Abstract
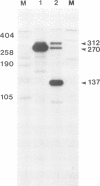
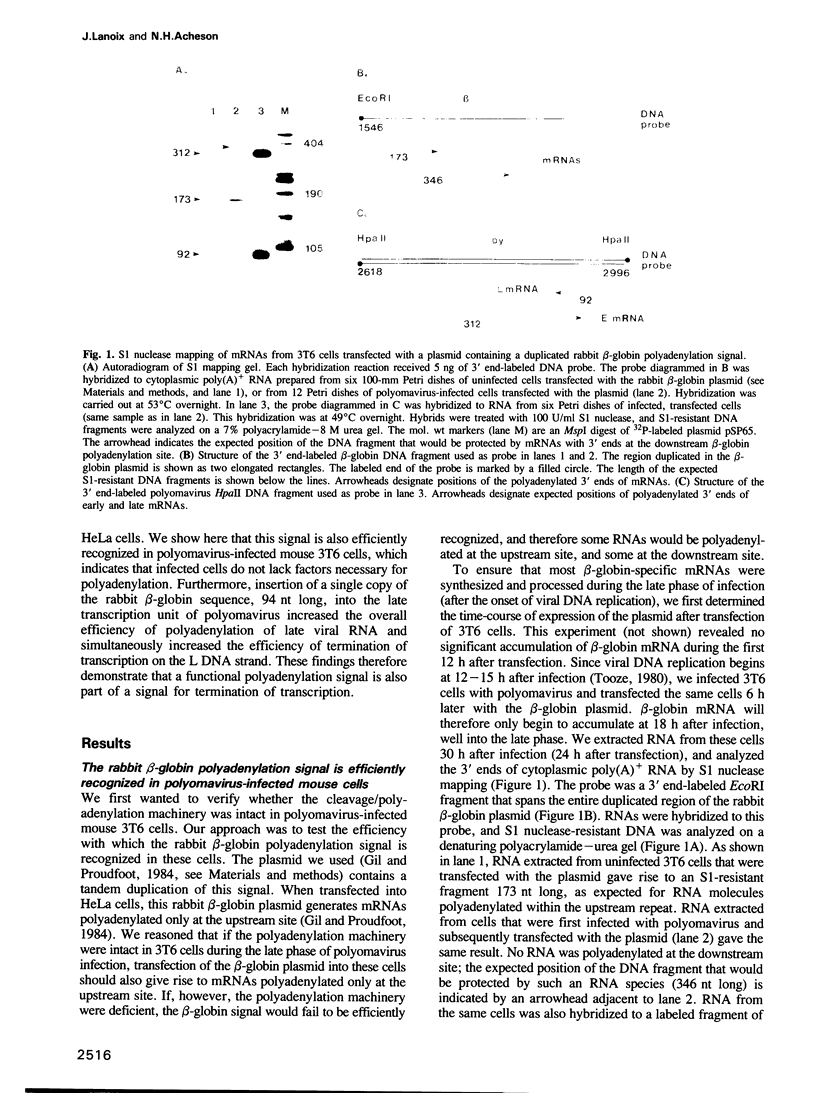
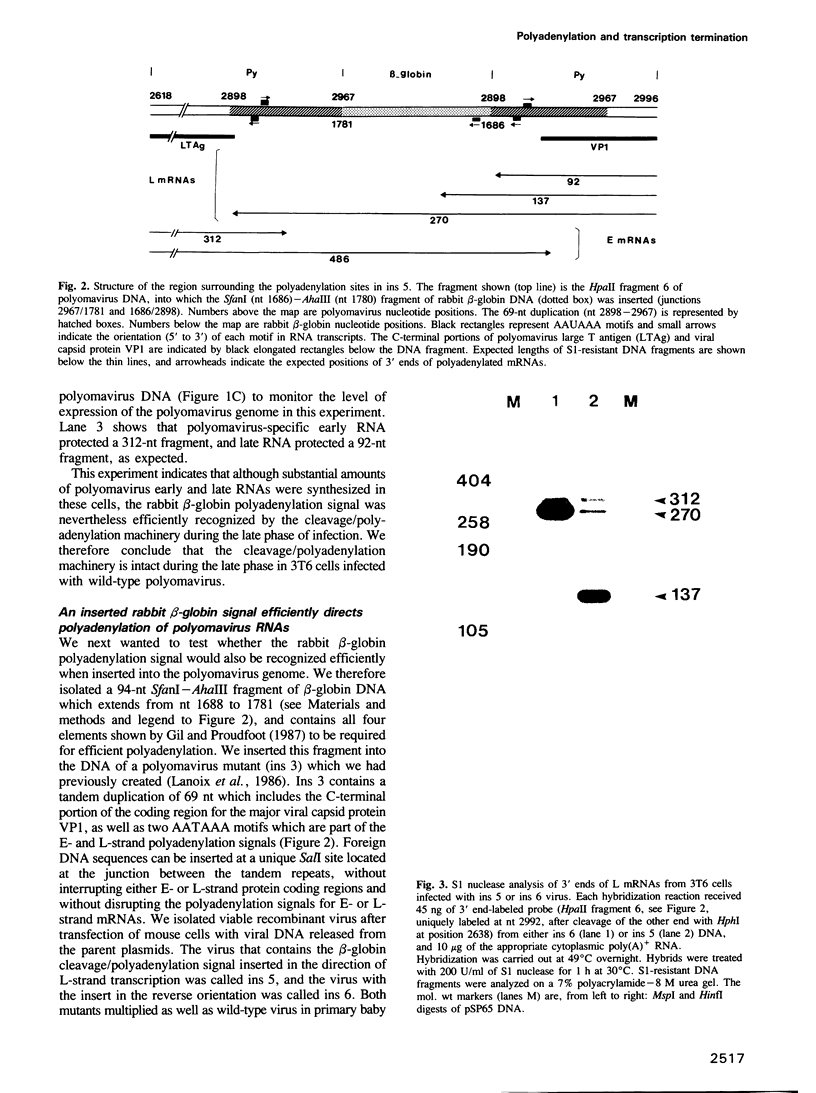
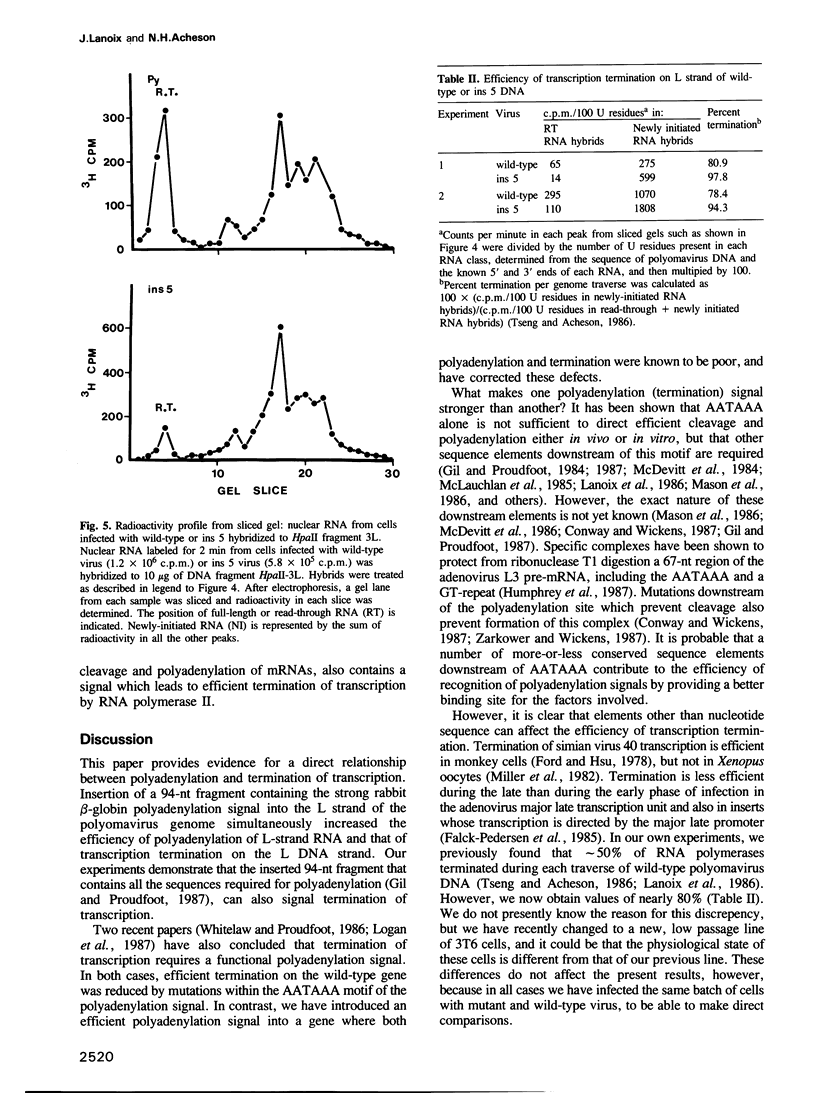
We constructed a viable insertion mutant (ins 5) of polyomavirus which contains, upstream of the L-strand polyadenylation signal, a 94-nt fragment of rabbit beta-globin DNA. Included in this fragment are all of the sequence elements required for efficient cleavage and polyadenylation of rabbit beta-globin RNA. The beta-globin signal was efficiently recognized by the cleavage/polyadenylation machinery in mouse 3T6 cells infected with ins 5, signalling greater than 90% of the polyadenylation events on L-strand RNAs. Furthermore, the presence of this efficient polyadenylation signal resulted in a 1.4- to 2.5-fold increase in the fraction of virus-specific RNAs that were polyadenylated. Most importantly, termination of transcription by RNA polymerase II on ins 5 DNA was also increased compared with wild-type virus; nearly 100% of polymerases terminated per traverse of the ins 5 genome. These findings demonstrate that the rabbit beta-globin insert, which contains a strong polyadenylation signal, also contains at least part of a signal for termination of transcription by RNA polymerase II. These results also show that the multiple, spliced leaders on polyomavirus L-strand mRNAs, which arise as a result of inefficient termination and polyadenylation, are not necessary for efficient virus replication.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Acheson N. H. Efficiency of processing of viral RNA during the early and late phases of productive infection by polyoma virus. J Virol. 1981 Feb;37(2):628–635. doi: 10.1128/jvi.37.2.628-635.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Kinetics and efficiency of polyadenylation of late polyomavirus nuclear RNA: generation of oligomeric polyadenylated RNAs and their processing into mRNA. Mol Cell Biol. 1984 Apr;4(4):722–729. doi: 10.1128/mcb.4.