SUMMARY
Streptococcal pyrogenic exotoxin B (SpeB) is an extracellular cysteine protease that is a critical virulence factor made by the major human pathogen group A Streptococcus (GAS). speB expression is dependent on the regulator of proteinase B (RopB) and is upregulated with increasing cell density and during infection. Because computer modeling suggested significant structural similarity between RopB and peptide-sensing regulatory proteins made by other Gram-positive bacteria, we hypothesized that speB expression is influenced by RopB-peptide interactions. Inactivation of the gene (vfr) encoding the virulence factor related (Vfr) protein resulted in increased speB transcript level during the exponential growth phase, whereas provision of only the amino-terminal region of Vfr comprising the secretion signal sequence in trans restored a wild-type speB expression profile. Addition of the culture supernatant from a Vfr signal peptide-expressing GAS strain restored wild-type speB transcript level to a vfr-inactivated isogenic mutant strain. A distinct peptide in the Vfr secretion signal sequence specifically bound to recombinant RopB. Finally, overexpression of the Vfr secretion signal sequence significantly decreased speB transcript level and attenuated GAS virulence in two mouse models of invasive infection. Taken together, these data delineate a previously unknown small peptide-mediated regulatory system that controls GAS virulence factor production.
Keywords: Virulence, gene regulation, SpeB, RopB, signal peptide
Introduction
Precise temporal regulation of virulence factor production is critical to microbial pathogenesis (Liu et al., 2011, Somerville & Proctor, 2009, Yoon et al., 2009). The genomic era has brought significant advances in understanding the range of genes influenced by specific transcriptional regulators (Goodman & Lory, 2004). However, information regarding the molecular mechanisms by which stand-alone transcription factors respond to environmental stimuli to fine tune expression of virulence-factor encoding genes is limited (Declerck et al., 2007, Withers et al., 2001). A better understanding of how pathogenic microbes regulate virulence factor production is critical to the goal of designing novel antimicrobials that can interfere with such signaling pathways (George et al., 2008, Hung et al., 2005, Kreikemeyer et al., 2003, Rasko et al., 2008).
Group A Streptococcus (GAS) is a Gram-positive bacterium that has long served as a model pathogen for investigating bacterial virulence factor regulation (Kreikemeyer et al., 2003, McIver, 2009). GAS causes a broad spectrum of human infections ranging from mild pharyngitis and impetigo to life threatening necrotizing fasciitis and streptococcal toxic shock syndrome (Olsen et al., 2009). Among the many virulence factors made by GAS, a secreted cysteine protease known as streptococcal pyrogenic exotoxin B (SpeB) is critical for virulence and dissemination of infection (Lukomski et al., 1998, Lukomski et al., 1999, Lukomski et al., 1997, Svensson et al., 2000). SpeB production is growth-phase dependent and increases as GAS transitions from the exponential to the stationary growth phase (Neely et al., 2003). Moreover, speB transcript is significantly increased during infection compared to growth in a standard laboratory medium (Loughman & Caparon, 2006). Given its significance in GAS pathogenesis, SpeB is subject to multiple layers of temporal and environmental regulation (Carroll & Musser, 2011).
Transcription of speB is directly controlled by a global regulator known as regulator of proteinase B (RopB) (Chaussee et al., 1999, Lyon et al., 1998, Carroll & Musser, 2011). RopB belongs to the Rgg-family of transcription regulators that are present in a diverse array of pathogenic low G+C Gram-positive bacteria (McIver, 2009). The speB promoter region has various cis-acting elements including two putative RopB binding sites with pseudo-palindromic sequences that are separated from each other by approximately ~120 bp (Fig. 1A) (Neely et al., 2003). RopB is required for activation of speB transcription, but attempts to activate speB expression at exponential phase of growth by ectopic provision of RopB failed to produce early onset of speB expression (Neely et al., 2003). Thus, it appears that additional signals are required for temporal regulation of SpeB production, but the identity and nature of such signals remain poorly understood (Neely et al., 2003, Chaussee et al., 1997).
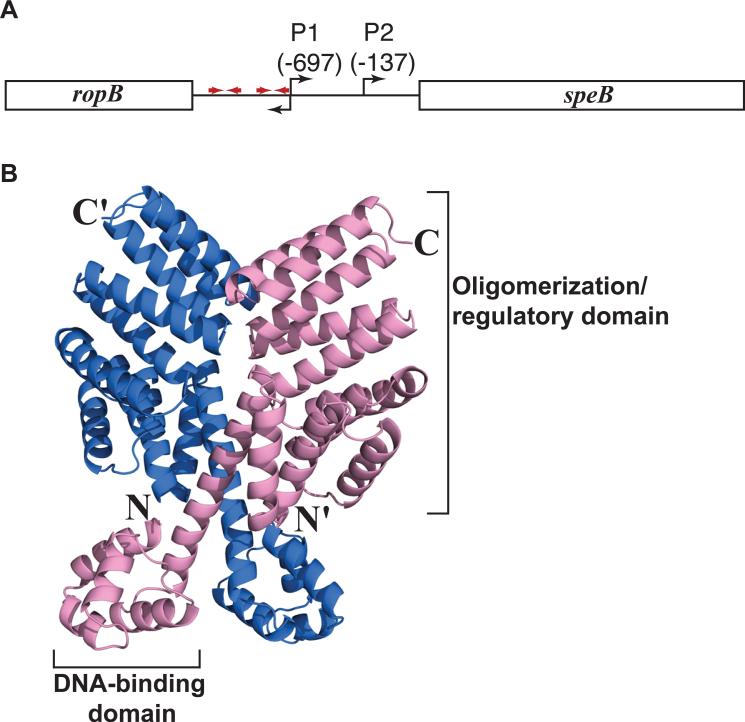
Figure 1. Organization of the speB gene region and RopB model.
(A) Organization of the speB gene region in strain MGAS10870. The divergently transcribed speB and ropB genes are shown as rectangle boxes and labeled. The bent arrows above the line indicate two transcription start sites of speB and bent arrow below denote the transcription start site of ropB. Red arrows indicate putative palindromic sites that have the RopB binding site. Data regarding transcription start sites and RopB binding sites are derived from (Neely et al., 2003). (B) RopB structure predicted by I-TASSER protein modeling server (http://zhanglab.ccmb.med.umich.edu/ITASSER). Ribbon representation of the predicted RopB homodimer is shown and the individual subunits of RopB dimer are colored in blue and pink. The amino- and carboxy-terminus of one subunit are indicated as N and C, respectively, and those of the second subunit are indicated with a prime ('). The putative DNA-binding domain in the amino-terminus and the oligomerization/regulatory domain in the carboxy-terminus of one subunit are labeled. The model shown is modified from Figure 2 from (Carroll et al., 2011).
To better understand the molecular mechanism of gene regulation by RopB, we recently performed three-dimensional computer modeling of RopB (Carroll et al., 2011). We discovered that RopB has significant predicted structural homology with PlcR from Bacillus cereus and PrgX from Enterococcus faecalis, the founding members of the Rap/Npr/PlcR/PrgX (RNPP)-family of quorum-sensing regulators (Fig. 1B) (Rocha-Estrada et al., 2010). Members of the RNPP-family use secreted peptides as regulatory signals and alter gene expression in response to changes in bacterial cell density (Mashburn-Warren et al., 2010, Rocha-Estrada et al., 2010, Fleuchot et al., 2011, Fontaine et al., 2010, Declerck et al., 2007). Given its predicted structural homology with PlcR and PrgX, it is possible that RopB uses secreted peptides as regulatory signals to control speB expression. In this regard, it is noteworthy that 15 years ago it was reported that inactivation of the GAS genes encoding oligopeptide permeases (Opp) or dipeptide permeases (Dpp) drastically reduced speB expression (Podbielski et al., 1996, Podbielski & Leonard, 1998) suggesting that peptide transport plays a role in speB regulation.
