Twin Cx9C proteins constitute a large protein family in the intermembrane space of mitochondria. One of these proteins, Cox19, serves as interaction partner of the copper transfer protein Cox11. The results suggest that a redox-regulated interplay of Cox19 and Cox11 is critical for copper insertion into cytochrome c oxidase.
Abstract
Members of the twin Cx9C protein family constitute the largest group of proteins in the intermembrane space (IMS) of mitochondria. Despite their conserved nature and their essential role in the biogenesis of the respiratory chain, the molecular function of twin Cx9C proteins is largely unknown. We performed a SILAC-based quantitative proteomic analysis to identify interaction partners of the conserved twin Cx9C protein Cox19. We found that Cox19 interacts in a dynamic manner with Cox11, a copper transfer protein that facilitates metalation of the Cu(B) center of subunit 1 of cytochrome c oxidase. The interaction with Cox11 is critical for the stable accumulation of Cox19 in mitochondria. Cox19 consists of a helical hairpin structure that forms a hydrophobic surface characterized by two highly conserved tyrosine-leucine dipeptides. These residues are essential for Cox19 function and its specific binding to a cysteine-containing sequence in Cox11. Our observations suggest that an oxidative modification of this cysteine residue of Cox11 stimulates Cox19 binding, pointing to a redox-regulated interplay of Cox19 and Cox11 that is critical for copper transfer in the IMS and thus for biogenesis of cytochrome c oxidase.
INTRODUCTION
To reach their functional states, newly synthesized polypeptides need to fold into their specific three-dimensional structures. Many proteins use the help of chaperones for surveillance of these folding reactions, for example, by preventing or reversing nonproductive folding intermediates (Bukau et al., 2006; Hartl et al., 2011). Members of the Hsp70 family play a pivotal role in protein folding and are present in most cellular compartments, including the cytosol, the mitochondrial matrix, and the endoplasmic reticulum (Kampinga and Craig, 2010). However, Hsp70 isoforms and other ATP-hydrolyzing chaperones appear to be absent from the intermembrane space (IMS) of mitochondria, and it is not known how IMS proteins are folded (Herrmann and Riemer, 2010, 2014; Vögtle et al., 2012; Hung et al., 2014).
About half of the ∼60 different proteins identified in the IMS of yeast mitochondria (Vögtle et al., 2012) contain structural disulfide bonds that are introduced during their folding process. Oxidative folding of these proteins coincides with their import across the outer membrane and is mediated by the essential oxidoreductase Mia40 (Chacinska et al., 2004; Naoe et al., 2004; Mesecke et al., 2005; Koch and Schmid, 2014). The conserved critical domain of Mia40 forms a helix-loop-helix fold in which the two antiparallel α-helices are connected by two parallel disulfide bonds (Banci et al., 2009; Kawano et al., 2009). These helices form on one side a hydrophobic dish-like cavity that is crucial for the binding and translocation of its substrates across the outer membrane. N-terminally adjacent to this binding region is a flexible arm that contains a cysteine-proline-cysteine (CPC) sequence mediating disulfide bond formation in bound substrates. Reoxidation of Mia40 is catalyzed by the sulfhydryl oxidase Erv1 (Lee et al., 2000; Allen et al., 2005; Mesecke et al., 2005; Tienson et al., 2009). This conserved flavoprotein generates disulfides de novo and transfers electrons via cytochrome c and cytochrome c oxidase to molecular oxygen, giving rise to the production of water (Farrell and Thorpe, 2005; Bihlmaier et al., 2007; Dabir et al., 2007; Fass, 2008).
Most Mia40 substrates belong to two groups of proteins that are referred to as “twin Cx3C” and “twin Cx9C” proteins. The twin Cx3C proteins are also called small Tim proteins owing to their essential role in protein import (Koehler et al., 1998; Sirrenberg et al., 1998; Chacinska et al., 2009). They bind to inner membrane carriers during their import into mitochondria and usher them from the translocase of the outer membrane of mitochondria pore in the outer membrane to their insertion site in the inner membrane. These small Tim proteins form stable hexameric complexes and exhibit chaperone activity in in vitro folding assays (Vial et al., 2002; Webb et al., 2006). However, in mitochondria, they presumably interact with a specific subset of precursors of hydrophobic membrane proteins and maintain them in a translocation-competent state rather than promote their folding.
The twin Cx9C proteins form a relatively large protein family (14 members in yeast, 29 in mammals), but the function of most of these factors is unclear (Gabriel et al., 2007; Longen et al., 2009; Cavallaro, 2010). Except for Mia40, all twin Cx9C proteins are small (7–14 kDa) and consist almost exclusively of the helix-loop-helix region. Mdm35 was found to serve as dynamic binding partner for the lipid transport protein Ups1 (Potting et al., 2010; Tamura et al., 2010). Dynamic interactions of Ups1 and Mdm35 were proposed to drive the regulated transfer of phosphatidic acid from the outer to the inner membrane (Connerth et al., 2012).
Here we screened for interaction partners for Cox19, a conserved twin Cx9C protein that is ubiquitously found in mitochondria of animals, fungi, and plants (Supplemental Figure S1). Cox19 was initially identified as an assembly factor for cytochrome c oxidase, but its specific function in this process remained unclear (Nobrega et al., 2002; Rigby et al., 2007). Owing to the observation that recombinant Cox19 can bind to copper in a 1:1 stoichiometry, it was suggested that Cox19 might serve as copper-binding protein. However, the four cysteine residues of Cox19 that are critical for metal binding in vitro are oxidized in vivo (Rigby et al., 2007; Bien et al., 2010; Fischer et al., 2013). Therefore a direct copper-binding activity of Cox19 in mitochondria is questionable (compare structure in Figure 1A).
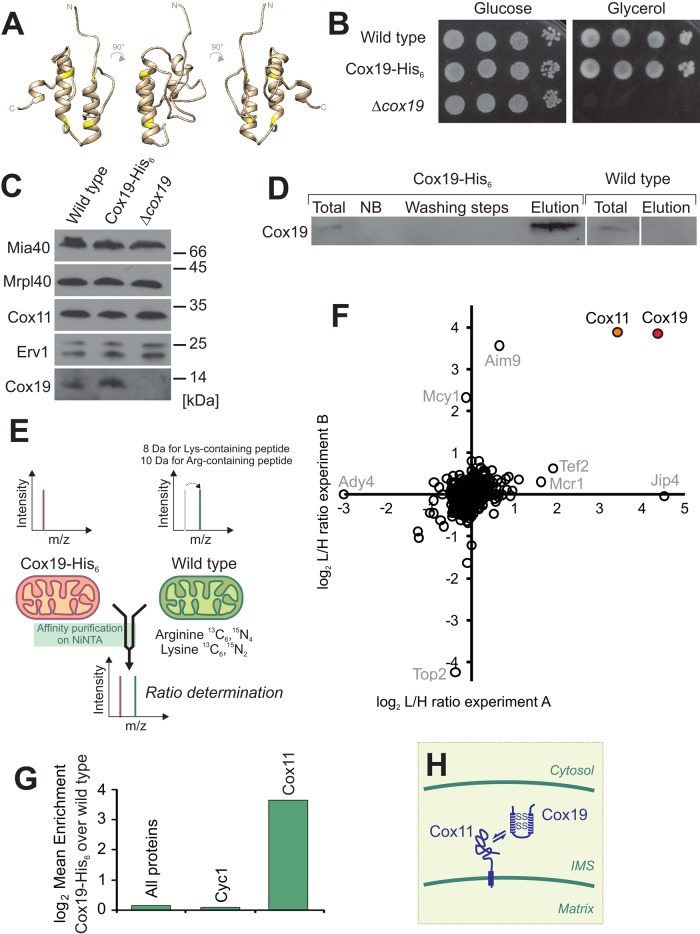
FIGURE 1:
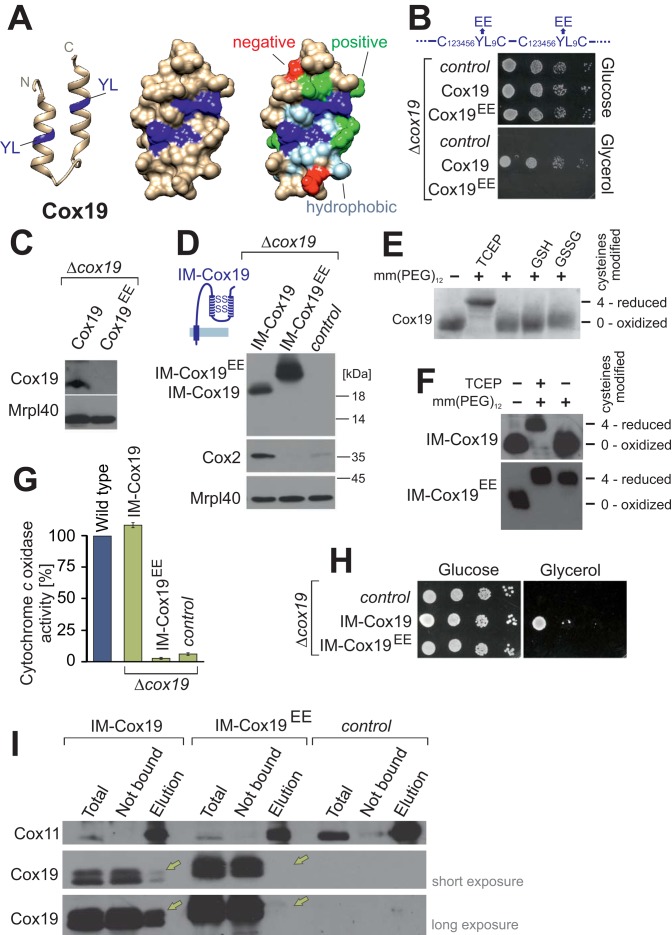
SILAC-based proteomic analysis identifies Cox11 as an interaction partner of Cox19. (A) The Cox19 structure was modeled on the basis of the yeast Mia40 structure (Protein Data Bank ID 2K3J; Banci et al., 2009) using the I-TASSER algorithm (Zhang, 2008) and was visualized with Chimera (Pettersen et al., 2004). Cysteine residues are shown in yellow. (B) Addition of the hexahistidine tag to Cox19 does not compromise its activity. The indicated strains were grown to log phase in galactose medium. Tenfold serial dilutions were dropped onto YPD (glucose) or YPG (glycerol) plates and incubated for 2 or 4 d, respectively. (C) Western blotting of mitochondria (100 μg) isolated from the indicated strains. (D) A 10-mg amount of mitochondria was lysed and either directly applied to SDS–PAGE (total) or used for affinity purification on Ni-NTA Sepharose. The proteins contained in washing steps and the elution after addition of 400 mM imidazole were precipitated by TCA and detected by Western blotting with Cox19-specific antibodies. The total and not-bound (NB) samples correspond to 7.5% of the material used for the other samples. (E) Principle of SILAC labeling to compare protein levels from Ni-NTA eluates of extracts from Cox19-His6 and wild-type mitochondria. (F, G) Enrichment factors from two independent experiments. (H) Model of the Cox19–Cox11 interaction.
