Summary
In mammals, DNA methylation is essential for protecting repetitive sequences from aberrant transcription and recombination. In some developmental contexts (e.g., preimplantation embryos) DNA is hypomethylated but repetitive elements are not dysregulated, suggesting that alternative protection mechanisms exist. Here we explore the processes involved by investigating the role of the chromatin factors DAXX and ATRX. Using genome-wide binding and transcriptome analysis, we found that DAXX and ATRX have distinct chromatin-binding profiles and are co-enriched at tandem repetitive elements in wildtype mouse ESCs. Global DNA hypomethylation further promoted recruitment of the DAXX/ATRX complex to tandem repeat sequences, including retrotransposons and telomeres. Knockdown of DAXX/ATRX in cells with hypomethylated genomes exacerbated aberrant transcriptional de-repression of repeat elements and telomere dysfunction. Mechanistically, DAXX/ATRX-mediated repression seems to involve SUV39H recruitment and H3K9 trimethylation. Our data therefore suggest that DAXX and ATRX safeguard the genome by silencing repetitive elements when DNA methylation levels are low.
Keywords: DAXX, ATRX, mouse embryonic stem cells, DNA methylation, DNMTs, repetitive sequences, telomeres, histone modification
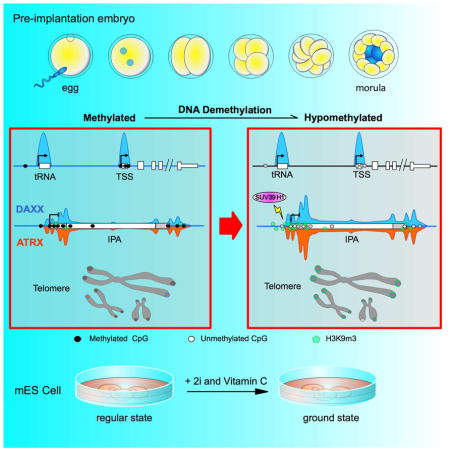
Graphical abstract
Introduction
DNA methylation is tightly regulated by the de novo DNA methyltransferases DNMT3a/3b and the maintenance DNA methyltransferase DNMT1, and required for somatic cell growth and survival in mammals (Jackson-Grusby et al., 2001; Tsumura et al., 2006). DNA methylation can inhibit gene transcription and facilitate the formation of compact and inactive chromatin or heterochromatin (e.g., repetitive sequences) to safeguard genome integrity and stability (Armour et al., 1996; Branzei and Foiani, 2010; Chan et al., 2006; Deepali Pathak 2012; Jurka et al., 2007; Ross et al., 2010; Sakaue et al., 2010; Treangen and Salzberg, 2012). Repeat elements such as telomeres and centromeres are located in specific regions and critical for maintaining the structure and integrity of chromosomes. The dysregulation of these sequences have been directly linked to genome instability and human diseases (Bzymek and Lovett, 2001; Heartlein, 1990; Mattick and Makunin, 2006). Other repeat elements such as long-terminal repeat (LTR) containing retrotransposons (or endogenous retroviruses (ERVs)) are scattered throughout the genome. Increasing evidence indicates that these sequence elements also possess the capacity to contribute to malignant transformation (Gao et al., 2008; Lee et al., 2012). It has been shown that ERVs are normally actively suppressed through chromatin maintenance mechanisms such as DNA methylation and histone modifications (Rebollo et al., 2012; Shalginskikh et al., 2013; Wolf et al., 2013), the disruption of which can have serious consequences and lead to diseases and cancer in humans (Bourc’his and Bestor, 2004; Dodge et al., 2005; Gaudet et al., 2003; Jackson-Grusby et al., 2001; Lewis et al., 2010; Lovejoy et al., 2012; Ross et al., 2010; Wilson et al., 2007).
Interestingly, embryonic stem (ES) cells can tolerate global loss of DNA methylation. Mammalian genomic DNA undergoes programmed genome-wide demethylation during specific developmental stages. For example, the vast majority of genomic DNA loses methylation due to restricted DNMT1 mobility and low DNMT3a/3b expression in preimplantation embryos (2–8 cell stage) and during primordial germ cell (PGC) specification (Cirio et al., 2008; Grohmann et al., 2005; Howell et al., 2001). Mouse ES (mES) cells deficient for DNMTs are also able to survive and maintain their self-renewal capacity (Jackson et al., 2004; Tsumura et al., 2006). Despite risks such as de-repression of repetitive elements, such genome-wide DNA demethylation does not lead to genomic instability (Baumann et al., 2010; Hutnick et al., 2010; Reik et al., 2001; Seisenberger et al., 2012), suggesting the existence of additional control mechanisms that ensure genome integrity and stability.
Recent studies have linked the DAXX/ATRX complex to DNA methylation. The SWI/SNF-like chromatin remodeling protein ATRX (α-thalassemia/mental retardation X-linked) can form a hetero-complex with the transcriptional co-repressor DAXX (death-domain associated protein). DAXX can also act as a chaperon for the chromatin deposition of histone H3 variant H3.3 (Wong et al., 2010; Xue et al., 2003). Knocking out (KO) either DAXX or ATRX in mice was embryonic lethal (Garrick et al., 2006; Michaelson et al., 1999). Previous work has indicated that both DAXX and ATRX can localize to pericentric heterochromatin and telomeres in somatic and ES cells (Baumann et al., 2008; Emelyanov et al., 2010; Wong et al., 2010). Recent genome-wide sequencing studies suggest that ATRX target regions (e.g., promoters and GC-rich tandem repeats) are enriched with the CpG dinucleotide (Law et al., 2010). In particular, ATRX associates with pericentric heterochromatin regions that are transcriptionally silenced by H3K9 trimethylation (H3K9me3) (McDowell et al., 1999). Mutations in ATRX were shown to cause changes in DNA methylation at repeat sequences including rDNA, interstitial heterochromatic repeats, and subtelomeric repeats (Gibbons et al., 2000). However, the precise role of DAXX and the DAXX-ATRX complex in these processes remains unclear.
In this study, we tested the hypothesis that the DAXX/ATRX complex participates in protecting repetitive elements in the absence of DNA methylation. To this end, we investigated genome-wide chromatin targeting of DAXX and ATRX in wildtype mES cells, and in mES cells that exhibit extensive loss of DNA methylation due to homozygous knockout of all three DNA methyltransferases (TKO cells) (Okano et al., 1999). Our data uncovered distinct genomic distribution patterns of DAXX vs. ATRX, and revealed significant changes in DAXX and ATRX targeting in cells with hypomethylated DNA. We showed that upon DNA hypomethylation DAXX and ATRX became preferentially targeted to tandem repetitive elements that are heavily methylated in wildtype mES cells, such as IAPs (intracisternal A particle) and telomeres. This chromatin re-distribution of DAXX/ATRX appeared to depend on DAXX, not ATRX. Our RNA-seq and quantitative RT-PCR experiments using TKO cells, mES cells maintained in ground-state conditions, and preimplantation embryos further support an essential role of the DAXX/ATRX complex in silencing these repetitive elements in cells undergoing global DNA demethylation. Additionally, we provide evidence that DAXX can recruit SUV39H1 and promote H3K9 trimethylation to silence tandem repetitive elements. Our findings demonstrate the functional link between the DAXX/ATRX complex and DNA methylation, and highlight the critical role of DAXX in silencing repetitive sequences in the absence of DNA methylation.