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acheson N. H. Polyoma virus giant RNAs contain tandem repeats of the nucleotide sequence of the entire viral genome. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4754–4758. doi: 10.1073/pnas.75.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adami G. R., Carmichael G. G. Polyomavirus late leader region serves an essential spacer function necessary for viability and late gene expression. J Virol. 1986 May;58(2):417–425. doi: 10.1128/jvi.58.2.417-425.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Citron B., Falck-Pedersen E., Salditt-Georgieff M., Darnell J. E., Jr Transcription termination occurs within a 1000 base pair region downstream from the poly(A) site of the mouse beta-globin (major) gene. Nucleic Acids Res. 1984 Nov 26;12(22):8723–8731. doi: 10.1093/nar/12.22.8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C. N., Santangelo G. M. Analysis in Cos-1 cells of processing and polyadenylation signals by using derivatives of the herpes simplex virus type 1 thymidine kinase gene. Mol Cell Biol. 1983 Feb;3(2):267–279. doi: 10.1128/mcb.3.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway L., Wickens M. Analysis of mRNA 3' end formation by modification interference: the only modifications which prevent processing lie in AAUAAA and the poly(A) site. EMBO J. 1987 Dec 20;6(13):4177–4184. doi: 10.1002/j.1460-2075.1987.tb02764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck-Pedersen E., Logan J., Shenk T., Darnell J. E., Jr Transcription termination within the E1A gene of adenovirus induced by insertion of the mouse beta-major globin terminator element. Cell. 1985 Apr;40(4):897–905. doi: 10.1016/0092-8674(85)90349-6. [DOI] [PubMed] [Google Scholar]