Although mutants in many of the twin Cx9C proteins lead to distinct phenotypes such as the absence of respiratory activity, the analysis of their biochemical function turned out to be difficult. In part, this might be due to the fact that most of these proteins are present only at very low amounts, so that they easily escape detection by proteomic analyses (Vögtle et al., 2012; Hung et al., 2014). We therefore combined affinity purification mass spectrometry with the stable isotope labeling by amino acids in cell culture (SILAC) strategy (Ong et al., 2002) to screen for proteins that associate with Cox19 in yeast mitochondria. In this way, Cox11 was identified as a prominent interaction partner of Cox19. Of interest, both proteins form a nonpersistent redox-dependent complex in the IMS. Cox11 binding to Cox19 depends on a hydrophobic region on Cox19 formed by the two disulfide-linked helices. This interaction is reminiscent of the binding of IMS proteins by Mia40, which was shown to serve, in addition to its role as oxidoreductase, as a chaperone for IMS proteins (Weckbecker et al., 2012). On the basis of our observations, we propose that twin Cx9C proteins serve as folding modulators in the IMS, which induce conformational changes of their client proteins. These folding modulators either interact with soluble IMS proteins, as in the case of the Mdm35-Ups1 interaction, or with inner membrane proteins, which expose larger structural domains into the IMS such as in the case of the Cox19–Cox11 interaction shown in this study.
RESULTS
SILAC-based proteomics identifies Cox11 as an interaction partner of Cox19
To identify potential interaction partners of the twin Cx9C protein Cox19 in the IMS, we generated a yeast strain in which Cox19 carries a C-terminal hexahistidine tag (Cox19-His6) by a chromosomal integration strategy to keep its expression under control of its endogenous promoter (Lafontaine and Tollervey, 1996). The resulting Cox19-His6 strain showed robust growth even on nonfermentable carbon sources, whereas a COX19-deletion strain (Δcox19) was unable to respire (Figure 1B). Moreover, the levels of the tagged Cox19 protein were indistinguishable from those of wild-type (wt) Cox19 (Figure 1C). The Cox19-His6 protein could be efficiently purified from isolated mitochondria on a nickel-nitriloacetic acid (Ni-NTA) resin, whereas the nontagged Cox19 protein was not recovered (Figure 1D).
We decided to use a highly sensitive, quantitative proteomics approach for the detection of Cox19 interactors to reduce the risk of false-positive hits. SILAC appeared to be a perfect method in this case (Ong et al., 2002), since protocols were established for yeast cultures that were successfully used to identify interaction partners of mitochondrial proteins (de Godoy et al., 2008; Harner et al., 2011; von der Malsburg et al., 2011; Gebert et al., 2012). We grew yeast cells in minimal media containing either light (Cox19-His6) or heavy (13C6 15N4-L-arginine, 13C6 15N2-L-lysine; wt) amino acids (Figure 1E). In two independent experiments, we isolated mitochondria from these strains, which were lysed and used for affinity chromatography on Ni-NTA resin. The eluates were mixed in a 1:1 ratio, tryptically digested, and analyzed by nano liquid chromatography–electrospray ionization tandem mass spectrometry on an Orbitrap instrument. Peptide identification and quantification was performed with the MaxQuant software package (Cox and Mann, 2008). Among the >300 proteins identified in the eluates (Supplemental Table S1), only two were consistently enriched in both experiments (Figure 1F): our bait protein Cox19, which was enriched by factors of 17.0 and 14.4 over background, and Cox11, which was enriched by factors of 14.7 and 12.4. None of the other proteins identified was enriched with Cox19-His6 in both of the purifications.
Cox11 is a conserved assembly factor for cytochrome c oxidase that is tethered to the inner membrane by an N-terminal membrane anchor (Figure 1H); it exposes a domain of 193 residues into the IMS, which is critical for copper insertion into subunit 1 of cytochrome c oxidase (Cox1; Tzagoloff et al., 1990; Carr et al., 2002; Banci et al., 2004; Horng et al., 2004).
Cox19 and Cox11 can be copurified from mitochondrial extracts
To verify a potential interaction of Cox11 and Cox19, we purified Cox19 from Triton X-100 extracts of Cox19-His6 mitochondria by affinity chromatography (Figure 2A). Cox11 was identified in the eluate fractions together with Cox19-His6, whereas neither Cox11 nor Cox19 was found upon control purification with wild-type mitochondrial extracts. This confirms the results of the SILAC data and shows that Cox11 is associated with Cox19-His6.
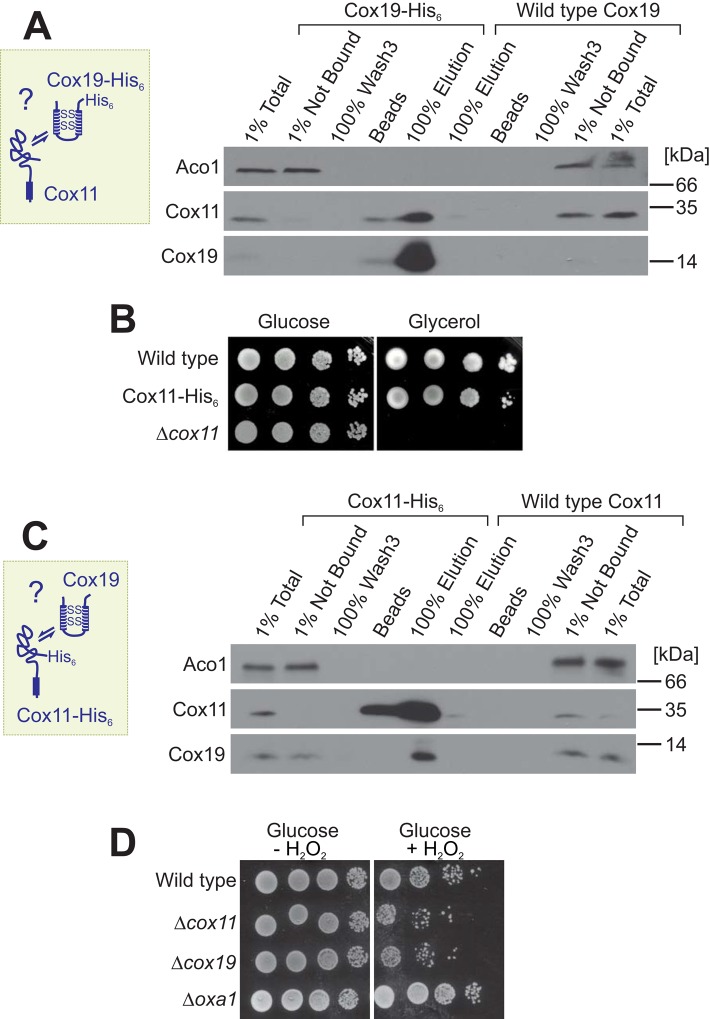
FIGURE 2:
The Cox19–Cox11 interaction is confirmed by reciprocal copurification experiments from mitochondrial extracts. (A) Affinity purification of extracts of mitochondria purified from Cox19-His6 and wild-type cells. Total and NB samples correspond to 1% of the material used for the other samples. (B) Mutants carrying a C-terminal hexahistidine tag on Cox11 are still able to grow on nonfermentable carbon source. (C) Cox19 can be copurified with Cox11-His6 on Ni-NTA material. (D) Mutants lacking Cox19 or Cox11 show increased sensitivity to hydrogen peroxide. Cells were grown to log phase in glucose medium and diluted to an OD of 0.5. After 2 h of incubation at 30°C with or without 6 mM hydrogen peroxide, cells were dropped onto YPD plates.
Next we asked whether the Cox11–Cox19 interaction is also observed upon purification of Cox11. To this end, we generated a Cox11 variant with a C-terminal hexahistidine tag (Cox11-His6). The tagged Cox11 is functional, as indicated by the ability of the Cox11-His6 yeast strain to grow on a nonfermentable carbon sources, whereas a COX11-deletion strain (Δcox11), similar to the COX19-deletion strain, was unable to respire (Figure 2B). Cox11-His6 was efficiently purified on the Ni-NTA resin from Cox11-His6 mitochondria, and a large proportion of Cox19 was copurified (Figure 2C). From this, we conclude that Cox11 physically interacts with Cox19 in the IMS of mitochondria.
The identification of Cox11 as a potential Cox19 interactor was highly interesting. Both proteins are essential for cytochrome c oxidase biogenesis, but an interaction of Cox11 and Cox19 had not previously been observed. We therefore tested how similar the phenotypes are that result from the deletion of Cox11 or Cox19. The absence of Cox11 leads to the accumulation of an assembly intermediate of cytochrome c oxidase that renders yeast cells highly sensitive toward exposure to hydrogen peroxide (Banting and Glerum, 2006; Khalimonchuk et al., 2007, 2010; Veniamin et al., 2011; Bode et al., 2013). We therefore tested whether the Δcox19 mutant shows comparable hydrogen peroxide sensitivity and indeed observed that the absence of Cox19 also renders cells particularly susceptible to oxidative damage, which was not found for Δoxa1 mutants, which also lack cytochrome c oxidase activity (Figure 2D). This comparable phenotype is consistent with a role of Cox11 and Cox19 in the same step of cytochrome c oxidase biogenesis.
Size exclusion chromatography indicates that Cox11 and Cox19 do not form a stable complex in the IMS
To analyze further the association of Cox11 and Cox19, we lysed isolated wild-type mitochondria with Triton X-100 and separated the protein complexes by size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (Figure 3, A and B). Cox11 migrated with an apparent mass of ∼170 kDa, whereas Cox19 eluted very late from the column in fractions with a peak of an apparent mass of ∼20 kDa. We did not observe Cox11 in the Cox19 fraction or Cox19 in the Cox11 fraction, suggesting that the interaction of Cox11 and Cox19 observed by Ni-NTA purification is rather labile and/or transient. In agreement with earlier studies, neither protein migrated with Cox2, nor was their migration altered in the absence of cytochrome c oxidase (Δcox6), indicating that they are not stable constituents of cytochrome c oxidase, as already shown by others (Nobrega et al., 2002; Khalimonchuk et al., 2010). The amounts of assembled cytochrome c oxidase were severely reduced in both Δcox19 and Δcox11 mitochondria, as indicated by low amounts of Cox2 the high–molecular weight elution fractions of the gel filtration compared with wild-type mitochondria (Figure 3A). These data, together with the inability of the deletion mutants to grow on nonfermentable carbon sources, confirm that both Cox11 and Cox19 play crucial roles in the biogenesis of cytochrome c oxidase.
