Abstract
Authentication of meat assumes significance in view of religious, quality assurance, food safety, public health, conservation and legal concerns. Here, we describe a PCR-RFLP (Polymerase Chain Reaction- Restriction Fragment Length Polymorphism) assay targeting mitochondrial cytochrome-b gene for the identification of meats of five most common food animals namely cattle, buffalo, goat, sheep and pig. A pair of forward and reverse primers (VPH-F & VPH-R) amplifying a conserved region (168–776 bp) of mitochondrial cytochrome-b (cytb) gene for targeted species was designed which yielded a 609 bp PCR amplicon. Further, restriction enzyme digestion of the amplicons with Alu1 and Taq1 restriction enzymes resulted in a distinctive digestion pattern that was able to discriminate each species. The repeatability of the PCR-RFLP assay was validated ten times with consistent results observed. The developed assay can be used in routine diagnostic laboratories to differentiate the meats of closely related domestic livestock species namely cattle from buffalo and sheep from goat.
Keywords: Meat, Adulteration, PCR-RFLP, Cytochrome-b gene, AluI, TaqI
Introduction
Authentication of the meat species assumes importance in view of the nagging problem of meat adulteration or substitution of meat with that of cheaper or less acceptable quality. Besides consumer satisfaction, certain social and religious concerns and possible health hazards associated with particular type of meat warrants honest labelling of the source of meat and meat products. The malpractice of fraudulent meat substitution or mislabelling is more common in countries with rather poor economy and high population with ever increasing demand for meat and meat products and their high cost. Furthermore, believers of Hinduism and Islam averse from inclusion of beef and pork in their diet, respectively, owing to their religious beliefs. In addition to socio-religious factors, food allergy due to consumption of particular type of meat or meat products has emerged as another major health concern implicating the beef (73 %), pork (58 %) and chicken (41 %) as the most common cause (Ayuso et al. 1999). Therefore, precise identification of origin of meat has become a vital element in food quality control procedures.
In the past, several analytical methods have been used to discriminate meats of different food animals, majority of which were based on the protein analysis either by electrophoretic (Vallejo et al. 2005), chromatographic (Toorop et al. 1997), or immunochemical assays (Chen and Hsieh 2000). However, all protein based methods share the common disadvantage of denaturation of proteins at high temperatures, resulting in changed antigenicity and electrophoretic mobility of molecules.
Recently, DNA-based methods have proved to be reliable tools in meat speciation studies. Given the relative thermostability of DNA molecule, these methods outweigh the protein-based techniques (Lenstra et al. 2001). Among the different DNA-based techniques including DNA hybridization (Buntjer and Lenstra 1998); PCR (Matsunaga et al. 1999); PCR-RFLP (Girish et al. 2005); RAPD-PCR (Sebastio et al. 2001); PCR-SSCP (Rehbein et al. 1997) and PCR product sequencing (Bartlett and Davidson 1992), PCR-RFLP has been one of the most widely used technique for meat species authentication (Meyer et al. 1995; Girish et al. 2005; Maede 2006).
In PCR-RFLP assay, a conserved region is amplified and the amplicons are subjected to restriction digestion using appropriate restriction enzymes resulting in a specific band pattern able to differentiate the target species. Comparative merit of PCR-RFLP lies in the fact that a universal PCR-primer system combined with few restriction enzymes (RE) could satisfactorily identify and discriminate various meat species and obviate the inclusion of a reference sample from the test protocol (Meyer et al. 1995). Moreover, careful selection of target genes and REs prevents ambiguous results caused due to intraspecies polymorphism (Wolf et al. 1999). Keeping in view the above indications, the present study was undertaken to develop a new primer based PCR-RFLP assay targeting mitochondrial cytb gene for differentiation of beef, carabeef, chevon, mutton and pork.
