The 24:0 sphingomyelin of nuclear lipid microdomains from normal cells shifts to 16:0 sphingomyelin in nuclear lipid microdomains from cancer cells. The narrower microdomains in the nucleus are associated with the changes to proteins involved in hepatocarcinogenesis.
Abstract
Lipid microdomains localized in the inner nuclear membrane are considered platforms for active chromatin anchoring. Stimuli such as surgery, vitamin D, or glucocorticoid drugs influence their gene expression, DNA duplication, and RNA synthesis. In this study, we used ultrafast liquid chromatography–tandem mass spectrometry to identify sphingomyelin (SM) species coupled with immunoblot analysis to comprehensively map differences in nuclear lipid microdomains (NLMs) purified from hepatocytes and hepatoma cells. We showed that NLMs lost saturated very-long-chain fatty acid (FA; C24:0) SM in cancer cells and became enriched in long-chain FA (C16:0) SM. We also found that signaling proteins, such as STAT3, Raf1, and PKCζ, were increased and vitamin D receptor was reduced in cancer cells. Because recent researches showed a shift in sphingolipid composition from C24:0 to C16:0 in relation to cell life, we performed a comparative analysis of properties among C16:0 SM, C24:0 SM, and cholesterol. Our results led us to hypothesize that the enrichment of C16:0 SM could determine enhanced dynamic properties of NLMs in cancer cells with an increased shuttling of protein signaling molecules.
INTRODUCTION
Sphingomyelin (SM) was described as a dominant sphingolipid in mammalian cell membranes indispensable for cell function (Taniguchi and Okazaki, 2014). It contains acyl chains that vary in length from long-chain fatty acids (LCFAs), such as C16:0 palmitic acid, to very-long-chain fatty acids (VLCFAs) with 20 or more carbons, including saturated fatty acids (FAs) such as 20:0 arachidic acid, 22:0 behenic acid, 24:0 lignoceric acid, and mono- and poli-VLCFAs (Ohno et al., 2010). VLCFAs are produced from certain LCFAs, provided through the diet or generated by FA synthase and elongated by FA elongase (Soupene and Kuypers, 2008).
SM and cholesterol (CHO) formed functional nanoscale-ordered domains, characteristic in particular of the external leaflet of cell membranes, whose thickness increased with the length of the acyl chain of SM (Róg and Vattulainen, 2014). Although a high-affinity interaction between SM and CHO was suggested (Radhakrishnan et al., 2000; Li et al., 2001), no compelling experimental evidence for the molecular basis of such a specific interaction has been reported in the literature, suggesting that the driving force for the formation of the lipidic domains is provided by the hydrophobic matching condition in membranes (Holopainen et al., 2004). However, it should be mentioned that interactions between SM and CHO were computationally studied in binary mixtures of CHO and SM via molecular dynamic simulation, with a focus on hydrogen bonding by showing that CHO formed more hydrogen bonds with SM than it formed with phosphatidylcholine (PC; Róg and Pasenkiewicz-Gierula, 2006). The studies on interactions among atoms of Aittoniemi et al. (2007) indicated that hydrogen bonding alone could not explain the higher affinity of CHO for SM but that one must also consider the contributions of van der Waals interactions between CHO and the choline groups of SM. In addition, CHO preferred saturated SM, with ordered acyl chains, to establish more favourable entropic interactions, since CHO might increase local entropy, and its interactions with disordered acyl chains in unsaturated phospholipids would lead to an ordering effect of CHO on acyl chains, thereby decreasing the local entropy (Slotte, 2013a). Measurements of sterol bilayer affinity demonstrated that palmitoyl SM was the optimal SM analogue for CHO (Slotte, 2013b). X-ray scattering data showed that 22:0, 23:0, and 24:0 SM included in a bilayer can lead to transbilayer interdigitation, that is, the distal part of a long acyl chain from a SM molecule in one leaflet penetrates into the opposing leaflet (Levin et al., 1985). CHO and SM are present in membranes as lipid microdomains called lipid rafts, extracted as liquid ordered–phase detergent-resistant membranes whose functional role has long been subject of discussion in the membrane biophysics community. Brown (2006) claimed that the detergent-resistant membrane fractions did not represent lipid rafts present in the cells before extraction, even if raft-targeting signals identified by detergent-resistant membrane analysis were often required for protein function. Frisz et al. (2013) demonstrated that sphingolipid domains in the plasma membranes of fibroblasts did not contain CHO but that the latest affected the sphingolipid organization via an indirect mechanism that involved the cytoskeleton. Then Honigmann et al. (2014) proposed that alternative interactions were responsible for the strong local trapping of sphingolipid analogue in living nonstimulated cells. However, Mollinedo and Gajate (2015) demonstrated that lipid rafts behaved in cell membranes as major modulators of membrane geometry and lateral movement of molecules and as a platform for traffic and signal transduction proteins. In addition, lipid rafts represented the major platforms for signaling regulation in cancer (Mollinedo and Gajate, 2015).
We have previously demonstrated that lipid microdomains, rich in SM and CHO, are present in the inner nuclear membrane (NM), and we called these nuclear lipid microdomains (NLMs). They played different roles in relation to cell function by acting as platform for vitamin D receptor (VDR) in embryonic hippocampal cells (Bartoccini et al., 2011) and for glucorticoid drugs in non-Hodgkin's T-cell human lymphoblastic lymphoma (Cataldi et al., 2014). In the liver, NLMs acted as a resting place for active chromatin (Chr) and transcription factors by regulating DNA (Albi et al., 2013) and RNA (Cascianelli et al., 2008; Albi and Villani, 2009) synthesis. No data have existed until now about the SM FA species in NLMs.
Recent research on sphingolipid FAs has been focused on their role in cell physiopathology. Sassa et al. (2012) described that a shift in sphingolipid composition from C24:0 to C16:0 increased susceptibility to apoptosis in HeLa cells. The role of C16:0 had already been suggested by a study of Zhang et al. (2004) demonstrating that C16:0 induced apoptosis in human hepatoma HepG2 cells.
Hepatoma is a leading primary malignancy of the liver, one of the most common cancers worldwide. New therapeutic strategies targeted anti-signal transducer and activator of transcription 3 (STAT3) protein, a key regulator of inflammation, cell survival, and tumorigenesis of liver cells (Hung et al., 2014), and Raf1, which prolongs cell survival and leads to cancer, even in the absence of oncogenic mutations (Gauthier and Mitchell, 2013). Protein kinase Cζ (PKCζ) was involved in the hepatocarcinogenic mechanism by controlling glycogen synthase kinase-3β (Desbois-Mouthon et al., 2002). PKCζ stimulates and vitamin D inhibits hepatocellular carcinoma development (Guo et al., 2013).