Results
DAXX and ATRX target to distinct chromatin regions in wildtype mouse ES cells
To better understand the function of DAXX and ATRX, we first determined their genome-wide targets by ChIP-seq using antibodies against endogenous DAXX and ATRX in wildtype mouse ES cells (J1) (supplemental Figure S1A–B). A total of 767 DAXX and 1,505 ATRX-binding sites were identified, including those in promoter regions, tRNA genes, and various repetitive elements (e.g., satellite and simple repeats) (Figure 1A–B). Enrichment of DAXX and ATRX at a number of the identified sites was further validated by ChIP Q-PCR (supplemental Figure S1C–D).
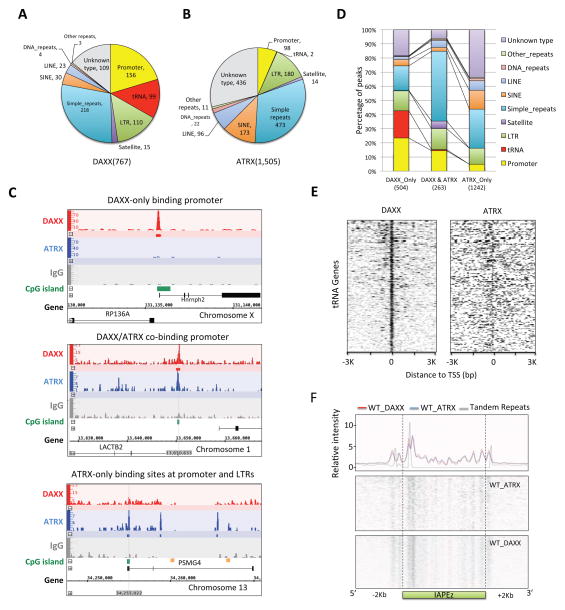
Figure 1. DAXX and ATRX exhibit distinct genome-wide distribution of binding sites in wildtype (WT) mES cells.
See also supplemental Figure S1. (A–B) ChIP-seq analysis of wildtype J1 mES cells was carried out using anti-DAXX and ATRX antibodies. The different types of binding sites identified throughout the genome are summarized here. (C) The identified binding sites at different gene loci were divided into DAXX only (top), DAXX & ATRX co-binding (middle), and ATRX only (bottom) sites. The distance to signal peaks and number of sites in each group were plotted on the x- and y-axis respectively, to show representative signal peak distributions of various DAXX/ATRX binding sites. Green bar, CpG island. Orange bar, LTR region. (D) Genomic features of the three groups of binding sites from (C) were plotted and further compared. (E) Comparison of the DAXX and ATRX binding profiles at tRNA genes. The x-axis indicates distance to the transcriptional start site (TSS) of tRNA genes. (F) The binding profiles of ATRX and DAXX at IAPEz regions (including ±2kb from these regions). Top panel, relative intensities of the peaks were plotted on the y-axis. The blue and red lines denote the average binding profiles of ATRX and DAXX respectively. The grey line indicates the distribution of tandem repetitive sequences. The bottom two panels are heat maps of ATRX and DAXX binding profiles.
Given that DAXX and ATRX can heterodimerize on chromatin, we were surprised by the relatively small percentage of shared binding sites (~34% for DAXX and ~18% for ATRX) (supplemental Figure S2A). Compared to ATRX, DAXX appeared to preferentially bind to promoter regions (156 vs. 98) (Figure 1C–D) and target tRNA genes (99 vs. 2) (Figure 1E). Many DAXX-only (504) and ATRX-only (1,242) binding sites were found at both gene promoters and repetitive sequences, supporting the notion that these two proteins carry out independent functions on different targets. Two major types of repetitive sequences (LTR and simple repeats) were highly enriched for both DAXX and ATRX, and satellite repeats were found exclusively in DAXX/ATRX co-binding sites, suggesting a role for these proteins in regulating repetitive sequences (Figure 1A–B, 1D). Consistent with DAXX and ATRX working as a complex on IAPs, the two proteins exhibit similar binding profiles and are enriched at the 5′ terminal regions of the tandem repeats (Figure 1F). These results underscore the unique functions of each protein and suggest related but distinct binding profiles/targets of DAXX vs. ATRX in mouse ES cells.
DNA hypomethylation impacts the genomic targeting of DAXX and ATRX
We reasoned that genome-wide alterations in DNA methylation should impact the targeting of DAXX and ATRX, given the importance of DNA methylation in silencing genes and repressing repeat sequences. To investigate this possibility, we carried out ChIP-Seq experiments using J1 cells knocked out for all three DNA methyltransferases (TKO cells) (supplemental Figure S1A–B). Compared to wildtype J1 cells, dramatic increases in the number of sites targeted by DAXX (2,280) and ATRX (2,191) were apparent in TKO cells, along with significant increases in both the number (263 vs. 863) and percentage (13% vs. 24%) of DAXX/ATRX co-occupied sites (Figure 2A, supplemental Figure S2B), indicating enhanced genomic targeting of individual DAXX and ATRX proteins and the DAXX/ATRX complex upon DNA hypomethylation (χ2: p=1.184−15). While most DAXX binding sites remained bound by DAXX in TKO cells (86.3%) with highly correlative binding intensities (coverage depth), only ~56% of the ATRX binding sites were retained in TKO cells, often with poor correlation in binding intensities (Figure 2A and B, supplemental Figure S2C–D). These results underline the different outcomes upon DNA hypomethylation: more chromatin sites became readily available for DAXX binding, whereas new potential ATRX target sites had to “compete” with existing sites for ATRX binding.
Figure 2. Loss of DNA methylation in TKO cells leads to preferential targeting of both DAXX and ATRX to repeat sequences.
See also supplemental Figure S1–3. TKO cells were used for ChIP-seq analysis using anti-DAXX and ATRX antibodies. (A) Comparison of DAXX and ATRX-binding sites in wildtype J1 (WT) vs. TKO cells. (B) A comparison of genome-wide DAXX and ATRX-binding sites in TKO vs. wildtype J1 (WT) cells. Left, all of the identified binding sites for DAXX and ATRX were plotted, where the x-axis is the distance to signal peaks and the y-axis represents the signaling intensity for each site. Right, the binding sites specific for TKO cells were classified and graphed as pie charts. (C) DAXX-binding sites that were found only in wildtype J1 (WT_Alone) or TKO (TKO_Alone) cells, or shared between the cells (Shared) were plotted based on various genomic features. (D) The DAXX-binding sites on LTR-containing sequences were further classified and similarly plotted as in (C). (E) The average distribution of DAXX, ATRX, and DNA methylation (±5kb from the binding peaks on LTRs) in wildtype J1 (WT) (blue) and TKO (red) cells was plotted and compared. The center vertical dotted lines indicate the summit of binding peaks.
