Abstract
We report the molecular characterization of a homeobox-containing gene that maps at 84A in the proximal region of the Antennapedia-complex. The structure and complete sequence are presented. Deletion analysis indicates that the cloned gene, F24, most likely corresponds to the labial (lab) gene. Northern blot experiments show a single approximately 3-kb transcript that is expressed at all embryonic stages from cellular blastoderm onwards and during larval development. The homeobox is split by an intron in the region which encodes the putative DNA-binding helix, a splicing position for homeobox-containing genes which is unique so far. The 5' part of the gene contains four M-repeat sequences (CAA/G repeats) in the protein-coding region. In situ hybridization to the transcripts during embryogenesis reveals two domains of expression. The anterior one is located in parts of the developing head, mainly in the hypopharyngeal organ and in anterior parts of the mandibular lobe, and is restricted to the ectoderm. The posterior domain is part of the posterior midgut primordium (endoderm), that invaginates and later contacts the endoderm cells from the anterior midgut invagination.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. E., Martinez-Arias A. The distribution of Ultrabithorax transcripts in Drosophila embryos. EMBO J. 1985 Jul;4(7):1689–1700. doi: 10.1002/j.1460-2075.1985.tb03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Beachy P. A., Helfand S. L., Hogness D. S. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985 Feb 14;313(6003):545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Bender W., Akam M., Karch F., Beachy P. A., Peifer M., Spierer P., Lewis E. B., Hogness D. S. Molecular Genetics of the Bithorax Complex in Drosophila melanogaster. Science. 1983 Jul 1;221(4605):23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Burke J. F. High-sensitivity S1 mapping with single-stranded [32P]DNA probes synthesized from bacteriophage M13mp templates. Gene. 1984 Oct;30(1-3):63–68. doi: 10.1016/0378-1119(84)90105-7. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Laymon R. A., McCutcheon M. A., Riley P. D., Scott M. P. The localization and regulation of Antennapedia protein expression in Drosophila embryos. Cell. 1986 Oct 10;47(1):113–122. doi: 10.1016/0092-8674(86)90372-7. [DOI] [PubMed] [Google Scholar]
- Carroll S. B., Scott M. P. Localization of the fushi tarazu protein during Drosophila embryogenesis. Cell. 1985 Nov;43(1):47–57. doi: 10.1016/0092-8674(85)90011-x. [DOI] [PubMed] [Google Scholar]
- Chadwick R., McGinnis W. Temporal and spatial distribution of transcripts from the Deformed gene of Drosophila. EMBO J. 1987 Mar;6(3):779–789. doi: 10.1002/j.1460-2075.1987.tb04820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
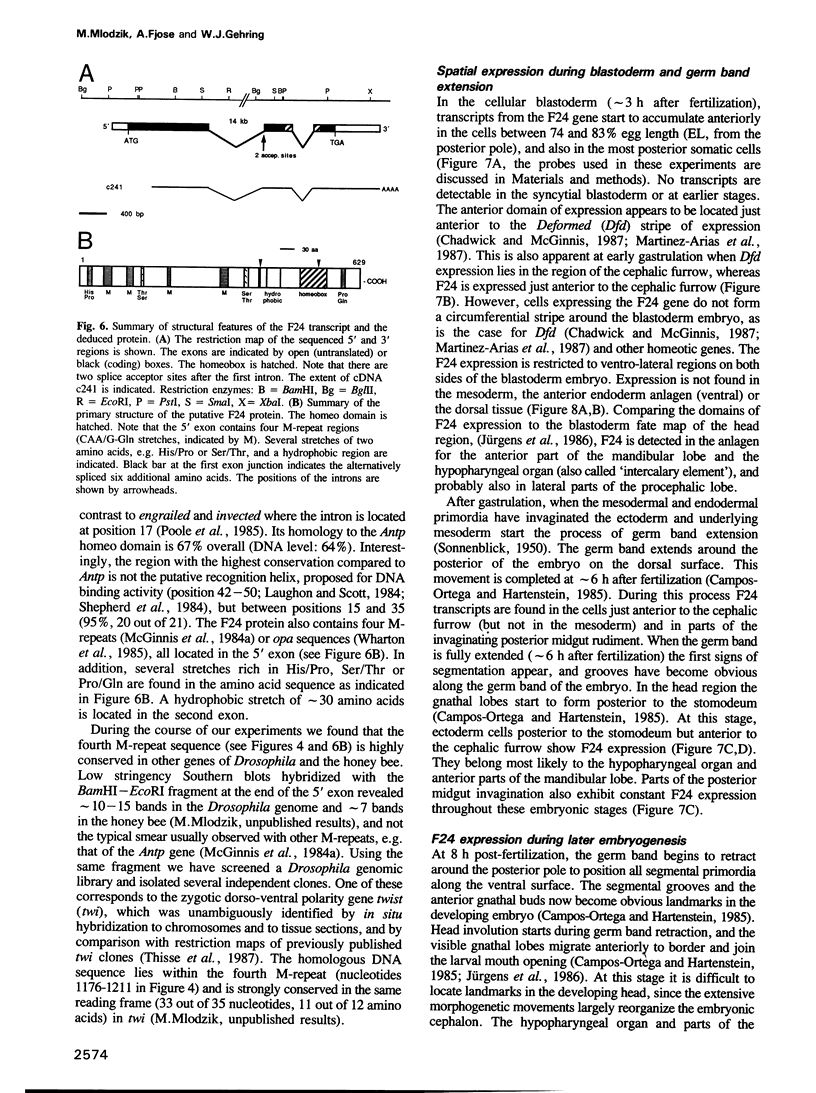
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- DiNardo S., Kuner J. M., Theis J., O'Farrell P. H. Development of embryonic pattern in D. melanogaster as revealed by accumulation of the nuclear engrailed protein. Cell. 1985 Nov;43(1):59–69. doi: 10.1016/0092-8674(85)90012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle H. J., Harding K., Hoey T., Levine M. Transcripts encoded by a homoeo box gene are restricted to dorsal tissues of Drosophila embryos. Nature. 1986 Sep 4;323(6083):76–79. doi: 10.1038/323076a0. [DOI] [PubMed] [Google Scholar]
