Previous work identified a single nonmuscle tropomyosin isoform, but genetics suggests the existence of additional tropomyosins essential for proper development. Three nonmuscle tropomyosins in Drosophila are identified, and one of the newly identified isoforms plays an unexpected role in chromosome segregation.
Abstract
Most eukaryotic cells express multiple isoforms of the actin-binding protein tropomyosin that help construct a variety of cytoskeletal networks. Only one nonmuscle tropomyosin (Tm1A) has previously been described in Drosophila, but developmental defects caused by insertion of P-elements near tropomyosin genes imply the existence of additional, nonmuscle isoforms. Using biochemical and molecular genetic approaches, we identified three tropomyosins expressed in Drosophila S2 cells: Tm1A, Tm1J, and Tm2A. The Tm1A isoform localizes to the cell cortex, lamellar actin networks, and the cleavage furrow of dividing cells—always together with myosin-II. Isoforms Tm1J and Tm2A colocalize around the Golgi apparatus with the formin-family protein Diaphanous, and loss of either isoform perturbs cell cycle progression. During mitosis, Tm1J localizes to the mitotic spindle, where it promotes chromosome segregation. Using chimeras, we identified the determinants of tropomyosin localization near the C-terminus. This work 1) identifies and characterizes previously unknown nonmuscle tropomyosins in Drosophila, 2) reveals a function for tropomyosin in the mitotic spindle, and 3) uncovers sequence elements that specify isoform-specific localizations and functions of tropomyosin.
INTRODUCTION
Eukaryotic cells construct and maintain multiple actin filament networks with different functions: cortical networks that provide mechanical rigidity; lamellipodial networks that drive plasma membrane protrusion; stress fibers that contribute to adhesion; and a contractile ring that divides one cell into two (Michelot and Drubin, 2011). How are these diverse networks, each with a different architecture and a different set of accessory factors, constructed from the same basic building material: the same actin filaments? One answer to this question may be provided by the tropomyosins, a large family of actin-binding proteins that can promote or block the interaction of actin filaments with a variety of regulatory proteins (Gunning et al., 2005).
Tropomyosins are evolutionally conserved, α-helical, coiled-coil proteins that bind along the sides of actin filaments in a highly cooperative manner and participate in a variety of cellular processes. In muscle cells, tropomyosin collaborates with the troponin complex to regulate interaction of “thick” myosin filaments with “thin” actin filaments (Gunning et al., 2008). In nonmuscle cells, tropomyosin isoforms help regulate many processes, including actin filament nucleation by formins and the Arp2/3 complex (Blanchoin et al., 2001), filament severing by cofilin (Ono and Ono, 2002), and force generation by myosin motors (Clayton et al., 2010).
Mammalian genomes contain four tropomyosin genes, which, with alternative splicing, generate more than 40 tropomyosin isoforms, all expressed at different levels in different tissues and developmental stages (Gunning et al., 2008). Individual mammalian cells express as many as seven tropomyosin isoforms at once, and some cellular processes require the participation of multiple, functionally distinct tropomyosins (Tojkander et al., 2011).
In contrast to vertebrates, almost nothing is known about the diversity of insect tropomyosins. The Drosophila genome contains two tropomyosin genes—Tm1 and Tm2—and perturbation of these genes produces defects in several developmental processes, including oskar mRNA localization, head morphogenesis, neuronal dendritic field specification, and border cell migration (Erdélyi et al., 1995; Tetzlaff et al., 1996; Li and Gao, 2003; Kim et al., 2011). Instabilities in the nomenclature and annotation of Drosophila tropomyosin genes, however, have made the literature difficult to follow. The Tm1 gene, for example, has variously been called TmII, cTm, mTmII, and TnH. Similarly, Tm2 has been called TmI, mTmI, and Ifm(3)3 (FlyBase.org). Remarkably, even though splicing predictions indicate the potential for expression of 18 tropomyosins in Drosophila, only one has thus far been identified in nonmuscle cells (Hanke et al., 1987). This protein, called cTm or Tm1A, is a product of the Tm1 gene and is generally regarded as the only nonmuscle tropomyosin in Drosophila. Results from several studies, however, strongly hint at the existence of additional, cryptic tropomyosin isoforms with important biological functions. Until now, however, the identities and properties of these cryptic tropomyosins have been a mystery.
Using a combination of molecular genetic, biochemical, and cell biological techniques, we identified three tropomyosin isoforms in Drosophila S2 cells: two encoded by the Tm1 gene (Tm1A and Tm1J) and one encoded by Tm2 (Tm2A). We found that each tropomyosin localizes to a different intracellular structure, together with a different set of actin-binding proteins. In interphase, Tm1A colocalizes with myosin-II to contractile networks (cortex and lamellum/convergence zone), whereas Tm1J and Tm2A colocalize with Diaphanous to the Golgi apparatus. During mitosis, Tm1A precedes myosin-II to the cleavage furrow, whereas Tm1J localizes to the mitotic spindle and plays a role in maintaining fidelity of chromosome segregation. In addition to identifying cryptic tropomyosins, our work reveals Drosophila to be an excellent model system with which to study the functional diversity of tropomyosin isoforms.
RESULTS
Drosophila S2 cells express three tropomyosin isoforms
Using three different anti-tropomyosin antibodies (E-17, ab11190, and TM311), we detected at least one “short” and one “long” tropomyosin in S2 cell lysate (Figure 1A; Schevzov et al., 2011). The electrophoretic mobility of the smaller species corresponds to the predicted size of Tm1A (32 kDa), a “short” tropomyosin isoform previously identified in Drosophila nonmuscle cells (Hanke and Storti, 1986). The larger species migrates with an apparent molecular weight of 38 kDa (Figure 1A).
FIGURE 1:
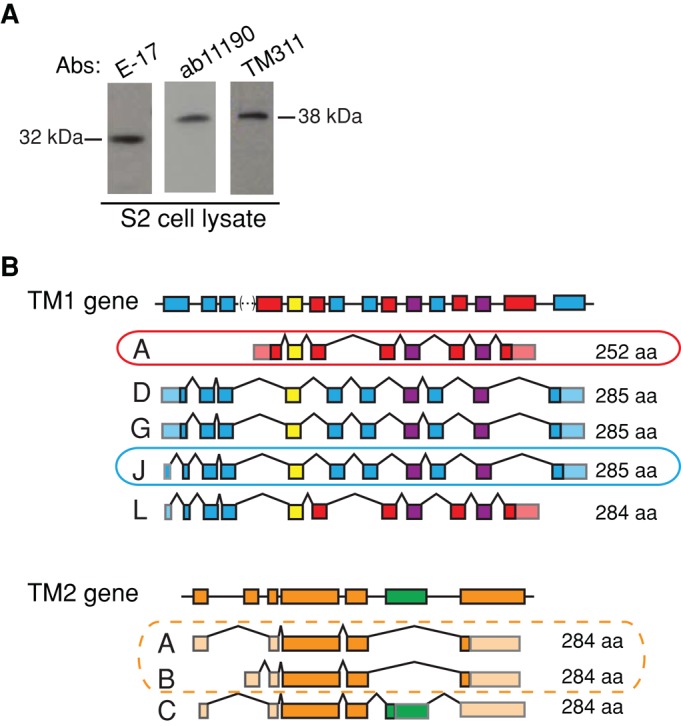
Drosophila S2 cells express three tropomyosin isoforms: Tm1A, Tm1J, and Tm2A/B. (A) Western blots show that at least two tropomyosin isoforms are present in S2 cell lysates: one 32 kDa in size and one 38 kDa in size. Three commercial antibodies were used: E-17 (goat; Santa Cruz Biotechnology), ab11190 (rabbit; Abcam), and TM311 (mouse; Sigma-Aldrich). (B) Schematic of Drosophila Tm1 and Tm2 gene structures. Predicted splice variants producing tropomyosin isoforms of 32 or 38 kDa in size (i.e., “canonical”) are displayed below and are based on FlyBase (FB2008_09, Dmel Release 5.12). Left, splice variant name. Right, amino acid length. Circled isoforms were confirmed through various methods (details in Supplemental Figure S1).
To identify specific isoforms, we analyzed cDNAs amplified by PCR from S2 cell mRNA using exon-specific primers (Supplemental Table S1) for Drosophila tropomyosin genes Tm1 and Tm2 (Supplemental Figure S1, A and B). From Tm1, we detected the entire coding sequences of two isoforms, which we call Tm1A and Tm1J (Figure 1B and Supplemental Figure S1C). We also amplified a full-length transcript from Tm2, which we call Tm2A/B (Tm2A and Tm2B transcripts encode the same protein but have different 5′ untranslated regions; Figure 1B and Supplemental Figure S1D). We were surprised by this result because Tm2 was previously believed to encode only muscle-specific tropomyosins (Basi et al., 1984).
We confirmed the presence of the Tm1J protein by mass spectrometry of endogenous tropomyosins partially purified from Drosophila S2 cells (see Materials and Methods). We detected peptides common to both Tm1J and Tm1A, as well as five unique peptides present only in Tm1J (Supplemental Figure S1E). Immunoblots using the E-17 antibody (which likely recognizes the C-terminus of Tm1A) indicate that potential isoform Tm1L—a chimera of Tm1J and Tm1A—is probably not expressed in S2 cells (Supplemental Figure S1F). We find no evidence for expression of “noncanonical” tropomyosins of the type found in indirect flight muscles (unpublished data; Kreuz et al., 1996). Taken together, our data argue strongly that Drosophila S2 cells express three cytoplasmic tropomyosins: Tm1A, Tm1J, and Tm2A/B (Figure 1B and Supplemental Table S2).