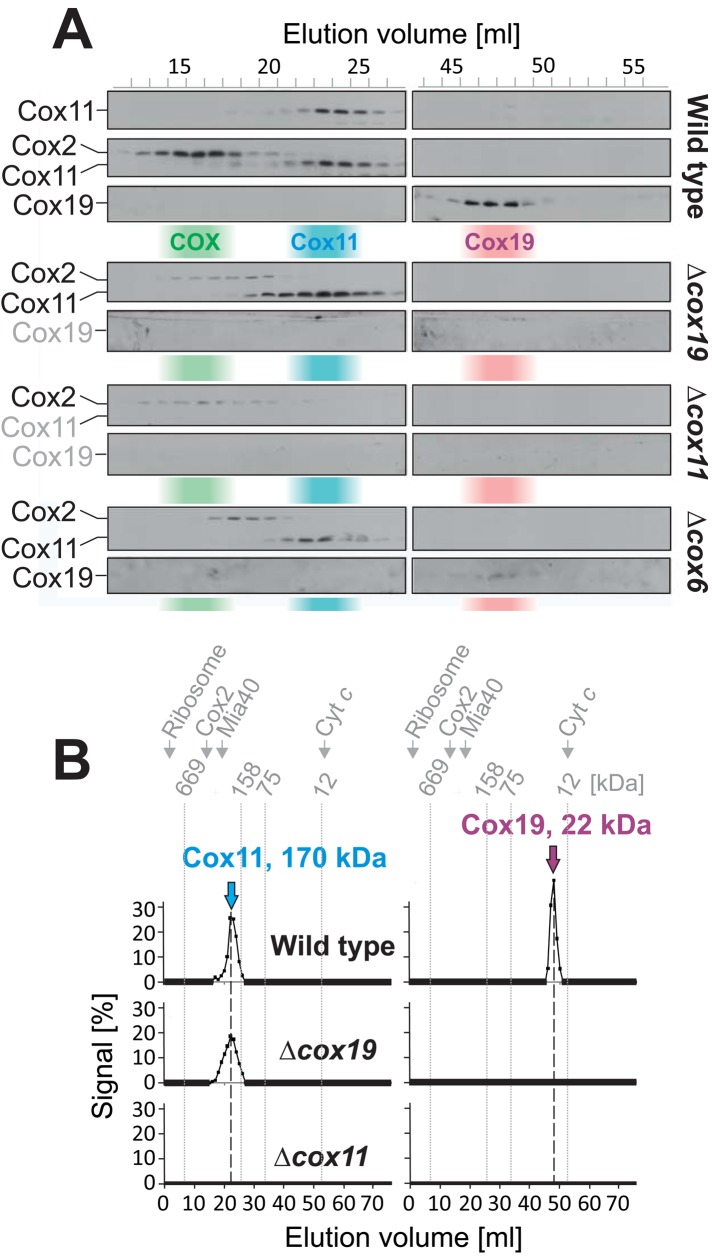
FIGURE 3:
Cox11 and Cox19 are not part of a common stable protein complex. (A) Mitochondria (3 mg) were isolated from the strains indicated and lysed in 500 μl of 25 mM Tris, pH 7.4, 250 mM NaCl, 1% Triton-X 100, and 2 mM PMSF. After a purifying centrifugation at 25,000 × g for 30 min at 4°C, the clarified extract was applied to size exclusion chromatography on a HiLoad 16/60 Superdex 200 column. Proteins were TCA precipitated from the resulting fractions and analyzed by Western blotting. Distribution of cytochrome c oxidase (COX, green), Cox11 (blue), and Cox19 (red) in wild-type extracts. (B) Quantification of the experiment in A.
The apparent molecular mass of the 170-kDa Cox11 complex was not changed in mitochondria from a Δcox19 strain, indicating that Cox19 is not essential for its formation or maintenance (Figure 3, A and B). However, it is of interest that no Cox19 was detectable in Δcox11 mutants, suggesting that Cox11 is required for the stable accumulation of Cox19 in the IMS of mitochondria.
Deletion of Cox11 leads to reduced levels of Cox19 in mitochondria
To better assess the relevance of Cox11 for the accumulation of Cox19 in mitochondria, we compared the steady-state protein levels of Cox11 and Cox19 in Δcox19 and Δcox11 mutant mitochondria (Figure 4A). We found that the levels of Cox11 are not considerably influenced by the deletion of Cox19. Moreover, overexpression of Cox19 also did not affect the amounts of the Cox11 protein in mitochondria. Hence Cox19 appears not to be critical for the biogenesis of Cox11. However, on the contrary, Cox19 was again almost undetectable in Δcox11 mitochondria. A comparable depletion of Cox19 was not observed in other cytochrome c oxidase–deficient mitochondria that we tested (Figure 4B).
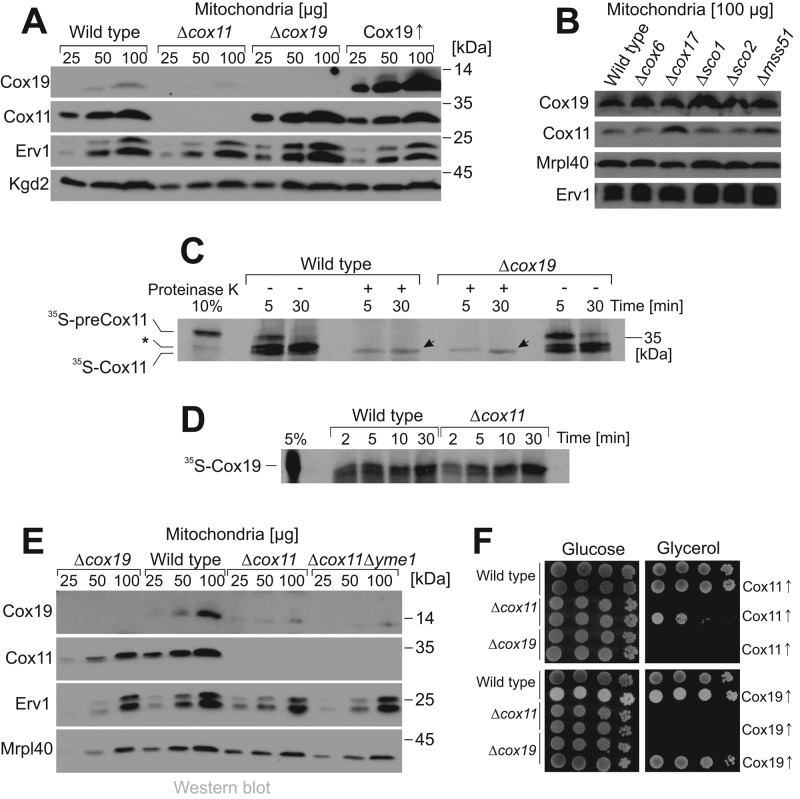
FIGURE 4:
Cox11 import does not depend on Cox19. (A, B) Mitochondrial proteins of the indicated strains were analyzed by Western blotting. (C) Radioactive 35S-preCox11 was synthesized in reticulocyte lysate and incubated with mitochondria purified from wild-type or Δcox19 cells. After 5 or 30 min, aliquots were taken and further incubated on ice in the absence or presence of 100 μg/ml proteinase K. Samples were applied to SDS–PAGE and analyzed by autoradiography. Arrowheads indicate the mature 35S-Cox11 protein that reached a protease-resistant location, indicating its complete import into the mitochondria. Processing of Cox11 presumably occurs in two steps, in which a higher-migrating intermediate is observed that is not fully imported (asterisk) and hence is not protease resistant. This might correspond to the two alternative N-termini of mature Cox11 that were reported on the basis of a proteomic study (Vögtle et al., 2009). (D) 35S-Cox19 was incubated with isolated mitochondria for the times indicated. All samples were treated with proteinase K to remove nonimported material. The samples were analyzed by autoradiography. (E) Mitochondrial proteins of the indicated strains were analyzed by Western blotting. (F) Complementation experiment of the indicated strains. Cells were transformed with plasmids overexpressing Cox11 (top) or Cox19 (bottom), serially diluted, and dropped on glucose and glycerol plates.
We next tested whether the interaction of Cox11 and Cox19 is relevant for the import of both components into mitochondria. To this end, we synthesized the precursor form of Cox11 in reticulocyte lysate in the presence of [35S]methionine and incubated the radioactive protein with isolated yeast mitochondria (35S-preCox11; Figure 4C). The in vitro–synthesized Cox11 precursor has an apparent molecular weight of 35 kDa, corresponding well to the calculated mass of 34.5 kDa. On incubation with mitochondria, 35S-preCox11 was converted to a faster-migrating form (35S-Cox11; Figure 4C), which was partially resistant to externally added proteinase K, indicating its translocation across the outer membrane (Figure 4C, arrowhead). Δcox19 mitochondria imported Cox11 with the same kinetics as wild-type mitochondria, indicating that Cox19 is not crucial for Cox11 import. Similarly, 35S-Cox19 was efficiently imported into Δcox11 mitochondria, suggesting that also Cox11 is not critical for the import of Cox19 (Figure 4D). It therefore appears unlikely that the reduced levels of Cox19 in Δcox11 mitochondria is due to reduced import rates of Cox19, but instead is the result of decreased stability of Cox19 in the IMS of Δcox11 mitochondria. However, Cox19 remained barely detectable even when the iAAA-protease Yme1, which is the major protease of the IMS (Leonhard et al., 1999; Schreiner et al., 2012), was deleted in the Δcox11 background (Figure 4E).
Next we tested whether overexpression of Cox11 or Cox19 suppresses the defects in Δcox19 and Δcox11 cells, respectively (Figure 4F). Overexpression of Cox11 from a multicopy plasmid largely complemented the respiration defect of Δcox11 cells but did not suppress the defects of Δcox19 mutants. Similarly, overexpression of Cox19 complemented a Δcox19 mutant but did not suppress the defects of Δcox11 strains. Hence we conclude that both factors are critical for biogenesis of cytochrome c oxidase, and their relevance is not simply explained by a potential stabilizing effect of one factor for the other.
Cox19 forms a hydrophobic cavity that is essential for its function
Cox19 is a conserved protein found in mitochondria of fungi, plants, and animals, including humans (Supplemental Figure S1). It is characterized by twin Cx9C signatures with conserved tyrosine-leucine (YL) residues at positions 7 and 8 between the cysteine residues (twin Cx6YLxC). Modeling of the Cox19 sequence onto the Mia40 structure suggests that these residues are part of a hydrophobic dish-like structure (Figure 5A). This structure is reminiscent of the hydrophobic substrate-binding pocket of Mia40 (Banci et al., 2009; Kawano et al., 2009; Sideris et al., 2009), but in Cox19, the hydrophobic cavity is surrounded by conserved charged residues (Figure 5A, green and red). In Mia40, mutation of conserved phenylalanine residues in this pocket to glutamate residues had been shown to compromise severely the Mia40–substrate interaction (Kawano et al., 2009). Following the same strategy, we generated a Cox19EE mutant in which the tyrosine-leucine residues were replaced by glutamate residues. Expression of this variant even from a multicopy plasmid did not rescue the respiration defect of a Δcox19 mutant, indicating that the Cox19EE mutant is not functional (Figure 5B). Because the conserved tyrosine-leucine residues are part of the two mitochondrial IMS-sorting signal (MISS)/IMS-targeting signal (ITS) signals of Cox19 (Sideris et al., 2009), we considered it as possible that the Cox19EE mutant might not be properly imported into the IMS. Indeed, the Cox19EE protein was not detectable in isolated mitochondria (Figure 5C).
FIGURE 5:
Conserved tyrosine-leucine residues are essential for Cox19 activity. (A) Structure of the hydrophobic cavity formed by the twin Cx6YLxC motif of Cox19. The structure was modeled as in Figure 1A, but only the helix-loop-helix region is shown. YL dipeptides, dark blue; on rightmost structure, other hydrophobic residues, light blue; negative residues, red; and positive residues, green. (B) Cells from a Δcox19 strain were transformed with an empty plasmid (control) or plasmids overexpressing Cox19 or Cox19EE. Growth was analyzed on SD (glucose) or SG (glycerol) plates. (C) Western blot analysis of mitochondria (100 μg) isolated from Cox19- and Cox19EE-expressing cells. (D) Fusion proteins of an IMS-targeting sequence with Cox19 and Cox19EE were overexpressed in Δcox19 cells. Mitochondria were isolated and analyzed by Western blotting. (E) Cox19 was incubated with 5 mM TCEP, GSH, or GSSG, reisolated by TCA precipitation, and treated with the alkylating agent mm(PEG)12. Modification with mm(PEG)12 leads to a slower electrophoretic mobility, which is indicative of the presence of reduced thiols. Cox19 was fully oxidized unless treated with the strong reductant TCEP. (F) Mitochondrial proteins of the indicated strains were TCA precipitated and treated with TCEP and/or mm(PEG)12. (G) Activities of cytochrome c oxidase were analyzed in mitochondria isolated from the indicated strains. (H) Drop dilution experiment of the strains analyzed in D. (I) Cox11 was purified by immunoprecipitation with Cox11-specific antibodies from extracts of mitochondria isolated from the indicated strains. Copurified Cox19 was detected by Western blotting and is indicated by yellow arrows. Note that no Cox11 can be detected in the pull downs with IM-Cox19EE.