Compared to wildtype J1 cells, the TKO cell-specific DAXX (1,665) and ATRX (1,354) binding sites were highly enriched for LTR-type sequences (24% vs. 14% for DAXX and 39% vs. 12% for ATRX) (Figure 2B–C), particularly simple repeats and the IAP and RLTR repeats of endogenous retroviruses family K (ERVK) (Figure 2D, supplemental Figure S2E). The binding intensity on these sites was also higher in TKO cells for both proteins (especially DAXX) (Figure 2E, supplemental Figure S2F). Taken together with our failure to detect similar enrichment at sites such as promoters and tRNA genes (data not shown), these results strongly suggest that DNA hypomethylation can specifically promote the binding of DAXX and ATRX to LTR-containing sequences. Because repetitive sequences are often methylated, we postulated that the additional DAXX/ATRX sites identified in TKO cells should be protected by DNA methylation in wildtype J1 cells. This was indeed the case when we calculated the relative intensities of DNA methylation on these sites using published DNA bisulfite sequencing data of TKO and wildtype J1 cells (Figure 2E) (Williams et al., 2011). Importantly, DNA methylation signals were only detected at IAP and RLTR sites targeted by DAXX and ATRX, reinforcing the link between the loss of DNA methylation and gain of DAXX and ATRX binding on repeat sequences.
The DAXX/ATRX complex is essential for transcriptional repression of repetitive elements in mES cells during DNA hypomethylation
To further determine the changes in DAXX and ATRX-mediated transcriptional repression in response to DNA demethylation, we generated J1 and TKO cells knocked out for DAXX and ATRX using the CRISPR-Cas9 technology (supplemental Figure S3) (Cong et al., 2013), and examined the transcription levels of IAP repeats in these cells. As shown in Figure 3A, knocking out DAXX or ATRX resulted in an increase of IAP transcription in both TKO and wildtype J1 cells. Again, the increase was more pronounced in TKO cells, consistent with the notion that loss of DNA methylation in TKO cells may have rendered the cells more dependent on DAXX and ATRX for repressing IAP transcription.
Figure 3. DAXX and ATRX are important for repressing repetitive sequences in cells undergoing global hypomethylation.
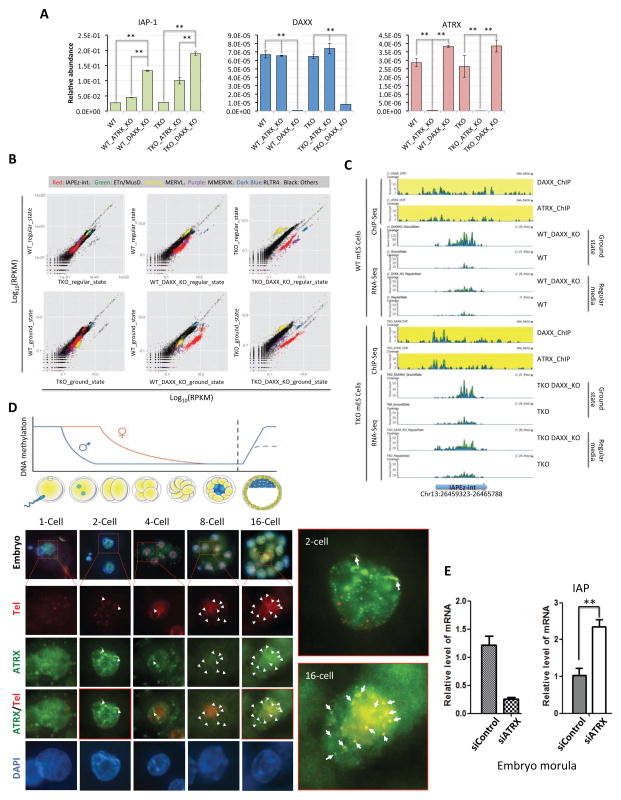
See also Figure S4. (A) RT-qPCR analysis of transcript levels of IAP-1, DAXX, and ATRX in wildtype J1 (WT) and TKO cells, as well as in WT and TKO cells knocked out for DAXX or ATRX. Error bars are standard deviation (n=3). The data were analyzed using the Student t-test. ** indicates p<0.01 and * indicates p<0.05. (B) Wildtype J1 (WT) and TKO cells as well as J1 and TKO cells knocked out for DAXX were grown in regular or ground-state (2i+vitamin C (VC)) media and examined for the transcriptional levels of five types of repetitive elements (color coded as indicated). RPKMs (Reads Per Kilobase of transcript per Million mapped reads) of various samples were plotted in pairs on log10 scale. In each paired comparison (plot), the expression of a particular repetitive element (represented by RPKMs) in different cells or conditions was plotted on the x- or y-axis as indicated. A shift from the diagonal line indicates differential expression between the two samples. IAPEz-int, intracisternal A particle interspersed repeats. ETn/MusD, the early transposon family of long repeated sequences, also known as MMET in Repbase database (Jurka et al., 2005). MERVL, mouse retroelement MuERV-L/MERVL. MMERVK, LTR family of mouse endogenous retrovirus K. RLTR4, Long Terminal Repeat for HERV3 endogenous retrovirus. (C) A snapshot of aligned ChIP-seq and RNA-seq data shows a representative case of IAP expression differences between samples. RNA-seq data from wildtype J1, wildtype DAXX KO, TKO, and TKO DAXX KO cells grown under regular vs. ground-state conditions were used. The relative coverage was normalized by the total number of aligned reads for each sample and the plot was drawn using GenomeBrowse (Golden Helix Inc.). Blue and green represent reads aligned to the plus and minus strand respectively. (D) Top, the global DNA demethylation process that occurs in early pre-implantation embryos. Bottom, different stages of mouse embryos were examined by immuno-FISH using a telomere FISH probe (red) and anti-ATRX antibodies (green). Arrowheads indicate co-localized foci. Magnified images of 2-cell and 16-cell embryos are shown on the right. (E) ATRX knockdown induces IAP de-repression in early mouse embryos. Mouse embryos were injected with control siRNA oligos or those against ATRX, and allowed to recover. Morula-stage embryos were then collected for quantitative RT-PCR analysis to determine the expression level of ATRX (left) and IAP (right) messages. Error bars indicate standard error. The data were analyzed using the Student t-test. ** indicates p<0.01.
It has been shown that culturing normal mES cells in ground-state conditions (e.g., addition of 2i (MEK/ERK and GSK3 inhibitors) (Ying et al., 2008) and vitamin C) could also lead to global DNA hypomethylation (Blaschke et al., 2013). We therefore carried out RNA-seq experiments using parental and DAXX KO J1 and TKO cells grown in ground-state conditions (Figure 3 and supplemental Figure S4). As illustrated in Figure 3B, we found no significant differences in the expression of repetitive elements between wildtype and TKO cells under different culturing conditions (leftmost plots), indicating that loss of DNA methylation alone is insufficient to derepress repetitive elements. Derepression of IAPs and MMERVK retrotransposons induced by DAXX loss was much more pronounced under ground-state conditions in J1 cells (center top vs. bottom). Consistent with our RT-PCR results, IAP and MMERVK transcription was significantly higher in TKO DAXX KO cells than in TKO cells (top right). In comparison, maintaining TKO DAXX KO cells in ground-state media produced no further IAP derepression (Figure 3B bottom right, supplemental Figure S4A), since such treatment of these cells likely yielded no additional DNA demethylation. A more detailed examination of one IAP locus highlights the expression pattern of IAPEZ in different cells and under different conditions (Figure 3C). Again, combined DNA hypomethylation and DAXX loss was required for IAP derepression in both WT and TKO cells. These data support our hypothesis that DAXX is critical for repressing repeat elements such as IAPs and LTRs, especially when cells undergo global DNA demethylation. The coordinated action of these two pathways mediates the regulation of repetitive element expression.