- Ford J. P., Hsu M. T. Transcription pattern of in vivo-labeled late simian virus 40 RNA: equimolar transcription beyond the mRNA 3' terminus. J Virol. 1978 Dec;28(3):795–801. doi: 10.1128/jvi.28.3.795-801.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3'-end formation. 1984 Nov 29-Dec 5Nature. 312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987 May 8;49(3):399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Grummt I., Rosenbauer H., Niedermeyer I., Maier U., Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986 Jun 20;45(6):837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Hagenbüchle O., Wellauer P. K., Cribbs D. L., Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984 Oct;38(3):737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- Humphrey T., Christofori G., Lucijanic V., Keller W. Cleavage and polyadenylation of messenger RNA precursors in vitro occurs within large and specific 3' processing complexes. EMBO J. 1987 Dec 20;6(13):4159–4168. doi: 10.1002/j.1460-2075.1987.tb02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig M., Hoffman E. P., Bertelson C. J., Monaco A. P., Feener C., Kunkel L. M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987 Jul 31;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- Lanoix J., Tseng R. W., Acheson N. H. Duplication of functional polyadenylation signals in polyomavirus DNA does not alter efficiency of polyadenylation or transcription termination. J Virol. 1986 Jun;58(3):733–742. doi: 10.1128/jvi.58.3.733-742.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMeur M. A., Galliot B., Gerlinger P. Termination of the ovalbumin gene transcription. EMBO J. 1984 Dec 1;3(12):2779–2786. doi: 10.1002/j.1460-2075.1984.tb02209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legon S., Flavell A. J., Cowie A., Kamen R. Amplification in the leader sequence of late polyoma virus mRNAs. Cell. 1979 Feb;16(2):373–388. doi: 10.1016/0092-8674(79)90013-8. [DOI] [PubMed] [Google Scholar]
- Legon S. The binding of ribosomes to polyoma virus RNA. Possible role of the leader region in initiation site recognition. J Mol Biol. 1979 Oct 25;134(2):219–240. doi: 10.1016/0022-2836(79)90033-0. [DOI] [PubMed] [Google Scholar]