We therefore constructed a variant in which the import of the Cox19EE protein is ensured by fusion to the N-terminal IMS targeting sequence of Mia40, which stably tethers the imported protein to the inner membrane (Figure 5D, inset). A similar strategy relying on a fusion to an inner membrane–spanning domain was successfully used previously to import Cox19 and other twin Cx9C proteins into the IMS (Maxfield et al., 2004; Rigby et al., 2007). The corresponding inner membrane (IM)-Cox19EE fusion protein was well detectable in isolated mitochondria. The slower migration of the IM-Cox19EE fusion compared with that of IM-Cox19 is presumably due to its more acidic nature. The abundance of the IM-Cox19EE protein was comparable to that of the IM-Cox19 fusion (Figure 5D). Cox19 contains two disulfide bonds, which are stable even in the presence of physiological concentrations of glutathione (Bien et al., 2010). This can be demonstrated by incubation of Cox19 with the alkylating agent methyl–polyethylene glycol maleimide 12 (mm(PEG)12), which adds ∼1.2 kDa to each reduced cysteine residue. As shown in Figure 5E, the four cysteine residues are inaccessible to mm(PEG)12 unless Cox19 was pretreated with the thiol-free reducing agent Tris-carboxyethyl phosphine (TCEP). The cysteine residues are also oxidized in IM-Cox19 but reduced in the IM-Cox19EE mutant (Figure 5F). This nicely illustrates that protein oxidation in the IMS depends on the presence of the MISS/ITS signal (Milenkovic et al., 2009; Sideris et al., 2009) and is not just the consequence of “oxidizing conditions” in this compartment.
Measurements of the cytochrome c oxidase activity with isolated mitochondria revealed that the expression of the IM-Cox19EE fusion protein was not able to suppress the cytochrome c oxidase deficiency of a Δcox19 mutant to any detectable extent (Figure 5G). Consistent with this, IM-Cox19EE was not able to complement the growth defect of a Δcox19 mutant (Figure 5H).
Immunoprecipitation from mitochondrial extracts with Cox11-specific antibodies proved that IM-Cox19 still binds to Cox11 (Figure 5I). In contrast, IM-Cox19EE was not recovered with Cox11, indicating that this mutant lost its ability to interact with Cox11. Thus the hydrophobic cavity of Cox19 presumably serves as a binding site for its partner protein Cox11. Comparable Cx6YLxC signatures are not present in other members of the twin Cx9C family (Longen et al., 2009) but are characteristic of Cox19 homologues, for which the tyrosine-leucine dipeptide is invariantly found in the second Cx9C helix (Supplemental Figure S1, red).
Cox19 binds to selective regions of the IMS domain of Cox11
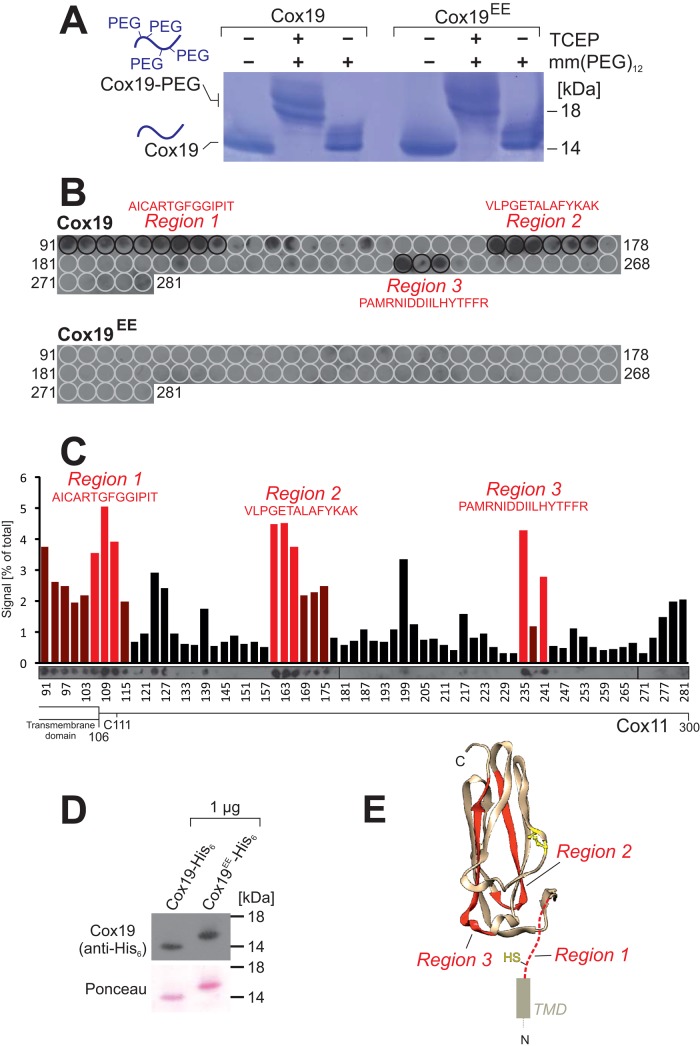
After identification of the nonfunctional Cox19EE mutant, we used this variant as a proper control to identify potential binding regions in Cox11 for Cox19. For Mia40, it was shown that the hydrophobic binding cleft interacts with relatively short helical stretches of 9–11 amino acid residues (Kawano et al., 2009; Milenkovic et al., 2009; Sideris et al., 2009), allowing for the identification of binding motifs by the use of peptide display arrays (Weckbecker et al., 2012; Longen et al., 2014). We therefore cloned the sequences of Cox19 and the Cox19EE mutant into an Escherichia coli expression plasmid and purified recombinant forms of both variants as fusion proteins from bacterial extracts. Because the recombinant proteins were synthesized in the bacterial cytosol, we first tested the redox state of their cysteine residues. To this end, purified Cox19 and Cox19EE were incubated with mm(PEG)12 (Figure 6A). Both proteins were not modified by mm(PEG)12 unless pretreated with TCEP, indicating that in Cox19 and Cox19EE, both disulfide bonds are formed upon expression in bacteria. Thus the introduced negative charges do not prevent cysteine oxidation per se but rather the recognition by Mia40 in the IMS.
FIGURE 6:
Cox19, but not Cox19EE, has potential binding affinity to three distinct regions in the IMS domain of Cox11. (A) Cox19-His6 and Cox19EE-His6 were recombinantly expressed in E. coli and purified. The protein was denatured and treated with TCEP and/or mm(PEG)12 and analyzed by nonreducing SDS–PAGE. Only after TCEP treatment are the cysteine residues in Cox19-His6 and Cox19EE-His6 accessible to alkylation, indicating that the recombinant protein is fully oxidized. (B, C) Peptides 20 residues in length corresponding to the IMS domain of Cox11 were spotted onto a nylon membrane and incubated with Cox19-His6 or Cox19EE-His6, respectively, as indicated. After extensive washing, bound protein was transferred to nitrocellulose and analyzed by immunoblotting with monoclonal antihexahistidine antibodies. The signals were quantified. The regions specifically bound to Cox19-His6, but none of the peptides was bound by Cox19EE-His6, although the protein was efficiently recognized by the antihexahistidine antibody (D). (E) The structure of yeast Cox11 was modeled on the basis of the Cox11 structure (PDB 1SP0) of S. meliloti (Banci et al., 2004). The three binding regions are shown in red.
Next we spotted two identical nylon membranes with 65 peptides of 20 amino acid residues in length, each representing the entire sequence of the IMS domain of Cox11 (residues 108–300). The peptides represented residues 91–110, 94–113, 97–116,…, 280–299, and 281–300. We incubated these membranes with recombinantly expressed purified Cox19-His6 and Cox19EE-His6, respectively (Figure 6, B and C). The membranes were washed, and Cox19/Cox19EE bound to the peptides was transferred to nitrocellulose and detected by immunoblotting with a monoclonal hexahistidine antibody that recognized the Cox19-His6 and the Cox19EE-His6 protein equally well (Figure 6D). Strong Cox19 signals were detected at three regions of Cox11, whereas Cox19EE did not bind to any of the peptides. This result again confirmed that Cox11 binding by Cox19 depends on the conserved YL residues of the Cx9C region. Chou–Fasman algorithms predict helical structures for all Cox11 peptides bound by Cox19 (Chou and Fasman, 1978). However, when we modeled the yeast Cox11 structure onto the published Cox11 structure of Sinorhizobium meliloti (Banci et al., 2004; Figure 6E), region 1 was directly N-terminal to the crystallized Cox11 fragment and contained a strictly conserved cysteine residue of Cox11. This cysteine is not part of the copper-binding site but is essential for cytochrome c oxidase biogenesis, although its specific function is unclear (Banci et al., 2004; Carr et al., 2005; Thompson et al., 2010).
Cox19 binds to Cox11 in a redox-sensitive manner
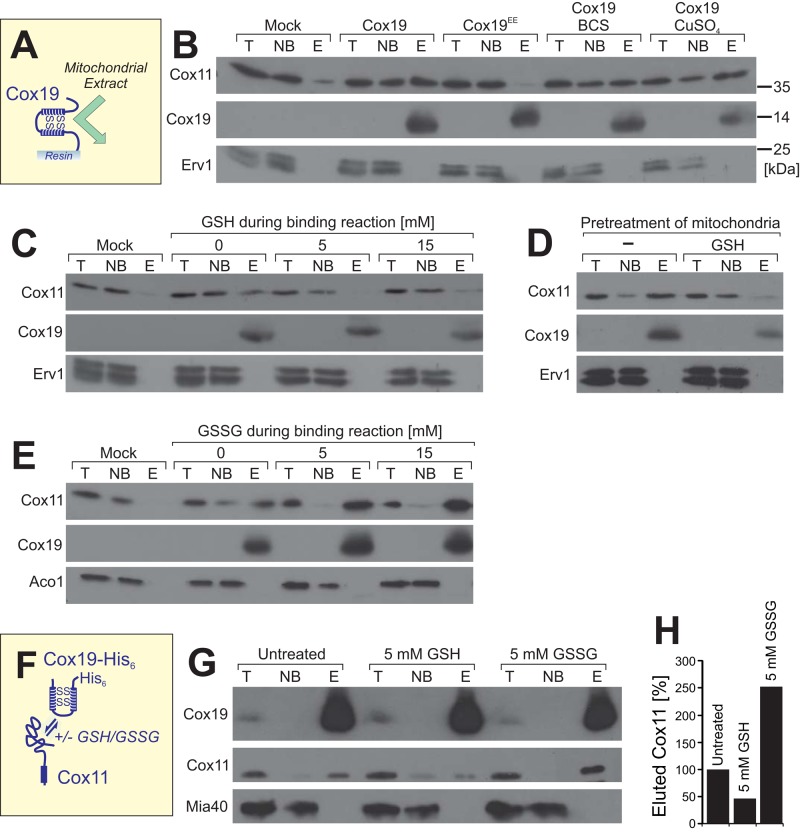
To better characterize the conditions under which Cox19 binds to Cox11, we developed an in vitro binding assay using immobilized recombinant Cox19-His6 (or Cox19EE-His6 for control) and mitochondrial extracts (Figure 7A). To this end, Cox19-His6 or Cox19EE-His6 was immobilized on Ni-NTA Sepharose beads and incubated with Triton X-100–lysed mitochondria. Subsequently, proteins in the nonbound (NB) and bound (E, eluate) fractions were analyzed by Western blotting (Figure 7B). We found that Cox11 was efficiently bound by Cox19-His6 on the resin, whereas Cox11 was not recovered with Cox19EE-His6 or with noncoated Sepharose beads. The interaction between Cox11 and Cox19 was not significantly influenced by the addition of copper sulfate or the copper chelator bathocuproine disulfonate (BCS), indicating that copper is not critical for the Cox11–Cox19 interaction. In contrast, the binding of Cox11 to Cox19-His6 was fully prevented in the presence of 5 mM reduced glutathione (GSH; Figure 7C). To test whether this GSH sensitivity is an effect of the reductant on Cox11 or on Cox19, we exclusively preincubated the mitochondria with GSH. Then we removed the reductant by extensive washing before the mitochondria were lysed. This pretreatment compromised the binding of Cox11 to the immobilized Cox19-His6, indicating that GSH treatment converts Cox11 into a species that does not efficiently interact with Cox19 (Figure 7D).