Early pre-implantation embryos undergo genome-wide epigenetic reprogramming through rapid and global DNA demethylation (Figure 3D) (Messerschmidt et al., 2014). Given the important role of the DAXX/ATRX complex in repressing retrotransposons in hypomethylated mES cells, we speculated that this complex might also be necessary for retrotransposon silencing in early pre-implantation embryos. We therefore determined the expression of IAPs by quantitative RT-PCR using one-cell stage embryos knocked down (KD) for ATRX. Consistent with the data obtained from mES cells, IAP expression was activated in ATRX KD embryos compared to controls (Figure 3E). These data strongly suggest that the DAXX/ATRX complex likely represents an alternative mechanism that cells utilize to protect retrotransposons in the event of DNA hypomethylation, adding support to our model that DNA hypomethylation can induce DAXX-dependent repression of IAPs/LTRs.
Loss of DNA methylation promotes the targeting of the DAXX/ATRX complex to telomere/subtelomeric regions in mouse ES cells and during early embryogenesis
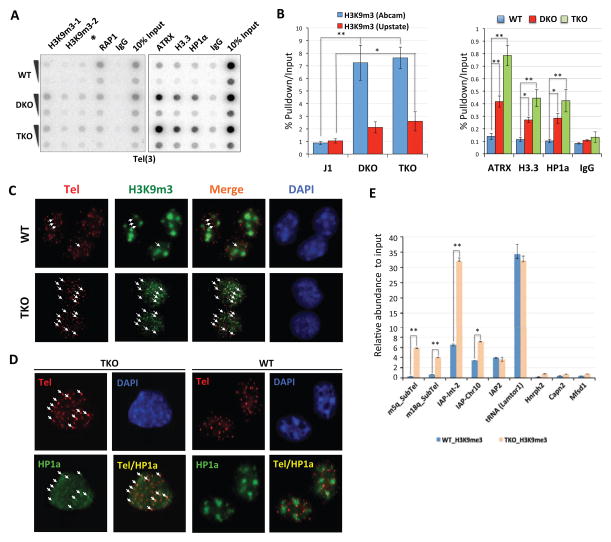
Both DAXX and ATRX are capable of binding telomeric/subtelomeric regions in mouse and human ES cells (Law et al., 2010; Lewis et al., 2010; Lovejoy et al., 2012). Loss of ATRX has been shown to lead to upregulated transcription, aberrant epigenetic changes, and DNA damage at telomeres (Heaphy et al., 2011; Lovejoy et al., 2012; Wong et al., 2010). We therefore examined how changes in DNA methylation would affect DAXX/ATRX targeting to these regions. Our ChIP-seq experiments suggest increased DAXX/ATRX targeting to telomeric repeat-containing sequences in TKO cells compared to J1 cells (Figure 4A). In support of the ChIP-seq results, telomere ChIP experiments using antibodies against endogenous DAXX and ATRX also brought down more telomeric DNA from TKO cells (Figure 4B–C). To rule out the possibility that this enrichment was due to longer telomeres in TKO cells (Gonzalo et al., 2006), we also performed telomere FISH-immunostaining assays. J1 cells knocked out for both Dnmt3a and Dnmt3b (DKO cells) were included as controls (supplemental Figure S1A–B). As expected, the majority of DAXX and ATRX localized to pericentric heterochromatin in wildtype J1 cells (Figure 4D–E). However, most DAXX and ATRX foci co-stained with telomeres in both DKO (~60%) and TKO (>80%) cells, which was accompanied by reduced staining for pericentric heterochromatin.
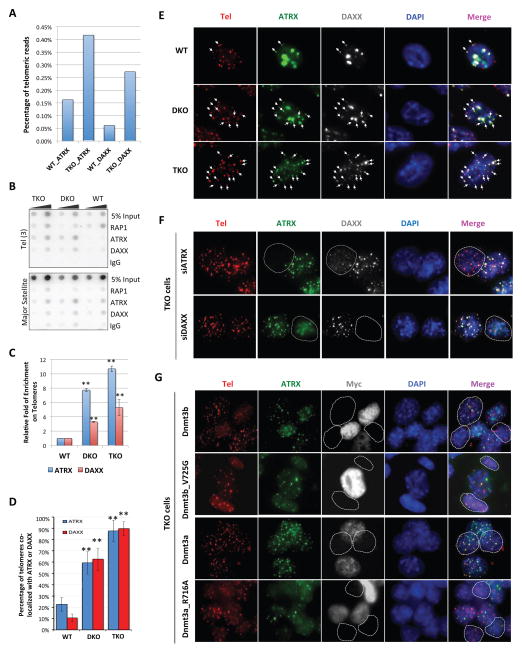
Figure 4. The DAXX/ATRX complex is targeted to subtelomeric/telomeric regions in response to genomic DNA demethylation in ES cells.
See also supplemental Figure S5–6. (A) We calculated and plotted the percentages of telomeric reads (TTAGGG(6)) out of total reads that were aligned to the mouse genome using our ChIP-seq data from wildtype J1 (WT) and TKO cells. (B) Telomere ChIP experiments were performed using antibodies against endogenous DAXX and ATRX. The telomeric protein RAP1 served as a positive control, and rabbit IgG was used as a negative control. The blots were probed with a radiolabeled TTAGGG(3) (Tel(3)) probe or a major satellite DNA probe. (C) Quantification of data from (B). Three independent experiments were performed. Error bars indicate standard deviation. The data were analyzed using the Student t-test. ** indicates p<0.01. (D) The percentages of telomeres occupied by ATRX and DAXX. Immuno-FISH experiments were carried out in wildtype J1 (WT), DKO, and TKO cells. ATRX and DAXX were detected by immunostaining using anti-ATRX and DAXX antibodies, while telomeres were detected using a Tel-RNP FISH probe. Error bars indicate standard deviation (n=3). The data were analyzed using the Student t-test. ** indicates p<0.01. (E) Representative IF-FISH images from (D). The white arrows indicate ATRX/DAXX occupied telomeres. (F) TKO cells depleted of DAXX or ATRX (siDAXX_3 and siATRX_1) were immunostained with antibodies against ATRX (green), DAXX (gray), and the Tel-RNP FISH probe that marks telomeres (red). Dotted circles indicate cells depleted of ATRX or DAXX due to successful knockdown. (G) TKO cells stably expressing Myc-tagged wildtype or mutant (Dnmt3a_R716A and Dnmt3b_V725G) Dnmt3a and Dnmt3b proteins were immunostained with anti-ATRX (green) and Myc (gray) antibodies. Telomeres were marked with a Tel-RNP FISH probe (red). Dotted circles indicate cells not expressing ectopic Dnmt3a/3b proteins.
Our immnuostaining data using TKO cells also suggest a central role for DAXX in the recruitment of DAXX/ATRX to telomere/subtelomere regions. Loss of ATRX had a minor effect on the telomeric localization of DAXX (Figure 4F, supplemental Figure S5 and S6A–B). However, knockdown of DAXX severely compromised the ability of ATRX to localize to telomeres. This was especially evident when cells with or without DAXX expression were compared to each other in the same image. When we ectopically expressed wildtype Dnmt3a or Dnmt3b in TKO cells (supplemental Figure S6C), these cells exhibited enhanced pericentric heterochromatin localization of DAXX and ATRX, with concomitant decreases in co-staining of DAXX/ATRX with telomere markers (Figure 4G). In contrast, ectopic expression of the enzymatically dead mutants of Dnmt3a/3b (Dnmt3a_R716A and Dnmt3b_V725G) failed to rescue the pericentric heterochromatin localization of DAXX and ATRX (Figure 4G). These data combined further support our hypothesis that DNA hypomethylation promotes binding/translocation of the DAXX/ATRX complex to tandem repetitive sequences such as telomeres.