Materials and methods
Sample collection
In the present study, species considered for sampling include cattle (Bos indicus, n = 15), buffalo (Bubalus bubalis, n = 15), goat (Capra hircus, n = 15), sheep (Ovis aries, n = 15) and pig (Sus domesticus, n = 15). Approximately, 50 g each of authentic meat samples were collected from local markets and abattoirs (Pantnagar and Rampur). However, in case of cattle, tissue samples were obtained from the postmortem hall and Veterinary Clinic of the College of Veterinary and Animal Sciences, G.B.P.U.A&T, Pantnagar, Uttarakhand as cow slaughter is banned in India. The meat samples were collected under aseptic conditions, brought to laboratory in icebox and kept at −20 °C till further use.
DNA Extraction
DNA was isolated from meat samples using Wizard® Genomic DNA purification kit (Promega, Madison, Wisconsin, USA) as per the manufacturer’s specifications. The DNA extracted from meat samples was electrophoresed in 1 % agarose gel containing 1 μg/ml ethidium bromide in 0.5 % Tris borate buffer (TBE) for 1 h at 50 V. Thereafter, the stained DNA bands were visualized under UV trans-illuminator and documented over a gel documentation system.
Design of oligonucleotide primers and selection of REs
The mitochondrial (cytb) sequences of cattle, buffalo, goat, sheep and pig were downloaded from the NCBI database and aligned using Megalign program (Lasergene software; DNAStar, Inc. Madison, Wisconsin, USA). A conserved region of 609 bp length was identified and a pair of forward (VPH-F, position 168–193) and reverse primer (VPH-R, position 752–776) was designed using Primer-Select program (Lasergene software; DNAStar, Inc.). The sequence and description of the primers used are summarized in Table 1. The selected primers were confirmed for species specificity using the PRIMER-BLAST programme of NCBI (http://ncbi.nlm.nih.gov/primerblast). The primers were custom synthesized from IDT, Inc., Coralville, Iowa, USA and were used for PCR amplification. Restriction endonucleases were selected by employing tabulation and comparison using NEBcutter V2.0, New England BioLabs (http://tools.neb.com/NEBcutter2/). The REs namely Alu1 and Taq1 were selected in this study and procured from Bangalore Genei, Bangalore, India.
Table 1.
Primers designed for amplification of a conserved region of mitochondrial cytochrome-b gene
| Sl. No. | Primer | Direction | Sequence | Target | Length | Tm °C | Amplicon |
|---|---|---|---|---|---|---|---|
| 1. | VPH-F | Forward | ATCCGACACAACAACAGCATTCTCCT | Mt cyt-b gene | 26 bp | 60.9 | 609 bp |
| 2. | VPH-R | Reverse | GCTGGGGTGTAGTTGTCTGGGTCTC | Mt cyt-b gene | 25 bp | 60.9 |
Standardization of PCR
The PCR was set up in a volume of 50 μL containing 5 μL of 10X assay buffer (160 mM (NH4)2SO4, 670 mM Tris-HCl, pH 8.8, 0.1 % tween-20, 25 mM MgCl2, Bioron-GmbH, Ludwigshafen, Germany), 1 μL (200 μM each) of dNTP mix (pH 7.5, Promega, Madison, Wisconsin, USA), 1 μL (25 pmol) each of forward and reverse primer (IDT, Iowa, USA), 2.5U Taq DNA polymerase (Bioron-GmbH, Ludwigshafen, Germany), 50 ng of purified DNA and nuclease free water (Merck, Darmstadt, Germany). The tubes were flash spun and the PCR was performed in a thermocycler (Gene AMP® PCR System 9700, Applied Biosystems, Foster City, California, USA). The cycling conditions consisted of an initial denaturation (95°C for 5 min) followed by 40 cycles of denaturation (95°C, 45 s), primer annealing (57°C, 45 s), extension (72°C, 45 s) and final extension (72°C, 10 min). The amplicons were separated by agarose gel electrophoresis (2 %, prepared in 0.5X TBE buffer, 50 V for 1.5 h) and their size determined by comparing with 100 bp DNA ladder (M/s Bangalore Genei, Bangalore, India). The amplicons were visualized using a gel documentation system (AlphaImager® HP, Alpha Innotech Corp., San Leandro, California, USA).