FIGURE 7:
Cox11 binds to Cox19 in a glutathione-dependent manner. (A, B) Purified Cox19-His6 or Cox19EE-His6 was immobilized on Sepharose beads and incubated with extracts of mitochondria expressing Cox11-HA. After washing, proteins of the total (T), not-bound (NB), and bound (E, eluate) fractions were analyzed by Western blotting. Cox11 was detected by use of an HA antibody. For the mock sample, naive Sepharose beads were used that carried neither Cox19-His6 nor Cox19EE-His6. In the samples labeled with BCS and CuSO4, 4 mM bathocuproine disulfonate or 100 μM copper sulfate, respectively, was added to 450 μg of wild-type mitochondria. After 15 min of incubation, the mitochondria were recovered by centrifugation and lysed. The clarified lysate was incubated with Cox19-coated Sepharose beads. (C,E) Pull-down experiments with naive beads (mock) or immobilized Cox19-His6 (other samples) as described in B, with the exception that different amounts of reduced glutathione (GSH) or oxidized glutathione disulfide (GSSG) were present during the binding reaction. (D) Mitochondria were preincubated with 5 mM reduced glutathione. The glutathione was removed by washing. Mitochondria were lysed, and the resulting extract was used for binding experiments as described in B. (F–H) Mitochondria expressing Cox19 with a hexahistidine tag were pretreated in the absence or presence of 5 mM GSH or GSSG for 15 min at 30°C. The mitochondria were washed and lysed in a buffer containing 1% Triton X-100, and the extract was clarified by centrifugation. Cox19-His6 was precipitated by 1 h incubation with Ni-NTA Sepharose and, after several washing steps, eluted with 250 mM imidazole. The T and NB samples correspond to 1% of the material used for the E sample. The samples were analyzed by Western blotting, and the signal intensities were quantified.
On the contrary, in the presence of oxidized glutathione disulfide (GSSG), the association of Cox11 and Cox19 was strongly increased, suggesting that oxidized glutathione converts Cox11 into a species that is competent for Cox19 binding (Figure 7E). This suggests that the redox states of the cysteines in Cox11 determine its affinity for Cox19.
Next we tested whether GSH and GSSG also influence the Cox11–Cox19 association in mitochondria (Figure 7F). To this end, we preincubated mitochondria in the absence or presence of 5 mM GSH or GSSG for 15 min at 30°C. After extensive washing to remove glutathione, we lysed the mitochondria and used the extracts for coimmunoprecipitation with Cox19 antibodies (Figure 7G). The amount of Cox19, as well as of coisolated Cox11 and Mia40 for control, was detected by Western blotting and quantified (Figure 7H). Consistent with the in vitro experiments, reduced GSH impaired the Cox19–Cox11 interaction, whereas preincubation with GSSG strongly increased the levels of Cox11 that were recovered with Cox19. This suggests that an oxidative modification of Cox11 increases its affinity for Cox19.
Cox11 contains a disulfide between C208 and C210 that depends on C111
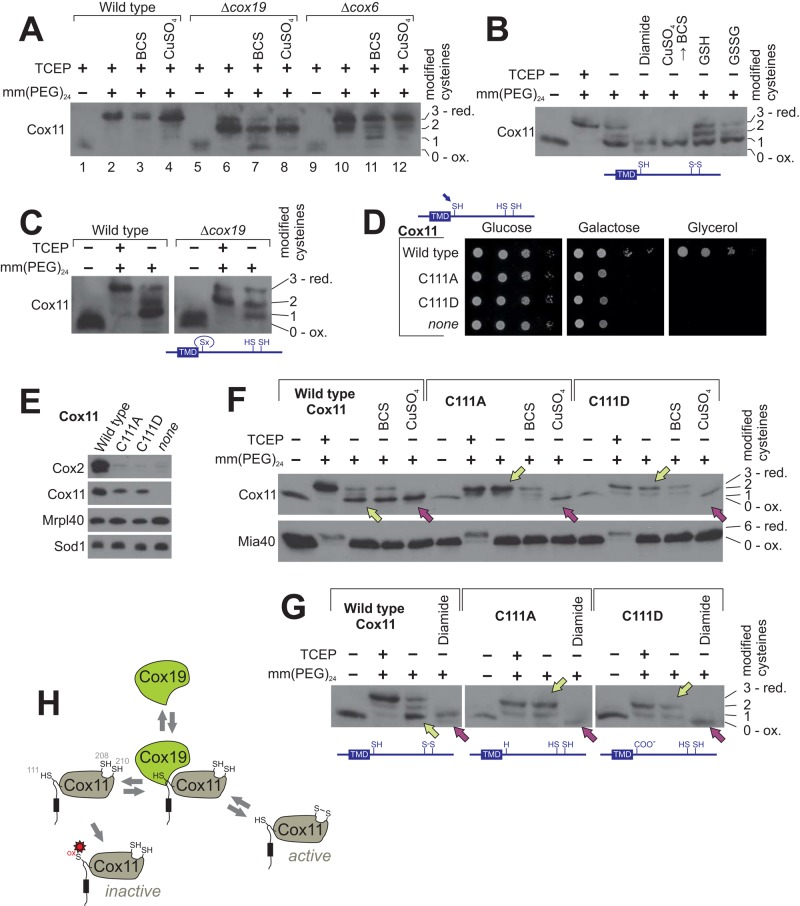
The strong influence of glutathione on the Cox11–Cox19 interaction inspired us to test whether the redox state of the three cysteine residues in Cox11 is altered in Δcox19 mutants. To this end, we precipitated mitochondrial proteins with trichloroacetic acid (TCA), since low pH prevents cysteine oxidation. Then we denatured the proteins and incubated them with mm(PEG)24 to assess the accessibility of cysteine residues (Figure 8A). In wild-type mitochondria, Cox11 was efficiently shifted up after pretreatment with the reductant TCEP due to the addition of presumably three mm(PEG)24 groups to the three cysteine residues of Cox11 (Figure 8A, lane 2). Treatment with BCS or copper sulfate did not considerably influence the accessibility of the cysteine residues to mm(PEG)24 (Figure 8A, lanes 3 and 4). In contrast, when extracts of Δcox19 mitochondria were analyzed, two different species were observed after treatment with mm(PEG)24, one that resembled the fully reduced species, which was also observed in wild-type mitochondria, and a second form in which only two mm(PEG)24 moieties were bound to Cox11 (Figure 8A, lanes 6–8). This indicates that a Cox11 species accumulates in Δcox19 mitochondria in which one cysteine residue does not react with the maleimide group of the mm(PEG)24, even after treatment with the strong reductant TCEP. This suggests that here one cysteine is overoxidized to the sulfinic or sulfonic acid state. The incomplete alkylation observed in Δcox19 mitochondria is not the consequence of the absence of cytochrome c oxidase in this mutant, as the variant in which three mm(PEG)24 moieties are bound to Cox11 was the predominant form in Δcox6 mitochondria (Figure 8A, lanes 10–12). Hence we conclude that Cox19 is critical to maintain the cysteine residues in Cox11 in a functional state.
FIGURE 8:
In Cox11, C208–C210 are partially oxidized in a C111-dependent manner. (A) Proteins of wild-type or Δcox19 mitochondria were TCA precipitated to prevent cysteine oxidation, denatured in SDS, and incubated with TCEP and mm(PEG)24 as indicated. Mitochondria were pretreated with or without 2 mM BCS or 100 μM copper sulfate for 10 min at 30°C before TCA precipitation. Samples were analyzed by Western blotting. (B) Proteins of wild-type mitochondria were TCA precipitated and incubated as indicated with or without TCEP, 5 mM diamide, 5 mM reduced (GSH) or oxidized (GSSG) glutathione, or first with 100 μM copper sulfate for 15 min before copper was again removed by addition of 2 mM BCS. Then free thiols were alkylated with mm(PEG)24. (C) The redox state of Cox11 in wild-type and Δcox19 mitochondria was determined as described in B. (D) Growth test for the indicated strains. (E) Protein levels in isolated mitochondria of the indicated strains as determined by Western blotting. (F, G) The redox state of Cox11 in mitochondria of the indicated mutants was determined as described in B. When indicated, mitochondria were pretreated with BCS to remove copper ions. Yellow arrowheads indicate steady-state redox conditions, and purple arrows point at samples in which disulfide bond formation was induced by oxidants. (H) Model for the Cox19–Cox11 interaction. Cox11 contains three cysteine residues. The regions in Cox11 that show affinity to Cox19 include the conserved cysteine C111 right after the transmembrane domain (shown here as a black box). Cox19 binding promotes disulfide bond formation in the copper-binding site of Cox11, potentially by preventing overoxidation of C111 (red asterisk).
Next we assessed the redox state of Cox11 in wild-type mitochondria. We again tested the accessibility to mm(PEG)24 but omitted the pretreatment with TCEP to monitor the endogenous redox state (Figure 8B). Surprisingly, we observed that in the prevalent fraction of Cox11, only one cysteine was accessible and two were blocked, suggesting that Cox11 contains a disulfide bond. Preincubation with oxidants such as diamide or copper sulfate shifted the entire Cox11 fraction into that form. Oxidized Cox11, albeit in lower amounts, were still found after pretreatment of mitochondria with physiological concentrations of glutathione (Figure 8B, GSH). The disulfide bond is presumably formed in the surface-exposed C208xC210 motif, which is part of the copper-binding site (Banci et al., 2004; Thompson et al., 2010). Of interest, in mitochondria isolated from Δcox19 cells, a considerably smaller fraction of Cox11 contained the disulfide bond, suggesting that Cox19 maintains Cox11 in an oxidation-competent form (Figure 8C).
Next we tested whether the cysteine in position 111 affects the redox state of C208 and C210 in Cox11. First we replaced the C111 residue by alanine or aspartate, the latter mimicking the negative charge of sulfenylated or glutathionylated cysteines. Both mutations completely compromised Cox11 activity, resulting in respiration-deficient mutants (Figure 8D) that do not contain detectable levels of cytochrome c oxidase (Figure 8E). When we assessed the thiol redox state in these Cox11 mutants, we found that C208 and C210 were accessible to mm(PEG)24 unless chemical oxidants such as copper sulfate (Figure 8F) or diamide (Figure 8G) were added. From this, we conclude that disulfide bond formation between C208 and C210 depends on the presence of the conserved cysteine residue at position 111, which is part of the presumed Cox19 binding site (Figure 8H).