To determine whether the localization of DAXX and ATRX also changes in pre-implantation embryos, we immunostained early embryos at different stages (zygotes to 16-cell morula) using anti-DAXX and ATRX antibodies (Figure 3E, supplemental Figure S6D). In zygotes, DAXX and ATRX exhibited nuclear staining and localized to pericentric heterochromatin. By morula stage, the majority of ATRX and DAXX appeared to have translocated to telomeres, which coincided with the drastic decrease in DNA methylation in embryos, indicating that DNA demethylation during early embryonic development can also induce the translocation of DAXX and ATRX to telomeres, where DAXX/ATRX likely play critical roles in telomere repression.
The DAXX/ATRX complex protects telomeres in the absence of DNA methylation
It has been shown that ATRX depletion leads to modestly increased telomere damage and de-repression of the telomere repeat containing non-coding RNA transcripts (TERRA) in mouse ES cells (Azzalin et al., 2007; Wong et al., 2010). When we knocked down DAXX or ATRX in TKO and wildtype J1 cells (supplemental Figure S6), we also observed increased TERRA expression, which was more pronounced in TKO cells (Figure 5A), suggesting a reliance on the DAXX/ATRX complex in repressing TERRA expression in the absence of DNA methylation. On hypomethylated telomeres, the DAXX/ATRX complex might function to inhibit telomeric sister-chromatid exchange events (T-SCE). Indeed, depleting ATRX or DAXX led to more cells showing T-SCE, and TKO cells again exhibited more severe phenotypes (Figure 5B), underlining the importance of the DAXX/ATRX complex in maintaining telomeres when DNA is hypomethylated. Additionally, double depletion of both DAXX and ATRX had no apparent additive or synergistic effects in cells.
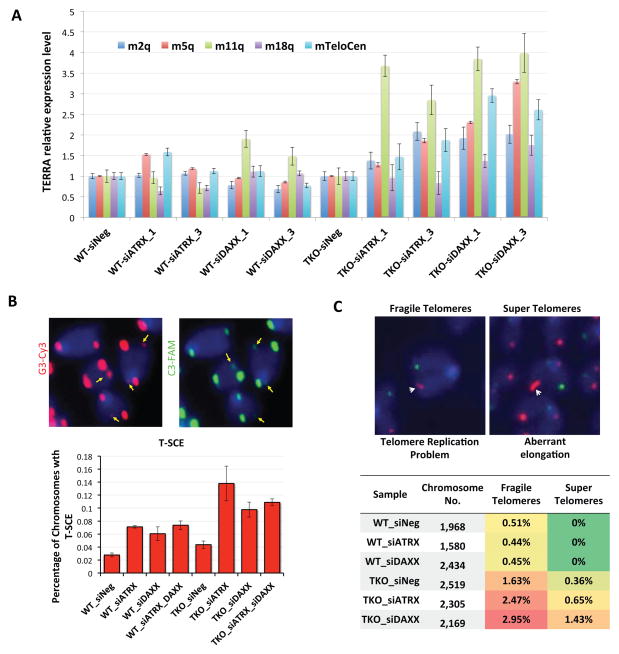
Figure 5. The DAXX/ATRX complex protects telomeres and ensures genome stability in response to DNA hypomethylation.
(A) Wildtype J1 (WT) and TKO cells knocked down for DAXX or ATRX were analyzed by RT-PCR for TERRA expression. Two different siRNA oligos each were used for DAXX and ATRX. Error bars indicate standard deviation (n=3). (B) The levels of telomeric sister-chromatid exchange (T-SCE) in wildtype J1 (WT) and TKO cells and those knocked down for DAXX and ATRX individually or together (siDAXX_3 and siATRX_1) were determined by CO-FISH. Leading and lagging strands were labeled by G3-Cy3 (TTAGGGTTAGGGTTAGGG) and C3-FAM (CCCTAACCCTAACCCTAA) respectively. Representative images are shown on top. The percentages of T-SCE in different cells were quantitated and plotted below. Error bars indicate standard deviation (n=3). (C) Metaphase spreads from wildtype J1 (WT) and TKO cells and those knocked down for DAXX or ATRX were prepared for FISH hybridization using the G3-Cy3 and C3-FAM oligo probes. Representative images of fragile and super telomeres are shown on top. The percentages of defective telomeres in different cells were quantitated and summarized below.
Telomere recombination may hinder DNA replication. We therefore carried out Q-FISH assays to examine abnormal fragile telomeres, a hallmark of defective telomere replication. While depleting DAXX or ATRX had little effect on wildtype J1 cells, TKO cells exhibited increased fragile telomeres, with smeared or double telomere signals on metaphase chromosomes (Figure 5C). Dysregulated telomere length control can result in abnormally long telomeres with average lengths of >1MB (super telomeres) (Chiodi et al., 2013). Again, we could observe super telomeres in TKO but not wildtype cells (Figure 5C). These observations help establish a role for DAXX and ATRX in protecting telomeres from damage and aberrant elongation in cells with hypomethylated DNA.
DAXX may recruit SUV39H1 to facilitate repression of repeat sequences
Both histone modifications and DNA methylation are crucial to transcriptional control. To better understand the epigenetic changes that occur with DNA hypomethylation, we investigated repressive epigenetic marks on telomeres in TKO and DKO cells. As shown in Figure 6A–B, antibodies against the repressive histone mark H3K9me3, the H3K9me3-binding protein HP1α (Kourmouli et al., 2005), and ATRX were able to bring down more telomeric DNA from DKO and TKO cells than from wildtype J1 cells. Furthermore, immunostaining analysis revealed extensive co-staining of H3K9me3 and HP1α with telomeres in TKO cells, but not in wildtype J1 cells (Figure 6C, 6D). ChIP-qPCR assays indicated that H3K9me3 was also enriched in TKO cells on DAXX/ATRX co-occupied repetitive sequences (e.g., subtelomeric regions and IAP), but not at promoter regions and tRNA genes (Figure 6E). These results provide evidence for the corresponding increases in repressive epigenetic marks following DNA hypomethylation at DAXX/ATRX target sites. Consistent with DAXX/ATRX-dependent H3.3 deposition, we also observed elevated accumulation of H3.3 on telomeres in DKO and TKO cells (Figure 6A–B).
Figure 6. DNA hypomethylation leads to increased recruitment of H3K9me3 and HP1 on telomeres and LTR-containing repeat elements.