- Logan J., Falck-Pedersen E., Darnell J. E., Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason P. J., Elkington J. A., Lloyd M. M., Jones M. B., Williams J. G. Mutations downstream of the polyadenylation site of a Xenopus beta-globin mRNA affect the position but not the efficiency of 3' processing. Cell. 1986 Jul 18;46(2):263–270. doi: 10.1016/0092-8674(86)90743-9. [DOI] [PubMed] [Google Scholar]
- McDevitt M. A., Hart R. P., Wong W. W., Nevins J. R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986 Nov;5(11):2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDevitt M. A., Imperiale M. J., Ali H., Nevins J. R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984 Jul;37(3):993–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3' termini. Nucleic Acids Res. 1985 Feb 25;13(4):1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T. J., Stephens D. L., Mertz J. E. Kinetics of accumulation and processing of simian virus 40 RNA in Xenopus laevis oocytes injected with simian virus 40 DNA. Mol Cell Biol. 1982 Dec;2(12):1581–1594. doi: 10.1128/mcb.2.12.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Tseng R. W., Acheson N. H. Use of a novel S1 nuclease RNA-mapping technique to measure efficiency of transcription termination on polyomavirus DNA. Mol Cell Biol. 1986 May;6(5):1624–1632. doi: 10.1128/mcb.6.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw E., Proudfoot N. Alpha-thalassaemia caused by a poly(A) site mutation reveals that transcriptional termination is linked to 3' end processing in the human alpha 2 globin gene. EMBO J. 1986 Nov;5(11):2915–2922. doi: 10.1002/j.1460-2075.1986.tb04587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D., Wickens M. Specific pre-cleavage and post-cleavage complexes involved in the formation of SV40 late mRNA 3' termini in vitro. EMBO J. 1987 Dec 20;6(13):4185–4192. doi: 10.1002/j.1460-2075.1987.tb02765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]