Restriction fragment length polymorphism (RFLP)
The amplicons were subjected to purification using GeneiTM Quick PCR Purification kit (Bangalore Genei, Bangalore, India) as per the standard protocol supplied with the kit. Purified PCR products (609 bp) were digested with AluI and TaqI restriction enzymes. Enzyme buffer mix was prepared by mixing 3 μL of recommended 10X buffer and 1.5 μL (AluI, 12 units and TaqI, 15 units) of restriction enzyme. The reaction mix was prepared by adding 20 μL of purified PCR product with 4.5 μL of enzyme buffer mix and the volume was made up to 30 μL using autoclaved Milli Q water. The reaction mixture was subsequently incubated at 37°C for AluI and 65°C for TaqI overnight. Thereafter, the enzyme activity was arrested by freezing reaction mixture at –20°C for 30 min. The digested PCR products were electrophoresed along with 100 bp DNA ladder in 2.5 % agarose gel at 60 V for 2 h. The bands were analyzed using gel documentation system (Alpha Innotech Corp., USA).
Results and discussion
Although, DNA is present in the nucleus as well as in mitochondria; mitochondrial DNA being more conserved is preferred for species identification. High mutation rate of mitochondrial DNA (ten times greater than nuclear DNA) helps point mutations to accumulate very quickly, thus allowing precise discrimination of closely related species. Using an appropriate primer pair, mitochondrial sequences have been amplified in many species, and the resulting differences used for species identification (Di Pinto et al. 2005). The commonly targeted mitochondrial genes for species identification include cytb (Kanuch et al. 2007), mt rRNA (5 s, 12 s, 16 s, 18 s etc) (Girish et al. 2004, 2005; Martin et al. 2007; Karabasanavar et al. 2010) and d-loop region (Fajardo et al. 2007; Kumar et al. 2011; Karabasanavar et al. 2011a). Among these, cytb gene is a preferred target for meat speciation and taxonomic/ phylogenetic studies (Verma and Singh 2003).
Different DNA-based techniques have been used for meat authentication, which include DNA hybridization (Janssen et al. 1998); PCR and its variants (Kumar et al. 2011; Karabasanavar et al. 2011a, b, c); PCR-RFLP (Wolf et al. 1999; Girish et al. 2005; Maede 2006); RAPD-PCR (Sebastio et al. 2001); PCR-SSCP (Asensio et al. 2001) and PCR-sequencing (Bartlett and Davidson 1992). Among these techniques, PCR-RFLP is considered as a highly discriminatory, cheaper and reliable (Meyer et al. 1995). Although, PCR-sequencing is considered to be more precise, the high set-up cost and lengthy procedure involved can be a hindrance where speedy results are expected.