(A) Telomere ChIP experiments were performed using wildtype J1 (WT), DKO, and TKO cells with the indicated antibodies. Precipitated DNA was dot-blotted and probed with the Tel(3) probe (TTAGGG(3)). IgG and anti-RAP1 antibodies were used as negative and positive controls respectively. Two different anti-H3K9me3 antibodies were used (H3K9me3-1 from Abcam and H3K9me3-2 from Upstate). *, non-relevant sample. (B) Three independent telomere ChIP experiments as described in (A) were performed and quantified. Error bars indicate standard deviation. The data were analyzed using the Student t-test. ** indicates p<0.01 and * indicates p<0.05. (C) Wildtype J1 (WT) and TKO cells were used for immunostaining analysis with an anti-H3K9me3 antibody (Abcam). Telomeres were marked with a telomere PNA probe. Arrows indicate co-staining of H3K9me3 with telomere foci. (D) Wildtype J1 (WT) and TKO cells were used for Immuno-FISH studies with antibodies against HP1α. Telomeres were marked with a Tel-RNP FISH probe. Arrows indicate co-staining of HP1α and telomere foci. (E) Wildtype J1 (WT) and TKO cells were used in ChIP-qPCR experiments with anti-H3K9me3 antibodies for LTR-containing repeat sites that are targeted by DAXX. Error bars indicate standard deviation (n=3). The data were analyzed using the Student t-test. ** indicates p<0.01 and * indicates p<0.05.
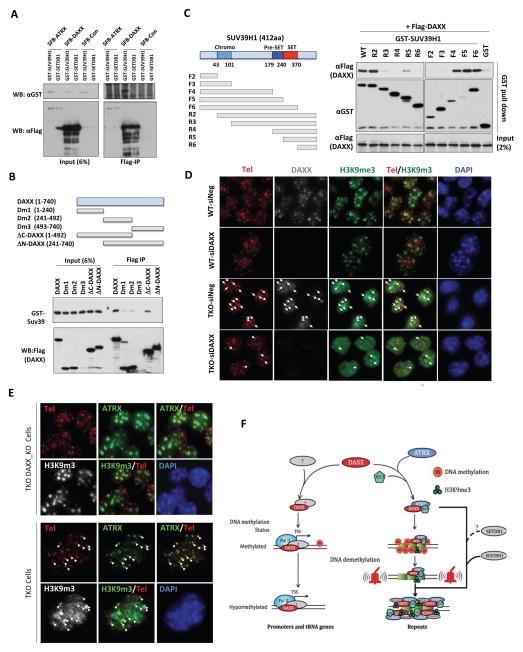
Heightened H3K9me3 marks could be a result of increased recruitment of histone modification enzymes. To investigate this possibility, we performed pull-down assays using SFB-tagged DAXX and ATRX, along with GST-tagged SUV39H1 and SETDB1, two major histone methyltransferases for H3K9. As shown in Figure 7A, DAXX was able to co-precipitate with SUV39H1, suggesting interaction between these two proteins. Further deletional mapping and Co-IP experiments using truncated DAXX and SUV39H1 proteins identified specific regions within DAXX and SUV39H1 that mediated the interaction (Figure 7B–C). In TKO cells, knockdown or knockout of DAXX resulted in the loss of telomeric accumulation of H3K9me3 (Figure 7D–E), suggesting DAXX-dependent H3K9me3 enrichment on telomeres in TKO cells. Taken together, our data support the model in which the DAXX/ATRX complex may upregulate H3K9me3 at its binding sites through interaction between DAXX and SUV39H1 (Figure 7F).
Figure 7. DAXX associates with SUV39H1 and facilitates H3K9me3 enrichment on telomeres in response to DNA demethylation.
See also supplemental Figure S7. (A) SFB-tagged DAXX or ATRX were co-expressed in mES cells with GST-tagged SUV39H1 or SETDB1. The cells were then harvested for immunoprecipitations (IP) using anti-FLAG antibodies. Co-precipitated proteins were detected by anti-GST antibodies. SFB-Con, vector alone. (B) GST-tagged full-length SUV39H1 was co-expressed with FLAG-tagged full-length or truncation mutants of DAXX in mES cells. The cells were then harvested for immunoprecipitations (IP) using anti-FLAG antibodies and probed with anti-GST antibodies. (C) FLAG-tagged full-length DAXX was co-expressed with GST-tagged full-length or truncation mutants of SUV39H1 in mES cells. The cells were then harvested for GST pull down and probed with anti-FLAG antibodies. (D) Wildtype J1 (WT) and TKO cells were transfected with control (siNeg) or DAXX (siDAXX_3) siRNAs and then analyzed by Immuno-FISH using anti-DAXX and H3K9me3 antibodies along with the telomere FISH probe TelG3-Cy3. Arrows indicate co-stained foci. (E) Parental and DAXX-knockout TKO cells were immunostained using anti-ATRX and H3K9me3 antibodies. Telomeres were marked by the telomere FISH probe TelG3-Cy3. Arrows indicate co-stained foci. (F) DAXX may act as a transcriptional regulator on gene promoters and tRNA regions that are normally low in DNA methylation. Global DNA demethylation may promote the recruitment/translocation of the DAXX/ATRX complex to tandem repeats (e.g. IAP, RLTR, and telomeres) to silence transcription and prevent instability. Through interaction with SUV39H1, DAXX may facilitate H3K9me3 recruitment to repetitive sequences for heterochromatin maintenance in the absence of DNA methylation.
Discussion
In this study, we generated genome-wide binding profiles of ATRX and DAXX and the transcriptomes of their target genes in both wildtype and DNA hypomethylated mouse ES cells. DAXX and ATRX clearly differ in their genomic binding distribution. The large number of DAXX-only binding sites, primarily located near promoters and tRNA genes, provides strong evidence for ATRX-less DAXX complexes, a conclusion also supported by findings of substantial DAXX-alone fractions in gel fractionation experiments (Lewis et al., 2010). Interestingly, inhibiting DAXX in mouse ES cells did not affect the transcription of DAXX-only target genes in general (supplemental Figure S4B), supporting differential function of DAXX on these two types of targets. Additionally, the lack of overlap between DAXX and H3.3 on DAXX-only targets also argues against a role for DAXX in H3.3 deposition in these cases (supplemental Figure S7).
Our study also revealed significant changes in DAXX and ATRX targeting when genomic DNA became hypomethylated. The DAXX/ATRX complex was preferentially recruited to the ERVK family of LTRs (e.g., IAP and RLTR) and telomeres under DNA hypomethylation conditions, and loss of DAXX and ATRX led to pronounced de-repression of these repeat sequences. Our findings have thus uncovered a pathway essential for repressing repetitive sequences in response to global DNA demethylation, which occurs during early embryogenesis, in ground-state mES cells, and when cells lose DNA methyltransferases (e.g., DKO and TKO cells). Indeed, our data with preimplantation embryos suggest that the DAXX/ATRX complex translocates to telomeres following global DNA demethylation. Given previous data in mammalian somatic cells that also found changes in DNA methylation distribution due to loss of ATRX (Gibbons et al., 2000), the dynamic modification and repression of repeat elements by both the ATRX/DAXX complex and DNA methylation pathways may be a general feature in most cell types. DAXX and ATRX are not responsible for H3.3 enrichment at ETn/MusD ERVs (Elsasser et al., 2015). We found derepression of ETn/MusD ERVs to be more apparent in DAXX KO cells under DNA hypomethylation (Figure 3B), suggesting that DAXX and DNA methylation work together to inhibit ETn/MusD ERVs. Our study underscores the role of the DAXX/ATRX complex in repressing retrotransposon transcription and safeguarding the genome in cells with hypomethylated DNA. While our manuscript was in revision, two groups published their work showing that ATRX and histone H3.3 were required for silencing specific sets of LTRs (IAPs) in ES cells (Elsasser et al., 2015; Sadic et al., 2015). In this study, we have further demonstrated overlapping yet independent functions of DAXX and ATRX in regulating repetitive sequences beyond IAPs.