The three tropomyosins have distinct localizations during interphase
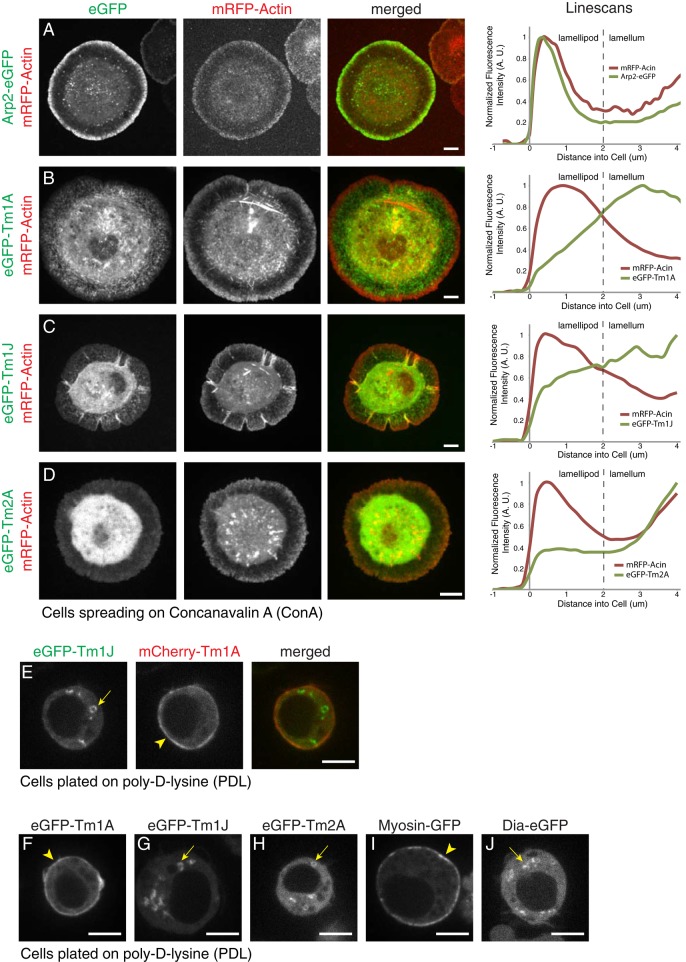
To determine the localization and dynamics of the three tropomyosins, we expressed each isoform in S2 cells as an N-terminal enhanced green fluorescent protein (eGFP) fusion protein under control of a copper-inducible pMT promoter (Invitrogen). We fused GFP to the N-terminus on the basis of previous work demonstrating that N-terminal fusions do not interfere with head-to-tail self-association of tropomyosin (Martin et al., 2010). We imaged the fluorescent fusion proteins in live cells, using spinning-disk confocal microscopy. On surfaces coated with concanavalin A (ConA), Drosophila S2 cells spread rapidly, projecting a flat, lamellar structure across the substrate (Rogers et al., 2003). This flat structure resembles the leading edge of migrating cells and is supported by three overlapping actin networks: a dense and highly branched lamellipodial network adjacent to the leading membrane; a less dense and less branched lamellar network, closer to the cell body; and a convergence zone at the boundary of the cell body, where actin filaments coalesce into bundles (Supplemental Figure S2A; Salmon et al., 2002). Based on comparisons with actin–monomeric red fluorescent protein (mRFP), both Tm1A and Tm1J are more concentrated in lamellar actin networks than with peripheral lamellipodia (Figure 2, B and C, and Supplemental Figure S2C). This contrasts with eGFP-labeled Arp2/3 complex, which localizes preferentially to lamellipodial networks and is excluded from the lamellum (Figure 2A and Supplemental Figure S2B), and agrees with our previous work on Tm1A in S2 cells (Iwasa and Mullins, 2007). Coexpression of GFP-labeled myosin-II and mCherry-Tm1A shows that Tm1A becomes progressively enriched in the lamellum network as it flows toward the cell body, where myosin-II and Tm1A colocalize in the convergence zone (Supplemental Figure S2D). Unlike the other tropomyosins, Tm2A shows no enrichment in the lamellum compared with soluble eGFP (compare Figure 2D and Supplemental Figure S2E). To further verify that our fluorescent tropomyosins incorporate into actin networks, we used latrunculin B to depolymerize filamentous actin in spreading S2 cells and observed relocalization of both eGFP-Tm1J and mCherry-Tm1A (Supplemental Figure S2F).
FIGURE 2:
Tm1A and Tm1J colocalize in spreading cells but do not colocalize in nonspreading cells. (A–D) Live-cell imaging of S2 cells spreading on ConA with mRFP-actin (red), a marker of the lamellipod, and eGFP-tagged proteins (green): Arp2 (A), Tm1A (B), Tm1J (C), and Tm2A (D). Arp2 localizes to the lamellipod with mRFP-actin, whereas Tm1A and Tm1J both localize to the lamellum and are excluded from the lamellipod. Tm2A shows no significant localization. Right, normalized average fluorescence intensity line scans. (E) Coexpression of eGFP-Tm1J (green) and mCherry-Tm1A (red) in S2 cells on PDL show that in the same cell, Tm1A localizes to the cortex, whereas Tm1J localizes to cytoplasmic ring-like structures. (F–J) Live-cell imaging of interphase S2 cells on PDL with eGFP-tagged proteins: Tm1A, Tm1J, Tm2A, myosin-II, and Diaphanous (Dia). Tm1A (F) and myosin-II (I) colocalize to the cortex (arrowheads), whereas Tm1J (G), Tm2A (H), and Dia (J) localize to cytoplasmic ring-like structures (arrows). mCherry-α-tubulin was coexpressed (not shown) to verify that cells were in interphase. See also Supplemental Figure S2.
On surfaces coated with poly-d-lysine (PDL), S2 cells adhere weakly and remain more or less spherical. In these nonspreading S2 cells, Tm1A and Tm1J do not colocalize (Figure 2E). Tm1A concentrates in the cortex at the cell periphery (Figure 2F), whereas both Tm1J and Tm2A form rings scattered through the cytoplasm (Figure 2, G and H). We compared these tropomyosins to other actin-regulatory proteins and found that the motor protein myosin-II (myosin-GFP) colocalizes with Tm1A to the cortex (Figure 2I), whereas the formin-family nucleation factor Diaphanous (Dia-eGFP) localizes to the same cytoplasmic rings (Figure 2J) as Tm1J and Tm2A.
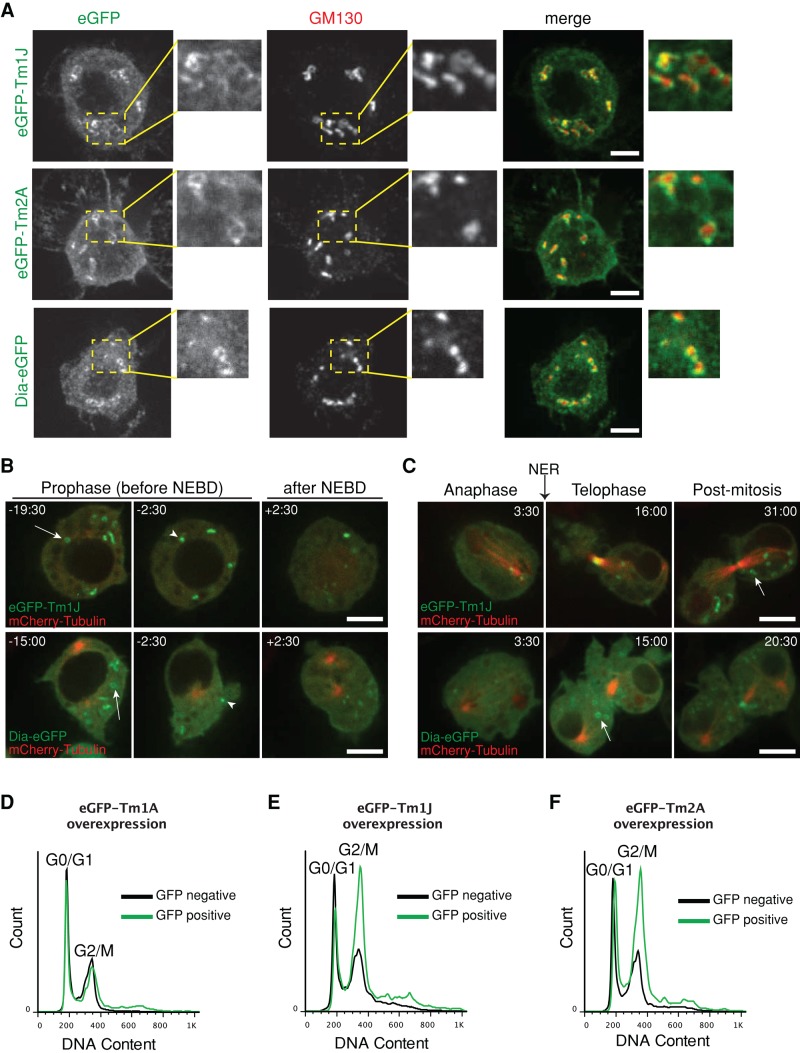
To determine whether cytoplasmic rings of Tm1J, Tm2A, and Dia are associated with organelles, we compared the localization of eGFP-Tm1J with markers for different intracellular compartments, including the endoplasmic reticulum (ER-tracker), mitochondria (Mito-tracker), and Golgi apparatus (Bodipy TR C5-ceramide; Invitrogen). We found that eGFP-Tm1J marginally overlaps with ER-tracker but associates closely with C5-ceramide, a marker of the Golgi apparatus (Supplemental Figure S3A). We confirmed that Tm1J/Tm2A/Dia rings encircle Golgi membranes by immunofluorescence, using an antibody against a resident Golgi protein, GM130 (Figure 3A). Of interest, when purifying endogenous tropomyosins from S2 cells, we found that the long isoforms (Tm1J and Tm2A) but not the short one (Tm1A) require high salt for solubilization (unpublished data), similar to other Golgi-associated proteins (Ivan et al., 2008).