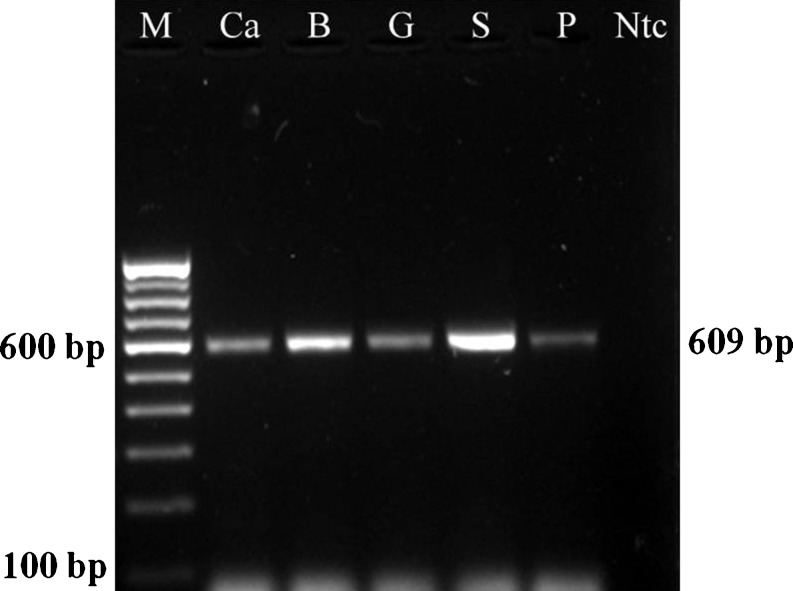
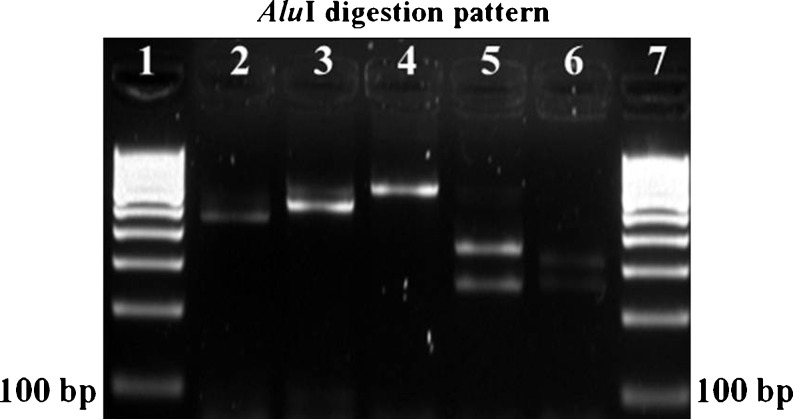
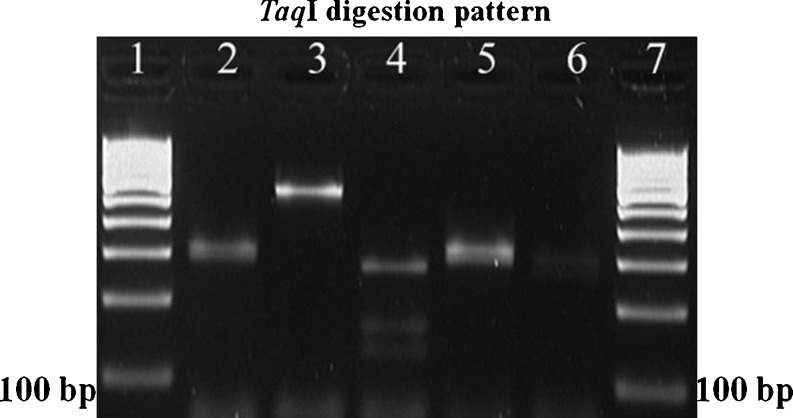
In the present study, we employed PCR-RFLP assay for identification of beef, carabeef, mutton, chevon and pork by amplifying a 609 bp region of cytb gene and subsequent digestion with AluI and TaqI restriction enzymes (Figs. 1, 2 and 3). AluI yielded three fragments of sizes 64, 92 and 453 bp in cattle, two fragments of 92 and 517 bp in buffalo, two fragments of 259 and 350 bp in sheep and four fragments of 17, 27, 243 and 322 bp in pig PCR products. However, the goat PCR product was not cleaved by AluI (Table 2 and Fig. 2). Similarly, the TaqI restriction enzyme yielded two fragments of 294 and 315 bp in cattle and sheep, two fragments of 70 and 539 bp in buffalo, four fragments of 43, 131, 163 and 272 bp in goat and three fragments of 43, 267 and 299 bp in pig PCR products (Table 2 and Fig. 3). Unique restriction pattern of AluI and TaqI for each species was found satisfactory to differentiate all five food animal species. However, digested PCR fragments of size less than 80 bp were not visible on gel electrophoresis, because of their small product size and rapid migration. Owing to their smaller size, they migrated faster through the gel while the larger fragments were still in the process of separation. Therefore, fragments <80 bp were neglected for result interpretation and similar considerations were also made by Wolf et al. (1999) previously. During the process of PCR standardization, annealing temperatures ranging from 53 °C to 60 °C were attempted to amplify the target sequence and 57 °C was selected as the optimum annealing temperature. Similarly, primer concentration of 25 pmol and PCR amplification at 40 cycles enhanced the intensity of amplification, sufficient to produce discrete bands on agarose gel.