Repressive epigenetic marks such as H3K9me3 are highly enriched on LTRs and telomeres. It is possible that H3K9me3 enrichment may prime telomeres for the recruitment of DAXX/ATRX, similar to what occurs at pericentric heterochromatin (Kourmouli et al., 2005). In fact, DAXX appears to play an active role in maintaining H3K9me3 at telomeres. Depletion of DAXX resulted in the loss of telomeric localization of both ATRX and H3K9me3. Furthermore, the interaction between DAXX and SUV39H1 offers a possible mechanism for DAXX-dependent establishment of H3K9me3 marks on telomeres. Previous reports have shown an association between the ADD domain of ATRX and H3K9me3 (Iwase et al., 2011) and a role of SUV39H1/2 in the maintenance and spreading of H3K9me3 on a subset of repeat elements (Bulut-Karslioglu et al., 2014). Our findings have added another layer of regulatory control to telomere chromatin. Recent studies suggest SETDB1 and KAP1 to be important for H3K9me3 of ERVs in mES cells (Elsasser et al., 2015), while SUV39H1/2 is partially responsible. Our model does not exclude factors such as SETDB1 from repetitive sequence repression. It is possible that SUV39H1, DAXX, H3K9me3, and ATRX can form a positive feedback loop to ensure the repression of repetitive sequences when DNA methylation level is low.
What are the mechanisms for increased recruitment of ATRX and DAXX to unmethylated regions? ATRX may be recruited through binding to H3K9me3 or tandem repetitive sequences that can adopt G-quadruplex structures. Our study here underscores the central role of DAXX in targeting DAXX/ATRX/H3K9me3 to repetitive sequences, where ATRX appears largely dispensable. The ChIP-seq and RNA-seq analyses have revealed that tandem repeat sequences are common in both DAXX only and DAXX/ATRX co-occupied sites. For instance, DAXX/ATRX clearly accumulate on IAP 5′ UTRs that are enriched in short tandem repeat sequences. These tandem repetitive DNA sequences (and not just ERV LTRs) are likely crucial to determining DAXX targeting as well.
Despite the ubiquitous expression of both DAXX and ATRX, co-localization of the DAXX/ATRX complex with telomeres was only observed in certain cell lines and tissues. Our study points to DNA methylation as a major determinant in the telomeric recruitment of DAXX/ATRX. Cells with reduced DNA methylation therefore may be more sensitive to DAXX/ATRX dysfunction. Mutations of DAXX and ATRX have been directly linked to chromosome instability, reduced cell survival, and activation of the ALT pathway in telomerase-negative cancers such as pancreatic neuroendocrine tumors (Bower et al., 2012; Marinoni et al., 2013; Zhang et al., 2013). Further investigation of DNA methylation status in ALT tumors should shed more light on the pathogenesis and progression of these cancers.
Experimental Procedures
Vectors and cell lines
Human ATRX (Dr. Junjie Chen, UT MDACC) and DAXX (GE Lifesciences) cDNAs were cloned into pDEST-27 and pCL-based vectors for GST, FLAG, or SFB tagging. Wildtype J1, DNMT3a/3b double KO (DKO), and DNMT1/DNMT3a/3b triple KO (TKO) mES cells were gifts from Dr. Margaret A. Goodell (Baylor College of Medicine). Ground-state cells were cultured in media with 2i (1 μM MEK inhibitor PD0325901 and 3 μM GSK3β inhibitor CHIR99021) and treated with vitamin C (100 μg/ml) (L-ascorbic acid 2-phosphate, A8960, Sigma) for 72 hours before analysis.
Cells were passaged 4 hours before transfection of siRNA oligos (supplemental Table S1), replenished with fresh media 4 hours after transfection and then harvested 48 hours later. CRISPRs/Cas9 gRNA vectors (supplemental Table S2) were generated using the pX330–hSpCas9+chimeric construct (Addgene) (Cong et al., 2013). Cells were transfected twice with the constructs and plated by limiting dilution four days after the second round of transfection. Deletion of ATRX and DAXX was also confirmed by western blots and immunostaining.
ChIP-seq and data analysis
ChIP-seq experiments and analysis were carried out essentially as previously described (Yang et al., 2011). ChIP-Seq data are available from the GEO database (GSE70850). DNA libraries were prepared from whole-genome ChIP using the DNA Sample Kit (Illumina), with ~10ng of precipitated DNA end-repaired and ligated to Illumina adaptors. The IgG sample served as a control. See Supplemental Materials for detailed analysis procedures. Published ChIP-seq data sets are listed in supplemental Table S3. Antibodies, ChIP-qPCR primers, and qRT-PCR primers used in this study can be found in supplemental Table S4, S5, and S6 respectively.
RNA-seq and data analysis
Cytoplasmic RNAs were extracted using the RNeasy Mini Kit (QIAGEN). mRNAs were captured using oligo-dT magnetic beads and fragmented. First-strand and second-strand cDNAs were generated using random primers and in the presence of dUTP respectively. The double-stranded cDNAs were end repaired, followed by the addition of a 3′A and Y-shaped adaptors to each end. The dUTP-containing strand was digested using UDG before PCR and sequencing on the Illumina HiSeq 2000. RNA-Seq data are available from the GEO database (GSE70850). Analysis follows published protocols with minor modifications (supplemental Materials) (Trapnell et al., 2012).
Supplementary Material
Highlights.
Loss of DNA methylation increases DNA binding by DAXX and ATRX
Knockdown of DAXX/ATRX leads to derepression of repetitive elements
DAXX/ATRX targeting also protects telomeres from recombination
DAXX/ATRX recruits SUV39H to promote H3K9 trimethylation
Acknowledgments
This work was supported by National Basic Research Program (973 Program 2012CB911201), National Natural Science Foundation of China (NSFC 91213302, 81330055, and 31371508), and the Program of International S&T Cooperation (2014DFB30020). We would also like to acknowledge the support of NIGMS GM095599, the Welch Foundation Q-1673, and the C-BASS Shared Resource at the Dan L. Duncan Cancer Center (P30CA125123). ChIP-seq and RNA-seq experiments were conducted at the Functional Genomic Core [FGC] facility at Baylor College of Medicine supported by shared Instrument Grant 1S10RR026550 from NIH.