To address the role of SM present in NLMs on cell function, we examined the presence of SM species in NLMs purified from hepatocytes (H) and H35 hepatoma cells (H35) in relation to signal proteins and vitamin D3 receptor (VDR).
RESULTS
Nuclear rafts of hepatocytes and hepatoma cells
Highly purified H and H35 nuclei were used to prepare NLMs. The purification level of the nuclear preparation was similar to that previously reported (Cascianelli et al., 2008). In the nuclei, after Barnes treatment, the activity of glucose-6-phosphatase was 7 ± 2 nmol/mg protein/min (H) and 6 ± 2 nmol/mg protein/min (H35). The NADH–cytochrome c reductase activity was undetectable in both preparations. The NLM fraction was obtained from triton solubilization. In H NLMs, the level of protein was 29.40 ± 2.21 μg/g liver, and in H35 NLMs, it was 1.31 ± 0.02 μg/106 cells, according to Cascianelli et al. (2008) and Bartoccini et al. (2011), respectively. No activity of glucose-6-phosphatase and NADH–cytochrome c reductase was detected in both preparations, indicating the absence of cytoplasmic contamination. These data were strongly supported by immunoblot analysis with giantin antibodies, a marker protein for Golgi membrane. The results showed that the band for giantin corresponding to an apparent molecular weight of 367 kDa was absent in H NLMs and H35 NLMs (Figure 1). The presence of STAT3, which is a marker of NLMs, demonstrates the purification of NLMs (Cascianelli et al., 2008) (Figure 1a), although it was expressed 3.43 times in H35 NLMs in comparison with H NLMs (Figure 1b). For highlighting the level of NLM purification and to exclude possible Chr and nuclear matrix (NMx) contamination, lamin B was analyzed as marker of NLMs, whereas Chr and NMx were used as controls. In samples prepared from both H and H35 nuclei, the results showed the absence of lamin B in NMx and the very low level of protein in Chr, indicated by the possible presence of small parts of inner NM in the sample, and a higher protein content in NLMs, as previously reported (Cascianelli et al., 2008; Figure 2). The level of CHO was 13.40 ± 0.22 μg/mg protein in H NLMs and 14.65 ± 0.38 μg/mg protein in H35 NLMs, similar to what was previously reported (Cascianelli et al., 2008; Bartoccini et al., 2011).
FIGURE 1:
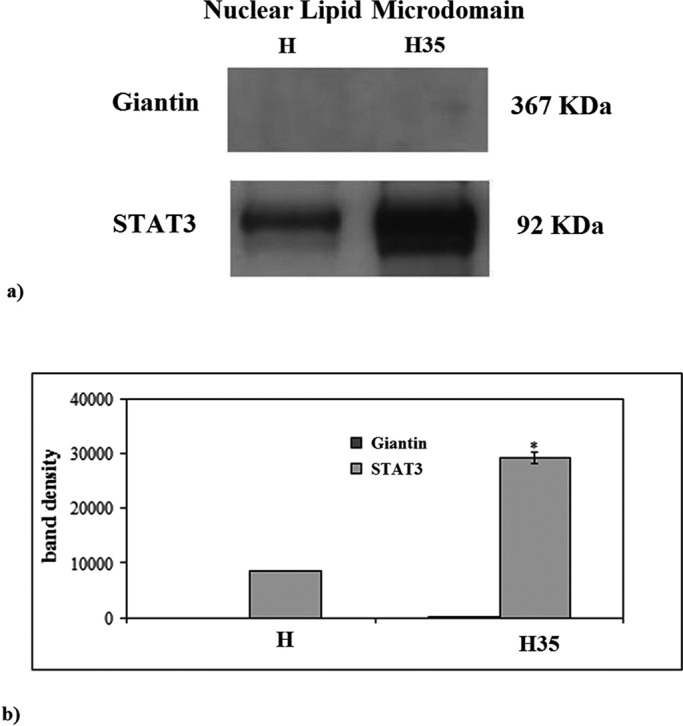
Giantin and STAT3 in NLMs purified from hepatocytes and hepatoma cells. (a) Immunoblot of proteins probed with specific antibodies. The position of giantin and STAT3 was indicated in relation to the position of molecular size standards. (b) The area density evaluated by densitometry scanning and analysis with Scion Image; the data represent the mean ± SD of three separate experiments. H, hepatocyte; H35, hepatoma cell line. *, p < 0.001 vs. H.
FIGURE 2:
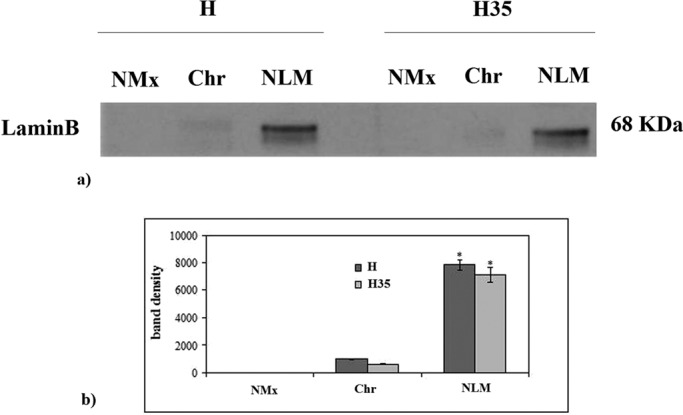
Lamin B in NMx, Chr, and NLM from hepatocytes and hepatoma cells. (a) Immunoblot of protein probed with specific antibody. The position of lamin B was indicated in relation to the position of the molecular size standard. (b) The area density evaluated by densitometry scanning and analysis with Scion Image; the data represent the mean ± SD of three separate experiments. H, hepatocyte; H35, hepatoma cell line. *, p < 0.001 vs. NMx and Chr.