Ma et al. (2009) recently reported that inactivation of the gene (vfr) encoding virulence factor related (Vfr) protein increased speB expression and decoupled speB expression from growth-phase dependency (Ma et al., 2009). Vfr lacks typical characteristics of a DNA binding protein and the molecular mechanism by which it alters SpeB production is not known. Vfr contains a putative amino-terminal secretion signal sequence (Ma et al., 2009). After processing by peptidases, signal peptides in the extracellular milieu can be imported to the cytosol where they interact with transcriptional regulators (Kozlowicz et al., 2006, Antiporta & Dunny, 2002, Lazazzera, 2001). In this study we tested the hypothesis that a peptide derived from the amino-terminal region of the Vfr secretion signal sequence directly interacts with RopB to regulate SpeB production. Our findings confirmed this hypothesis and elucidated a new mechanism of GAS virulence factor regulation.
Results
Kinetics of vfr, ropB and speB transcript levels in a serotype M3 GAS strain
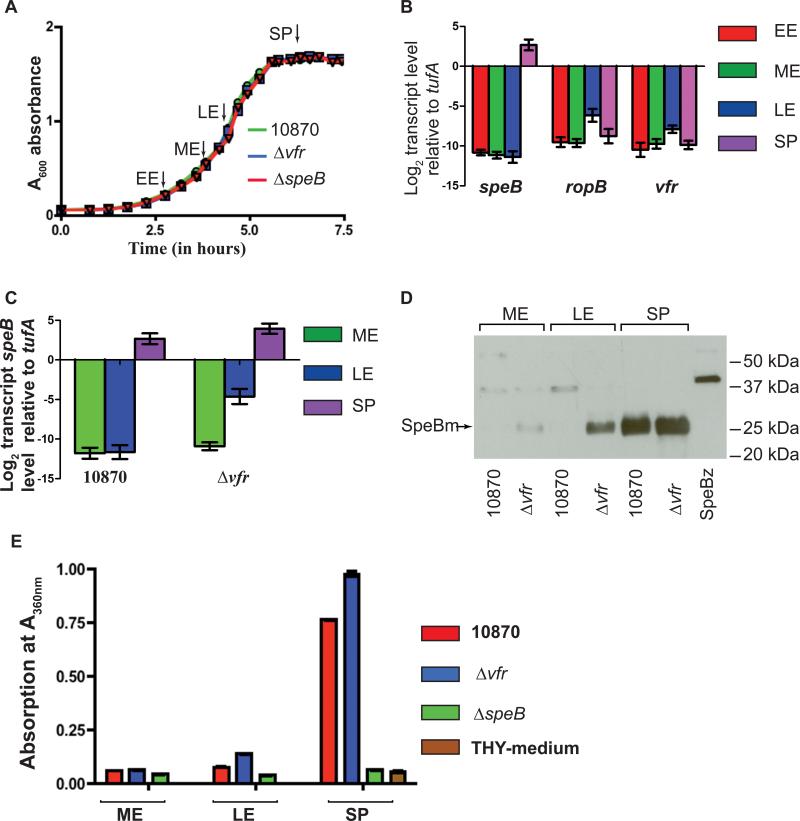
The only previous study of the vfr gene reported that its transcript peaks in the exponential growth phase and gradually decreases in stationary phase (Ma et al., 2009). Given that there is significant strain-to-strain variation in gene expression profiles among GAS isolates (Dmitriev et al., 2008), we first measured the transcript levels of vfr, ropB and speB in a serotype M3 strain (MGAS10870) at different growth stages (Fig. 2A). MGAS10870 was chosen for this study because it is an invasive isolate that has been fully sequenced, and it contains the most common RopB isoform (Beres et al., 2010, Carroll et al., 2011). Consistent with previous observations (Carroll et al., 2011, Neely et al., 2003), in standard laboratory medium ropB transcript reached a maximum level in late-exponential phase, and the speB transcript level peaked in the stationary phase (Fig. 2B). The kinetics of the vfr and ropB transcripts paralleled one another, reaching their highest levels at late exponential phase and returning to a basal level in stationary phase (Fig. 2B). These data suggest that the presence of Vfr in the late exponential growth phase may be inhibiting RopB-mediated speB transcription, and this inhibition is subsequently relieved in the stationary phase of growth.
Figure 2. vfr inactivation alters speB expression profile.
(A) Growth curve of indicated strains in THY broth. Samples were collected at early-exponential (EE, (A600 ~ 0.3), mid-exponential (ME, A600 ~ 0.6), late-exponential (LE, A600 ~ 1.0) and stationary phase (SP, A600 ~ 1.7) for transcript and protein analysis. Arrows indicated the sampling time points. (B) Transcript levels of speB, ropB and vfr in strain MGAS10870 measured by Taqman qRT-PCR. (C) speB transcript level in strains MGAS10870 and isogenic mutant strain Δvfr. For (B) and (C) duplicate biological replicates were grown on two separate occasions and analyzed in duplicate. Data graphed are mean ± standard deviation. (D) Representative western immunoblot analysis of SpeB in filtered growth media from strain MGAS10870 and isogenic mutant strain Δvfr. Each experiment was performed on three different occasions with essentially identical results. Growth media collected at indicated sampling points from strains MGAS10870 and Δvfr were analyzed using anti-SpeB polyclonal rabbit antibody and chemiluminescence. The bands corresponding to the mature form of SpeB are labeled (SpeBm) and indicated by an arrow. (E) Assay of SpeB protease activity by azocasein hydrolysis assay. Filtered growth media collected at indicated growth phases from strains MGAS10870 and Δvfr were assayed for secreted SpeB protease activity. Azocasein hydrolysis by SpeB was determined by measuring the absorption at A360 nm.
Vfr negatively influences speB expression in the late-exponential growth phase
To investigate the role of vfr in regulation of speB expression, we used non-polar insertional mutagenesis to construct an isogenic mutant derivative of strain MGAS10870 in which the entire vfr gene was replaced by a spectinomycin-resistance cassette. For the purpose of clarity, strain 10870Δvfr will be referred to hereafter as Δvfr. The growth kinetics of strains MGAS10870 and Δvfr did not differ significantly in rich laboratory media (Fig. 2A). Next, we compared speB transcript levels of the wild-type and mutant strains at different phases of growth. Very little speB transcript was expressed in the mid-exponential growth phase by either strain MGAS10870 or strain Δvfr (Fig. 2C). However, compared to wild-type, strain Δvfr had an approximately 64-fold increase in the speB transcript level at the late-exponential growth phase (Fig. 2C). The speB transcript level in mutant strain Δvfr remained elevated in the stationary-growth phase, but the difference at this point was only about two-fold compared to wild-type (Fig. 2C).
To determine if these differences in transcript level resulted in variation in the amount of SpeB made by the strains, we assessed the presence of secreted SpeB in the extracellular medium by Western immunoblotting and SpeB protease activity via by an azocasein-cleavage assay. Consistent with the transcript data, immunoreactive SpeB was detected only in the growth medium of strain Δvfr at the late-exponential growth phase, whereas SpeB was detected in the growth medium of both strains at the stationary phase (Fig. 2D). Next, we measured the SpeB protease activity by a chromogenic azocasein hydrolysis assay. Strain Δvfr had increased SpeB protease activity in the late-exponential and stationary growth phase compared to wild-type, although the differences observed at the stationary growth phase were more pronounced compared to those observed at the late-exponential growth phase (Fig. 2E). Taken together, these data indicate that, directly or indirectly, Vfr negatively influences speB expression and SpeB production, with the majority of the regulatory effect occurring in the late-exponential growth phase.