Footnotes
Contributions
Z.S. and Q.H. conceived the project and organized components; Q.H., H.K., R.H., W.L., M.T., F.S., and D. Y. performed experiments and data analysis; Q.H., D.L., and Z.S. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
Accession number
Our ChIP-seq and RNA-seq data are available in the GEO database under the accession number GSE70850.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Armour JA, Anttinen T, May CA, Vega EE, Sajantila A, Kidd JR, Kidd KK, Bertranpetit J, Paabo S, Jeffreys AJ. Minisatellite diversity supports a recent African origin for modern humans. Nat Genet. 1996;13:154–160. doi: 10.1038/ng0696-154. [DOI] [PubMed] [Google Scholar]
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- Baumann C, Schmidtmann A, Muegge K, De La Fuente R. Association of ATRX with pericentric heterochromatin and the Y chromosome of neonatal mouse spermatogonia. BMC molecular biology. 2008;9:29. doi: 10.1186/1471-2199-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C, Viveiros MM, De La Fuente R. Loss of maternal ATRX results in centromere instability and aneuploidy in the mammalian oocyte and pre-implantation embryo. PLoS Genet. 2010;6:e1001137. doi: 10.1371/journal.pgen.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martinez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bower K, Napier CE, Cole SL, Dagg RA, Lau LM, Duncan EL, Moy EL, Reddel RR. Loss of wild-type ATRX expression in somatic cell hybrids segregates with activation of Alternative Lengthening of Telomeres. PloS one. 2012;7:e50062. doi: 10.1371/journal.pone.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature reviews Molecular cell biology. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galan C, Winter GE, Engist B, Gerle B, et al. Suv39hdependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Molecular cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- Bzymek M, Lovett ST. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc Natl Acad Sci U S A. 2001;98:8319–8325. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Zhang X, Bernatavichute YV, Jacobsen SE. Two-step recruitment of RNA-directed DNA methylation to tandem repeats. PLoS biology. 2006;4:e363. doi: 10.1371/journal.pbio.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodi I, Belgiovine C, Zongaro S, Ricotti R, Horard B, Lossani A, Focher F, Gilson E, Giulotto E, Mondello C. Super-telomeres in transformed human fibroblasts. Biochimica et biophysica acta. 2013;1833:1885–1893. doi: 10.1016/j.bbamcr.2013.03.030. [DOI] [PubMed] [Google Scholar]
- Cirio MC, Ratnam S, Ding F, Reinhart B, Navara C, Chaillet JR. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. BMC developmental biology. 2008;8:9. doi: 10.1186/1471-213X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepali Pathak SA. Repetitive DNA: A Tool to Explore Animal Genomes/Transcriptomes. Functional Genomics. 2012:157–180. [Google Scholar]
- Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, Ueda Y, Dyson N, Li E. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem. 2005;280:17986–17991. doi: 10.1074/jbc.M413246200. [DOI] [PubMed] [Google Scholar]
- Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015 doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov AV, Konev AY, Vershilova E, Fyodorov DV. Protein complex of Drosophila ATRX/XNP and HP1a is required for the formation of pericentric beta-heterochromatin in vivo. J Biol Chem. 2010;285:15027–15037. doi: 10.1074/jbc.M109.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2:e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H, Higgs DR. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Gonzalo S, Jaco I, Fraga M, Chen T, Li E, Esteller M, Blasco M. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nature cell biology. 2006;8:416–440. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- Grohmann M, Spada F, Schermelleh L, Alenina N, Bader M, Cardoso MC, Leonhardt H. Restricted mobility of Dnmt1 in preimplantation embryos: implications for epigenetic reprogramming. BMC developmental biology. 2005;5:18. doi: 10.1186/1471-213X-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaphy CM, de Wilde RF, Jiao Y, Klein AP, Edil BH, Shi C, Bettegowda C, Rodriguez FJ, Eberhart CG, Hebbar S, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heartlein MW. Chromosome instability associated with human repetitive sequences and DNA amplification. Progress in clinical and biological research. 1990;340B:415–424. [PubMed] [Google Scholar]
- Howell CY, Bestor TH, Ding F, Latham KE, Mertineit C, Trasler JM, Chaillet JR. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104:829–838. doi: 10.1016/s0092-8674(01)00280-x. [DOI] [PubMed] [Google Scholar]
- Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Xiang B, Ghosh S, Ren T, Lewis P, Cochrane J, Allis C, Picketts D, Patel D, Li H, et al. ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nature structural & molecular biology. 2011;18:769–845. doi: 10.1038/nsmb.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, Chevassut T, Forrester L, Ansell J, Ramsahoye B. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Molecular and cellular biology. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annual review of genomics and human genetics. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenetic and genome research. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kourmouli N, Sun YM, van der Sar S, Singh PB, Brown JP. Epigenetic regulation of mammalian pericentric heterochromatin in vivo by HP1. Biochemical and biophysical research communications. 2005;337:901–907. doi: 10.1016/j.bbrc.2005.09.132. [DOI] [PubMed] [Google Scholar]
- Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, de Lange T, De S, Petrini JH, Sung PA, Jasin M, et al. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS genetics. 2012;8:e1002772. doi: 10.1371/journal.pgen.1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni I, Kurrer AS, Vassella E, Dettmer M, Rudolph T, Banz V, Hunger F, Pasquinelli S, Speel EJ, Perren A. Loss of DAXX and ATRX are Associated with Chromosome Instability and Reduced Survival of Patients with Pancreatic Neuroendocrine Tumors. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- McDowell TL, Gibbons RJ, Sutherland H, O’Rourke DM, Bickmore WA, Pombo A, Turley H, Gatter K, Picketts DJ, Buckle VJ, et al. Localization of a putative transcriptional regulator (ATRX) at pericentromeric heterochromatin and the short arms of acrocentric chromosomes. Proc Natl Acad Sci U S A. 1999;96:13983–13988. doi: 10.1073/pnas.96.24.13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812–828. doi: 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JS, Bader D, Kuo F, Kozak C, Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes & development. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Rebollo R, Miceli-Royer K, Zhang Y, Farivar S, Gagnier L, Mager DL. Epigenetic interplay between mouse endogenous retroviruses and host genes. Genome biology. 2012;13:R89. doi: 10.1186/gb-2012-13-10-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science (New York, NY) 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Ross JP, Rand KN, Molloy PL. Hypomethylation of repeated DNA sequences in cancer. Epigenomics. 2010;2:245–269. doi: 10.2217/epi.10.2. [DOI] [PubMed] [Google Scholar]
- Sadic D, Schmidt K, Groh S, Kondofersky I, Ellwart J, Fuchs C, Theis FJ, Schotta G. Atrx promotes heterochromatin formation at retrotransposons. EMBO reports. 2015 doi: 10.15252/embr.201439937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue M, Ohta H, Kumaki Y, Oda M, Sakaide Y, Matsuoka C, Yamagiwa A, Niwa H, Wakayama T, Okano M. DNA methylation is dispensable for the growth and survival of the extraembryonic lineages. Current biology : CB. 2010;20:1452–1457. doi: 10.1016/j.cub.2010.06.050. [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Molecular cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalginskikh N, Poleshko A, Skalka AM, Katz RA. Retroviral DNA methylation and epigenetic repression are mediated by the antiviral host protein Daxx. Journal of virology. 2013;87:2137–2150. doi: 10.1128/JVI.02026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nature reviews Genetics. 2012;13:36–46. doi: 10.1038/nrg3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes to cells : devoted to molecular & cellular mechanisms. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochimica et biophysica acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Wolf G, Nielsen AL, Mikkelsen JG, Pedersen FS. Epigenetic marking and repression of porcine endogenous retroviruses. The Journal of general virology. 2013;94:960–970. doi: 10.1099/vir.0.049288-0. [DOI] [PubMed] [Google Scholar]
- Wong L, McGhie J, Sim M, Anderson M, Ahn S, Hannan R, George A, Morgan K, Mann J, Choo K. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome research. 2010;20:351–411. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Gibbons R, Yan Z, Yang D, McDowell TL, Sechi S, Qin J, Zhou S, Higgs D, Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, Songyang Z. Human telomeric proteins occupy selective interstitial sites. Cell research. 2011;21:1013–1027. doi: 10.1038/cr.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Francois R, Iyer R, Seshadri M, Zajac-Kaye M, Hochwald SN. Current understanding of the molecular biology of pancreatic neuroendocrine tumors. Journal of the National Cancer Institute. 2013;105:1005–1017. doi: 10.1093/jnci/djt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.