24:0 SM shifts to 16:0 SM in nuclear lipid rafts (NLRs) of cancer cells
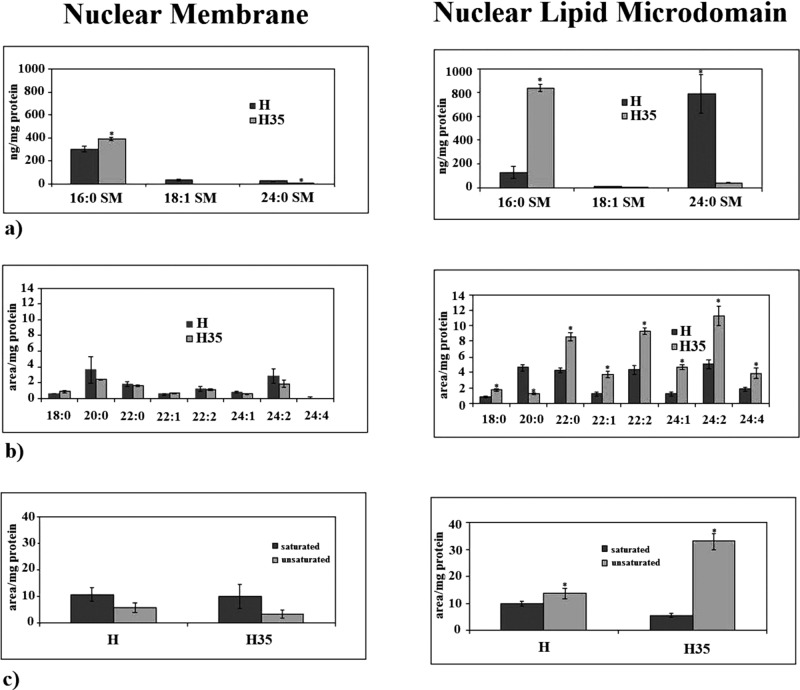
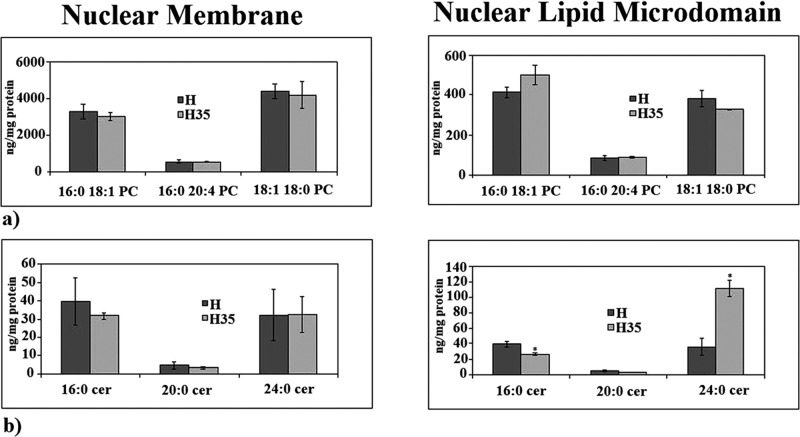
We analyzed SM species in NLMs by using 16:0 SM, 18:1 SM, and 24:0 SM external calibrators; the results were compared with those of total NM. The results highlighted that, in NLMs prepared from cancer cells, the value of 16:0 SM increased 6.5 times and that of 24:0 SM decreased 18.65 times in comparison with NLMs from normal cells (Figure 3a). In NM, 16:0 SM increased 1.29 times and 24:0 SM decreased 4.6 times (Figure 3a). As NM contained NLMs, it is possible that the low variations present in NM reflected the changes observed in NLMs. To have a deeper insight of SM species containing saturated or unsaturated FAs, we evaluated the areas of all the peaks identified on the basis of their molecular weights and we analyzed their values in relation to protein content. A total of 24 species were investigated: 16:1 SM, 18:0 SM, 18:2 SM, 20:0 SM, 20:1 SM, 20:2 SM, 20:3 SM, 22:0 SM, 22:1 SM, 22:2 SM, 22:3 SM, 22:4 SM, 24:1 SM, 24:2 SM, 24:3 SM, 24:4 SM, 24:5 SM, 26:0 SM, 26:1 SM, 26:2 SM, 26:3 SM, 26:4 SM, 26:5 SM, 26:6 SM. Seven peaks were detected (Figure 3b). Significant differences in the levels of various lipid molecular species were found between H NLMs and H35 NLMs. H NLMs were richer in 20:0 SM content than H35 NLMs, and H35 NLMs were richer in 22:0 SM, 22:1 SM, 22:2 SM, 24:1 SM, 24:2 SM, and 24:4 SM content than H NLMs. In the intermediate-length acyl chains (20:0 and 22:0), there was an opposite effect to that observed for 16:0 and 24:0. However, 20:0 decreased 3.72 times and 22: 0 increased 2.07 times, similar to other FAs. Thus we focused our attention on FAs with higher variations, such as 16: 0 and 24:0. In NM, the changes in SM content between normal and cancer cells were not statistically significant. It is possible that the variations observed in NLMs were not high enough to effect changes in the total NM. We then compared the changes in the total levels of SM species containing saturated and unsaturated FAs. As reported in Figure 3c, the SM saturated FAs were 1.77 times lower and SM unsaturated FAs were 2.42 times higher in NLMs of cancer cells than in NLMs of normal cells. Thus the saturated/unsaturated FA ratio was 0.71 in H NLMs and 0.17 in H35 NLMs. Among unsaturated FAs, the monounsaturated FAs increased 3.10 times (22:1 SM) and 3.89 (24:1 SM), the di-unsaturated and tetra-unsaturated FAs increased in a range between 2.16 and 2.42. No significant changes were seen in NM (Figure 3c). To verify the specificity of SM changes, we analyzed PC and ceramide species by using 16:0 18:1 PC, 16:0 24:0 PC, 18:1 18:0 PC, 16:0 ceramide, 20:0 ceramide, and 24:0 ceramide as external calibrators. When the total SM species (Figure 3a) was compared with the total PC species (Figure 4a), it could be seen that PC was ∼23 times higher than SM in NM, whereas both lipids had similar value in NLMs, as previously reported (Cascianelli et al., 2008). The results showed no changes of PC species in both NM and NLMs prepared from normal and cancer cells (Figure 4a). Also, no changes were present in ceramide species in NM, whereas 16:0 ceramide decreased 1.5 times and 24:0 ceramide increased 3.13 times in NLMs (Figure 4b). It is possible that the variations of ceramide species were the results of the changes of substrates for sphingomyelinase, such as SM 16:0 and SM 24:0 (Figure 3a).
FIGURE 3:
SM in NM and NLMs purified from hepatocytes and hepatoma cells. (a) SM species studied by using 16:0 SM, 18:1 SM, and 24:0 SM external calibrators. Data are expressed as nmol/mg protein and represent the mean ± SD of three separate experiments. (b) SM species studied by evaluating the areas of all the peaks identified on the basis of their molecular weight. Data are expressed as area/mg protein and represent the mean ± SD of three separate experiments. (c) Total saturated and unsaturated FAs. Data are expressed as area/mg protein and represent the mean ± SD of three separate experiments. H, hepatocyte; H35, hepatoma cell line. *, p < 0.001 vs. H.