Overexpression of Vfr negatively influences speB expression
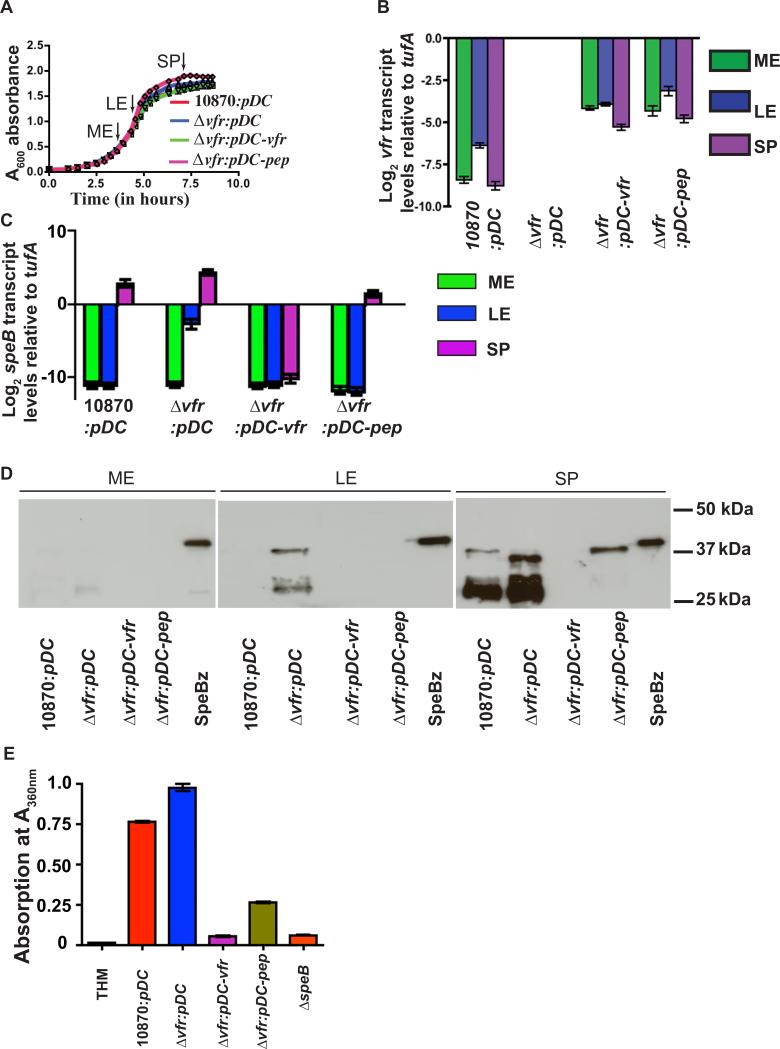
To test the hypothesis that ectopic expression of vfr by a plasmid would complement the vfr-inactivated isogenic mutant strain, we constructed plasmid pDC-vfr containing the entire vfr gene and its promoter region. The complemented mutant strain (strain Δvfr:pDC-vfr), strain Δvfr with empty vector (strain Δvfr:pDC), and the wild-type strain with empty vector (strain 10870: pDC) did not differ significantly in growth (Fig. 3A). To ensure that vfr in the pDC-vfr construct constituted a functional transcriptional unit, we measured the level of vfr transcript in the complemented strain by qRT-PCR. The pattern of the relative abundance of vfr transcripts at different growth stages was preserved in the complemented strain. We also found an overall increase (≥16 fold) in vfr transcript level in the complemented strain relative to wild-type strain MGAS10870, indicating that the complemented strain overexpressed vfr transcript (Fig. 3B). Importantly, the presence of the vfr-overexpressing plasmid in the vfr-inactivated strain restored a speB phenotype similar to that of wild-type at the late-exponential growth phase (Fig. 3C). Suppression of speB transcription by pDC:vfr was extended well into the stationary phase of growth, as the increased speB transcript level characteristic of stationary phase was absent in strain Δvfr:pDC-vfr (Fig. 3C). Consistent with the qRT-PCR data, no detectable immunoreactive SpeB was made by the complemented mutant strain (Fig. 3D). Similarly, negligible SpeB protease activity was detected in culture supernatants prepared from strain Δvfr:pDC-vfr samples with levels being comparable to those of the isogenic mutant SpeB-inactivated strain (strain 10870ΔspeB) (Fig. 3E). Thus, the speB phenotype of the Vfr-inactivated strain can be reversed by trans-complementation with vfr. In addition, the negative regulatory influences of Vfr on speB can be extended into stationary growth phase by vfr overexpression.
Figure 3. The secretion signal sequence of Vfr restores a wild-type speB expression profile to strain Δvfr.
(A) Growth pattern of indicated strains in THY broth. Samples were collected at mid-exponential (ME A600 ~ 0.6), late-exponential (LE A600 ~ 1.0) and stationary phase (SP A600 ~ 1.7) for transcript and protein analysis at time points designated by arrows. (B) vfr transcript levels as measured by Taqman qRT-PCR at indicated growth phase. (C) speB transcript levels in complemented strains at indicated growth phases. For (B) and (C), biological replicates were analyzed in duplicate on two separate occasions with data graphed being mean ± standard deviation. (D) Western immunoblot analysis of SpeB in filtered growth media prepared from indicated strains at indicated growth phases. (E) Assay of SpeB protease activity in the culture supernatant by azocasein hydrolysis. Azocasein hydrolysis by SpeB was assessed by absorption at A360 nm. Samples were analyzed in triplicates on two separate occasions with data graphed are mean ± standard deviation.
Overexpression of only the amino-terminal secretion signal sequence of Vfr negatively influences speB expression
To test the hypothesis that the element in Vfr responsible for regulating SpeB resides in the amino-terminal secretion signal sequence, we cloned the vfr gene region that encodes only the amino-terminal 39 amino acid residues of Vfr and the vfr promoter region into pDC123 to create plasmid pDC-pep. Consistent with our hypothesis, the complemented strain Δvfr:pDC-pep had a speB transcript level profile essentially identical to that observed for the wild-type strain containing empty vector (Fig. 3C). The decreased speB transcript level observed in the late-exponential growth phase for strain Δvfr:pDC-pep was comparable to that of full-length vfr complementation. These results indicate that the secretion signal peptide of Vfr alone is sufficient to negatively influence speB expression at the late-exponential growth phase (Fig. 3C). Similarly, results from western immunblotting analysis showed that SpeB production in strain Δvfr:pDC-pep was comparable to wild-type in the late exponential phase but there was decreased production of the mature SpeB isoform in strain Δvfr:pDC-pep in the stationary phase (Fig. 3D). The azocasein cleavage assay was too insensitive to reveal differences in SpeB protease activity in exponential-phase samples (Fig. 2E). Thus, we performed the assay only with stationary-phase samples. Compared to the vfr-inactivated strain containing empty vector, SpeB protease activity was significantly reduced at stationary phase in both complemented strains, with full-length complementation causing more pronounced regulation (Fig. 3E). In the aggregate, these data show that the amino-terminal secretion signal sequence of Vfr is sufficient for the negative influence of Vfr on speB expression in the late exponential phase of growth.
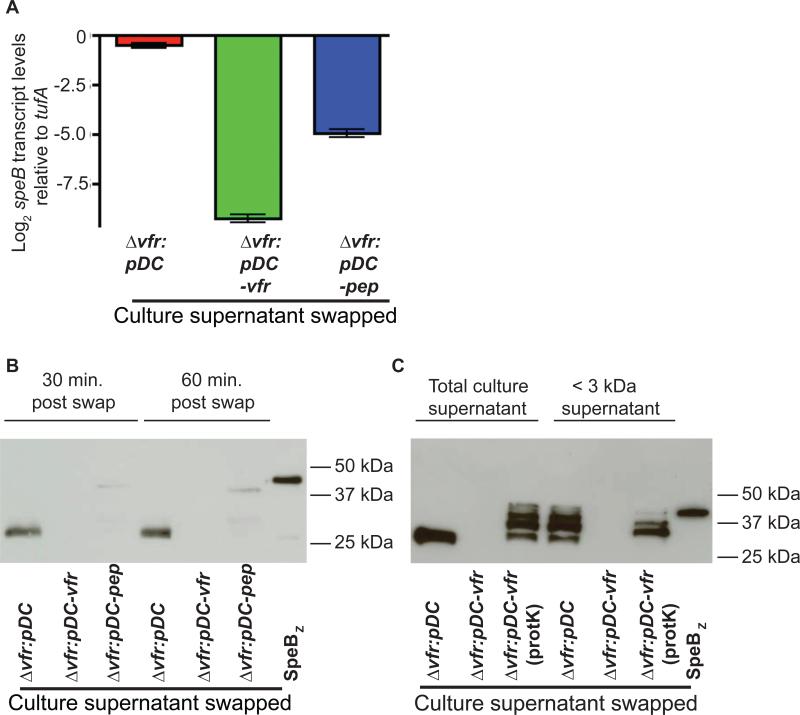
A factor in the culture supernatant of a Vfr-expressing strain restores a wild-type speB phenotype to strain Δvfr
Multiple attempts to synthesize peptides corresponding to regions of the Vfr secretion signal sequence failed to yield synthetic peptide of reliable sequence, high purity, high yield, and solubility, likely due to marked hydrophobicity of the peptides. Thus, we could not investigate whether direct addition of a synthetic peptide containing the Vfr secretion signal sequence to the growth medium inhibited speB expression. Rather, we performed culture supernatant-swapping experiments designed to determine if the Vfr secretion signal peptide is present in the extracellular environment and then internalized. Strain Δvfr was grown to mid-exponential phase (A600 ~0.6), cells were collected by centrifugation, and the cell pellet was suspended in the filtered growth media prepared from late-exponential phase cultures (A600 ~0.8) from one of the following strains: Δvfr:pDC, strain Δvfr:pDC-vfr, or Δvfr:pDC-pep. No immunoreactive SpeB was present in the pre-swap samples (data not shown). Samples were collected 1 h post-swapping (A600 ~1.0), and speB transcript level and secreted SpeB were measured by qRT-PCR and Western immunoblotting, respectively. Consistent with the hypothesis that the secretion signal sequence of Vfr is present in the extracellular milieu and subsequently imported into the cytoplasm to mediate speB regulation, significantly less speB transcript level was observed in strain Δvfr grown in the medium derived from strain Δvfr:pDC-vfr or strain Δvfr:pDC-pep compared to the same strain exposed to the medium derived from strain Δvfr-pDC (Fig. 4A and 4B). Similarly, culture supernatant derived from strain Δvfr:pDC-vfr or strain Δvfr:pDC-pep, but not strain Δvfr:pDC, drastically reduced the production of immunoreactive SpeB (Fig. 4B). Thus, speB expression is negatively influenced by Vfr or a product derived from it that is present in the culture supernatant.