DISCUSSION
Proteins of the mitochondrial IMS play important roles in transport of molecules between mitochondria and the cytosol, biogenesis of the respiratory chain, and redox homeostasis (Herrmann and Riemer, 2010). Despite their relevance, surprisingly little is known about the biogenesis and function of most of these proteins. The iAAA protease Yme1 is the only characterized ATP-hydrolyzing enzyme of the IMS (Leonhard et al., 1999; Schreiner et al., 2012), and it is largely unclear how protein functions in this compartment are controlled. During evolution, the IMS was derived from the bacterial periplasm, which lacks ATP and, hence, ATP-hydrolyzing enzymes (Merdanovic et al., 2011; Goemans et al., 2014). The ATP-independent strategies to regulate protein activities in the periplasm might therefore also be used by eukaryotic cells to control protein functions in the IMS of mitochondria.
Twin Cx9C proteins constitute the largest group of IMS proteins: in yeast, 14 of the 31 soluble IMS proteins, and in humans, 29 of the 127 IMS proteins, belong to this family (Gabriel et al., 2007; Longen et al., 2009; Cavallaro, 2010; Vögtle et al., 2012; Hung et al., 2014). Although they are numerous, evolutionarily conserved, and of similar overall structure, we know almost nothing about their molecular functions. We therefore used an unbiased quantitative proteomic approach to identify interactors of the conserved twin Cx9C protein Cox19 that may shed light on the molecular function of this conserved IMS protein.
Cox19 has a helix-turn-helix structure with a hydrophobic surface on one side that is characterized by two adjacent leucine-tyrosine residues. As reported here, these residues are essential for Cox19 function and critical for its ability to bind to the IMS domain of Cox11. We identified three regions in Cox11 that showed affinity for Cox19. All Cox19-binding peptides are predicted to form α-helical structures with one hydrophobic surface and contain the aromatic residues phenylalanine or tyrosine. However, the published bacterial Cox11 structure suggests that in the context of the folded protein, region 2 is part of a β-sheet. Peptides of regions 2 and 3 lack cysteine residues, but the helical nature and the hydrophobic residues are similar to, although still distinct from, the MISS/ITS signals that are recognized by the hydrophobic substrate-binding region of Mia40 (Milenkovic et al., 2009; Sideris et al., 2009). Cox19 apparently has a similar protein-binding activity as Mia40; however, in the case of Cox19, this activity is not required for the import of its binding partner Cox11 but rather for Cox11 function.
The precise role of Cox11 during biogenesis of cytochrome c oxidase is not clear, despite some considerable interest in the protein since several genome-wide association studies recently reported that a single-nucleotide polymorphism in the human Cox11 gene is an important risk factor for breast cancer (Ahmed et al., 2009; Fasching et al., 2012). Cox11 was shown to be required for the incorporation of the Cu(B) center into Cox1. Cox11 can accept copper from the copper-binding protein Cox17 (Horng et al., 2004), suggesting that it serves as a shuttle mediating copper transfer from Cox17 to Cox1. Similar copper transfer factors are known for the Cu(A) site of cytochrome c oxidase and the copper/zinc superoxide dismutase (Sod1), for which metalation is mediated by Sco1 and Ccs1, respectively (Rae et al., 1999; Leary et al., 2007; Banci et al., 2010; Gross et al., 2011).
The IMS domain of Cox11, which exhibits the main activity of the protein, contains three conserved cysteine residues, all of which are essential for its function (Banting and Glerum, 2006). The surface-exposed C208xC210 motif in Cox11 (compare Figure 6E) presumably constitutes the copper-binding site. The function of cysteine C111, which is located directly C-terminal of the transmembrane region, is not clear, but it was suggested that C111 is involved in the metalation of Cox1 (Thompson et al., 2010). It was also speculated that C111 mediates homodimerization of Cox11 by the formation of an intermolecular disulfide (Banci et al., 2004), but at least in the bacterium Rhodobacter sphaeroides, no evidence for homodimers was found (Thompson et al., 2010). Similarly, we did not observe Cox11 dimers under different conditions tested (unpublished results). The alkylation experiments in this study suggest that Cox11 can form an intramolecular disulfide between C208 and C210 that is less efficiently formed in Δcox19 mitochondria and absent if C111 is mutated. In addition, we observe a TCEP-resistant modification of presumably C111 in Δcox19 mitochondria, suggesting that Cox19 protects C111 of Cox11 against overoxidation to a sulfenic, sulfinic, or sulfonic acid form (Figure 8H).
Hence the absence of Cox19 might block the reaction cycle of Cox11 at a stage that makes the protein particularly redox sensitive and therefore leads to overoxidation and inactivation of Cox11.
Our Cox11–Cox19 in vitro binding assay showed that pretreatment of Cox11 with glutathione disulfide strongly stimulated its binding by Cox19, and, on the contrary, reduced glutathione basically abolished the Cox19–Cox11 interaction. Hence the redox state of the cysteine residues in Cox11 obviously determines its interaction with Cox19 (Figure 7G). It is unclear whether the observed cysteine oxidation step is directly linked to the copper transfer, but the observation that addition or removal of copper did not affect the Cox19–Cox11 interaction suggests that the binding of Cox19 to Cox11 is not triggered by copper binding. Of interest, it was recently shown that in human cells, Cox19 can be released from the IMS to regulate copper efflux from the Golgi-localized, ATP7A copper-transporting ATPase (Leary et al., 2013). Cox19 release thereby correlates with the redox state of cysteine residues in Sco1 and Sco2, the copper chaperones for the Cu(A) site of cytochrome c oxidase. A direct interaction of Cox19 with Sco1 or Sco2 was not reported in that study, nor did we find Sco1 or Sco2 in our Cox19 interactome. It is conceivable that perturbation of Sco1 function impairs the Cox11–Cox19 interaction and thereby allows Cox19 to be exported back from the IMS to the cytosol.
The interaction of Cox19 with Cox11 in the IMS is reminiscent of the recently observed interaction of the twin Cx9C protein Mdm35 with the lipid-binding protein Ups1 (Potting et al., 2010; Tamura et al., 2010). Ups1 binds phosphatidic acid at the IMS side of the mitochondrial outer membrane and transfers it to the inner membrane (Connerth et al., 2012). In this process, Mdm35 is critical to maintaining Ups1 in a transfer-competent state, although its precise role in this reaction is not entirely clear. The overall structure of Mdm35 is similar to that of Cox19, and it even has the same conserved tyrosine residues at the same positions (twin Cx6Yx2C). Hence the principles by which Mdm35 supports Ups1-mediated lipid transfer might be similar to those by which Cox19 mediates Cox11-mediated copper transfer in the IMS. Possibly, twin Cx9C proteins might play a general function as folding modulators in the IMS that drive reactions in this compartment by a dynamic interaction with their client proteins. Thereby the function of IMS proteins might be controlled in an ATP-independent manner by modulations of conformational states triggered by the association or dissociation of twin Cx9C proteins. It will be exciting to develop an in vitro assay in which the Cox11-mediated copper transfer from Cox19 to Cox1 can be studied to dissect the precise functions of Cox19 in facilitating the redox-dependent copper transfer by Cox11.
MATERIALS AND METHODS
Yeast strains and media
The strains used for SILAC analysis were derived from YPH499 Δarg4 (von der Malsburg et al., 2011). To generate YPH499 Δarg4 Cox19-His6, the Schizosaccharomyces pombe HIS5 cassette from pFA6a-His3MX6 was amplified with primers with homologous ends to the 3′ untranslated region replacing the stop codon of COX19. The forward primer contained the sequence for six histidine residues followed by a stop codon. The resulting product was transformed into YPH499 Δarg4 and integrated into the DNA by homologous recombination.
All other yeast strains were derived from the wild-type strain W303 (Sherman et al., 1986). His6 tagging of Cox11 and Cox19 was done as described. For addition of a 3× hemagglutinin (3xHA) tag to Cox11, a genomic integration strategy was used employing the Kluyveromyces lactis TRP1 cassette with a 3xHA tag from a pYM22 vector (Janke et al., 2004).
For deletion of COX19, the open reading frame was replaced by homologous recombination with the spHIS5 cassette from pFA6a-His3MX6 (Bähler et al., 1998). The Δcox11 strain was made by replacing the COX11 reading frame by a KanMX4 cassette. For deletion of YME1, the open reading frame was replaced by URA3 from Saccharomyces cerevisiae.
Cox11 and Cox19 overexpression plasmids were made by amplifying the coding regions of the genes with additional 300 base pairs upstream and 150 base pairs downstream and inserting them via HindIII and BamHI into pRS426 (Sikorski and Hieter, 1989). The pRS426 plasmid containing COX19 was used for QuikChange Site-Directed Mutagenesis (Stratagene, Santa Clara, CA) to generate in the first step the mutations Y37E and L38E and in the second step Y59E and L60E. For generation of IM-Cox19 and IM-Cox19EE, the coding sequences of Cox19 and Cox19EE were amplified from the respective pRS426 plasmids and inserted using BamHI and XhoI into pYX223 (Novagen, Darmstadt, Germany), which contained the sequence for the first 75 amino acids of Mia40 between the EcoRI and BamHI sites.
pGEM plasmids (Promega, Fitchburg, WI) were used to synthesize radiolabeled Cox11 and Cox19. COX11 was inserted into pGEM3 via HindIII and BamHI. pGEM4-Cox19 was previously described (Bien et al., 2010).
Yeast strains were grown in YP (1% yeast extract and 2% peptone) or minimal medium with 2% glucose or galactose as carbon sources at 30°C (Altmann et al., 2007). For the SILAC labeling, the carbon source of the medium was glycerol. The wild type (YPH499 Δarg4) was grown in medium containing [13C6/15N4]l-arginine and [13C6/15N2]l-lysine, and the strain expressing Cox19-His6 was grown in medium with [12C/14N]arginine and lysine (von der Malsburg et al., 2011).
Purification of recombinant Cox19
The coding sequence without the stop codon of Cox19 and Cox19EE was amplified from pRS426-Cox19 or Cox19EE and cloned into pET22b (Novagen) upstream of a hexahistidine sequence using the restriction sites BamHI and XhoI. These plasmids were transformed into E. coli Rosetta 2(DE3) cells (Merck, Billerica, MA). The cells were grown at 37°C to an OD600 of 0.6 before expression was induced by the addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG) for 4 h at 37°C. The native purification of the recombinant proteins was performed as described in protocols 9 and 12 of the QIAexpressionist Handbook (Qiagen, Venlo, Netherlands).
Generation of antibodies against Cox11 and Cox19
The sequence corresponding to the IMS domain of Cox11 (bases 301–903) was cloned downstream of the sequence for a hexahistidine tag of the pET16b plasmid (Novagen) using the restriction sites Nde1 and BamH1. The plasmid was transformed into E. coli BL21-(DE3) cells (Novagen), and positive cells were grown at 37°C in LBAmp (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 100 mg/l ampicillin) to an OD600 of 0.6. Subsequently, gene expression was induced by the addition of 0.5 mM IPTG. After 4 h of induction at 37°C cells, were centrifuged. Cells were lysed for 10 min with vortexing and subsequent sonication in a buffer containing 8 M urea, 10 mM Tris, and 100 mM NaH2PO4, pH 8.0. Cell debris was removed by centrifugation, and Ni-NTA resin (Qiagen) was added to the supernatant. This was incubated for 1.5 h at room temperature under constant agitation. After washing the column twice with 8 M urea, 10 mM Tris, and 100 mM NaH2PO4, pH 6.3, and twice with 20 mM Tris, pH 7.4, we eluted recombinant His6-Cox11 with 20 mM Tris, 0.1% Triton X-100, and 250 mM imidazole, pH 7.4.