FIGURE 4:
PC and ceramide in NM and NLMs purified from hepatocytes and hepatoma cells. (a) PC species studied by using 16:0 18:1 PC, 16:0 24:0 PC, and 18:1 18:0 PC external calibrators. (b) Ceramide species studied by using 16:0 ceramide, 20:0 ceramide, and 24:0 ceramide standards. Data are expressed as nmol/mg protein and represent the mean ± SD of three separate experiments. H, hepatocyte; H35, hepatoma cell line. *, p < 0.001 vs. H.
Comparative analysis of properties among C16:0 SM, C24:0 SM, and CHO
To understand the possible meaning of the FA change in NLM SM of cancer cells, we studied the properties of C16:0 SM, C24:0 SM, and CHO, using molecular modeling calculations. Accordingly, tridimensional models of C16:0 SM, C24:0 SM, and CHO were generated in silico as detailed in the Materials and Methods. These models were instrumental in calculating surface descriptors, such as the polar surface area (Å2) and total van der Waals surface area (Å2), and geometrical descriptors, including the topological radius (Å) and topological diameter (Å). Specifically, the topological radius and diameter are respectively defined as the minimum vertex and the maximum vertex of a molecular graph representing the molecule, thereby providing the dimensions of the lipid (Todeschini et al., 2000).
Based on our results, 16:0 SM (Figure 5a) is composed of a PC headgroup, a sphingosine moiety of 18 carbon atoms, and an N-linked FA chain of 16 saturated carbon atoms. Its polar surface area is 118 Å2, representing 10% of the total van der Waals surface area (1173 Å2). The topological radius of 16:0 SM is 17 Å, whereas the topological diameter is 33 Å. We found that 24:0 SM (Figure 5b) contains a PC headgroup, a sphingosine moiety of 18 carbon atoms, and an N-linked FA chain of 24 saturated carbon atoms. Its polar surface area is also 118 Å2, representing 8.5% of the total van der Waals surface area (1384 Å2) of the lipid. The topological radius of 24:0 SM is 21 Å, whereas the topological diameter is 41 Å. CHO (Figure 5c) is composed of a steroid nucleus with a polar hydroxyl group at C3 and a lipophilic side chain. Its polar surface area is 20 Å2, representing 3% of the total van der Waals surface area (607 Å2) of the lipid. The topological radius of CHO is 8 Å, whereas its topological diameter is 15 Å.
FIGURE 5:
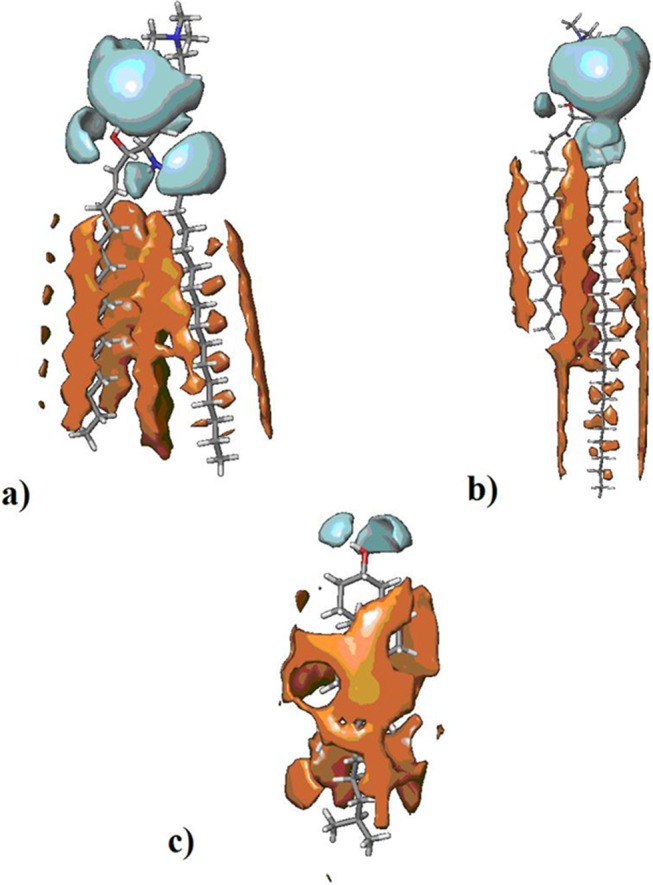
Models of 16:0 SM, 24:0 SM, and CHO. (a) Polar surface area (cyan) and lipophilic surface area (orange) of 16:0 SM. (b) Polar surface area (cyan) and lipophilic surface area (orange) of 24:0 SM. (c) Polar surface area (cyan) and lipophilic surface area (orange) of CHO.
Membrane thickness values are reported in the literature for 18:0 SM (46–47 Å) and 24:0 SM (52–56 Å) (Maulik and Shipley, 1995, 1996). Because membrane thickness is related to lipid chain length, we compared the topological diameter of lipids taken as a value of lipid chain length to infer the thickness of lipid rafts formed by 16:0 SM and 24:0 SM. This approximation resulted in a range of thickness values of 43–44 Å for the membrane composed of 16:0 SM and of 52–56 Å for the membrane composed of 24:0 SM in combination with CHO.
Signal proteins and VDR located in NLRs are different between hepatocytes and hepatoma cells
We have tested the possibility that the changes of SM species between H NMs and H35 NMs might be associated with variations of functional protein content. Raf1, PKCζ, and VDR expression have been analyzed by immunoblotting with specific antibodies in whole cells and in NLMs. Both samples showed immunoreactivity in correspondence to the bands with apparent molecular weights corresponding to 80 kDa (Raf1), 78 kDa (PKCζ), and 50 kDa (VDR) (Figure 6a). In whole cells, the band density increased 1.15 and 1.89 times for Raf1 and PKCζ, respectively, and decreased 1.92 times for VDR in cancer cells in comparison with normal cells. An increase of band density of 1.69 and 2.39 times for Raf1 and PKCζ, respectively, and a decrease of 2.87 times for VDR appeared in H35 NLMs in comparison with H NLMs (Figure 6b).