Figure 4. Vfr represses SpeB production by a product present in the culture supernatant.
(A) speB transcript level analysis of mutant strain Δvfr grown in the culture supernatant the indicated strains for 1 hr as described in Materials and Methods. Two biological replicates were performed on two separate days and analyzed in duplicate with data graphed being mean ± standard deviation. P value is derived from a two-sample t-test. (B) Western immunoblot analysis of SpeB in the growth medium of mutant strain Δvfr grown for 30 minutes and 60 minutes in the culture supernatant derived from indicated strains as described in (A). (C) Western immunoblot analysis of SpeB in the growth medium of mutant strain Δvfr grown for 60 minutes in either the total culture supernatant or culture supernatant filtered with YM-3 (< 3 kDa) membrane derived from indicated strains. Proteinase K treated culture supernatants (Prot K) used for swapping are indicated.
To determine the chemical nature and the likely molecular mass of the regulatory factor, we performed a swap assay with culture supernatants of Δvfr:pDC-vfr that is sieved through YM-3 (~3 kDa molecular weight cut-off filter) membrane filter and/or treated with proteinase K. In accordance with the hypothesis that the secreted regulatory factor has low molecular mass, strain Δvfr grown in the low molecular weight component (≤ 3 kDa) of the culture supernatant obtained from Δvfr:pDC-vfr strain had negligible levels of secreted SpeB compared to that of strain Δvfr grown in the low molecular weight component of supernatant derived from strain Δvfr:pDC (Fig. 4C). Consistent with the hypothesis that the regulatory factor present in the culture supernatant of Δvfr:pDC-vfr is proteinaceous, strain Δvfr grown in the proteinase K treated culture supernatant of Δvfr:pDC-vfr failed to repress SpeB production (Fig. 4C). Together, these data indicate that the secreted factor present in the culture supernatant of Δvfr:pDC-vfr that negatively regulates speB expression is a low molecular weight peptide.
A distinct region of the amino-terminal secretion signal sequence of Vfr interacts with RopB
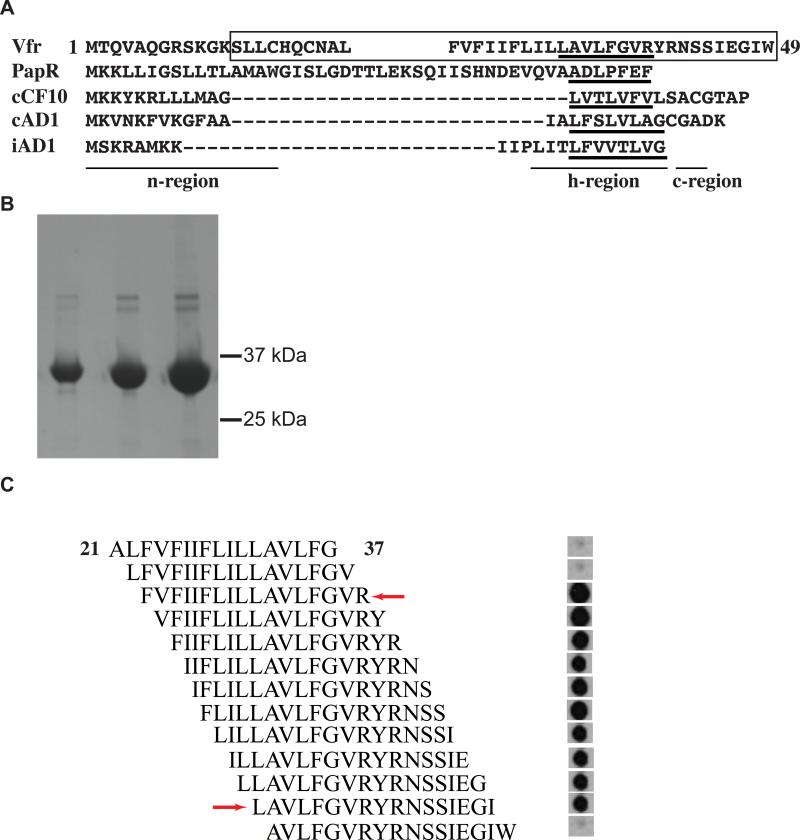
Inasmuch as the predicted structural homologs of RopB interact with secreted peptides to mediate gene regulation (Shi et al., 2005, Declerck et al., 2007), we next investigated whether a distinct region of the amino-terminal signal sequence of Vfr directly interacts with RopB using a synthetic peptide array. Overlapping 17-amino acid synthetic peptides that span the entire predicted signal sequence of Vfr (amino acid residues 1-49) were generated by SPOT synthesis on a cellulose membrane. The resulting synthetic peptide array encompassed all three components commonly found in secretion signal sequences, namely a basic n-region, a hydrophobic h-region, and a c-region with the protease cleavage site (Fig. 5A). Interaction between the synthetic peptides and purified recombinant hexa-histidine-tagged RopB (Fig. 5B) was assessed. The data show that RopB interacts with a distinct region of the amino-terminal secretion signal sequences of Vfr (Fig. 5C). The amino acids required for RopB binding are located in the h-region and c-region fragment of the Vfr signal sequence (Fig. 5C). Omission of amino acid residues R39 at the carboxy-terminus and L32 at the amino-terminus markedly decreased RopB binding, indicating the likely boundaries of the RopB binding site in the Vfr signal sequence (Fig. 5C). Thus, we conclude that a region of the amino-terminal secretion signal sequence of Vfr interacts directly with RopB suggesting a mechanism by which Vfr alters speB expression.
Figure 5. Recombinant RopB binds the Vfr amino-terminus signal sequence.
(A) Primary sequence alignment of Vfr signal sequence with characterized Gram-positive bacterial pheromones, PapR from Bacillus cereus; and cCF10, cAD1, and iAD1 from Enterococcus faecalis. Regions corresponding to the three major components of signaling peptides, n-region, h-region, and c-region, are underlined and labeled. Amino acid sequences corresponding to mature peptides of each pheromone and the putative Vfr mature peptide were underlined. Amino acid sequences of Vfr included in the peptide array are boxed. (B) SDS-polyacrylamide gel with increasing concentrations of purified recombinant hexa-histidine tagged RopB. The corresponding positions of molecular weight markers (in kilodaltons) are indicated. (C) Peptide array of overlapping 17-mer peptides that spans the entire putative Vfr signal sequence was generated using SPOT synthesis on a cellulose membrane. Membrane was incubated with purified recombinant hexa-histidine tagged RopB, and bound protein was detected with anti-hexahistidine tag antibodies and chemiluminescence. The red arrows indicate the likely amino-terminal and carboxy-terminal boundaries of the RopB binding site in the Vfr secretion signal sequence, located at L32 and R39 amino acid residues, respectively. The numbering of amino acid residues indicates their position within the Vfr secretion signal sequence.