FIGURE 3:
Tm1J, Tm2A, and Diaphanous localize to cytoplasmic rings surrounding the Golgi in a cell cycle–dependent manner. (A) The ring-shaped patterns of tropomyosin isoforms, eGFP-Tm1J and eGFP-Tm2A, and Dia-eGFP encircle the immunofluorescence cis-Golgi marker, GM130 (red). Areas outlined by yellow dashed box in left-hand images are enlarged in the right-hand images. (B) eGFP-Tm1J and Dia-eGFP rings (arrows) localize to the Golgi in late G2 phase but during prophase begin to disassemble (arrowheads) minutes before nuclear envelope breakdown (NEBD) at the onset of mitosis and fully disperse after NEBD. Times indicated are relative to NEBD. See also Supplemental Figure 3 Videos 1 and 2. (C) At the end of mitosis, eGFP-Tm1J and Dia-eGFP rings (arrows) reform at the Golgi during telophase after the nuclear envelope reforms (NER). Times indicated are relative to the onset of anaphase. (D–F) Cell cycle analysis using flow cytometry shows that overexpression of eGFP-tagged Golgi-related TMs Tm1J (E) or Tm2A (F) but not Tm1A (D) causes an increase in the population of cells in G2/M phase. After >24 h of induction at 1 mM CuSO4, S2 cells were fixed and stained with propidium iodide (a DNA marker). Graphs depict histograms of DNA content of GFP-negative (black) and GFP-positive (green) cells (displayed as overlays).
The presence of tropomyosin and Diaphanous suggests the existence of a network of actin filaments surrounding the Golgi apparatus (Figures 2J and 3A). We saw no accumulation of eGFP-actin around the Golgi, but this is not particularly surprising, given that formin-family proteins (like Diaphanous) cannot use GFP-actin as a substrate (Chen et al., 2012). We tested four live-cell actin filament probes in S2 cells and found only one—Utr230-eGFP a truncated actin-binding domain from utrophin (Belin et al., 2013)—that clearly forms rings around the Golgi apparatus (Supplemental Figure S3B). Similarly, Percival et al. (2004) found a vertebrate tropomyosin, Tm5NM-2, localized to Golgi-associated actin filaments that could only be detected by a single, specialized antibody, one that reacts preferentially with filament ends. These results are also consistent with our recent study of commonly used actin probes (Belin et al., 2015), each of which turns out to bind a different subset of actin filaments in live cells.
Golgi-associated Diaphanous and tropomyosin influence cell cycle progression
The architecture of the Golgi apparatus is linked to cell cycle progression (Wang and Seemann, 2011). In mammalian cells, pericentriolar Golgi cisternae become separated in G2 phase and begin to fragment into small tubules and vesicles throughout G2 and into mitosis (Villeneuve et al., 2013). In late anaphase/telophase, the dispersed Golgi vesicles begin to fuse and reassemble into stacks as the nuclear envelope reforms. Of interest, preventing Golgi fragmentation actually blocks entry into mitosis (Rabouille and Kondylis, 2007). In S2 cells, Golgi ministacks are paired by an Abi-Arp2/3 actin network during G1 phase and must be unpaired and dispersed in order for cells to enter mitosis (Kondylis et al., 2007).
To track the fate of Golgi-associated Diaphanous/tropomyosin networks through the cell cycle, we performed time-lapse imaging of eGFP fusions of Dia, Tm1J, and Tm2A in cells coexpressing mCherry-tubulin. In contrast to Abi and Arp2/3, which disappear during S phase, Dia/Tm1J/Tm2A rings remain associated with the Golgi apparatus through G2 and into prophase of mitosis (arrows, Figure 3B and Supplemental Figure S3C). During prophase, however, Dia/Tm1J/Tm2A rings rapidly disassemble (arrowheads, Figure 3B and Supplemental Figure S3C) and are almost fully dissolved by the point of nuclear envelope breakdown (Supplemental Figure 3 Videos 1 and 2). At the end of mitosis, as the nuclear envelope reforms, Dia, Tm1J, and Tm2A all return to the reassembling Golgi (Figure 3C and Supplemental Figure S3D). We observed similar dynamics in cells expressing an eGFP-tagged Golgi-resident protein, fringe (Supplemental Figure S3E and Supplemental Figure 3 Video 1).
The dynamics of Golgi-associated tropomyosins suggests that perhaps the rings must be disassembled before Golgi cisternae can disperse during mitosis. If true, depletion of Golgi-associated tropomyosins might accelerate entry into mitosis, whereas overexpression of these tropomyosins would delay mitotic entry and slow progression through G2. To test this idea, we depleted Tm1J and Tm2A, singly and together, in S2 cells using RNA interference (RNAi) and monitored cell cycle progression by flow cytometry. Surprisingly, depletion of either Tm1J or Tm2A increases the fraction of cells in G1/G0 relative to G2/M cells (Supplemental Figure S3, G and H). Depletion of Tm2A seems to especially increase the percentage of cells in S phase, whereas depleting both proteins produces an additive effect on the G2:G1 ratio (Supplemental Figure S3I). In contrast, depletion of Tm1A has no measurable effect on the G2:G1 ratio (Supplemental Figure S3F).
We next overexpressed eGFP fusions of Golgi-associated tropomyosins Tm1J and Tm2A and looked at the effect on cell cycle progression. We chose growth conditions that allow wild-type S2 cells to pass through mitosis and accumulate in G1 phase (Rizzino and Blumenthal, 1978). Under these conditions, overexpressing eGFP-Tm1A had no effect on cell cycle distribution (Figure 3D), whereas overexpressing either eGFP-Tm1J or eGFP-Tm2A shifted the majority of GFP-positive cells into the G2 phase (Figure 3, E and F). Together these results suggest that Tm1J and Tm2A stabilize a Golgi-associated actin network that helps maintain structural integrity of the Golgi apparatus and must be disassembled before entry into mitosis. The obvious difference between cell cycle distributions of the control populations in knockdown versus overexpression protocols is due to the different growth conditions used in these experiments. The RNAi knockdown experiments required incubating cells for several days with double-stranded RNA in serum-free medium. Overexpression studies required less time, and so we grew S2 cells for approximately one cell cycle under growth conditions chosen to promote synchrony in G1 phase (Rizzino and Blumenthal, 1978).
Tm1A dynamically colocalizes with myosin-II during interphase
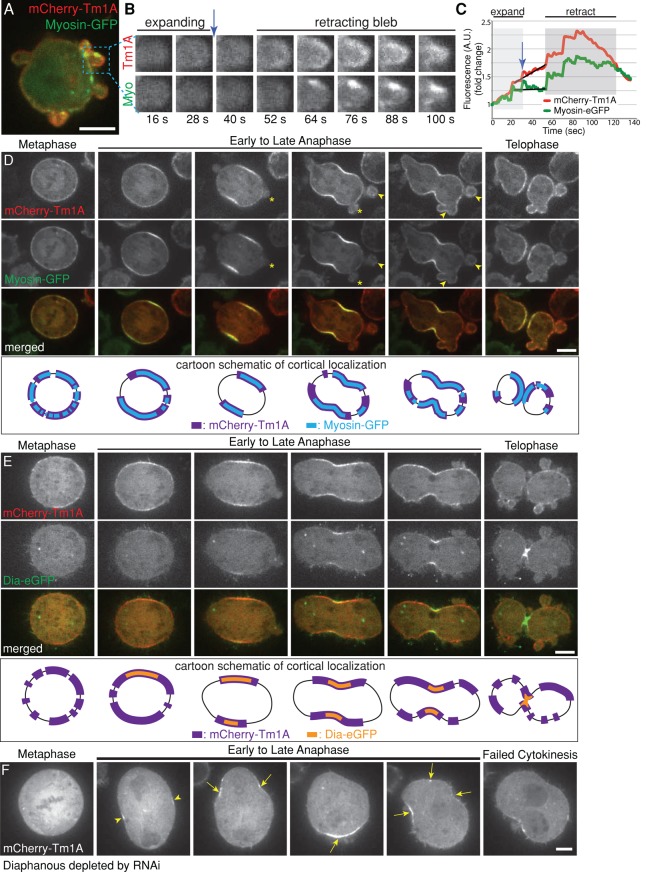
Tropomyosins are known to regulate myosin motors, so we compared the localization of Tm1A with myosin-II throughout the cell cycle. In interphase, mCherry-Tm1A and myosin-GFP colocalize throughout the cortex (Figure 4A and Supplemental Figure 4 Video 1), but, when part of the plasma membrane detaches from the underlying cytoskeleton to form a bleb, both Tm1A and myosin-II are lost. The moment when the bleb stops expanding, however, both Tm1A and myosin-II reappear and become concentrated as the bleb begins to retract (Figure 4A). Whereas mCherry-Tm1A appears all along the edge of the retracting bleb, myosin-GFP appears only in patches (retracting bleb, Figure 4B). Furthermore, Tm1A arrives in the bleb just before myosin-II, just before the bleb begins to retract (blue arrows, Figure 4, B and C). This timing is similar to that observed by Charras et al. (2006), who found that tropomyosin arrives more or less simultaneously with actin, 10 s before myosin-II. These authors did not investigate isoform specificity of bleb-associated tropomyosins, whereas we found that only Tm1A localizes to retracting blebs in Drosophila S2 cells (Supplemental Figure S4, A and B). In agreement with past studies of mDia1 (Charras et al., 2006), we found that Dia-eGFP also fails to localize to retracting blebs (Supplemental Figure S4C). Together the spatial and temporal dynamics of Tm1A and myosin-II suggest that they specifically function together to induce or maintain cortical contractility in S2 cells.