FIGURE 6:
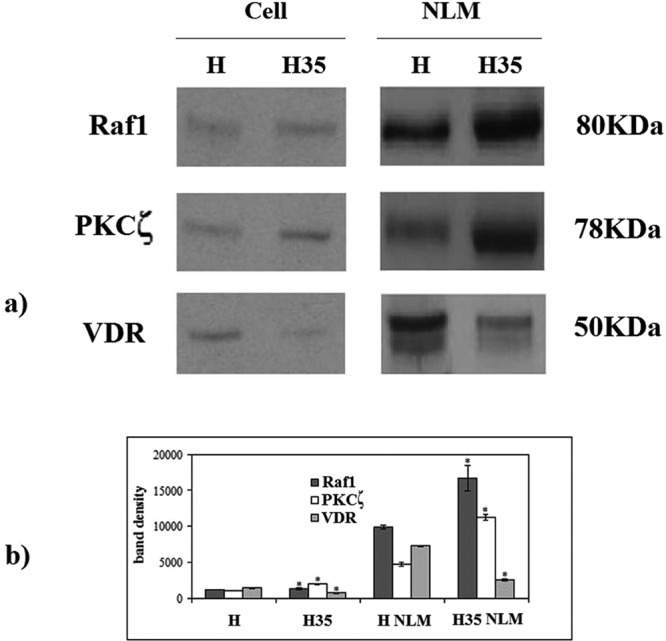
Raf1, PKCζ, and vitamin D3 receptor (VDR) in whole cells and in NLM purified from hepatocytes and hepatoma cells. (a) Immunoblot of proteins probed with specific antibodies, The position of proteins is indicated in relation to the position of molecular size standards. (b) The area density evaluated by densitometry scanning and analysis with Scion Image; the data represent the mean ± SD of three separate experiments. H, hepatocyte; H35, hepatoma cell line. *, p < 0.001 vs. H.
DISCUSSION
Considerable evidence suggests the implication of sphingolipids in cancer (Adan-Gokbulut et al., 2013). Nowadays, researchers focus attention on the different species of sphingolipid molecules containing long and very long FAs. In has been demonstrated that cancer cells incorporate and remodel exogenous 16:0 into structural and oncogenic glycerophospholipids, sphingolipids, and ether lipids (Louie et al., 2013). In addition, supplementation of culture medium with 16:0 modifies the FA composition of Reuber H35 hepatoma cells (Martínez-Cayuela et al., 2000). The shift of sphingolipid composition from very long FAs (24:0) to long FAs (16:0) changes cell function (Sassa et al., 2012).
We have demonstrated that NLMs act as a platform for duplication and transcription of active Chr (Cascianelli et al., 2008; Albi et al., 2013) and VDR (Bartoccini et al., 2011), and as a site for dexamethasone regulation of gene expression (Cataldi et al., 2014).
In this study, we demonstrated for the first time that, in NLMs of cancer cells, the FAs of SM change from very long FAs (24:0 SM) to long FAs (16:0 SM) in comparison with normal cells. Taking into account the calculated properties of 16:0 SM and 24:0 SM (van der Waals surface area and topological radius), it is possible to envisage that 24:0 SM is stronger than 16:0 SM in forming lipid microdomains with CHO. This hypothesis is supported by the larger van der Waals surface area of 24:0, which would allow a better accommodation of CHO underneath the polar headgroup of SM. It has been reported that SM forms bilayers with different thicknesses depending on the size of its FA chain, such as 46–47 Å for 18:0 SM and 52–56 Å for 24:0 SM (Maulik and Shipley, 1995, 1996). Comparing the topological diameters of 16:0 (33 Å), 18:0 (35 Å), 20:0 (37 Å), and 24:0 (41 Å) SM, the thickness of lipid rafts formed by 16:0 and 24:0 SM in combination with CHO can be in a range of 43–44 Å and 52–56 Å, respectively. In addition, our results indicate that H35 NLMs are enriched in unsaturated FAs, which is known to increase the area of a lipid and, consequently, membrane fluidity (Ziegelhöffer et al., 2012). Rapidly growing evidence reinforces the notion that lipid rafts in membranes participate in the recruitment of proteins and lipid signaling molecules (Brown and London, 2000; Róg and Vattulainen, 2014). Therefore, considering our results, we hypothesize that the narrower thickness of NLMs composed of 16:0 SM and the increase of unsaturated FAs might determine enhanced dynamic properties of the NLRs in hepatoma cells, with an increased shuttling of protein molecules. We demonstrate here, in H35 NLMs, an increase of proteins involved in hepatocarcinogenesis, such as STAT3 (Hung et al., 2014), Raf1 (Gauthier and Mitchell, 2013), and PKCζ (Desbois-Mouthon et al., 2002) in spite of the reduction of VDR, probably for the reduction of vitamin D3, which inhibits hepatocellular carcinoma development (Guo et al., 2013). The changes are higher in NLMs than in whole cells, supporting the idea that the modifications of NLMs in cancer cells might influence the content of functional proteins in these microdomains that act as platform for active Chr (Cascianelli et al., 2008).
In conclusion, we show changes in SM and functional proteins of NLMs in cancer cells. This underlines the importance of focusing attention on NLMs instead of the global NM when cancer cells are studied.
MATERIALS AND METHODS
Animals and cells
Thirty-day-old Sprague Dawley rats of either sex (Harlan Nossan, Milan, Italy) kept at normal light–dark periods were used. They had free access to pelleted food and water before being killed between 9 and 10 a.m. All treatments were made according to the international regulations of the National Institutes of Health. H35 hepatoma cells were obtained from the European Collection of Animal Cell Cultures (Salisbury, UK).
Materials
DMEM, bovine serum albumin (BSA), dithiothreitol (DTT), fetal bovine serum (FBS), phenylmethylsulfonylfluoride (PMSF), methanol, 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide, 2-propanol, metyl-tert-butyl ether, formic acid, chloroform, and CHO were obtained from Sigma-Aldrich (St. Louis, MO); lipid standards 16:0 SM, 18:1 SM, 24:0 SM, 16:0 18:1 PC, 16:0 24:0 PC, 18:1 18:0 PC, 16:0 ceramide, 20:0 ceramide, and 24:0 ceramide were purchased from Avanti (Avanti Polar, Alabaster, AL); anti-giantin, anti-STAT3, anti-Raf1, anti-PKCζ, and anti-VDR were obtained from Santa Cruz Biotechnology (Santa Cruz, CA); anti-lamin B was obtained from Oncogene (Boston, MA)
Rat liver
Rat liver was homogenized in 10 mM Tris-HCl buffer (pH 7.4) containing 0.25 M sucrose, 1 mM EDTA, 0.1% ethanol, 0.1 M PMSF, and 0.2 M DTT by using the Thomas homogenizer; the homogenate was filtered through two layers of surgical gauze and was used for hepatocyte nuclei isolation.
Cell culture
H35 hepatoma cells were seeded in 25 cm2 flasks and were grown in monolayer in DMEM enriched with 10% FBS, 2 mM of l-glutamine, 100 IU/ml of penicillin, 100 μg/ml of streptomycin, and 250 μg/ml of amphotericin B. Cells were maintained at 37°C in a saturating humidity atmosphere containing 95% air, 5% CO2 and were used for hepatoma nuclei isolation.