Overexpression of the vfr secretion signal sequence significantly decreases GAS virulence in mouse models of invasive infection
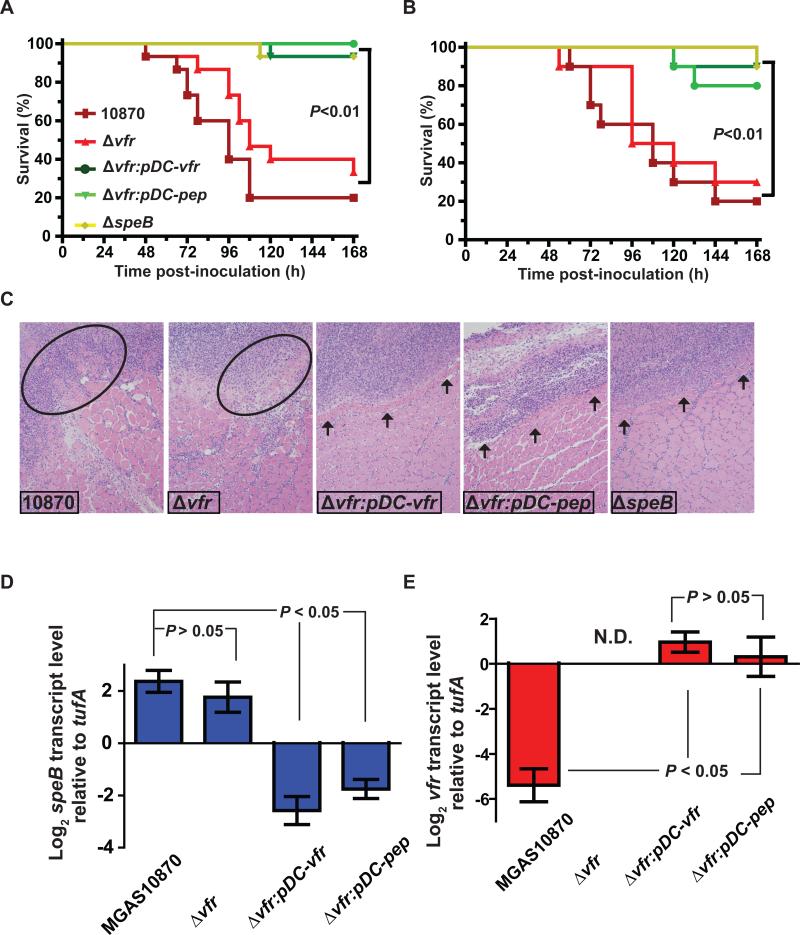
To test the hypothesis that the vfr gene and its influence on speB expression contribute to GAS virulence, we compared strains MGAS10870, ΔspeB, Δvfr, Δvfr:pDC-vfr, and Δvfr:pDC-pep in two different mouse models of invasive infection. First, mice were injected intraperitoneally with 1 × 107 colony-forming units (CFU) of each strain, and the ability to cause near mortality was monitored for seven days. The strains could be divided into two groups based on virulence phenotype. Wild-type strain MGAS10870 and mutant strain Δvfr caused significantly higher mortality compared to strains Δvfr:pDC-vfr, Δvfr:pDC-pep, or ΔspeB (P < 0.01 when comparing any two strains in different groups) (Fig. 6A). There was no significant difference in mortality among strains within either of the two groups.
Figure 6. Overexpression of the vfr signal sequence decreases GAS virulence in mouse models of infection.
(A) Fifteen CD-1 mice were inoculated with each indicated strain intraperitoneally and the near mortality was monitored. Kaplan-Meier survival curve with P values derived by log rank test. (B) Ten CD-1 mice were injected intramuscularly with 1 × 107 CFU of each strain. Kaplan-Meier survival curve with P values derived by log rank test. (C) Histopathologic analysis of muscular lesions from mice infected with each indicated strain. Areas of host tissue damage are circled, whereas confined, less destructive lesions are indicated by arrows. (D) and (E) TaqMan qRT-PCR analysis of speB and vfr gene transcript levels from mouse tissue samples infected with indicated strains. Samples were collected from the lesions of three mice per infecting strain and analyzed in duplicate. Resulting data were graphed as mean ± standard deviation with P values derived from two-sample t-test.
GAS cause a wide range of human infections with different virulence factors having varying impact depending on the site of infection (Engleberg et al., 2004, Olsen et al., 2009). Therefore, we also tested the role of vfr in GAS virulence using an intramuscular infection model that mimics human necrotizing fasciitis. Consistent with the results obtained from the intraperitoneal injection experiments, strains MGAS10870 and Δvfr were significantly more virulent than strains Δvfr:pDC-pep, Δvfr:pDC-vfr, and ΔspeB (P < 0.01 when comparing any two strains in different groups) (Fig. 6B). To better understand the role of vfr in GAS pathogenesis at the cellular level, we performed histopathologic analysis of intramuscular tissue lesions. The strains from the two groups caused contrasting pathologic characteristics. Lesions from the mice infected with strains MGAS10870 and Δvfr showed severe host-tissue damage and disseminated infection (Fig. 6C). In comparison, lesions from mice infected with strains Δvfr:pDC-pep, Δvfr:pDC-vfr, and ΔspeB were small, circumscribed and bordered by healthy muscle tissue (Fig. 6C).
To gain insight into the molecular mechanism by which overexpression of the Vfr secretion signal sequence reduces GAS virulence in vivo, we performed qRT-PCR analysis for speB and vfr transcript levels in infected mouse tissue. Similar to our observations for the stationary phase of growth in standard laboratory medium (Fig. 2C), there was no significant difference in in vivo speB transcript level between wild-type strain MGAS10870 and mutant strain Δvfr (Fig. 6D). In contrast, there was a significant decrease in speB transcript level in strains Δvfr:pDC-vfr and Δvfr:pDC-pep (Fig. 6D). As expected, no detectable vfr transcript was made by strain Δvfr, whereas vfr transcripts were present at a relatively high level in both complemented strains, Δvfr:pDC-vfr and Δvfr:pDC-pep (Fig. 6E). Interestingly, vfr transcript level was nearly undetectable in strain MGAS10870 suggesting strong repression of vfr expression during infection (Fig. 6E). Together, these data indicate that inactivation of Vfr does not significantly affect GAS virulence but overexpression of the Vfr secretion signal sequence significantly reduces speB transcript level in the host and attenuates GAS virulence.
Discussion
Many bacterial pathogens sense changes in population density in their environment via secreted molecules and respond by altering expression of genes involved in diverse cellular processes such as competence, conjugation, biofilm formation, and pathogen-host interaction (Bassler & Losick, 2006). Virulence gene expression at high cell density can result in host tissue damage and promote microbial dissemination and survival (Bassler, 1999, Van Delden & Comte, 2001, De Kievit & Iglewski, 2000). Conversely, pathogens also have mechanisms to downregulate virulence gene expression at low cell density to avoid premature elicitation of host defense responses which could promote bacterial clearance from the site of infection (Kozlowicz et al., 2006). The data presented herein provide new insights into virulence gene regulation in response to changes in the GAS growth phase. We have shown that at low cell density and in a growth-phase dependent manner, the amino-terminal secretion signal sequence of Vfr negatively influences expression of a key virulence factor, SpeB, by interfering with RopB-dependent speB transcription.
A previous study reported that genetic inactivation of vfr relieves growth-phase-dependent repression of speB expression (Ma et al., 2009). It was proposed that the Vfr influence occurs through protein-protein interactions with molecules in other regulatory networks (Ma et al., 2009). Our data show that the SpeB-related phenotype in Vfr inactivated strain can be reversed with only the region of vfr encoding the secretion signal sequence. This finding indicates that the speB regulatory function of vfr resides in the amino-terminal 40 amino acids of Vfr. Known regulatory peptides in Gram-positive bacteria are generally derived from either a small peptide-encoding open reading frame or from the amino-terminal secretion signal sequence of lipoproteins (Lazazzera, 2001, Antiporta & Dunny, 2002). In this regard, we note that the Vfr propeptide region has many of the characteristics of Gram-positive pheromones including a basic amino acid-rich amino-terminus region (n-region) and a hydrophobic central region (h-region). However, the Vfr secretion signal sequence lacks a carboxy-terminal cysteine residue commonly required for signal peptidase II cleavage and secretion (Paetzel et al., 2002, Von Heijne, 1990) (Fig. 5A). Interestingly, recently identified pheromone propeptides from S. mutans and S. thermophilus also lack the characteristic cysteine residue at the cleavage site and are believed to be processed and secreted by unknown secretion systems (Fontaine et al., 2010, Mashburn-Warren et al., 2010). Additionally, Chang et.al recently reported that short-hydrophobic peptides (SHP2 and SHP3) interact with RopB-like regulators (designated Rgg2 and Rgg3 in that study) in GAS and mediate gene regulation (Chang et al., 2011). The Vfr-RopB interaction seems fundamentally distinct from those of the SHP-Rgg as shp2 and shp3 are encoded immediately upstream of the respective rgg gene and are not part of a larger open reading frame as is the case with the Vfr N-terminal signal sequence. Thus, in concert with recently published data, our findings add to the rapidly expanding appreciation of the role of the diverse role of small peptide molecules in GAS pathophysiology.