Fig. 1.
PCR amplification of cytb gene of different food animals. M-100 bp DNA ladder, Ca- Cattle, B- Buffalo, G-Goat, S-Sheep, P-Pig, Ntc- negative control
Fig. 2.
RFLP profile of cytb gene. PCR amplicons were subjected to restriction digestion with Alu1 producing following restriction products: Lane 1: 100 bp DNA ladder, Lane 2: Cattle + Alu1 (64, 92, 453), Lane 3: Buffalo + Alu1 (92,517), Lane 4: Goat + Alu1 (609), Lane 5: Sheep + Alu1 (259,350), Lane 6: Pig + Alu1 (17, 27, 243, 322) and Lane 7: 100 bp DNA ladder
Fig. 3.
RFLP profile of cytb gene. PCR amplicons were subjected to restriction digestion with Taq1 producing following restriction products: Lane 1: 100 bp DNA ladder, Lane 2: Cattle + taq1 (294,315), Lane 3: Buffalo + taq1 (70,539), Lane 4: Goat + taq1 (43,131,163,272), Lane 5: Sheep + taq1 (294,315), Lane 6: Pig + taq I (43,267,299) and Lane 7: 100 bp DNA ladder
Table 2.
Restriction enzymes with their detailed description of the fragments that could discriminate the most commonly used meat animal species
| Species | Restriction enzyme that digests the 609 bp PCR product (size in bp) | |||||
|---|---|---|---|---|---|---|
| Alu I | Taq I | |||||
| RE site (s) No. & position | Fragments (bp) | RE site (s) No. & position | Fragments (bp) | |||
| Expected | Diagnostic | Expected | Diagnostic | |||
| Cattle | (2) 92 | (3) 64 | (1) 453 | (1) 315 | (2) 294 | (2) 294 |
| 545 | 92 | 315 | 315 | |||
| 453 | ||||||
| Buffalo | (1) 92 | (2) 92 | (1) 517 | (1) 70 | (2) 70 | (1) 539 |
| 517 | 539 | |||||
| Goat | – | – | – | (3) 43 | (4) 43 | (3) 131 |
| 315 | 131 | 163 | ||||
| 446 | 163 | 272 | ||||
| 272 | ||||||
| Sheep | (1) 350 | (2) 259 | (2) 259 | (1) 315 | (2) 294 | (2) 294 |
| 350 | 350 | 315 | 315 | |||
| Pig | (3) 17 | (4) 17 | (2) 243 | (2) 43 | (3) 43 | (2) 267 |
| 260 | 27 | 322 | 342 | 267 | 299 | |
| 287 | 243 | 299 | ||||
| 322 | ||||||
In past, few investigators have used this approach to identify food animal species targeting 12S rRNA gene (Girish et al. 2005), satellite DNA (Verkaar et al. 2002) and cytb gene (Maede 2006; Murugaiah et al. 2009) etc. However, these studies used comparatively more number of REs. The improvement of our work lies in the use of only two REs to differentiate five food animal species in a two-step process of PCR amplification followed by RFLP analysis. In a similar report, Meyer et al. (1995) amplified a 359-bp fragment of cytb gene followed by restriction digestion with four enzymes (AluI, RsaI, TaqI and HinfI) to identify meats of pig, cattle, wild boar, buffalo, sheep, goat, horse, chicken and turkey. Girish et al. (2005) reported differentiation of cattle, buffalo, sheep and goat meats by amplifying a conserved region (456-bp) of the mitochondrial 12S rRNA gene using AluI, HhaI, ApoI and BspTI enzymes.