FIGURE 4:
Tm1A colocalizes with myosin-II but only partially overlaps with Diaphanous in the cleavage furrow, where Diaphanous is required for retention, but not recruitment, of Tm1A. (A–C) During interphase, mCherry-Tm1A and myosin-GFP localize to retracting, but not expanding, blebs at the cell periphery. See also Supplemental Figure 4 Video 1. (B) Zoomed-in single-color frames from time-lapse imaging. (C) Graph of total fluorescence intensity in the example bleb over time during bleb expansion (light gray) and bleb retraction (darker gray). (D) During mitosis, Tm1A and myosin-II colocalize at the equatorial cortex (arrow) during early anaphase and to retracting blebs (arrowhead), but not expanding blebs (asterisks), at the cell poles during late anaphase and telophase. Bottom, cartoon schematic illustrating localization of Tm1A (purple) and myosin (blue) in corresponding upper images. See also Supplemental Figure 4 Video 2. (E) During metaphase, Tm1A localizes to the cortex before Dia. As the cell progresses through mitosis, Tm1A occupies a larger, expanding region of the cortex around the periphery of the cell, whereas in contrast, Dia occupies a smaller region in the middle of the cell that seems to concentrate at the site of cleavage furrow ingression. Bottom, schematic diagram illustrating differential localizations of Tm1A (purple) and Dia (orange). See also Supplemental Figure 4 Video 3. (F) Dia is required for retention, not recruitment, of Tm1A to the equatorial cortex during anaphase. mCherry-Tm1A in a dividing cell after depletion of Dia with RNAi. Tm1A still initially localizes to the equatorial cortex (arrowheads), but it does not remain there. As the cell proceeds through anaphase, Tm1A localizes to sites of contraction (arrows) around the periphery of the cell during shape instability. See also Supplemental Figure 4 Video 4.
Distinct localizations of tropomyosin isoforms during mitosis
In yeast and vertebrate cells, tropomyosins help assemble the contractile ring (Thoms et al., 2008; Laporte et al., 2010). To determine whether any of the three Drosophila tropomyosins plays a similar role in cell division, we coexpressed each as an eGFP fusion, together with mCherry-α-tubulin. We plated cells on poly-d-lysine–coated coverslips to promote complete division and identified early mitotic cells by overall morphology and arrangement of microtubules. We then imaged the cells through the entire process of division.
Tm1A and myosin-II concentrate at the cleavage furrow and retracting polar blebs
During prophase and metaphase, Tm1A localizes throughout the cortex, but, early in anaphase, it disappears from the regions nearest the spindle poles, remaining only in the equatorial region of the cortex (arrowheads, Supplemental Figure S4D). As the cell elongates during anaphase, eGFP-Tm1A localization spreads throughout the cortex along the sides of the elongating cell, remaining generally absent from the cell poles (asterisks, Supplemental Figure S4D). As in interphase, the Tm1A pattern mirrors that of myosin-II (Supplemental Figure S4E) during mitosis (Figure 4D). When the cell elongates late in anaphase, membrane blebs form at the cell poles (Paluch et al., 2006) and, as in interphase, both Tm1A and myosin-II colocalize to polar blebs during the retraction phase (arrowheads, Figure 4D; Supplemental Figure 4 Video 2).
Diaphanous does not recruit Tm1A to the cleavage furrow but helps maintain Tm1A there
Formins help construct actin filaments in the contractile rings of many cells, including yeast and Drosophila S2 cells (Laporte et al., 2010). In Schizosaccharomyces pombe, Cdc12p (a homologue of Dia) localizes to the site of cell division, where it generates actin filaments that subsequently recruit a tropomyosin (Cdc8p) and myosin-II (Skau et al., 2009). To determine whether Tm1A coordinates with Dia during cell division, we coexpressed Dia-eGFP with mCherry-Tm1A (Figure 4E). Although both Tm1A and Dia localize to the equatorial cortex (site of cleavage furrow ingression) during early anaphase, Tm1A arrives before Dia (schematic, Figure 4E). This is the opposite of the temporal relationship between the Diaphanous-family formin (Cdc12p) and tropomyosin (Cdc8p) in S. pombe. In addition, whereas the region of the cortex occupied by mCherry-Tm1A expands as the cell elongates and anaphase progresses, Dia-eGFP localization contracts to a smaller region near the site of division (schematic, Figure 4E; Supplemental Figure 4 Video 3).
To verify that Tm1A localization to the equatorial cortex does not depend on Dia, we depleted Dia using RNAi in cells stably expressing mCherry-Tm1A and eGFP-Tm1J. As in previous studies (Rogers et al., 2003), depletion of Dia produces a multinucleate phenotype as cells fail to complete cytokinesis (Figure 4F). During cell division, Tm1A is still initially recruited to the equatorial cortex (arrowheads, Figure 4F), but, in the absence of Diaphanous, its localization is not stable, and Tm1A repeatedly appears and disappears in randomly located patches around the cell cortex. These patches often undergo transient contraction, which relaxes when the Tm1A disappears (arrows, Figure 4F; Supplemental Figure 4 Video 4), further supporting the idea that Tm1A recruits myosin-II or locally promotes its activity. These results likely explain results from a previous study reporting that Dia is not necessary for initial recruitment of myosin-II to the equatorial cortex but is required to stabilize myosin-II at the furrow (Dean et al., 2005).
Tm1J localizes to centrosomes, kinetochores, and the central spindle
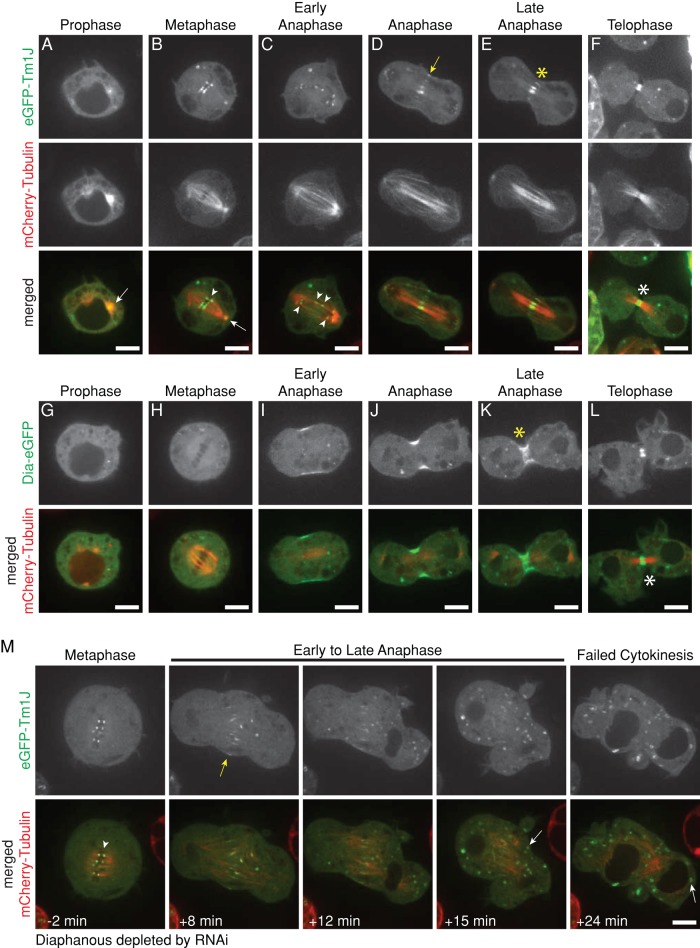
During prophase, most Tm1J remains associated with the Golgi ministacks, but a fraction of the protein localizes to the centrosomes (Figure 5A). Upon nuclear envelope breakdown, Golgi-associated Tm1J rings completely disperse, and the protein immediately concentrates at kinetochores (Figure 5B). Tm1J remains associated with both centrosomes and kinetochores through metaphase, as the chromosomes are captured and aligned. During anaphase, as chromosomes begin to separate, Tm1J leaves kinetochores and localizes to the central spindle, midway between the separating chromosomes (Figure 5, C– E, and Supplemental Figure 5 Video 1). Later in mitosis, as the ingressing cleavage furrow compresses central spindle microtubules into a bundle, Tm1J becomes concentrated at the central spindle and midbody (Figure 5F). Unlike canonical midbody proteins that remain concentrated in the remnants of the central spindle long after division is complete, Tm1J deserts the midbody for the Golgi-associated rings as the nuclear envelope begins to reform during telophase (Figure 3C).
