inducible enzyme systems in Acidiphilium spp. J Appl Microbiol. 2002;92:315–321. doi: 10.1046/j.1365-2672.2002.01535.x. [DOI] [PubMed] [Google Scholar]
- 52.Coupland K, Johnson DB. Evidence that the potential for dissimilatory ferric iron reduction is widespread among acidophilic heterotrophic bacteria. FEMS Microbiol Lett. 2008;279:30–35. doi: 10.1111/j.1574-6968.2007.00998.x. [DOI] [PubMed] [Google Scholar]
- 53.Osorio H, Mangold S, Denis Y, Nancucheo I, Esparza M, Johnson DB, Bonnefoy V, Dopson M, Holmes DS. Anaerobic sulfur metabolism coupled to dissimilatory iron reduction in the extremophile Acidithiobacillus ferrooxidans. Appl Environ Microbiol. 2013;79:2172–2181. doi: 10.1128/AEM.03057-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leimbach A: bac-genomics-scripts. [https://github.com/aleimba/bac-genomics-scripts]
- 55.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124–133. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallberg KB, Johnson DB. Biodiversity of acidophilic prokaryotes. Adv Appl Microbiol. 2001;49:37–84. doi: 10.1016/S0065-2164(01)49009-5. [DOI] [PubMed] [Google Scholar]
- 57.García-Moyano A, González-Toril E, Aguilera A, Amils R. Prokaryotic community composition and ecology of floating macroscopic filaments from an extreme acidic environment, Río Tinto (SW, Spain) Syst Appl Microbiol. 2007;30:601–614. doi: 10.1016/j.syapm.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 58.Kay C, Rowe O, Rocchetti L, Coupland K, Hallberg K, Johnson DB. Evolution of microbial “streamer” growths in an acidic, metal-contaminated stream draining an abandoned underground copper mine. Life. 2013;3:189–211. doi: 10.3390/life3010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Touvinen OH, Kelly DP. Studies on the growth of Thiobacillus ferrooxidans. I. Use of membrane filters and ferrous iron agar to determine viable numbers, and comparison with 14CO2-fixation and iron oxidation as measures of growth. Arch Mikrobiol. 1973;88:285–298. doi: 10.1007/BF00409941. [DOI] [PubMed] [Google Scholar]
- 60.Kishimoto N, Fukaya F, Inagaki K, Sugio T, Tanaka H, Tano T. Distribution of bacteriochlorophyll a among aerobic and acidophilic bacteria and light-enhanced CO2-incorporation in Acidiphilium rubrum. FEMS Microbiol Ecol. 1995;16:291–296. doi: 10.1111/j.1574-6941.1995.tb00293.x. [DOI] [Google Scholar]
- 61.Gest H. Photosynthetic and quasi-photosynthetic bacteria. FEMS Microbiol Lett. 1993;112:1–6. doi: 10.1111/j.1574-6968.1993.tb06414.x. [DOI] [Google Scholar]
- 62.Küsel K, Dorsch T, Acker G, Stackebrandt E. Microbial reduction of Fe(III) in acidic sediments: isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose. Appl Environ Microbiol. 1999;65:3633–3640. doi: 10.1128/aem.65.8.3633-3640.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