Similar to many other bacterial transcription regulators, RopB may function as a dimer, and two RopB binding sites with inverted repeats have been identified in the speB promoter region that are separated from each other by ~130 bp (Fig. 1B) (Neely et al., 2003). For both PrgX and PlcR (structural homologs of RopB) peptide-binding triggers structural changes in their respective effector molecule that leads to altered DNA-binding and/or oligomerization, thereby altering gene expression (Kozlowicz et al., 2006, Shi et al., 2005, Declerck et al., 2007). PlcR-promoter interactions and the consequent transcription activation by PlcR occur only in the presence of the activating peptide PapR (Slamti & Lereclus, 2002). On the other hand, binding of either inhibitory peptide, iCF10 or activating peptide, cCF10 to PrgX controls the regulatory properties of PrgX. Interaction between PrgX and iCF10 promote PrgX tetramerization, resulting in the occupation of both PrgX binding sites by DNA looping and repression (Kozlowicz et al., 2006). Conversely, interactions between cCF10 and PrgX disrupt the tetramerization interface and the dimeric PrgX activates gene expression (Shi et al., 2005). Thus, the Vfr regulatory peptide may influence speB expression by modulating oligomerization of RopB, thereby interfering with or otherwise altering DNA-binding at one or both of the speB promoter binding sites.
The negative regulation of speB expression by Vfr is more pronounced during the exponential phase of growth, with repression being relieved in the stationary growth phase. Our data indicate that removal of Vfr-mediated negative regulation of speB expression by downregulation of vfr gene expression at stationary phase contributes to transcriptional activation of speB (Fig. 2B). Similarly, repression of vfr transcript level during infection may help to explain the high levels of speB transcript that are typically observed following GAS infection (Fig. 6D, 6E). However, inactivation of vfr alone cannot fully account for the rapid increase in speB expression observed in MGAS10870 at stationary-growth phase as the Vfrinactivated strain had the same speB transcript level as strain MGAS10870 at the mid-exponential growth phase (Fig. 2C). Moreover, the speB transcript level in strain Δvfr at late-exponential phase remained significantly less than that observed for strain MGAS10870 at the stationary phase of growth. Thus it is likely that an activation element, possibly a secreted peptide that is produced at high cell density is responsible for the activation of speB expression observed at stationary phase. Similar dynamics between inhibiting and activating peptides were observed in the regulation of PrgX function (Shi et al., 2005, Kozlowicz et al., 2006). The finding of such an activation peptide would explain the long-standing observation that elimination of transporter systems involved in the uptake of small peptides reduces SpeB production and might help explain how GAS is able to increase SpeB production during infection (Podbielski et al., 1996, Podbielski & Leonard, 1998).
In conclusion, we show that the amino-terminal secretion signal sequence of Vfr is an integral part of the system used by GAS to regulate expression of the gene encoding SpeB, a critical virulence factor. These data describe the existence of a pheromone-like regulatory circuit that controls virulence gene expression in a growth-phase depending manner. Complete elucidation of peptide-controlled regulatory networks such as this could serve as the platform for the development of novel intervention strategies.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are listed in Table 1. Strain MGAS10870 is a previously-described invasive serotype M3 isolate whose genome has been fully sequenced (Beres et al., 2010). MGAS10870 is representative of serotype M3 strains that cause invasive infections and has a wild-type sequence for all major regulatory genes including RopB (Beres et al., 2010). GAS was grown routinely on Trypticase Soy agar containing 5% sheep blood (BSA; Becton Dickinson) or in Todd-Hewitt broth containing 0.2% (w/v) yeast extract (THY; DIFCO). When required, spectinomycin or chloramphenicol was added to a final concentration of 150 μg/ml and 5 μg/ml, respectively. All GAS growth experiments were done in triplicate on three separate occasions for a total of nine replicates. Overnight cultures were inoculated into fresh medium to achieve an initial OD600 of 0.03. Growth was monitored by measuring A600. The Escherichia coli strain used for protein overexpression was grown in Luria-Bertani broth (LB broth; Fisher Scientific) and when appropriate ampicillin was added to a final concentration of 40 μg/ml.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Description | Reference |
|---|---|---|
| Strains | ||
| MGAS10870 | Invasive isolate, serotype M3 | (Beres et al., 2010) |
| MGAS10870:pDC | MGAS10870 with empty vector, Cm+ | This study |
| Δ vfr | MGAS10870Δvfr::aad9 | This study |
| Δvfr:pDC | MGAS10870Δvfr::aad9, empty vector, Cm+ | This study |
| Δvfr-pDC:vfr | MGAS10870Δvfr::aad9, vfr+, Cm+ | This study |
| Δvfr-pDC:pep | MGAS10870Δvfr::aad9, Vfr peptide+, Cm+ | This study |
| MGAS10870ΔspeB | MGAS 10870ΔspeB::aad9 | This study |
| Plasmids | ||
| pSL60-1 | Vector containing aad9 gene encoding spectinomycin resistance | (Lukomski et al., 2000) |
| pDC123 | Low-copy number plasmid capable of replication in GAS and Escherichia coli, Cm+ | (Li et al., 1997) |
| pDC:vjr | pDC123 with entire vfr gene plus promoter, Cm+ | This study |
| pDC:pep | pDC123 with Vfr N-terminal secretion signal plus promoter, Cm+ | This study |
Creation of vfr- and speB-inactivated mutant strains
Insertion inactivation of the speB and vfr genes in wild-type strain MGAS10870 was performed by methods described previously (Kuwayama et al., 2002, Lukomski et al., 2000). Briefly, a PCR fragment containing a spectinomycin-resistance (spc) cassette with the fragment of gene to be deleted on either side was generated in a three-step PCR process. Subsequently, the plasmid with the spc gene-disruption cassette was introduced into the parent strain by electroporation and the gene was disrupted through homologous recombination. The isogenic mutant strains were selected by growth on spectinomycin-containing medium. Inactivation of the gene was confirmed by DNA sequencing. Primers used for the construction of strain 10870ΔspeB and Δvfr are listed in supplemental Table S1.
Construction of Vfr full-length and Vfr-peptide complementation plasmids
To complement isogenic mutant strain Δvfr, the coding sequence of the full-length vfr gene or the region encoding the amino-terminus secretion signal sequence (amino acid residues 1-39) were cloned into the E. coli-GAS shuttle vector pDC123 (Chaffin & Rubens, 1998): Using the primers listed in Table S1, the respective fragments were amplified by PCR from GAS genomic DNA, digested with BglII and NdeI, and ligated into digested vector pDC123. The inserts were verified by DNA sequencing and electroporated into the appropriate mutant derivatives of strain MGAS10870.
Measurement of transcript levels by quantitative RT-PCR
GAS strains were grown to the indicated OD600 and incubated with two volumes of RNAprotect (Qiagen) for 10 min at room temperature. Bacteria were harvested by centrifugation and the cell pellets were snap frozen with liquid nitrogen. RNA isolation and purification were performed using an RNeasy kit (Qiagen). Purified RNA was analyzed for quality and concentration with an Agilent 2100 Bioanalyzer. cDNA was synthesized from the purified RNA using Superscript III (Invitrogen) and Taqman quantitative RT-PCR was performed with an ABI 7500 Fast System (Applied Biosystems). Comparison of transcript levels was done using ΔCT method of analysis using tufA as the endogenous control gene (Virtaneva et al., 2005). The Taqman primers and probes used are listed in Table S1.