Ni-NTA affinity purification
A 10-mg amount of isolated mitochondria was lysed in 25 mM Tris, 250 mM NaCl, 20 mM imidazole, 1% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride (PMSF), pH 7.4. After a clarifying spin, 1% of the supernatant was taken as total fraction. Ni-NTA resin (Qiagen) was equilibrated in lysis buffer and added to the samples, which were subsequently incubated for 1 h at 4°C with constant tumbling. The resin was centrifuged down, and 1% of the supernatant was taken as the not-bound fraction. After three washing steps with lysis buffer without Triton X-100 and PMSF, the proteins were eluted by 10-min incubation at 4°C with of 25 mM Tris, 250 mM NaCl, and 250 mM imidazole, pH 7.4. The complete supernatants of the elution and the third washing steps were TCA precipitated and used for SDS–PAGE.
Mass spectrometry
Protein pellets were digested and desalted as described (Sommer et al., 2014). Peptides were resuspended in 12 μl of loading buffer (2% acetonitrile, 0.4% acetic acid), and 3-μl samples were analyzed in triplicate using a nanoAQUITY UPLC (Waters, Milford, MA) coupled to an LTQ-Orbitrap XL (Thermo, Waltham, MA) running in data-dependent acquisition mode as described (Mühlhaus et al., 2011), with the following modifications: resolution for MS1 scans was set to 60,000, the seven most intense precursor ions from the MS1 scan were selected for MS2 analysis, and each precursor was selected twice for MS2 analysis before it was put on a dynamic exclusion list for 30 s. The HPLC gradient ramped from 2 to 35% acetonitrile within 90 min, then to 80% acetonitrile within 5 min, followed by a 10-min wash with 80% acetonitrile and 17-min reequilibration with 2% acetonitrile.
Peptide and protein identification and quantitation were done using MaxQuant software, version 1.2.0.18 (Cox and Mann, 2008; Cox et al., 2011), and Saccharomyces Genome Database (www.yeastgenome.org; yeast orf trans all, 03-Feb-2011). For heavy labels, Arg-10 and Lys-8 was chosen, up to three tryptic miscleavages were allowed, and protein N-terminal acetylation and Met oxidation were specified as variable modifications and Cys carbamidomethylation as fixed modification. The false discovery rate was set at 1% for peptides, proteins, and sites, and minimal peptide length was seven amino acids.
Size exclusion chromatography
A 3-mg amount of isolated mitochondria was lysed in 500 μl of 25 mM Tris, 250 mM NaCl, 1% Triton-X 100, and 2 mM PMSF, pH 7.4. After a purifying centrifugation at 25,000 × g for 30 min at 4°C, the clarified extract was applied to a HiLoad 16/60 Superdex 200 column (GE Healthcare, Chalfont St Giles, United Kingdom) and separated with 25 mM Tris, 250 mM NaCl, and 0.1% Triton-X 100, pH 7.4. Proteins were precipitated from the collected 1-ml fractions by addition of TCA and analyzed by Western blotting.
Protein import into mitochondria
Radiolabeled Cox11 and Cox19 were synthesized in vitro using the TNT T7 Quick Coupled Transcription/Translation Kit (Promega) from the plasmids pGEM3-Cox11 and pGEM4-Cox19. To avoid premature protein oxidation, protein synthesis of Cox19 was performed under hypoxic conditions. The import reactions and their analyses were performed as described previously (Mesecke et al., 2005). The import reaction for Cox19 also contained 5 mM GSH. To remove nonimported protein, mitochondria were treated with 100 μg/ml proteinase K for 30 min on ice after the import reactions.
COX activity assay
The assay was performed as previously described (Stehling et al., 2007).
Immunoprecipitation
A 1-mg amount of mitochondria was lysed in 100 mM NaH2PO4, 100 mM NaCl, 0.1% Triton X-100, and 2 mM PMSF, pH 8.0. After a clarifying spin, 5% of the supernatant was taken as total fraction. In lysis buffer, equilibrated protein A–Sepharose beads (Expedeon, Harston, United Kingdom) were added, to which antibodies against Cox11 were bound. The samples were incubated for 1 h at 4°C with constant tumbling. The beads were centrifuged down, and 5% of the supernatant was taken as the not-bound fraction. After two washing steps with lysis buffer and one with lysis buffer without Triton X-100 and PMSF, the proteins were eluted by 5-min incubation at 96°C with Laemmli buffer. The complete supernatant was used as the elution fraction.
Determination of the redox state of Cox11 and Cox19
For Cox11, 200 μg of isolated mitochondria was added to 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, with 2 mM BCS, 100 μM CuSO4, or no additional chemicals. After 10 min of incubation at 30°C, the samples were precipitated by addition of 12% TCA and incubated overnight at −20°C. The samples were centrifuged for 15 min at 4°C and 20,000 × g and washed with ice-cold acetone; the resulting pellet was dried and then resolved in modification buffer (80 mM Tris, pH 7 (HCl), 10% glycerol, 2% SDS, bromocresol purple). For Cox19, 12.5 μg of the isolated protein was directly added to the modification buffer. mm(PEG)24 (Thermo Scientific) was used for the alkylation of Cox11, and mm(PEG)12 was used for Cox19. To alkylate all cysteine residues in the proteins (“maximum shift”), samples were pretreated at 96°C with modification buffer containing 20 mM of thiol-free reductant TCEP before 15 mM methyl-PEG12/24-maleimide was added. As unmodified control (“minimum shift”), one sample was mock treated with modification buffer without mm(PEG)12/24. After addition of mm(PEG)12/24, all samples were incubated for 1 h in darkness.
Peptide scan
Twenty-six peptides of 20 residues each shifted by three amino acids to cover the sequence of the IMS domain of Cox11 were spotted two times onto cellulose membranes (Frank and Overwin, 1996; Kramer and Schneider-Mergener, 1998). The membranes were incubated at room temperature for 2 min in methanol, washed for 2 min with H2O, and equilibrated for 20 min with binding buffer (20 mM Tris-HCl, pH 7.0, 20 mM NaCl). Then one membrane was incubated with binding buffer containing 12.5 μM purified His6-tagged Cox19 and the other with Cox19EE. After washing twice for 10 min with binding buffer and twice for 10 min with 10 mM Tris-HCl, pH 7.4, and 150 mM NaCl, the bound Cox19 was transferred onto a nitrocellulose membrane used for Western blotting with hexahistidine-specific antibodies (Weckbecker et al., 2012).
In vitro binding assay with recombinant Cox19
A 450-μg amount of isolated mitochondria was lysed in 500 μl lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.1% Triton X-100, 1 mM PMSF, pH 8.0) during incubation at 4°C for 10 min. After a clarifying spin at 20,000 × g for 10 min, 5% of the supernatant was taken as the total fraction. The rest was added to 20 μl Ni-NTA agarose beads (Qiagen) that were equilibrated with binding buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0). A 10-μg amount of recombinant His6-tagged Cox19 or Cox19EE was added. The samples were incubated for 1 h at 4°C with constant agitation. The beads were centrifuged down, and 5% of the supernatant was taken as the not-bound fraction. The beads were washed four times with binding buffer before they were incubated for 5 min at 30°C with 25 μl of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0). The beads were centrifuged down, and the supernatant was taken as the elution fraction.
Supplementary Material
Acknowledgments
We thank Rodrigo Cuevas Arenas for help with some experiments, Jan Riemer and Bruce Morgan for critical reading of the manuscript, and Sabine Knaus for technical assistance. The work was supported by grants from the Deutsche Forschungsgemeinschaft (J.M.H., R.Z., M.S.), the International Graduate School IRTG1830 (J.M.H., R.Z.), Sonderforschungsbereich 746 (M.v.d.L.), the Excellence Initiative of the German Federal and State Governments (EXC294 BIOSS; M.v.d.L.), and the BioComp Landesschwerpunkt Rheinland-Pfalz (J.M.H., M.S.).
Abbreviations used:
- BCS
bathocuproine disulfonate
- GSH
glutathione
- GSSG
glutathione disulfide
- IM
inner membrane
- IMS
intermembrane space
- ITS
IMS-targeting signal
- MISS
mitochondrial IMS-sorting signal
- mm(PEG)
methyl-polyethylene glycol maleimide
- SILAC
stable isotope labeling by amino acids in cell culture
- TCA
trichloroacetic acid
- TCEP
Tris-carboxyethyl phosphine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1526) on April 29, 2015.