FIGURE 5:
Tm1J localizes to centrosomes, kinetochores, the central spindle, and midbody during mitosis and requires Diaphanous for localization to the central spindle during anaphase. (A–F) eGFP-Tm1J localizes to the spindle during mitosis. Tm1J is at the centrosomes (arrow) during prophase (A); at kinetochores (arrowhead) and centrosomes (arrow) during metaphase (B); remains associated with separating kinetochores (arrowheads) during early anaphase (C); shifts to the spindle midzone (yellow arrow) and the central spindle (yellow asterisk) during anaphase (D) and late anaphase (E); and becomes tightly focused on the microtubule bundle at the midpoint of the central spindle (white asterisk) during telophase (F). Representative images from two different cells. See also Supplemental Figure 5 Video 1. (G–L) Dia-eGFP localizes to the cleavage furrow during early anaphase (I). However, during late anaphase (K) and telophase (L), Dia localizes to the central spindle (yellow asterisk) and midbody (white asterisk), respectively, similar to Tm1J localization. (M) Diaphanous is required for Tm1J localization to the central spindle and midpoint in dividing cells but not for Tm1J localization to kinetochores or Golgi. After Diaphanous is depleted by RNAi, eGFP-Tm1J still localizes to kinetochores (arrowhead) during metaphase and the spindle midzone (yellow arrow) during early anaphase but does not localize to the central spindle or midpoint during late anaphase. Tm1J instead relocates from the spindle midzone directly to Golgi (white arrows) during late anaphase. Times indicated are relative to anaphase onset. See also Supplemental Figure 5 Video 2.
Diaphanous is required to localize Tm1J to the central spindle but not to kinetochores
The localization pattern of Tm1J reminded us of the Diaphanous-family formins. In mammalian cells, a Diaphanous-family formin (mDia1) helps organize the Golgi apparatus (Zilberman et al., 2011). During mitosis, a second Diaphanous-family formin (mDia2) localizes to the central spindle and midbody (Watanabe et al., 2010) and a third (mDia3) localizes to kinetochores (Cheng et al., 2011). Drosophila has only one Diaphanous-family formin, Dia, which colocalizes with Tm1J and Tm2A at the Golgi rings in interphase (Figure 3, A and B). Late in anaphase, Dia and Tm1J both localize to the central spindle and midbody (Figure 5, K and L). In contrast to Tm1J, however, Dia fails to localize to centrosomes or kinetochores (Figure 5, G and H). These results suggest that Dia may collaborate with Tm1J and Tm2A at the Golgi apparatus during interphase and with Tm1J at the central spindle and midbody late in anaphase.
To test whether Dia is required to localize Tm1J, we depleted Dia from S2 cells using RNAi and imaged eGFP-Tm1J together with either mCherry-Tm1A or mCherry-tubulin. In the absence of Dia, Tm1J still forms Golgi-associated rings in interphase (Supplemental Figure S5A) and concentrates on metaphase kinetochores during mitosis (arrowhead, Figure 5M). Tm1J does not, however, localize to the central spindle or the midbody in later phases of mitosis of Dia-depleted cells (Figure 5M and Supplemental Figure 5 Video 2). Instead, Tm1J relocates directly from kinetochores to the Golgi apparatus during cytokinesis (white arrow, Figure 5M).
Unlike the other tropomyosins, Tm2A displays no obvious localization pattern during mitosis (Supplemental Figure S5B). It becomes dispersed throughout the cytoplasm early in mitosis and returns to the Golgi rings upon exit from mitosis (Supplemental Figure S3D).
Molecular determinants of tropomyosin localization: the C-termini of Tm1A and Tm1J specify their localization during mitosis
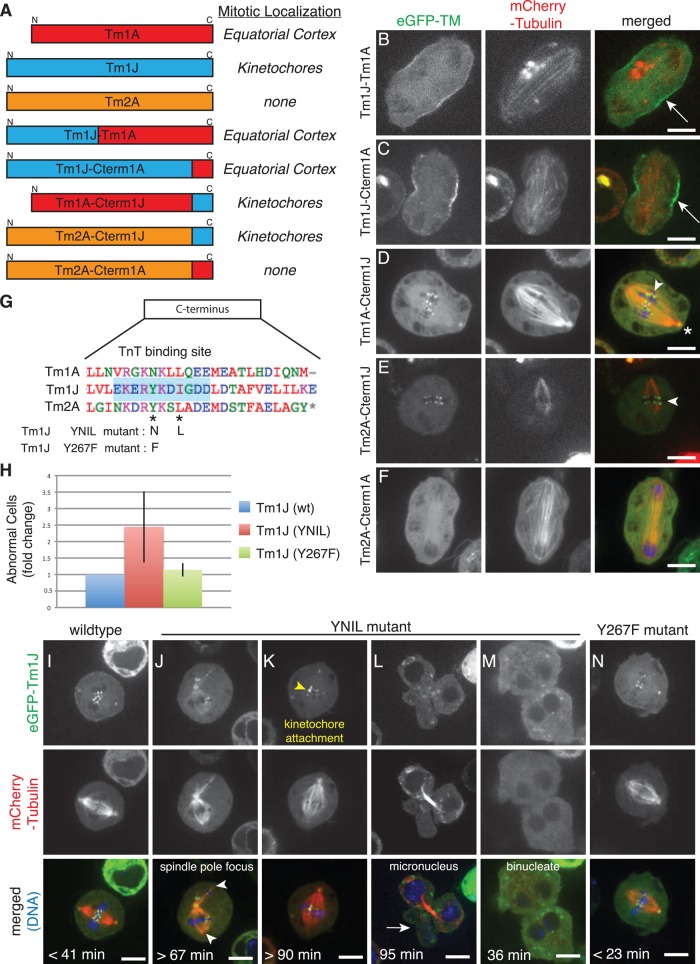
To understand why different tropomyosins localize to different places in the cell, we fused sequences from the three isoforms, creating chimeras. To maintain the coiled-coil structure and avoid shifting the register of residues that contact actin, we spliced sequences only at exon boundaries. The size of each chimera is, therefore, the same as that of either a canonical long or short isoform in Drosophila.
These chimeras reveal that sequences in the C-terminal region of tropomyosin regulate its localization during mitosis. For example, fusion of N-terminal regions of Tm1J to C-terminal regions of Tm1A (producing constructs such as Tm1L and Tm1J-Cterm1A) results in a Tm1A-like localization to the equatorial cortex and cleavage furrow (Figures 6, A–C). Conversely, fusing the C-terminus of Tm1J to the N-terminal portion of Tm1A (Tm1A-Cterm1J) produces a chimera that localizes, like Tm1J, to kinetochores (Figure 6, A and D). In addition, when we replaced the C-terminus of Tm2A, a tropomyosin with a diffuse localization during mitosis, with the C-terminal region of Tm1J (Tm2A-Cterm1J), this chimera also accumulated at kinetochores (Figure 6, A and E). Of interest, Tm2A-Cterm1A fails to localize to the equatorial cortex during mitosis (Figure 6, A and F). Together these results show that the C-terminus of Tm1A is necessary for localization to the equatorial cortex, whereas the C-terminus of Tm1J is both necessary and sufficient for localization of tropomyosin to kinetochores during mitosis.
FIGURE 6:
The C-terminus determines mitotic localization of tropomyosin isoforms, and mutational analysis shows that Tm1J participates in chromosome segregation and spindle morphology during mitosis. (A–F) Protein sequences in the C-terminus determine the localization of TM isoforms during mitosis. (A) Left, names and diagrams of TM isoform chimera constructs imaged during mitosis. Right, chart of results indicating whether each TM chimera localizes to “kinetochores” during metaphase, the “equatorial cortex” during anaphase, or no significant localization (“none”). (B–F) Images of dividing S2 cells expressing the indicated TM chimera tagged with eGFP (left) or mCherry-tubulin (middle) and merged images (eGFP, green; tubulin, red). Tm1J-Tm1A (B) and Tm1J-Cterm1A (C) both localize to the equatorial cortex (arrows). Tm1A-Cterm1J (D) and Tm2A-Cterm1J (E) both localize to kinetochores (arrowheads) and centrosomes (asterisk), whereas Tm2A-Cterm1A (F) shows no significant localization. (G) Alignments of the C-terminal regions of all three nonmuscle TM isoforms show that Tm1J, but not Tm1A or Tm2A, contains a consensus binding site for troponin T (blue shaded box). The “YNIL” mutant mutates two of the amino acids of this binding site, from Y267 to N and I270 to L, and has been shown to decrease binding to troponin T, whereas “Y267F” mutates Y267 to F. (H) Expression of the Tm1J YNIL mutation leads to an increase in the percentage of abnormal mitoses relative to cells expressing wild-type Tm1J, but expression of Tm1J-Y267F does not. (I–N) Representative images of dividing S2 cells expressing Tm1J wild type (I), YNIL mutant (J–M), or Y267F mutant (N). Supplemental Figure 6 Video 1 depicts cell shown in J. Hoechst 33258 was used to visualize DNA (blue). Time from NEBD to anaphase onset is noted in bottom left corner. Descriptions of mutant phenotypes are indicated on specific example cells.