- Fjose A., McGinnis W. J., Gehring W. J. Isolation of a homoeo box-containing gene from the engrailed region of Drosophila and the spatial distribution of its transcripts. Nature. 1985 Jan 24;313(6000):284–289. doi: 10.1038/313284a0. [DOI] [PubMed] [Google Scholar]
- Frasch M., Hoey T., Rushlow C., Doyle H., Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987 Mar;6(3):749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei E., Baumgartner S., Edström J. E., Noll M. Cloning of the extra sex combs gene of Drosophila and its identification by P-element-mediated gene transfer. EMBO J. 1985 Apr;4(4):979–987. doi: 10.1002/j.1460-2075.1985.tb03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei E., Schuh R., Baumgartner S., Burri M., Noll M., Jürgens G., Seifert E., Nauber U., Jäckle H. Molecular characterization of spalt, a homeotic gene required for head and tail development in the Drosophila embryo. EMBO J. 1988 Jan;7(1):197–204. doi: 10.1002/j.1460-2075.1988.tb02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio G., Burri M., Bopp D., Baumgartner S., Noll M. Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell. 1986 Dec 5;47(5):735–746. doi: 10.1016/0092-8674(86)90516-7. [DOI] [PubMed] [Google Scholar]
- Garber R. L., Kuroiwa A., Gehring W. J. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 1983;2(11):2027–2036. doi: 10.1002/j.1460-2075.1983.tb01696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Bellido A. Genetic control of wing disc development in Drosophila. Ciba Found Symp. 1975;0(29):161–182. doi: 10.1002/9780470720110.ch8. [DOI] [PubMed] [Google Scholar]
- Gehring W. J., Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- Hafen E., Kuroiwa A., Gehring W. J. Spatial distribution of transcripts from the segmentation gene fushi tarazu during Drosophila embryonic development. Cell. 1984 Jul;37(3):833–841. doi: 10.1016/0092-8674(84)90418-5. [DOI] [PubMed] [Google Scholar]
- Hafen E., Levine M., Garber R. L., Gehring W. J. An improved in situ hybridization method for the detection of cellular RNAs in Drosophila tissue sections and its application for localizing transcripts of the homeotic Antennapedia gene complex. EMBO J. 1983;2(4):617–623. doi: 10.1002/j.1460-2075.1983.tb01472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding K., Rushlow C., Doyle H. J., Hoey T., Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science. 1986 Aug 29;233(4767):953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Hoey T., Doyle H. J., Harding K., Wedeen C., Levine M. Homeo box gene expression in anterior and posterior regions of the Drosophila embryo. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4809–4813. doi: 10.1073/pnas.83.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 1988 Jan;7(1):189–196. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa A., Kloter U., Baumgartner P., Gehring W. J. Cloning of the homeotic Sex combs reduced gene in Drosophila and in situ localization of its transcripts. EMBO J. 1985 Dec 30;4(13B):3757–3764. doi: 10.1002/j.1460-2075.1985.tb04145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer P. R., Waldrop A. A., Ward D. C. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Boulet A. M., Bermingham J. R., Jr, Laymon R. A., Scott M. P. Structure of transcripts from the homeotic Antennapedia gene of Drosophila melanogaster: two promoters control the major protein-coding region. Mol Cell Biol. 1986 Dec;6(12):4676–4689. doi: 10.1128/mcb.6.12.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Scott M. P. Sequence of a Drosophila segmentation gene: protein structure homology with DNA-binding proteins. Nature. 1984 Jul 5;310(5972):25–31. doi: 10.1038/310025a0. [DOI] [PubMed] [Google Scholar]
- Levine M., Hafen E., Garber R. L., Gehring W. J. Spatial distribution of Antennapedia transcripts during Drosophila development. EMBO J. 1983;2(11):2037–2046. doi: 10.1002/j.1460-2075.1983.tb01697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Ingham P., Struhl G. Isolation, structure, and expression of even-skipped: a second pair-rule gene of Drosophila containing a homeo box. Cell. 1986 Dec 5;47(5):721–734. doi: 10.1016/0092-8674(86)90515-5. [DOI] [PubMed] [Google Scholar]
- Macdonald P. M., Struhl G. A molecular gradient in early Drosophila embryos and its role in specifying the body pattern. Nature. 1986 Dec 11;324(6097):537–545. doi: 10.1038/324537a0. [DOI] [PubMed] [Google Scholar]
- Mahaffey J. W., Kaufman T. C. Distribution of the Sex combs reduced gene products in Drosophila melanogaster. Genetics. 1987 Sep;117(1):51–60. doi: 10.1093/genetics/117.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Ingham P. W., Scott M. P., Akam M. E. The spatial and temporal deployment of Dfd and Scr transcripts throughout development of Drosophila. Development. 1987 Aug;100(4):673–683. doi: 10.1242/dev.100.4.673. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A., Lawrence P. A. Parasegments and compartments in the Drosophila embryo. Nature. 1985 Feb 21;313(6004):639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Garber R. L., Wirz J., Kuroiwa A., Gehring W. J. A homologous protein-coding sequence in Drosophila homeotic genes and its conservation in other metazoans. Cell. 1984 Jun;37(2):403–408. doi: 10.1016/0092-8674(84)90370-2. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Levine M. S., Hafen E., Kuroiwa A., Gehring W. J. A conserved DNA sequence in homoeotic genes of the Drosophila Antennapedia and bithorax complexes. 1984 Mar 29-Apr 4Nature. 308(5958):428–433. doi: 10.1038/308428a0. [DOI] [PubMed] [Google Scholar]
- Mlodzik M., Fjose A., Gehring W. J. Isolation of caudal, a Drosophila homeo box-containing gene with maternal expression, whose transcripts form a concentration gradient at the pre-blastoderm stage. EMBO J. 1985 Nov;4(11):2961–2969. doi: 10.1002/j.1460-2075.1985.tb04030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlodzik M., Gehring W. J. Expression of the caudal gene in the germ line of Drosophila: formation of an RNA and protein gradient during early embryogenesis. Cell. 1987 Feb 13;48(3):465–478. doi: 10.1016/0092-8674(87)90197-8. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Frohnhöfer H. G., Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987 Dec 18;238(4834):1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980 Oct 30;287(5785):795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Regulski M., McGinnis N., Chadwick R., McGinnis W. Developmental and molecular analysis of Deformed; a homeotic gene controlling Drosophila head development. EMBO J. 1987 Mar;6(3):767–777. doi: 10.1002/j.1460-2075.1987.tb04819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley P. D., Carroll S. B., Scott M. P. The expression and regulation of Sex combs reduced protein in Drosophila embryos. Genes Dev. 1987 Sep;1(7):716–730. doi: 10.1101/gad.1.7.716. [DOI] [PubMed] [Google Scholar]
- Rushlow C., Frasch M., Doyle H., Levine M. Maternal regulation of zerknüllt: a homoeobox gene controlling differentiation of dorsal tissues in Drosophila. Nature. 1987 Dec 10;330(6148):583–586. doi: 10.1038/330583a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneuwly S., Kuroiwa A., Baumgartner P., Gehring W. J. Structural organization and sequence of the homeotic gene Antennapedia of Drosophila melanogaster. EMBO J. 1986 Apr;5(4):733–739. doi: 10.1002/j.1460-2075.1986.tb04275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J., Hazelrigg T. I., Polisky B. A., Pirrotta V., Scalenghe F., Kaufman T. C. The molecular organization of the Antennapedia locus of Drosophila. Cell. 1983 Dec;35(3 Pt 2):763–776. doi: 10.1016/0092-8674(83)90109-5. [DOI] [PubMed] [Google Scholar]
- Scott M. P., Weiner A. J. Structural relationships among genes that control development: sequence homology between the Antennapedia, Ultrabithorax, and fushi tarazu loci of Drosophila. Proc Natl Acad Sci U S A. 1984 Jul;81(13):4115–4119. doi: 10.1073/pnas.81.13.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd J. C., McGinnis W., Carrasco A. E., De Robertis E. M., Gehring W. J. Fly and frog homoeo domains show homologies with yeast mating type regulatory proteins. Nature. 1984 Jul 5;310(5972):70–71. doi: 10.1038/310070a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stroeher V. L., Jorgensen E. M., Garber R. L. Multiple transcripts from the Antennapedia gene of Drosophila melanogaster. Mol Cell Biol. 1986 Dec;6(12):4667–4675. doi: 10.1128/mcb.6.12.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B., el Messal M., Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987 Apr 24;15(8):3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B. T., Turner F. R., Kaufman T. C. Defects in embryogenesis in mutants associated with the antennapedia gene complex of Drosophila melanogaster. Dev Biol. 1984 Mar;102(1):147–172. doi: 10.1016/0012-1606(84)90182-9. [DOI] [PubMed] [Google Scholar]
- Wharton K. A., Yedvobnick B., Finnerty V. G., Artavanis-Tsakonas S. opa: a novel family of transcribed repeats shared by the Notch locus and other developmentally regulated loci in D. melanogaster. Cell. 1985 Jan;40(1):55–62. doi: 10.1016/0092-8674(85)90308-3. [DOI] [PubMed] [Google Scholar]
- White R. A., Wilcox M. Protein products of the bithorax complex in Drosophila. Cell. 1984 Nov;39(1):163–171. doi: 10.1016/0092-8674(84)90202-2. [DOI] [PubMed] [Google Scholar]
- Wirz J., Fessler L. I., Gehring W. J. Localization of the Antennapedia protein in Drosophila embryos and imaginal discs. EMBO J. 1986 Dec 1;5(12):3327–3334. doi: 10.1002/j.1460-2075.1986.tb04647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]