Western immunoblot analysis of SpeB in the culture supernatant
Cells were grown to the indicated A600 and harvested by centrifugation. The culture supernatant was filtered and the filtrate was concentrated two-fold by speed-vac drying. Equal volumes of the samples were resolved on a 15% SDS-PAGE gel, transferred to a nitrocellulose membrane, and probed with polyclonal anti-SpeB rabbit antibodies (Shelburne et al., 2010). SpeB was detected by secondary antibody conjugated with horseradish peroxidase and visualized by chemiluminescence using SuperSignal West Pico Rabbit IgG detection kit (Thermo Scientific).
Casein hydrolysis assay for SpeB protease activity
Cysteine protease activity in the culture supernatant was analyzed by methods described previously (Collin & Olsén, 2000). Briefly, GAS strains were grown to indicated OD600 and harvested by centrifugation. The culture supernatant was filtered with a 0.22 μM membrane filter and the filtrate was used for the assay. Culture supernatant (200 μl) was mixed with an equal volume of activation buffer (0.1 M acetate buffer pH 5.0, 1 mM EDTA, and 20 mM DTT) and incubated at 40° C for 30 min. The activated sample was incubated with 400 μl of 2% azocasein (Sigma) in activation buffer for 60 min at 40° C. The reaction was stopped by adding trichloroacetic acid (TCA) to a final concentration of 10% v/v. After 5 min incubation at room temperature, the samples were vortexed and centrifuged at 15,000 rpm for 5 min. Casein hydrolysis was assessed by measuring the absorbance at 360 nm. A control sample in which TCA was added to the activated reaction before the addition of azocasein was used as blank.
Culture supernatant swapping experiments
Overnight cultures of strains Δvfr, Δvfr:pDC, Δvfr:pDC-vfr, and Δvfr:pDC-pep were inoculated into fresh THY media. The complemented strains were grown to A600 ~0.8, cells were pelleted by centrifugation, and culture supernatants were prepared by filtering through a 0.22 μM membrane filter. The cell pellets of strain Δvfr grown to mid-exponential phase (A600 ~0.6) were suspended in the culture supernatants prepared from one of the three strains tested, Δvfr:pDC or Δvfr:pDC-vfr or Δvfr:pDC-pep and incubated at 37°C for 1 h (A600 ~1.0). RNA analysis and Western immunoblotting were then performed as described above.
To characterize the regulatory factor present in the culture supernatant of Δvfr:pD-Cvfr, the complemented strain was grown at 37°C to late exponential phase (A600 ~0.8) and the culture supernatant was collected by centrifugation and prepared as described below. To obtain the low molecular weight component of the culture supernatant, the samples were filtered using YM-3 (3 k Da cut-off filter) membrane and the flow through was used for the culture supernatant swap experiments. Proteinase K treated culture supernatants were prepared by incubating the filtered growth medium with proteinase K (0.15 mg/ml) for 1 hour at 37°C and the enzyme was subsequently removed by filtering through YM-10 (10 k Da cut-off filter) membrane. The filtered samples were used for the swapping. Swapping experiments were performed as described above.
Protein overexpression and purification
The ropB gene of strain MGAS10870 was cloned into plasmid pET-15b and protein was overexpressed in E. coli strain BL21* (DE3). Cells were grown at 37°C till the A600 reaches 0.5 and induced with 0.5 mM IPTG at 13°C overnight. Cell pellets were suspended in 50 ml of buffer A (20 mM Tris HCl pH 8.5, 100 mM NaCl, 10 % glycerol, and 1 mM Tris 2-carboxyethyl phosphine hydrochloride (TCEP)) supplemented with one protease inhibitor cocktail pellet and DNaseI to a final concentration of 5 μg/ml. Cells were lysed by a microfluidizer M-110L device (Microfluidics) and cell debris was removed by centrifugation at 15,000 rpm for 30 min. RopB was purified by affinity chromatography using a Ni-NTA agarose column followed by size exclusion chromatography with Superdex 200G column. The protein was purified to >95 % homogeneity by Ni-affinity chromatography (Fig. 5B).
SPOT-peptide array
Cellulose-bound peptides were prepared by automated SPOT synthesis (MultiPep RS, Intavis, Bergisch Gladbach, Germany) as described previously (Frank & Overwin, 1996, Kramer et al., 1999). The SPOT membranes were rehydrated in Tris-buffered saline containing 0.05% Tween-20 (TBS-T) and blocked overnight at 4°C with 4% non-fat dry milk in TBS-T. After two washes with TBS-T, the membrane was incubated with purified hexa-histidine-tagged purified RopB at a final concentration of 2 μg/ml for 1 h at room temperature. RopB-peptide binding was assessed by chemiluminescence with anti-hexa-histidine tag antibodies conjugated to horseradish peroxidase (Clontech). Binding of the probing antibodies to the peptides was eliminated by control reactions with anti-hexahistidine tag antibody alone (data not shown).
Mouse infection studies
Virulence of the isogenic mutant GAS strains was assessed using two mouse models. For intraperitoneal infection, fifteen female 3-4 week-old CD1 mice (Harlan Laboratories) were used for each GAS strain. Animals were inoculated intraperitoneally with 1 × 107 CFUs and survival was monitored daily. Data were graphically displayed as a Kaplan-Meier survival curve and analyzed using the log-rank test. For intramuscular infection, 10 female 3-4 week-old CD1 mice (Harlan Laboratories) were inoculated in the right hindlimb with 1 × 107 CFU of each strain and monitored for near mortality. Results were graphically displayed as a Kaplan-Meier survival curve and analyzed using the log-rank test. For histopathology, infected hindlimbs were examined at 24 and 48 h post-inoculation. Tissues from excised lesions were fixed in 10% phosphate-buffered formalin, decalcified, serially sectioned, and embedded in paraffin using automated standard instruments. Hematoxylin and eosin and Gram's stained sections were examined in a blinded fashion with a BX5 microscope and photographed using a DP70 camera (Olympus). Micrographs of tissue taken from the inoculation site that showed pathology characteristic of each strain were selected for publication. All animal protocols were approved by the Institutional Animal Care and Use Committee at The Methodist Hospital Research Institute.
Transcript analysis from infected tissue
To assay in vivo transcript levels, skin lesions from three mice per infecting strain were collected 24 h post-infection and the tissue samples were incubated with RNAlater (Qiagen). Samples were snap frozen with liquid nitrogen and stored at −80°C until use. RNA was isolated and purified using an RNeasy fibrous tissue mini kit (Qiagen). The quality and concentration of RNA were assessed with an Agilent 2100 Bioanalyzer. cDNAs were prepared using Superscript III (Invitrogen) and transcript levels were measured by Taqman qRT-PCR. Data were analyzed using the ΔCT method.
Supplementary Material
Acknowledgements
We thank Gary Dunny for critical reading of the manuscript. This work was supported by an American Heart Association grant 09GRNT2280109 (S.A.S) and a National Institute Allergy and Infectious Diseases K08 Career Development Award AI-064564 (S.A.S.).