Mutation of the troponin-binding site in Tm1J causes defects in chromosome segregation and spindle morphology during mitosis
To understand how the C-terminal regions of tropomyosins specify their localization and function during mitosis, we compared the sequences of Tm1A, Tm1J, and Tm2A. Because the C-terminus of Tm1J plays an essential role in binding kinetochores, we looked first for residues near the C-terminus of Tm1J that are not conserved in the other isoforms. Somewhat surprisingly, we found that this region of Tm1J contains the consensus sequence for high-affinity binding of the actin:tropomyosin regulatory protein troponin T (Murakami et al., 2008), whereas key residues in this motif are absent from Tm1A and Tm2A (Figure 6G).
To determine whether troponin T (TnT) might play a role in the mitotic localization and/or function of Tm1J, we mutated two residues in the putative TnT-binding site (Y267 and I270) to their analogues in Tm1A (N267 and L270, respectively) to form the mutant Tm1J-YNIL. The equivalent mutations in skeletal muscle tropomyosin greatly reduce the affinity of skeletal muscle TnT for tropomyosin (Murakami et al., 2008). Like wild-type Tm1J, the Tm1J-YNIL mutant localizes to centrosomes, kinetochores, and the central spindle (Supplemental Figure S5C). Remarkably, however, expression of Tm1J-YNIL produces significant defects in chromosome segregation and spindle morphology, even in the presence of endogenous Tm1J. We quantified the severity of these defects by time-lapse imaging of dividing cells (Figure 6H) and found that more than twice as many Tm1J-YNIL-expressing cells exhibit abnormal mitoses (n = 58 cells) than cells expressing wild-type Tm1J (n = 54 cells). Cells expressing the Tm1J-YNIL mutant exhibited kinetochore attachment defects, unfocused spindle poles, and delayed anaphase progression (Figure 6, J– L, and Supplemental Figure 6 Video 1). Cells expressing Tm1J-YNIL that do eventually enter anaphase often have defects in chromosome segregation, leading to cell division failure or micronuclei surrounding unaligned and/or missegregated chromosomes (Figure 6, L and M).
To control for the possibility that the Y-N mutation in Tm1J abolishes an important regulatory phosphorylation site, we mutated this tyrosine (267) to phenylalanine, which is structurally similar but cannot be phosphorylated. We found that Tm1J-Y267F still localizes to centrosomes, kinetochores, and the central spindle during mitosis (Supplemental Figure S5C), but, unlike Tm1J-YNIL, expression of the nonphosphorylatable Tm1J-Y267F mutant caused no significant increase in abnormal mitoses (Figure 6, H and N). These results show that Tm1J plays roles in spindle morphology and chromosome segregation and confirms the importance of the C-terminus in mediating kinetochore-specific functions. Furthermore, the fact that mutation of the TnT consensus binding motif does not abolish Tm1J localization to the spindle but clearly affects Tm1J function at the spindle raises the possibility that the troponin regulatory complex may also participate in chromosome segregation and maintenance of spindle morphology.
DISCUSSION
Previous evidence of multiple nonmuscle tropomyosin isoforms
Our discovery of additional nonmuscle Drosophila tropomyosins distinct from the previously described Tm1A isoform likely explains some puzzling results from previous studies of Drosophila embryogenesis. These studies suggested that unknown tropomyosin isoforms might be required to 1) drive oskar mRNA localization (Erdélyi et al., 1995), 2) specify neuronal dendritic field size (Li and Gao, 2003), and 3) promote border cell migration (Kim et al., 2011). On the basis of the location of P-element insertions in the Tm1 gene of Drosophila lines used in some of these studies, together with our observation that both Tm1A and Tm1J localize to dynamic lamellar actin networks in S2 cells, we suggest that Tm1J and Tm1A work together in many of these processes.
Challenges to identifying nonmuscle tropomyosins in Drosophila
Confusion over tropomyosin isoform expression has also limited the ability of RNAi screens to identify cellular functions of tropomyosin. Most RNAi screens target a limited set of isoforms encoded by one gene. This approach is not well suited to untangling overlapping contributions from multiple isoforms, encoded by multiple genes. Specifically, five sequences have been used to knock down expression of tropomyosins encoded by the Tm1 gene in a large number of RNAi screens (∼100 screens; Drosophila RNAi Screening Center [www.flyrnai.org/]). Only one of these sequences, however, actually targets the previously identified nonmuscle tropomyosin, Tm1A. The other sequences appear to have been optimized for “efficiency” and/or the ability to deplete multiple possible isoforms. We believe that this is why previous RNAi screens in Drosophila S2 cells failed to identify many of the functions of tropomyosin.
Localization and function of tropomyosin isoforms through the cell cycle
Each Drosophila tropomyosin shifts localization several times during the cell cycle, and the choreography of these shifts provides insight into the biological functions of these molecules (Figure 7). The Tm1A isoform, for example, is linked spatially and temporally to myosin-II throughout the cell cycle, suggesting that Tm1A may help recruit myosin-II to actin filaments and/or promote myosin motor activity. Previous studies also suggested that nonmuscle tropomyosins may help recruit myosin-II to actin filaments in other systems (Bryce et al., 2003; Huckaba et al., 2006; Tojkander et al., 2011).
FIGURE 7:
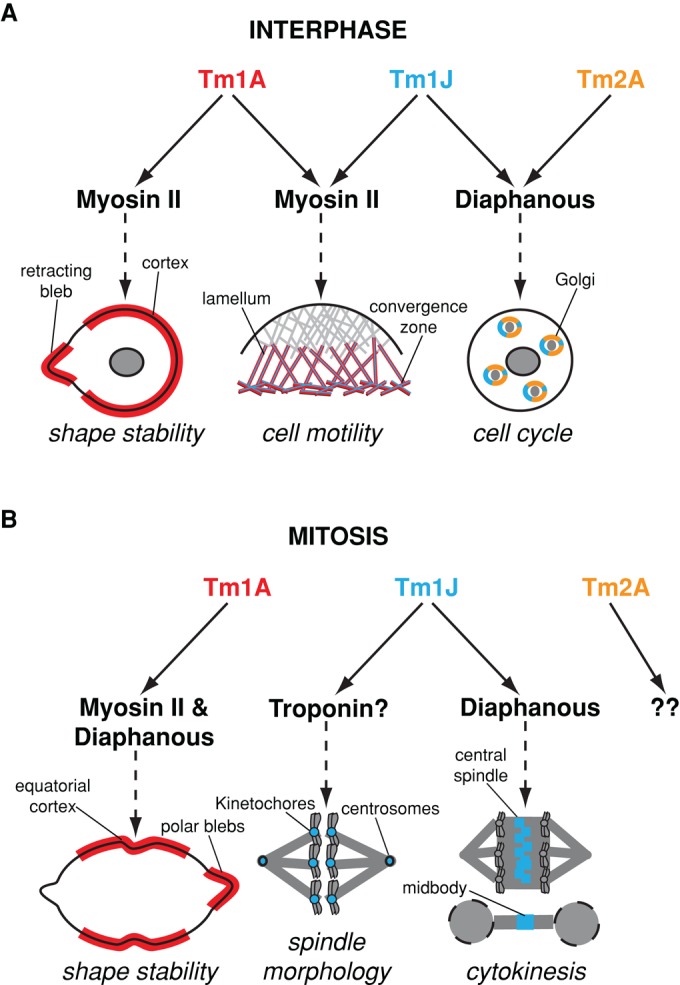
Model of Drosophila tropomyosin functions throughout the cell cycle. (A) During interphase, Tm1A colocalizes with myosin-II to the cell cortex and retracting membrane blebs, where they function to maintain cortical contractility and shape stability. In spreading cells, Tm1A and Tm1J localize with myosin-II to the lamellum and convergence zone, two networks previously shown to be important for cell motility (Salmon et al., 2002). Tm1J also colocalizes with Tm2A and Diaphanous to actin networks surrounding Golgi ministacks. There they help maintain Golgi architecture and, in so doing, influence cell cycle progression by preventing premature exit from G2 phase into mitosis. (B) During mitosis, Tm1A and myosin-II (Dean et al., 2005) initially colocalize to the equatorial cortex independent of Diaphanous but are maintained at the equator through a Diaphanous-dependent mechanism. Mislocalization of Tm1A causes shape instability and cytokinesis failure. Tm1J, on the other hand, localizes to the mitotic spindle. During metaphase, Tm1J localizes to kinetochores and centrosomes and may function with the troponin regulatory complex to influence spindle morphology, anaphase progression, and chromosome segregation. During anaphase, through a Diaphanous-dependent mechanism, Tm1J localization shifts to the central spindle and midbody, two structures previously shown to be important for cytokinesis (Giansanti et al., 1998).
The Tm1J isoform is linked to the formin-family protein Dia throughout the cell cycle. Both Tm1J and Dia localize to Golgi-associated rings during interphase and to the central spindle and midzone during mitosis. In addition, localization of Tm1J to the central spindle and midzone actually requires Dia. The only lapse in colocalization of these proteins is early in mitosis, when Tm1J localizes to centrosomes and kinetochores without Dia. It is possible that Dia also localizes to kinetochores early in mitosis but that we are unable to detect it there, possibly due to interference caused by fusion to a fluorescent protein. Of interest, one mammalian homologue of Dia, mDia3, localizes to kinetochores early in mitosis and relocates to the central spindle and midbody late in anaphase (Yasuda et al., 2004). In addition, expression of a dominant-active mDia3 causes misalignment of chromosomes and produces multinucleate cells (Cheng et al., 2011), similar to the effect of expressing the YNIL mutant of Tm1J we observed in Drosophila S2 cells. The mitotic dynamics of Tm1J in S2 cells and mDia3 in mammals resemble those of the chromosomal passenger complex (CPC). The CPC is a highly dynamic protein complex that regulates multiple aspects of mitosis, including spindle architecture, kinetochore–microtubule attachment, mitotic progression, and completion of cytokinesis (Ruchaud et al., 2007). One subunit of the CPC, the kinase Aurora B, actually phosphorylates mDia3 and regulates its activity at kinetochores (Cheng et al., 2011) and so may also affect Tm1J activity at kinetochores.