Hepatocyte and hepatoma cell nuclei isolation
H nuclei were isolated from liver homogenate according to Bresnick et al. (1967) in the presence of 1 mM PMSF, as previously described (Albi et al., 1994). Briefly, liver homogenate was centrifuged at 700 × g for 10 min at 4°C. The procedure was repeated twice, and the final pellet was resuspended in 2.4 M sucrose containing 1 mM MgCl2; this was followed by centrifugation at 50,000 × g for 60 min at 4°C. The pellet was washed with 0.25 M sucrose containing 1 mM MgCl2 and centrifuged at 2000 × g for 10 min. This method yielded a homogeneous population of hepatocyte nuclei with no contamination from other types of nuclei (Albi et al., 1994).
H35 nuclei were isolated as previously reported (Albi et al., 2005). Briefly, the homogenized cells were treated with 1% Triton X-100 in hypotonic buffer (0.5:1 vol/vol); the cellular suspension was stirred on a vortex mixer for 30 s; the buffer containing 1.5 M sucrose was added (0.25:1 vol/vol), and the solution was centrifuged at 2000 × g for 10 min.
The H and H35 cell nuclei were then washed twice with Barnes solution (0.085 M KCl, 0.0085 NaCl, 0.0025 M MgCl2, 0.005 M trichloroacetic acid [TRA]-HCl), as previously reported (Rossi et al., 2007). This treatment, during which the nuclei were sedimented at 2000 × g, removed mitochondrial and microsomal contaminations. The nuclei were checked for possible mitochondria and microsome contamination by evaluating the activity of a microsomal marker (NADH–cytochrome c reductase) and glucose-6-phosphatase, as previously reported (Albi et al., 2005).
Purification of NM
NMs were purified from H and H35 isolated nuclei as previously reported (Albi et al., 1997, 1999).
Purification of Chr
Chr was purified from H and H35 isolated nuclei as previously reported (Albi et al., 2003b).
Purification of NMx
NMx was purified from H and H35 isolated nuclei as previously reported (Albi et al., 2003a).
Purification of NLMs
NLMs were purified from H and H35 cell nuclei from normal and hepatectomized rats (Cascianelli et al., 2008; Bartoccini et al., 2011). The extraction was carried out with Triton X-100 dissolved in distilled water (10% vol/vol) on ice. This solution was added to the purified nuclei to a final detergent concentration of 1% (vol/vol). The extract was placed in a cushion of 80% sucrose with a gradient of 15–40% sucrose on top. After overnight centrifugation, floating fractions were carefully collected with a pipette, diluted five times with 25 mM HEPES-HCl, 150 mM NaCl (pH 7.1), and centrifuged at 100,000 × g for 120 min to obtain the pellet containing rafts. For testing the purity of NLRs, the absence of giantin, as a marker protein for Golgi membrane (Satoh et al., 2005), and the presence of STAT3, as a specific NLR marker (Cascianelli et al., 2008), were evaluated by immunoblot analysis.
Lipid extraction
Lipid extraction was performed according to Matyash et al. (2008) as reported by Lazzarini et al. (2014) with modifications. The pellets of H and H35 NLRs were suspended in Tris 10 mM, pH 7.4, and diluted with 1 ml methanol. Three milliliters ultra pure water and 3 ml MTBE were added. Each sample was vortexed for 1 min and centrifuged at 3000 × g for 5 min. The supernatant was recovered. The extraction with MTBE was repeated on the pellet and the supernatant was added to the first. The organic phase was dried under nitrogen flow and resuspended in 500 μl of methanol.
Ultrafast liquid chromatography–tandem mass spectrometry
The 16:0 SM, 18:1 SM, 24:0 SM, 16:0 18:1 PC, 16:0 24:0 PC, 18:1 18:0 PC, 16:0 ceramide, 20:0 ceramide, and 24:0 ceramide standards were prepared according to Matyash et al. (2008). Standards were dissolved in chloroform/methanol (9:1 vol/vol) at 10 μg/ml final concentration. The stock solutions were stored at −20°C. Working calibrators were prepared by diluting stock solutions with methanol to 500:0, 250:0, 100:0, and 50:0 ng/ml final concentrations. Twenty microliters of standards or lipids extracted from serum was injected after purification with specific nylon filters (0.2 μm)
Analyses were carried out according to Rabagny et al. (2011) by using the Ultra Performance Liquid Chromatography system tandem mass spectrometer (Applied Biosystems, Italy). The lipid species were separated, identified, and analyzed as previously reported (Garcia-Gil et al., 2014). The samples were separated on a Phenomenex Kinetex phenyl-hexyl 100 A column (50 × 4.60-mm diameter, 2.6-μm particle diameter) with a precolumn security guard Phenomenex ULTRA phenyl-hexyl 4.6. For SM, column temperature was set at 50°C and flow rate at 0.9 ml/min. Solvent A was 1% formic acid; solvent B was 100% isopropanol containing 0.1% formic acid. The run was performed for 3 min in 50% solvent B and then in a gradient to reach 100% solvent B in 5 min. The system needed to be reconditioned for 5 min with 50% solvent B before the next injection. The SM species were identified by using positive turbo-ion spray and modality multipole-reaction monitoring. The identification and analysis of CHO was conducted by atmospheric pressure chemical ionization in positive ionization conditions and multipole-ion scan modality.
Protein content
Total protein concentration was determined spectrophotometrically at 750 nm by using bovine BSA as a standard, as previously reported (Albi et al., 2008).
Electrophoresis and Western blot analysis
Thirty micrograms of protein from H and H35 NLRs was submitted to SDS–PAGE electrophoresis in an 8% polyacrylamide slab gel for giantin detection and a 10% gel for STAT3, Raf1, PKCζ, VDR, and lamin B according to Laemmli (1970). For the electrophoresis image analysis, the gel was stained with Coomassie blue. The transfer of protein was carried out onto nitrocellulose in 90 min according to Towbin et al. (1979). The membranes were blocked for 30 min with 5% nonfat dry milk in PBS (pH 7.5) and incubated overnight at 4°C with specific antibodies. The blots were treated with horseradish-conjugated secondary antibodies for 90 min. Visualization was performed with the enhanced chemiluminescence kit from Amersham Pharmacia Biotech (Rainham, Essex, UK).
Properties of 16:0 SM, 24:0 SM, and CHO
Three-dimensional chemical structures of 16:0 SM, 24:0 SM, and CHO were generated using Maestro v9.5 (Schrödinger, NY). Geometries were optimized using semiempirical calculations with AM1 method and RHF wavefunction. Surface and geometrical properties, including polar surface area (Å2), total van der Waals surface area (Å2), topological radius (Å), and topological diameter (Å), were calculated using Canvas v1.7 (Schrödinger, NY).