References
- Antiporta MH, Dunny GM. ccfA, the Genetic Determinant for the cCF10 Peptide Pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2:582–587. doi: 10.1016/s1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially Speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA. 2010;107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll RK, Musser JM. From transcription to activation: how group A streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol. 2011 Jun; doi: 10.1111/j.1365-2958.2011.07709.x. epub. [DOI] [PubMed] [Google Scholar]
- Carroll RK, Shelburne SA, III, Olsen RJ, Suber B, Sahasrabhojane P, Kumaraswami M, Beres SB, Shea PR, Flores AR, Musser JM. Naturally occurring single amino acid replacements in a regulatory protein alter streptococcal gene expression and virulence in mice. J Clin Invest. 2011;121:1956–1968. doi: 10.1172/JCI45169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin DO, Rubens CE. Blue/white screening of recombinant plasmids in Gram-positive bacteria by interruption of alkaline phosphatase gene (phoZ) expression. Gene. 1998;219:91–99. doi: 10.1016/s0378-1119(98)00396-5. [DOI] [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jiminez JC, Aggarwal C, Federle MJ. Two Group A Streptococcal Peptide Pheromones Act through Opposing Rgg Regulators to Control Biofilm Development. PLoS Pathog. 2011;7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MS, Ajdic D, Ferretti JJ. The rgg Gene of Streptococcus pyogenes NZ131 Positively Influences Extracellular SpeB Production. Infect Immun. 1999;67:1715–1722. doi: 10.1128/iai.67.4.1715-1722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussee MS, Phillips ER, Ferretti JJ. Temporal production of streptococcal erythrogenic toxin B (streptococcal cysteine proteinase) in response to nutrient depletion. Infect Immun. 1997;65:1956–1959. doi: 10.1128/iai.65.5.1956-1959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin M, Olsén A. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol Microbiol. 2000;36:1306–1318. doi: 10.1046/j.1365-2958.2000.01942.x. [DOI] [PubMed] [Google Scholar]
- De Kievit TR, Iglewski BH. Bacterial Quorum Sensing in Pathogenic Relationships. Infect Immun. 2000;68:4839–4849. doi: 10.1128/iai.68.9.4839-4849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci USA. 2007;104:18490–18495. doi: 10.1073/pnas.0704501104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev AV, McDowell EJ, Chaussee MS. Inter-and intraserotypic variation in the Streptococcus pyogenes Rgg regulon. FEMS Microbiol Letters. 2008;284:43–51. doi: 10.1111/j.1574-6968.2008.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleberg NC, Heath A, Vardaman K, DiRita VJ. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect Immun. 2004;72:623–628. doi: 10.1128/IAI.72.2.623-628.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleuchot B, Gitton C, Guillot A, Vidic J, Nicolas P, Besset C, Fontaine L, Hols P, Leblond-Bourget N, Monnet V, Gardan R. Rgg proteins associated with internalized small hydrophobic peptides: a new quorum-sensing mechanism in streptococci. Mol Microbiol. 2011;80:1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x. [DOI] [PubMed] [Google Scholar]
- Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. A Novel Pheromone Quorum-Sensing System Controls the Development of Natural Competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol. 2010;192:1444–1454. doi: 10.1128/JB.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R, Overwin H. SPOT synthesis. Methods Mol Biol. 1996;66:149–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- George EA, Novick RP, Muir TW. Cyclic peptide inhibitors of staphylococcal virulence prepared by Fmoc-based thiolactone peptide synthesis. J Am Chem Soc. 2008;130:4914–4924. doi: 10.1021/ja711126e. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Lory S. Analysis of regulatory networks in Pseudomonas aeruginosa by genomewide transcriptional profiling. Curr Opin Microbiol. 2004;7:39–44. doi: 10.1016/j.mib.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Hung DT, Shakhnovich EA, Pierson E, Mekalanos JJ. Small-Molecule Inhibitor of Vibrio cholerae Virulence and Intestinal Colonization. Science. 2005;310:670–674. doi: 10.1126/science.1116739. [DOI] [PubMed] [Google Scholar]
- Kozlowicz BK, Shi K, Gu Z-Y, Ohlendorf DH, Earhart CA, Dunny GM. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol. Microbiol. 2006;62:958–969. doi: 10.1111/j.1365-2958.2006.05434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Reineke U, Dong L, Hoffmann B, Hoffm,ller U, Winkler D, Volkmer engert R, Schneider mergener J. Spot synthesis: observations and optimizations. J Pept Res. 1999;54:319–326. doi: 10.1034/j.1399-3011.1999.00108.x. [DOI] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 2002;30:e2. doi: 10.1093/nar/30.2.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazazzera BA. The intracellular function of extracellular signaling peptides. Peptides. 2001;22:1519–1527. doi: 10.1016/s0196-9781(01)00488-0. [DOI] [PubMed] [Google Scholar]
- Li J, Kasper DL, Ausubel FM, Rosner B, Michel JL. Inactivation of the alpha C protein antigen gene, bca, by a novel shuttle/suicide vector results in attenuation of virulence and immunity in group B Streptococcus. Proc Natl Acad Sci U S A. 1997;94:13251–13256. doi: 10.1073/pnas.94.24.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang M, Peterfreund GL, Tsou AM, Selamoglu N, Daldal F, Zhong Z, Kan B, Zhu J. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci USA. 2011;108:810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughman JA, Caparon M. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J Bacteriol. 2006;188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Burns EH, Jr., Wyde PR, Podbielski A, Rurangirwa J, Moore-Poveda DK, Musser JM. Genetic Inactivation of an Extracellular Cysteine Protease (SpeB) Expressed by Streptococcus pyogenes Decreases Resistance to Phagocytosis and Dissemination to Organs. Infect Immun. 1998;66:771–776. doi: 10.1128/iai.66.2.771-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Hoe NP, Abdi I, Rurangirwa J, Kordari P, Liu M, Dou SJ, Adams GG, Musser JM. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Montgomery CA, Rurangirwa J, Geske RS, Barrish JP, Adams GJ, Musser JM. Extracellular Cysteine Protease Produced by Streptococcus pyogenes Participates in the Pathogenesis of Invasive Skin Infection and Dissemination in Mice. Infect Immun. 1999;67:1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser JM. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Gibson CM, Caparon MG. A role for Trigger Factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Bryant AE, Salmi DB, McIndoo E, Stevens DL. vfr, a Novel Locus Affecting Cysteine Protease Production in Streptococcus pyogenes. J Bacteriol. 2009;191:3189–3194. doi: 10.1128/JB.01771-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIver KS. Stand-alone response regulators controlling global virulence networks in Streptococcus pyogenes. Contrib Microbiol. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- Neely MN, Lyon WR, Runft DL, Caparon M. Role of RopB in Growth Phase Expression of the SpeB Cysteine Protease of Streptococcus pyogenes. J Bacteriol. 2003;185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RJ, Shelburne SA, Musser JM. Molecular mechanisms underlying group A streptococcal pathogenesis. Cell Microbiol. 2009;11:1–12. doi: 10.1111/j.1462-5822.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. Signal Peptidases. Chem Rev. 2002;102:4549–4580. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- Podbielski A, Leonard BAB. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol Microbiol. 1998;28:1323–1334. doi: 10.1046/j.1365-2958.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- Podbielski A, Pohl B, Woischnik M, Körner C, Schmidt KH, Rozdzinski E, Leonard BAB. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol Microbiol. 1996;21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Moreira CG, Li DR, Reading NC, Ritchie JM, Waldor MK, Williams N, Taussig R, Wei S, Roth M, Hughes DT, Huntley JF, Fina MW, Falck JR, Sperandio V. Targeting QseC Signaling and Virulence for Antibiotic Development. Science. 2008;321:1078–1080. doi: 10.1126/science.1160354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha-Estrada J, Aceves-Diez AE, Guarneros G, de la Torre M. The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Applied Microbiol & Biotech. 2010:1–11. doi: 10.1007/s00253-010-2651-y. [DOI] [PubMed] [Google Scholar]
- Shelburne SA, Olsen RJ, Suber B, Sahasrabhojane P, Sumby P, Brennan RG, Musser JM. A combination of independent transcriptional regulators shapes bacterial virulence gene expression during infection. PLoS Pathog. 2010;6:e1000817. doi: 10.1371/journal.ppat.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Brown CK, Gu ZY, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natil Acad Sci USA. 2005;102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slamti L, Lereclus D. A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 2002;21:4550–4559. doi: 10.1093/emboj/cdf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville GA, Proctor RA. At the Crossroads of Bacterial Metabolism and Virulence Factor Synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson MD, Scaramuzzino DA, Sjöbring U, Olsén A, Frank C, Bessen DE. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol Microbiol. 2000;38:242–253. doi: 10.1046/j.1365-2958.2000.02144.x. [DOI] [PubMed] [Google Scholar]
- Van Delden C, Comte R. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins LD, Romero RA, Corn GJ, Gardner DJ, Bailey JR, Parnell MJ, Musser JM. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Heijne G. The signal peptide. J Mem Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- Withers H, Swift S, Williams P. Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria. Curr Opin Microbiol. 2001;4:186–193. doi: 10.1016/s1369-5274(00)00187-9. [DOI] [PubMed] [Google Scholar]
- Yoon H, McDermott JE, Porwollik S, McClelland M, Heffron F. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 2009;5:e1000306. doi: 10.1371/journal.ppat.1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.