The connection between specific tropomyosin isoforms and Diaphanous-family formins is likely to be highly conserved. Drosophila express only one Diaphanous-family formin, whereas mammals express three: mDia1, mDia2, and mDia3. The three mammalian proteins appear to reflect the separation of several functions carried out by the more generalist Drosophila Dia. For example, mDia1 localizes to the Golgi apparatus in interphase (Zilberman et al., 2011), whereas mDia3 localizes to kinetochores early in mitosis (Yasuda et al., 2004), and mDia2 localizes to the central spindle and midzone late in mitosis (Watanabe et al., 2010).
Tm1J, Tm2A, and cell cycle progression
Knockdown of Tm1J and Tm2A probably influences cell cycle progression indirectly, via effects on Golgi architecture. The Golgi apparatus plays host to important cell cycle checkpoints, including one that involves the Golgi-associated kinase myt1. Similar to the effect of knocking down Tm1J and Tm2A, the depletion of myt1 causes S2 cells to pile up in the G1 phase of the cell cycle (Cornwell et al., 2002), and overexpression of myt1 (Wells et al., 1999) has the same effect as overexpressing Tm1J or Tm2A, causing cells to accumulate in the G2/M phase. Perturbing the architecture of the Golgi-associated Tm1J/Tm2A/actin network likely leads to misregulation of Golgi-associated cell cycle checkpoints.
Previous evidence that tropomyosin-actin contributes to mitotic spindle function
Mainstream models of mitotic spindle assembly do not include actin or its binding partners as central players, but growing evidence suggests that actin filaments are somehow involved in the process (Sandquist et al., 2011). Kwon et al. (2008) found, for example, that spindle defects caused by depleting the microtubule-based motor kinesin-14 (ncd) are exacerbated by inhibition of actin assembly by latrunculin A or depletion of the actin-based motor Myo10A.
Tropomyosin had not previously been reported on the mitotic spindle, but previous studies indicated that tropomyosin and troponin are required for proper spindle assembly and function. Thoms et al. (2008) found that overexpressing tropomyosin isoform Tm1 in U373G astrocytoma cells perturbs both spindle architecture and DNA segregation. In an RNAi screen, Kwon et al. (2008) found that depletion of troponin I (wupA) produces supernumerary centrosomes in Drosophila S2 cells, and Sahota et al. (2009) found that Drosophila troponin I (wupA) and tropomyosin mutant embryos display abnormal spindle orientation and defects in chromosome segregation. Many of these authors assumed that tropomyosin-associated mitotic defects were secondary to perturbation of cytokinesis, but our results suggest a more direct connection.
There are many ways in which tropomyosin–actin filaments could contribute to spindle function, but our results suggest that one key site of actin activity is on the kinetochore. Our results also raise the possibility that kinetochore-associated tropomyosin–actin filaments might interact with troponin T to form calcium-regulated “thin filaments” similar to those found in skeletal muscle sarcomeres.
MATERIALS AND METHODS
Immunoblotting and immunofluorescence
Immunoblotting and immunofluorescence were performed as previously described (Kondylis and Rabouille, 2003; Dean et al., 2005) using anti-tropomyosin antibodies E-17 (goat; Santa Cruz Biotechnology, Santa Cruz, CA), TM311 (mouse; Sigma-Aldrich, St. Louis, MO), and ab11190 (rabbit; Abcam) and anti-GM130 antibody ab30637 (rabbit; Abcam, Cambridge, MA). TM311 is a well-characterized monoclonal antibody that recognizes long tropomyosin (TM) isoforms in many species (Schevzov et al., 2011).
Purification of endogenous tropomyosin(s) and mass spectrometry analysis
Endogenous tropomyosin protein was partially purified from 1 l of S2 cells using a combination of heat denaturation, ammonium sulfate precipitation, pH precipitation, and hydroxyapatite column chromatography similar to past studies purifying exogenously expressed tropomyosin from Escherichia coli (Hitchcock-DeGregori and Heald, 1987). Individual protein bands were excised from SDS–PAGE gels stained with Coomassie blue and sent for mass spectrometry identification by the Burlingame lab at the University of California, San Francisco. To identify previously unknown TM isoforms, we added the protein sequences for all TM isoforms predicted by FlyBase to the mass spectrometry search database. Five unique protein peptides present in Tm1J but not present in Tm1A or Tm2A were found.
Cell culture, transfection, and RNAi
Drosophila Schneider S2 cells were cultured as previously described (Rogers et al., 2003; Iwasa and Mullins, 2007). Gateway cloning technology (Invitrogen, Carlsbad, CA) was used to create vectors for expression of fluorescently tagged proteins under the copper-inducible pMT promoter. Vectors for the expression of Dia-eGFP and myosin-GFP were a kind gift from the Vale lab (Uehara et al., 2010). S2 cells were transfected with 1 μg of total DNA including pCoHygro plasmid (Invitrogen) using Effectene transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. S2 cells were grown in the presence of hygromycin for 2 wk to select for stably transfected cells. RNAi and cell cycle analysis were performed as previously described (Bettencourt-Dias and Goshima, 2009). In brief, RNAi was performed on S2 cells cultured in six-well tissue culture plates for 7 d according to the methods of Rogers et al. (2003) using PCR products flanked at their 5′ and 3′ ends by T7 promoter sequences. Primers that would amplify tropomyosin isoform-specific sequences were designed and evaluated using the E-RNAi Web application (Horn and Boutros, 2010). Primer sequences can be found in Supplemental Table 3. Double-strand RNA (dsRNA) was produced by in vitro transcription using T7 Megascript kits (Ambion, Austin, TX) according to the manufacturer's instructions. S2 cells were incubated with dsRNA for 7 d and then processed for microscopy or cell cycle analysis. These conditions resulted in full depletion of the Tm1A isoform but only partial depletion of the Tm1J and Tm2A isoforms (assessed by immunoblots).
Tropomyosin overexpression and cell cycle analysis
eGFP-tagged TM isoforms under the pMT promoter were overexpressed by >24-h induction with 1 mM CuSO4 and processed for flow cytometry and cell cycle analysis essentially as described (Davidson and Duronio, 2011).
Live-cell imaging and analysis
For live-cell imaging, S2 cell lines were induced with 100 μM CuSO4 overnight and then transferred to 96-well glass-bottom plates (Matrical, Spokane, WA) coated with ConA or PDL (Sigma-Aldrich) and allowed to spread or adhere for 1–2 h before imaging. To depolymerize filamentous actin, S2 cells were treated with 10 μM latrunculin B. We used a Yokagawa CSU22 spinning-disk confocal microscope (Nikon Ti-E microscope) equipped with perfect focus and an automated piezo-driven stage (Nikon, Melville, NY) and controlled by Micromanager software (Edelstein et al., 2010) for all imaging. Single confocal planes are shown for all imaging. All scale bars are 5 μm.
Dividing cells were followed using multiposition time-lapse imaging. For analysis of cell division defects, we identified cells near the beginning of division on the basis of morphology of the microtubule cytoskeleton and imaged them throughout mitosis. We then scored the mitoses blind as either “normal” or “abnormal.” We defined as abnormal any mitosis that 1) took >45 min to progress from metaphase to anaphase or 2) exhibited obvious chromosome segregation defects, including those that produce multiple cleavage furrows or result in the formation of micronuclei or multinucleate cells. On each imaging day, S2 cells expressing eGFP-tagged wild-type Tm1J were imaged as controls, and graphs depict fold change relative to wild-type cells.
Supplementary Material
Acknowledgments
This work was supported by grants to R.D.M. from the National Institutes of Health (3R01GM061010) and the Howard Hughes Medical Institute. R.D.M. acknowledges J. R. L., F. R. Nada, and C. R. Amit for streamlining the operation. We are grateful to Juan Oses in the Burlingame lab at the University of California, San Francisco, for his assistance with mass spectrometry analysis; Kurt Thorn in the Nikon Imaging Center at University of California, San Francisco/QB3, where all confocal images were acquired; Eric Griffis and Thomas Huckaba in the Vale lab for advice and reagents for S2 cell culture work; members of the Mullins lab for their assistance and advice, especially Elena Ingerman for assistance with Matlab, Jenny Hsiao for help with data processing, and Chris Rivera for detailed discussions on cellular mechanisms; D Casimere and members of the FantFour Group, including Eric Chow, Catherine Foo, and Ashley Robinson, for valuable discussions throughout this project; Ron Vale and Henry Bourne for helpful discussion; and Peter Gunning for numerous reagents and advice on tropomyosin isoforms. L.G.C. would like to dedicate this paper to her mother, “Professor Goins.”