Statistical analysis
Data are expressed as mean ± SD, and a t test was used for statistical analysis.
Abbreviations used:
- BSA
bovine serum albumin
- CHO
cholesterol
- Chr
chromatin
- DTT
dithiothreitol
- FBS
fetal bovine serum
- H
hepatocyte
- H35
H35 hepatoma cells
- LCFA
long-chain fatty acid
- NLM
nuclear lipid microdomain
- NLR
nuclear lipid rafts
- NM
nuclear membrane
- NMx
nuclear matrix
- PC
phosphatidylcholine
- PKCζ
protein kinase Cζ
- PMSF
phenylmethylsulfonylfluoride
- SM
sphingomyelin
- STAT-3
signal transducer and activator of transcription 3
- VDR
vitamin D receptor
- VLCFA
very-long-chain fatty acids.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-02-0071) on May 13, 2015.
REFERENCES
- Adan-Gokbulut A, Kartal-Yandim M, Iskender G, Baran Y. Novel agents targeting bioactive sphingolipids for the treatment of cancer. Curr Med Chem. 2013;20:108–122. [PubMed] [Google Scholar]
- Aittoniemi J, Niemela PS, Hyvonen MT, Karttunen M, Vattulainen I. Insight into the putative specific interactions between cholesterol, sphingomyelin, and palmitoyl-oleoyl phosphatidylcholine. Biophys J. 2007;92:1125–1137. doi: 10.1529/biophysj.106.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albi E, Cataldi S, Rossi G, Magni MV. A possible role of cholesterol-sphingomyelin/phosphatidylcholine in nuclear matrix during rat liver regeneration. J Hepatol. 2003a;38:623–628. doi: 10.1016/s0168-8278(03)00074-6. [DOI] [PubMed] [Google Scholar]
- Albi E, La Porta CA, Cataldi S, Magni MV. Nuclear sphingomyelin-synthase and protein kinase C delta in melanoma cells. Arch Biochem Biophys. 2005;438:156–161. doi: 10.1016/j.abb.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Albi E, Lazzarini A, Lazzarini R, Floridi A, Damaskopoulou E, Curcio F, Cataldi S. Nuclear lipid microdomain as place of interaction between sphingomyelin and DNA during liver regeneration. Int J Mol Sci. 2013;14:6529–6541. doi: 10.3390/ijms14046529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albi E, Lazzarini R, Viola Magni M. Phosphatidylcholine/sphingomyelin metabolism crosstalk inside the nucleus. Biochem J. 2008;410:381–389. doi: 10.1042/BJ20070758. [DOI] [PubMed] [Google Scholar]
- Albi E, Mersel M, Leray C, Tomassoni ML, Viola-Magni MP. Rat liver chromatin phospholipids. Lipids. 1994;29:715–719. doi: 10.1007/BF02538916. [DOI] [PubMed] [Google Scholar]
- Albi E, Peloso I, Magni MV. Nuclear membrane sphingomyelin-cholesterol changes in rat liver after hepatectomy. Biochem Biophys Res Commun. 1999;262:692–695. doi: 10.1006/bbrc.1999.1188. [DOI] [PubMed] [Google Scholar]
- Albi E, Pieroni S, Viola Magni MP, Sartori C. Chromatin sphingomyelin changes in cell proliferation and/or apoptosis induced by ciprofibrate. J Cell Physiol. 2003b;196:354–361. doi: 10.1002/jcp.10314. [DOI] [PubMed] [Google Scholar]
- Albi E, Tomassoni ML, Viola-Magni M. Effect of lipid composition on rat liver nuclear membrane fluidity. Cell Biochem Funct. 1997;15:181–190. doi: 10.1002/(SICI)1099-0844(199709)15:3<181::AID-CBF737>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Albi E, Villani M. Nuclear lipid microdomains regulate cell function. Commun Integr Biol. 2009;2:23–24. doi: 10.4161/cib.2.1.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoccini E, Marini F, Damaskopoulou E, Lazzarini R, Cataldi S, Cascianelli G, Garcia-Gil M, Albi E. Nuclear lipid microdomains regulate nuclear vitamin D3 uptake and influence embryonic hippocampal cell differentiation. Mol Biol Cell. 2011;22:3022–3031. doi: 10.1091/mbc.E11-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E, Lanclos K, Sage J, Schwartz A, Yawn DH, Bush H, Hunuma T. Isolation and ribonucleic acid synthesis in nuclei of rat fetal liver. Exp Cell Res. 1967;46:396–411. doi: 10.1016/0014-4827(67)90076-6. [DOI] [PubMed] [Google Scholar]
- Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiol. 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- Cascianelli G, Villani M, Tosti M, Marini F, Bartoccini E, Magni MV, Albi E. Lipid microdomains in cell nucleus. Mol Biol Cell. 2008;19:5289–5295. doi: 10.1091/mbc.E08-05-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldi S, Codini M, Cascianelli G, Tringali S, Tringali AR, Lazzarini A, Floridi A, Bartoccini E, Garcia-Gil M, Lazzarini R, et al. Nuclear lipid microdomain as resting place of dexamethasone to impair cell proliferation. Int J Mol Sci. 2014;15:9832–9846. doi: 10.3390/ijms151119832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois-Mouthon C, Blivet-Van Eggelpoël MJ, Beurel E, Boissan M, Delélo R, Cadoret A, Capeau J. Dysregulation of glycogen synthase kinase-3β signaling in hepatocellular carcinoma cells. Hepatology. 2002;36:1528–1536. doi: 10.1053/jhep.2002.37192. [DOI] [PubMed] [Google Scholar]
- Frisz JF, Klitzing HA, Lou K, Hutcheon ID, Weber PK, Zimmerberg J, Kraft ML. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J Biol Chem. 2013;288:16855–16861. doi: 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gil M, Lazzarini A, Lazzarini R, Floridi E, Cataldi S, Floridi A, Albi E. Serum deprivation alters lipid profile in HN9.10e embryonic hippocampal cells. Neurosci Lett. 2014;589:83–87. doi: 10.1016/j.neulet.2014.12.059. [DOI] [PubMed] [Google Scholar]