The new set of primers elucidates more differentiating features between the closely related species such as cattle/buffalo and sheep/goat. AluI generated fragments of 453 bp and 517 bp for cattle and buffalo, respectively, with a difference of more than 50 bp that was satisfactorily visible on agarose gel. Similarly, TaqI generated fragments of 294 and 315 bp for cattle and a single fragment of 539 bp in buffalo. In case of sheep and goat, AluI resulted in fragments of 259 and 350 bp in the former; however, it did not digest the amplicon of the latter. Further, TaqI generated fragments of 131, 163 and 272 bp in goat and 294 and 315 bp in sheep. These differences in the band pattern were found adequate for unambiguous differentiation of closely related species.
Although, Partis et al. (2000) comprehensively differentiated animal species by using two REs, inclusion of buffalo in our study and the ability of our assay to accurately differentiate cattle and buffalo DNA offer a unique advantage, especially in the Asian and African continent, where buffalo meat is often adulterated with cattle meat. Further, the repeatability of the developed PCR-RFLP assay was validated ten times and consistent results were recorded with the constant cycling conditions and reagents. The confirmation of the results could however be accomplished by the use of species-specific PCRs or by the sequence analysis of resultant fragments, if desired; otherwise, the technique described in this study provides sufficient discrimination to identify the most common red meat animal species, i.e., cattle, buffalo, goat, sheep and pig.
Conclusion
In the present study, a PCR-RFLP assay was developed targeting mitochondrial cytb gene for the differentiation of beef, carabeef, chevon, mutton and pork with high specificity. The restriction digestion pattern of the two restriction enzymes (AluI and TaqI) was able to clearly distinguish the targeted species upon gel electrophoresis. Significantly, this assay could prove to be a useful tool for differentiation of closely related species such as cattle/ buffalo and sheep/ goat. Though, it takes about 24 h to yield results, the precision and specificity of the method justifies its application in meat speciation studies.
Acknowledgment
We sincerely thank The Dean, College of Veterinary and Animal Sciences, G.B.P.U.A&T, Uttarakhand, India for providing necessary facilities for smooth conduction of research. Authors would like to thank Dr. M. Suman Kumar, IVRI, Izatnagar for critical reading of the manuscript.
References
- Asensio L, González I, Fernández A, Rodríguez MA, Hernández PE, García T, Martín R. PCR-SSCP: a simple method for the authentication of grouper (Epinephelus guaza), wreck fish (Polyprion americanus), and Nile Perch (Lates niloticus) fillets. J Agric Food Chem. 2001;49:1720–1723. doi: 10.1021/jf001185w. [DOI] [PubMed] [Google Scholar]
- Ayuso R, Lehrer SB, Tanaka L, Ibanez MD, Pascual C, Burks AW, Sussman GL, Goldberg B, Lopez M, Reese G. Ig-E antibody response to vertebrate meat proteins including tropomyosin. Ann Allergy Asthma Immunol. 1999;83:399–405. doi: 10.1016/S1081-1206(10)62837-2. [DOI] [PubMed] [Google Scholar]
- Bartlett SE, Davidson WS. FINS (Forensically Informative Nucleotide Sequencing): a procedure for identifying the animal origin of biological specimens. Biotechniques. 1992;12:408–411. [PubMed] [Google Scholar]