REFERENCES
- Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, et al. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J Mol Biol. 2005;353:937–944. doi: 10.1016/j.jmb.2005.08.049. [DOI] [PubMed] [Google Scholar]
- Altmann K, Dürr M, Westermann B. Saccharomyces cerevisiae as a model organism to study mitochondrial biology. In: Leister D, Herrmann JM, editors. Mitochondria. Practical Protocols. Vol. 372. Totowa, NJ: Humana Press; 2007. pp. 81–90. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Gonnelli L, Mangani S. Solution structure of Cox11, a novel type of beta-immunoglobulin-like fold involved in CuB site formation of cytochrome c oxidase. J Biol Chem. 2004;279:34833–34839. doi: 10.1074/jbc.M403655200. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K. Mia40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat Struct Mol Biol. 2009;16:198–206. doi: 10.1038/nsmb.1553. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, Palumaa P. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- Banting GS, Glerum DM. Mutational analysis of the Saccharomyces cerevisiae cytochrome c oxidase assembly protein Cox11p. Eukaryot Cell. 2006;5:568–578. doi: 10.1128/EC.5.3.568-578.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proof read by glutathione. Mol Cell. 2010;37:516–528. doi: 10.1016/j.molcel.2010.01.017. [DOI] [PubMed] [Google Scholar]
- Bihlmaier K, Mesecke N, Terzyiska N, Bien M, Hell K, Herrmann JM. The disulfide relay system of mitochondria is connected to the respiratory chain. J Cell Biol. 2007;179:389–395. doi: 10.1083/jcb.200707123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode M, Longen S, Morgan B, Peleh V, Dick TP, Bihlmaier K, Herrmann JM. Inaccurately assembled cytochrome c oxidase can lead to oxidative stress-induced growth arrest. Antioxid Redox Signal. 2013;18:1597–1612. doi: 10.1089/ars.2012.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Carr HS, George GN, Winge DR. Yeast Cox11, a protein essential for cytochrome c oxidase assembly, is a Cu(I)-binding protein. J Biol Chem. 2002;277:31237–31242. doi: 10.1074/jbc.M204854200. [DOI] [PubMed] [Google Scholar]
- Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. J Biol Chem. 2005;280:22664–22669. doi: 10.1074/jbc.M414077200. [DOI] [PubMed] [Google Scholar]
- Cavallaro G. Genome-wide analysis of eukaryotic twin CX9C proteins. Mol Biosyst. 2010;6:2459–2470. doi: 10.1039/c0mb00058b. [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuan Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 2004;23:3735–3746. doi: 10.1038/sj.emboj.7600389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338:815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Dabir DV, Leverich EP, Kim SK, Tsai FD, Hirasawa M, Knaff DB, Koehler CM. A role for cytochrome c and cytochrome c peroxidase in electron shuttling from Erv1. EMBO J. 2007;26:4801–4811. doi: 10.1038/sj.emboj.7601909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–1254. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- Farrell SR, Thorpe C. Augmenter of liver regeneration: a flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry. 2005;44:1532–1541. doi: 10.1021/bi0479555. [DOI] [PubMed] [Google Scholar]
- Fasching PA, Pharoah PD, Cox A, Nevanlinna H, Bojesen SE, Karn T, Broeks A, van Leeuwen FE, van't Veer LJ, Udo R, et al. The role of genetic breast cancer susceptibility variants as prognostic factors. Hum Mol Genet. 2012;21:3926–3939. doi: 10.1093/hmg/dds159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass D. The Erv family of sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:557–566. doi: 10.1016/j.bbamcr.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Fischer M, Horn S, Belkacemi A, Kojer K, Petrungaro C, Habich M, Ali M, Kuttner V, Bien M, Kauff F, et al. Protein import and oxidative folding in the mitochondrial intermembrane space of intact mammalian cells. Mol Biol Cell. 2013;24:2160–2170. doi: 10.1091/mbc.E12-12-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R, Overwin H. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol Biol. 1996;66:149–169. doi: 10.1385/0-89603-375-9:149. [DOI] [PubMed] [Google Scholar]
- Gabriel K, Milenkovic D, Chacinska A, Muller J, Guiard B, Pfanner N, Meisinger C. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J Mol Biol. 2007;365:612–620. doi: 10.1016/j.jmb.2006.10.038. [DOI] [PubMed] [Google Scholar]
- Gebert M, Schrempp SG, Mehnert CS, Heisswolf AK, Oeljeklaus S, Ieva R, Bohnert M, von der Malsburg K, Wiese S, Kleinschroth T, et al. Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J Cell Biol. 2012;197:595–604. doi: 10.1083/jcb.201110047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goemans C, Denoncin K, Collet JF. Folding mechanisms of periplasmic proteins. Biochim Biophys Acta. 2014;1843:1517–1528. doi: 10.1016/j.bbamcr.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Gross DP, Burgard CA, Reddehase S, Leitch JM, Culotta VC, Hell K. Mitochondrial Ccs1 contains a structural disulfide bond crucial for the import of this unconventional substrate by the disulfide relay system. Mol Biol Cell. 2011;22:3758–3767. doi: 10.1091/mbc.E11-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. The intermembrane space of mitochondria. Antioxid Redox Signal. 2010;13:1341–1358. doi: 10.1089/ars.2009.3063. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Riemer J. Three approaches to one problem: protein folding in the periplasm, the endoplasmic reticulum, and the intermembrane space. Antioxid Redox Signal. 2014;21:438–456. doi: 10.1089/ars.2014.5841. [DOI] [PubMed] [Google Scholar]
- Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome c oxidase. J Biol Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Yamano K, Naoe M, Momose T, Terao K, Nishikawa S, Watanabe N, Endo T. Structural basis of yeast Tim40/Mia40 as an oxidative translocator in the mitochondrial intermembrane space. Proc Natl Acad Sci USA. 2009;106:14403–14407. doi: 10.1073/pnas.0901793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol. 2010;30:1004–1017. doi: 10.1128/MCB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Bird A, Winge DR. Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J Biol Chem. 2007;282:17442–17449. doi: 10.1074/jbc.M702379200. [DOI] [PubMed] [Google Scholar]
- Koch JR, Schmid FX. Mia40 targets cysteines in a hydrophobic environment to direct oxidative protein folding in the mitochondria. Nat Commun. 2014;5:3041. doi: 10.1038/ncomms4041. [DOI] [PubMed] [Google Scholar]
- Koehler CM, Jarosch E, Tokatlidis K, Schmid K, Schweyen RJ, Schatz G. Import of mitochondrial carrier proteins mediated by essential proteins of the intermembrane space. Science. 1998;279:369–373. doi: 10.1126/science.279.5349.369. [DOI] [PubMed] [Google Scholar]
- Kramer A, Schneider-Mergener J. Synthesis and screening of peptide libraries on continuous cellulose membrane supports. Methods Mol Biol. 1998;87:25–39. doi: 10.1385/0-89603-392-9:25. [DOI] [PubMed] [Google Scholar]
- Lafontaine D, Tollervey D. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 1996;24:3469–3472. doi: 10.1093/nar/24.17.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary SC, Cobine PA, Kaufman BA, Guercin GH, Mattman A, Palaty J, Lockitch G, Winge DR, Rustin P, Horvath R, Shoubridge EA. The human cytochrome c oxidase assembly factors SCO1 and SCO2 have regulatory roles in the maintenance of cellular copper homeostasis. Cell Metab. 2007;5:9–20. doi: 10.1016/j.cmet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Leary SC, Cobine PA, Nishimura T, Verdijk RM, de Krijger R, de Coo R, Tarnopolsky MA, Winge DR, Shoubridge EA. COX19 mediates the transduction of a mitochondrial redox signal from SCO1 that regulates ATP7A-mediated cellular copper efflux. Mol Biol Cell. 2013;24:683–691. doi: 10.1091/mbc.E12-09-0705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hofhaus G, Lisowsky T. Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett. 2000;477:62–66. doi: 10.1016/s0014-5793(00)01767-1. [DOI] [PubMed] [Google Scholar]
- Leonhard K, Stiegler A, Neupert W, Langer T. Chaperone-like activity of the AAA domain of the yeast Yme1 AAA protease. Nature. 1999;398:348–351. doi: 10.1038/18704. [DOI] [PubMed] [Google Scholar]
- Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J. Systematic analysis of the twin cx9c protein family. J Mol Biol. 2009;393:356–368. doi: 10.1016/j.jmb.2009.08.041. [DOI] [PubMed] [Google Scholar]
- Longen S, Woellhaf MW, Petrungaro C, Riemer J, Herrmann JM. The disulfide relay of the intermembrane space oxidizes the ribosomal subunit Mrp10 on its transit into the mitochondrial matrix. Dev Cell. 2014;28:30–42. doi: 10.1016/j.devcel.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Maxfield AB, Heaton DN, Winge DR. Cox17 is functional when tethered to the mitochondrial inner membrane. J Biol Chem. 2004;279:5072–5080. doi: 10.1074/jbc.M311772200. [DOI] [PubMed] [Google Scholar]
- Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. Protein quality control in the bacterial periplasm. Annu Rev Microbiol. 2011;65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell. 2005;121:1059–1069. doi: 10.1016/j.cell.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Ramming T, Muller JM, Wenz LS, Gebert N, Schulze-Specking A, Stojanovski D, Rospert S, Chacinska A. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol Biol Cell. 2009;20:2530–2539. doi: 10.1091/mbc.E08-11-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlhaus T, Weiss J, Hemme D, Sommer F, Schroda M. Quantitative shotgun proteomics using a uniform (1)(5)N-labeled standard to monitor proteome dynamics in time course experiments reveals new insights into the heat stress response of Chlamydomonas reinhardtii. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.004739. M110.004739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoe M, Ohwa Y, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. Identification of Tim40 that mediates protein sorting to the mitochondrial intermembrane space. J Biol Chem. 2004;279:47815–47821. doi: 10.1074/jbc.M410272200. [DOI] [PubMed] [Google Scholar]
- Nobrega MP, Bandeira SC, Beers J, Tzagoloff A. Characterization of COX19, a widely distributed gene required for expression of mitochondrial cytochrome oxidase. J Biol Chem. 2002;277:40206–40211. doi: 10.1074/jbc.M207348200. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Potting C, Wilmes C, Engmann T, Osman C, Langer T. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 2010;29:2888–2898. doi: 10.1038/emboj.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- Rigby K, Zhang L, Cobine PA, George GN, Winge DR. Characterization of the cytochrome c oxidase assembly factor Cox19 of Saccharomyces cerevisiae. J Biol Chem. 2007;282:10233–10242. doi: 10.1074/jbc.M610082200. [DOI] [PubMed] [Google Scholar]
- Schreiner B, Westerburg H, Forne I, Imhof A, Neupert W, Mokranjac D. Role of the AAA protease Yme1 in folding of proteins in the intermembrane space of mitochondria. Mol Biol Cell. 2012;23:4335–4346. doi: 10.1091/mbc.E12-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks J. Methods in Yeast Genetics: A Laboratory Course. Cold Spring Harbor Laboratory Press. 1986 [Google Scholar]
- Sideris DP, Petrakis N, Katrakili N, Mikropoulou D, Gallo A, Ciofi-Baffoni S, Banci L, Bertini I, Tokatlidis K. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J Cell Biol. 2009;187:1007–1022. doi: 10.1083/jcb.200905134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C, Endres M, Fölsch H, Stuart RA, Neupert W, Brunner M. Carrier protein import into mitochondria mediated by the intermembrane proteins Tim10/Mrs11p and Tim12/Mrs5p. Nature. 1998;391:912–915. doi: 10.1038/36136. [DOI] [PubMed] [Google Scholar]
- Sommer F, Muhlhaus T, Hemme D, Veyel D, Schroda M. Identification and validation of protein-protein interactions by combining co-immunoprecipitation, antigen competition, and stable isotope labeling. Methods Mol Biol. 2014;1188:245–261. doi: 10.1007/978-1-4939-1142-4_17. [DOI] [PubMed] [Google Scholar]
- Stehling O, Smith PM, Biederbick A, Balk J, Lill R, Muhlenhoff U. Investigation of iron-sulfur protein maturation in eukaryotes. Methods Mol Biol. 2007;372:325–342. doi: 10.1007/978-1-59745-365-3_24. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Iijima M, Sesaki H. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 2010;29:2875–2887. doi: 10.1038/emboj.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AK, Smith D, Gray J, Carr HS, Liu A, Winge DR, Hosler JP. Mutagenic analysis of Cox11 of Rhodobacter sphaeroides: insights into the assembly of Cu(B) of cytochrome c oxidase. Biochemistry. 2010;49:5651–5661. doi: 10.1021/bi1003876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tienson HL, Dabir DV, Neal SE, Loo R, Hasson SA, Boontheung P, Kim SK, Loo JA, Koehler CM. Reconstitution of the mia40-erv1 oxidative folding pathway for the small tim proteins. Mol Biol Cell. 2009;20:3481–3490. doi: 10.1091/mbc.E08-10-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Capitanio N, Nobrega MP, Gatti D. Cytochrome oxidase assembly in yeast requires the product of COX11, a homolog of the P. denitrificans protein encoded by ORF3. EMBO J. 1990;9:2759–2764. doi: 10.1002/j.1460-2075.1990.tb07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veniamin S, Sawatzky LG, Banting GS, Glerum DM. Characterization of the peroxide sensitivity of COX-deficient yeast strains reveals unexpected relationships between COX assembly proteins. Free Radic Biol Med. 2011;51:1589–1600. doi: 10.1016/j.freeradbiomed.2011.06.024. [DOI] [PubMed] [Google Scholar]
- Vial S, Lu H, Allen S, Savory P, Thornton D, Sheehan J, Tokatlidis K. Assembly of Tim9 and Tim10 into a functional chaperone. J Biol Chem. 2002;277:36100–36108. doi: 10.1074/jbc.M202310200. [DOI] [PubMed] [Google Scholar]
- Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C. Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics. 2012;11:1840–1852. doi: 10.1074/mcp.M112.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Webb CT, Gorman MA, Lazarou M, Ryan MT, Gulbis JM. Crystal structure of the mitochondrial chaperone TIM9–10 reveals a six-bladed alpha-propeller. Mol Cell. 2006;21:123–133. doi: 10.1016/j.molcel.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Weckbecker D, Longen S, Riemer J, Herrmann JM. Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J. 2012 doi: 10.1038/emboj.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.