- Gauthier A, Mitchell H. The role of sorafenib in the treatment of advanced hepatocellular carcinoma: an update. Hepatol Res. 2013;43:147–154. doi: 10.1111/j.1872-034X.2012.01113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Ma Z, Ma Q, Wu Z, Fan P, Zhou X, Chen L, Zhou S, Goltzman D, Miao D, Wu E. 1, 25(OH)2D3 inhibits hepatocellular carcinoma development through reducing secretion of inflammatory cytokines from immunocytes. Curr Med Chem. 2013;20:4131–4141. doi: 10.2174/09298673113209990248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holopainen JM, Metso AJ, Mattila JP, Jutila A, Kinnunen PK. Evidence for the lack of a specific interaction between cholesterol and sphingomyelin. Biophys J. 2004;86:1510–1520. doi: 10.1016/S0006-3495(04)74219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honigmann A, Mueller V, Ta H, Schoenle A, Sezgin E, Hell SW, Eggeling C. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nat Commun. 2014;5:5412. doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- Hung MH, Tai WT, Shiau CW, Chen KF. Downregulation of signal transducer and activator of transcription 3 by sorafenib: a novel mechanism for hepatocellular carcinoma therapy. World J Gastroenterol. 2014;20:15269–15274. doi: 10.3748/wjg.v20.i41.15269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structure proteins during the assembly of bacteriophage T4. Nature. 1970;227:680–683. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazzarini A, Floridi A, Pugliese L, Villani M, Cataldi S, Codini M, Lazzarini R, Beccari T, Ambesi-Impiombato FS, Curcio F, Albi E. Analysis of serum sphingomyelin species by UFLC-MS/MS in patients affected with monoclonal gammopathy. J Chromat Separation Techniq. 2014;5:1000239. [Google Scholar]
- Levin IW, Thompson TE, Barenholz Y, Huang C. Two types of hydrocarbon chain interdigitation in sphingomyelin bilayers. Biochemistry. 1985;24:6282–6286. doi: 10.1021/bi00343a036. [DOI] [PubMed] [Google Scholar]
- Li XM, Momsen MM, Smaby JM, Brockman HL, Brown RE. Cholesterol decreases the interfacial elasticity and detergent solubility of sphingomyelins. Biochemistry. 2001;40:5954–5963. doi: 10.1021/bi002791n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie SM, Roberts LS, Mulvihill MM, Luo K, Nomura DK. Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochim Biophys Acta. 2013;1831:1566–1572. doi: 10.1016/j.bbalip.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cayuela M, García-Pelayo MC, Linares A, García-Peregrín E. Metabolism of palmitic and docosahexaenoic acids in Reuber H35 hepatoma cells. J Biochem. 2000;128:545–551. doi: 10.1093/oxfordjournals.jbchem.a022786. [DOI] [PubMed] [Google Scholar]
- Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulik PR, Shipley GG. X-ray diffraction and calorimetric study of N-lignoceryl sphingomyelin membranes. Biophys J. 1995;69:1909–1916. doi: 10.1016/S0006-3495(95)80061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulik PR, Shipley GG. Interactions of N-stearoyl sphingomyelin with cholesterol and dipalmitoylphosphatidylcholine in bilayer membranes. Biophys J. 1996;70:2256–2265. doi: 10.1016/S0006-3495(96)79791-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinedo F, Gajate C. Lipid rafts as major platforms for signaling regulation in cancer. Adv Biol Regul. 2015;57:130–146. doi: 10.1016/j.jbior.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Ohno Y, Suto S, Yamanaka M, Mizutani Y, Mitsutake S, Igarashi Y, Sassa T, Kihara A. ELOVL1 production of C24 acyl-CoAs is linked to C24 sphingolipid synthesis. Proc Natl Acad Sci USA. 2010;107:18439–18444. doi: 10.1073/pnas.1005572107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabagny Y, Herrmann W, Geisel J, Kirsch SH, Obeid R. Quantification of plasma phospholipids by ultra performance liquid chromatography tandem mass spectrometry. Anal Bioanal Chem. 2011;401:891–899. doi: 10.1007/s00216-011-5154-5. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Anderson TG, McConnell HM. Condensed complexes, rafts, and the chemical activity of cholesterol in membranes. Proc Natl Acad Sci USA. 2000;97:12422–12427. doi: 10.1073/pnas.220418097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róg T, Pasenkiewicz-Gierula M. Cholesterol–sphingomyelin interactions: a molecular dynamics simulation study. Biophys J. 2006;91:3756–3767. doi: 10.1529/biophysj.106.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Róg T, Vattulainen I. Cholesterol, sphingolipids, and glycolipids: what do we know about their role in raft-like membranes. Chem Phys Lipids. 2014;184:82–104. doi: 10.1016/j.chemphyslip.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Rossi G, Magni MV, Albi E. Sphingomyelin-cholesterol and double stranded RNA relationship in the intranuclear complex. Arch Biochem Biophys. 2007;459:27–32. doi: 10.1016/j.abb.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Sassa T, Suto S, Okayasu Y, Kihara A. A shift in sphingolipid composition from C24 to C16 increases susceptibility to apoptosis in HeLa cells. Biochim Biophys Acta. 2012;1821:1031–1037. doi: 10.1016/j.bbalip.2012.04.008. [DOI] [PubMed] [Google Scholar]
- Satoh A, Beard M, Warren G. Preparation and characterization of recombinant golgin tethers. Methods Enzymol. 2005;404:279–296. doi: 10.1016/S0076-6879(05)04026-7. [DOI] [PubMed] [Google Scholar]
- Slotte JP. Molecular properties of various structurally defined sphingomyelins—correlation of structure with function. Prog Lipid Res. 2013a;52:206–219. doi: 10.1016/j.plipres.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res. 2013b;52:424–437. doi: 10.1016/j.plipres.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Soupene E, Kuypers FA. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med. 2008;233:507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Okazaki T. The role of sphingomyelin and sphingomyelin synthases in cell death, proliferation and migration—from cell and animal models to human disorders. Biochim Biophys Acta. 2014;1841:692–703. doi: 10.1016/j.bbalip.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Todeschini R, Consonni V. Handbook of Molecular Descriptors, Methods and Principles in Medicinal Chemistry 11. Weinheim, Germany: Wiley-VCH; 2000. pp. 366–510. [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ji J, Zhu XY, Wu YY, Yu H, Zhang B, Li XL, Sun XZ. Palmitic acid induces apoptosis in human hepatoma cell line, HepG2 cells. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2004;26:671–676. [PubMed] [Google Scholar]
- Ziegelhöffer A, Waczulíková I, Ferko M, Šikurová L, Mujkošová J, Ravingerová T. Involvement of membrane fluidity in endogenous protective processes running on subcellular membrane systems of the rat heart. Physiol Res 61 (Suppl 2) 2012:S11–S21. doi: 10.33549/physiolres.932361. [DOI] [PubMed] [Google Scholar]