- Buntjer JB, Lenstra JA. Mammalian species identification by interspersed repeat PCR fingerprinting. J Ind Microbiol Biotechnol. 1998;21:121–127. doi: 10.1038/sj.jim.2900540. [DOI] [Google Scholar]
- Chen FC, Hsieh YH. Detection of pork in heat-processed meat products by monoclonal antibody-based ELISA. J Assoc Off Anal Chem. 2000;83:79–85. [PubMed] [Google Scholar]
- Di Pinto A, Forte VT, Conversano MC, Tantillo GM. Duplex polymerase chain reaction for detection of pork meat in horse meat fresh sausages from Italian retail sources. Food Control. 2005;16:391–394. doi: 10.1016/j.foodcont.2004.04.004. [DOI] [Google Scholar]
- Fajardo V, Gonzalez I, Calleja L, Martin I, Rojas M, Garcia T, Hernandez PE, Martin R. PCR amplification of meats from Chamois (Rupicapra ruicapra), Pyrenean ibex (Capra pyrenaica), and Mouflon (Ovis ammon) targeting specific sequences from the mitochondrial D-loop gene. Meat Sci. 2007;76:644–652. doi: 10.1016/j.meatsci.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Girish PS, Anjaneyulu ASR, Viswas KN, Anand M, Rajkumar N, Shivakumar BM, Sharma B. Sequence analysis of mitochondrial 12S rRNA gene can identify meat species. Meat Sci. 2004;66:551–556. doi: 10.1016/S0309-1740(03)00158-X. [DOI] [PubMed] [Google Scholar]
- Girish PS, Anjaneyulu ASR, Viswas KN, Shivakumar BM, Anand M, Patel M, Sharma B. Meat species identification by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) of mitochondrial 12S rRNA gene. Meat Sci. 2005;70:107–112. doi: 10.1016/j.meatsci.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Janssen FW, Hagele GH, Buntjer JB, Lenstra JA. Species identification in meat using PCR-generated satellite probes. J Ind Microbiol Biotechnol. 1998;21:115–120. doi: 10.1038/sj.jim.2900541. [DOI] [Google Scholar]
- Kanuch P, Hajkova P, Zdenek R, Bryja J. A rapid PCR-based test for species identification of two cryptic bats Pipistrellus pipistellus and P. pygmaeus and its application on museum and dropping samples. Acta Chiropterol. 2007;9:277–282. doi: 10.3161/1733-5329(2007)9[277:ARPTFS]2.0.CO;2. [DOI] [Google Scholar]
- Karabasanavar NS, Singh SP, Shebannavar SN, Kotresh AM, Girish PS, Umapathi PV. Identification of avian species using polymerase chain reaction and sequence analysis of mitochondrial 12SrRNA gene. Vet Arhiv. 2010;80:653–661. [Google Scholar]
- Karabasanavar NS, Singh SP, Umapathi V, Girish PS, Shebannavar SN, Kumar D (2011a) Authentication of carabeef (water buffalo, Bubalus bubalis) using highly specific polymerase chain reaction. Eur Food Res Technol. doi:10.1007/s00217-011-1583-9
- Karabasanavar NS, Singh SP, Umapathi V, Kumar D, Shebannavar SN. Identification of goat meat using highly species-specific polymerase chain reaction. J Food Qual. 2011;34:142–149. doi: 10.1111/j.1745-4557.2011.00376.x. [DOI] [Google Scholar]
- Karabasanavar NS, Singh SP, Umapathi V, Kumar D, Patil G, Shebannavar SN. A highly specific PCR assay for identification of raw and heat treated mutton (Ovis aries) Small Rumin Res. 2011;100:153–158. doi: 10.1016/j.smallrumres.2011.07.009. [DOI] [Google Scholar]
- Kumar D, Singh SP, Singh R, Karabasanavar NS. A highly specific PCR assay for identification of goat (Capra hircus) meat. Small Rumin Res. 2011;97:76–78. doi: 10.1016/j.smallrumres.2011.01.013. [DOI] [Google Scholar]
- Lenstra JA, Buntjer JB, Janssen FW. On the origin of meat- DNA techniques for species identification in meat products. Vet Sci Tomorrow. 2001;2:1–15. [Google Scholar]