Abbreviations used:
- ConA
concanavalin A
- CPC
chromosomal passenger complex
- Dia
Diaphanous
- eGFP
enhance green fluorescent protein
- NEBD
nuclear envelope breakdown
- NER
nuclear envelope reformation
- PDL
poly-d-lysine
- TM
tropomyosin
- TnT
troponin T
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-12-1619) on May 13, 2015.
REFERENCES
- Basi GS, Boardman M, Storti RV. Alternative splicing of a Drosophila tropomyosin gene generates muscle tropomyosin isoforms with different carboxy-terminal ends. Mol Cell Biol. 1984;4:2828–2836. doi: 10.1128/mcb.4.12.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell. 2013;24:982–994. doi: 10.1091/mbc.E12-09-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Goins LM, Mullins RD. Comparative analysis of tools for live imaging of actin network architecture. BioArchitecture. 2015 doi: 10.1080/19490992.2014.1047714. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Goshima G. RNAi in Drosophila S2 cells as a tool for studying cell cycle progression. Methods Mol Biol. 2009;545:39–62. doi: 10.1007/978-1-60327-993-2_3. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol. 2001;11:1300–1304. doi: 10.1016/s0960-9822(01)00395-5. [DOI] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP. Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell. 2003;14:1002–1016. doi: 10.1091/mbc.E02-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras GT, Hu C-K, Coughlin M, Mitchison TJ. Reassembly of contractile actin cortex in cell blebs. J Cell Biol. 2006;175:477–490. doi: 10.1083/jcb.200602085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, Rozier L, Yu H, Deng H, Mao Y. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20:342–352. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Nag S, Pollard TD. Formins filter modified actin subunits during processive elongation. J Struct Biol. 2012;177:32–39. doi: 10.1016/j.jsb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010;20:1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Cornwell WD, Kaminski PJ, Jackson JR. Identification of Drosophila Myt1 kinase and its role in Golgi during mitosis. Cell Signal. 2002;14:467–476. doi: 10.1016/s0898-6568(01)00276-5. [DOI] [PubMed] [Google Scholar]
- Davidson JM, Duronio RJ. Using Drosophila S2 cells to measure S phase-coupled protein destruction via flow cytometry. Methods Mol Biol. 2011;782:205–219. doi: 10.1007/978-1-61779-273-1_15. [DOI] [PubMed] [Google Scholar]
- Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci USA. 2005;102:13473–13478. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using μManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit 14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdélyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M. Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 1998;12:396–410. doi: 10.1101/gad.12.3.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev. 2008;88:1–35. doi: 10.1152/physrev.00001.2007. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 2005;15:333–341. doi: 10.1016/j.tcb.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Hanke PD, Lepinske HM, Storti RV. Characterization of a Drosophila cDNA clone that encodes a 252-amino acid non-muscle tropomyosin isoform. J Biol Chem. 1987;262:17370–17373. [PubMed] [Google Scholar]
- Hanke P, Storti R. Nucleotide sequence of a cDNA clone encoding a Drosophila muscle tropomyosin II isoform. Gene. 1986;45:211–214. doi: 10.1016/0378-1119(86)90256-8. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Heald RW. Altered actin and troponin binding of amino-terminal variants of chicken striated muscle alpha-tropomyosin expressed in Escherichia coli. J Biol Chem. 1987;262:9730–9735. [PubMed] [Google Scholar]
- Horn T, Boutros M. E-RNAi: a web application for the multi-species design of RNAi reagents–2010 update. Nucleic Acids Res. 2010;38:W332–W339. doi: 10.1093/nar/gkq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckaba TM, Lipkin T, Pon LA. Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J Cell Biol. 2006;175:957–969. doi: 10.1083/jcb.200609155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V, de Voer G, Xanthakis D, Spoorendonk KM, Kondylis V, Rabouille C. Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell. 2008;19:4352–4365. doi: 10.1091/mbc.E08-03-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa JH, Mullins RD. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr Biol. 2007;17:395–406. doi: 10.1016/j.cub.2007.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Cho A, Yin H, Schafer DA, Mouneimne G, Simpson KJ, Nguyen K-V, Brugge JS, Montell DJ. Psidin, a conserved protein that regulates protrusion dynamics and cell migration. Genes Dev. 2011;25:730–741. doi: 10.1101/gad.2028611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V, van Nispen tot Pannerden HE, Herpers B, Friggi-Grelin F, Rabouille C. The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell. 2007;12:901–915. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Kreuz AJ, Simcox A, Maughan D. Alterations in flight muscle ultrastructure and function in Drosophila tropomyosin mutants. J Cell Biol. 1996;135:673–687. doi: 10.1083/jcb.135.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Théry M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Zhao R, Wu J-Q. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol. 2010;21:892–898. doi: 10.1016/j.semcdb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Gao F-B. Actin filament-stabilizing protein tropomyosin regulates the size of dendritic fields. J Neurosci. 2003;23:6171–6175. doi: 10.1523/JNEUROSCI.23-15-06171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Schevzov G, Gunning P. Alternatively spliced N-terminal exons in tropomyosin isoforms do not act as autonomous targeting signals. J Struct Biol. 2010;170:286–293. doi: 10.1016/j.jsb.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Curr Biol. 2011;21:R560–R569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K, Stewart M, Nozawa K, Tomii K, Kudou N, Igarashi N, Shirakihara Y, Wakatsuki S, Yasunaga T, Wakabayashi T. Structural basis for tropomyosin overlap in thin (actin) filaments and the generation of a molecular swivel by troponin-T. Proc Natl Acad Sci USA. 2008;105:7200–7205. doi: 10.1073/pnas.0801950105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–1076. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch E, van der Gucht J, Sykes C. Cracking up: symmetry breaking in cellular systems. J Cell Biol. 2006;175:687–692. doi: 10.1083/jcb.200607159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival JM, Hughes JAI, Brown DL, Schevzov G, Heimann K, Vrhovski B, Bryce N, Stow JL, Gunning PW. Targeting of a tropomyosin isoform to short microfilaments associated with the Golgi complex. Mol Biol Cell. 2004;15:268–280. doi: 10.1091/mbc.E03-03-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Kondylis V. Golgi ribbon unlinking: an organelle-based G2/M checkpoint. Cell Cycle. 2007;6:2723–2729. doi: 10.4161/cc.6.22.4896. [DOI] [PubMed] [Google Scholar]
- Rizzino A, Blumenthal AB. Synchronization of Drosophila cells in culture. In Vitro. 1978;14:437–442. doi: 10.1007/BF02616105. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sahota VK, Grau BF, Mansilla A, Ferrús A. Troponin I and Tropomyosin regulate chromosomal stability and cell polarity. J Cell Sci. 2009;122:2623–2631. doi: 10.1242/jcs.050880. [DOI] [PubMed] [Google Scholar]
- Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandquist JC, Kita AM, Bement WM. And the dead shall rise: actin and myosin return to the spindle. Dev Cell. 2011;21:410–419. doi: 10.1016/j.devcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Whittaker SP, Fath T, Lin JJ, Gunning PW. Tropomyosin isoforms and reagents. BioArchitecture. 2011;1:135–164. doi: 10.4161/bioa.1.4.17897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Neidt EM, Kovar DR. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell. 2009;20:2160–2173. doi: 10.1091/mbc.E08-12-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetzlaff MT, Jäckle H, Pankratz MJ. Lack of Drosophila cytoskeletal tropomyosin affects head morphogenesis and the accumulation of oskar mRNA required for germ cell formation. EMBO J. 1996;15:1247–1254. [PMC free article] [PubMed] [Google Scholar]
- Thoms JAI, Loch HM, Bamburg JR, Gunning PW, Weinberger RP. A tropomyosin 1 induced defect in cytokinesis can be rescued by elevated expression of cofilin. Cell Motil Cytoskeleton. 2008;65:979–990. doi: 10.1002/cm.20320. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21:539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Uehara R, Goshima G, Mabuchi I, Vale RD, Spudich JA, Griffis ER. Determinants of myosin II cortical localization during cytokinesis. Curr Biol. 2010;20:1080–1085. doi: 10.1016/j.cub.2010.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve J, Scarpa M, Ortega-Bellido M, Malhotra V. MEK1 inactivates Myt1 to regulate Golgi membrane fragmentation and mitotic entry in mammalian cells. EMBO J. 2013;32:72–85. doi: 10.1038/emboj.2012.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Okawa K, Miki T, Sakamoto S, Morinaga T, Segawa K, Arakawa T, Kinoshita M, Ishizaki T, Narumiya S. Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol Biol Cell. 2010;21:3193–3204. doi: 10.1091/mbc.E10-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells NJ, Watanabe N, Tokusumi T, Jiang W, Verdecia MA, Hunter T. The C-terminal domain of the Cdc2 inhibitory kinase Myt1 interacts with Cdc2 complexes and is required for inhibition of G(2)/M progression. J Cell Sci. 1999;112:3361–3371. doi: 10.1242/jcs.112.19.3361. [DOI] [PubMed] [Google Scholar]
- Yasuda S, Oceguera-Yanez F, Kato T, Okamoto M, Yonemura S, Terada Y, Ishizaki T, Narumiya S. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nat Cell Biol. 2004;428:767–771. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
- Zilberman Y, Alieva NO, Miserey-Lenkei S, Lichtenstein A, Kam Z, Sabanay H, Bershadsky A. Involvement of the Rho-mDia1 pathway in the regulation of Golgi complex architecture and dynamics. Mol Biol Cell. 2011;22:2900–2911. doi: 10.1091/mbc.E11-01-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.