- Maede D. A strategy for molecular species detection in meat and meat products by PCR-RFLP and DNA sequencing using mitochondrial and chromosomal genetic sequences. Eur Food Res Technol. 2006;224:209–217. doi: 10.1007/s00217-006-0320-2. [DOI] [Google Scholar]
- Martin I, Garcia T, Fajardo V, Calleja L, Rojas M, Hernandez PE, Gonzalez I, Martin R. Mitochondrial markers for the identification of four duck species and the specific identification of Muscovy duck in meat mixtures using the polymerase chain reaction. Meat Sci. 2007;76:721–729. doi: 10.1016/j.meatsci.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Matsunaga T, Chikuni K, Tanabe R, Muroya S, Shibata K, Yamada J, Shinmura Y. A quick and simple method for the identification of meat species and meat products by PCR assay. Meat Sci. 1999;51:143–148. doi: 10.1016/S0309-1740(98)00112-0. [DOI] [PubMed] [Google Scholar]
- Meyer R, Hoefelein C, Luethy J, Candrian U. Polymerase chain reaction-restriction fragment length polymorphism analysis: a simple method for species identification in food. J Assoc Off Anal Chem. 1995;78:1542–1551. [PubMed] [Google Scholar]
- Murugaiah C, Noor ZM, Mastakim M, Bilung LM, Selamat J, Radu S. Meat species identification and Halal authentication analysis using mitochondrial DNA. Meat Sci. 2009;83:57–61. doi: 10.1016/j.meatsci.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Partis L, Croan D, Guo Z, Clark R, Coldham T, Murby J. Evaluation of a DNA fingerprinting method for determining the species origin of meats. Meat Sci. 2000;54:369–376. doi: 10.1016/S0309-1740(99)00112-6. [DOI] [PubMed] [Google Scholar]
- Rehbein H, Kress G, Schmidt T. Application of PCR-SSCP to species identification of fishery products. J Sci Food Agric. 1997;74:35–41. doi: 10.1002/(SICI)1097-0010(199705)74:1<35::AID-JSFA765>3.0.CO;2-2. [DOI] [Google Scholar]
- Sebastio P, Zanelli P, Neri TM. Identification of anchovy (Engraulis encrasicholus L.) and gilt sardine (Sardinella aurita) by polymerase chain reaction, sequence of their mitochondrial cytochrome b gene, and restriction analysis of polymerase chain reaction products in semipreserves. J Agric Food Chem. 2001;49:1194–1199. doi: 10.1021/jf000875x. [DOI] [PubMed] [Google Scholar]
- Toorop MR, Murch SJ, Ball RO. Methodology and development of prediction equations for the determination of pork substitution in veal. Food Res Int. 1997;30:629–636. doi: 10.1016/S0963-9969(98)00013-1. [DOI] [Google Scholar]
- Vallejo B, Gonzalez AF, Mazorra MA, Rodrıguez R. Capillary electrophoresis for the analysis of meat authenticity. J Sep Sci. 2005;28:826–836. doi: 10.1002/jssc.200500013. [DOI] [PubMed] [Google Scholar]
- Verkaar ELC, Nijman IJ, Boutaga K, Lenstra JA. Differentiation of cattle species in beef by PCR-RFLP of mitochondrial and satellite DNA. Meat Sci. 2002;60:365–369. doi: 10.1016/S0309-1740(01)00144-9. [DOI] [PubMed] [Google Scholar]
- Verma SK, Singh L. Novel universal primers establish identity of enormous number of animal species for forensic application. Mol Ecol Notes. 2003;3:28–31. doi: 10.1046/j.1471-8286.2003.00340.x. [DOI] [Google Scholar]
- Wolf C, Rentsch J, Hubner P. PCR-RFLP analysis of mitochondrial DNA: a reliable method for species identification. J Agric Food Chem. 1999;47:1350–1355. doi: 10.1021/jf9808426. [DOI] [PubMed] [Google Scholar]