The mechanisms governing the spatial organization of endocytosis and exocytosis are ill defined. A quantitative imaging screen and high-density single-vesicle tracking are used to identify mutants that are defective in endocytic and exocytic vesicle organization. The screen identifies a role for the exocyst complex in connecting the two pathways.
Abstract
The coupling of endocytosis and exocytosis underlies fundamental biological processes ranging from fertilization to neuronal activity and cellular polarity. However, the mechanisms governing the spatial organization of endocytosis and exocytosis require clarification. Using a quantitative imaging-based screen in budding yeast, we identified 89 mutants displaying defects in the localization of either one or both pathways. High-resolution single-vesicle tracking revealed that the endocytic and exocytic mutants she4∆ and bud6∆ alter post-Golgi vesicle dynamics in opposite ways. The endocytic and exocytic pathways display strong interdependence during polarity establishment while being more independent during polarity maintenance. Systems analysis identified the exocyst complex as a key network hub, rich in genetic interactions with endocytic and exocytic components. Exocyst mutants displayed altered endocytic and post-Golgi vesicle dynamics and interspersed endocytic and exocytic domains compared with control cells. These data are consistent with an important role for the exocyst in coordinating endocytosis and exocytosis.
INTRODUCTION
Stimulus-driven exocytosis, such as that occurring after fertilization of Xenopus laevis and sea urchin eggs, results in the fusion of >15,000 secretory granules with the plasma membrane within a brief period of 15 min (Whalley et al., 1995). Were it not for compensatory endocytosis, plasma membrane homeostasis would not be maintained. The coupling of endocytosis and exocytosis is also evident in stimulated neurotransmitter release at neuromuscular junctions (Ceccarelli et al., 1973; Heuser and Reese, 1973). Here the spatial coordination of endocytosis and exocytosis facilitates the rapid recycling of vesicle components that would otherwise become depleted upon sustained stimulation (Gundelfinger et al., 2003). The efficiency of the two trafficking pathways may be optimized by proximity, such as in Drosophila melanogaster nerve terminals, where endocytic sites surround the exocytic active zone (Roos and Kelly, 1999).
The spatial coordination of endocytosis and exocytosis also plays a role in regulating cell polarity, the asymmetric distribution of cellular components (Mostov et al., 2003). For example, in plants, PIN proteins transport auxins in a polarized manner, thus regulating directed auxin fluxes that contribute to plant growth and development (Hazak et al., 2010). PINs are transported to the center of the apical plasma membrane, where they cluster, exhibiting constrained lateral diffusion on the membrane. PIN proteins that escape the cluster are retrieved via polarized endocytosis; thus exocytic trafficking and endocytosis contribute to PIN polarity (Kleine-Vehn et al., 2011).
Processes requiring high rates of polarized growth, including pollen tube extension and yeast hyphal growth, also display spatial coordination of endocytosis and exocytosis (Kohli et al., 2008; Taheri-Talesh et al., 2008; Zonia and Munnik, 2008; Echauri-Espinosa et al., 2012; Riquelme et al., 2014). In Aspergillus nidulans and Ashbya gossypii, exocytic vesicles are targeted to the hyphal tip, where soluble N-ethylmaleimide–sensitive factor attachment protein receptor (SNARE)–mediated vesicle fusion delivers the membrane and proteins required for apical growth. Endocytic activity is concentrated in a trailing collar that tracks the exocytic pole, providing an opportunity to recycle vesicle-SNAREs for future rounds of exocytosis (Kohli et al., 2008; Taheri-Talesh et al., 2008).
The biological systems discussed so far share a common characteristic: all place a heavy demand on vesicle delivery, which may require specialized organization of endocytic and exocytic trafficking domains. However, even in slower-growing yeast such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, endocytic and exocytic vesicles display striking spatial organization (Chesneau et al., 2004; Gachet and Hyams, 2005). Previous work demonstrated that sites of endocytosis form a ring at the cortex around the time when growth becomes polarized to form a new cell, the bud (Kilmartin and Adams, 1984; Amberg, 1998; Layton et al., 2011). Simultaneous imaging of endocytic and exocytic markers subsequently demonstrated that endocytic sites surround a central exocytic zone (McCusker et al., 2012; Jose et al., 2013; Slaughter et al., 2013). This bull's-eye organization is reminiscent of the same trafficking domains in Drosophila neuromuscular junctions (Roos and Kelly, 1999). Based on high-speed, evanescent-field imaging and mathematical modeling in budding yeast, a mechanism of robust polarity establishment has been proposed in which endocytic activity focuses the exocytic pole via a corralling mechanism into a vertex of exocytic activity from which the newly formed bud emerges (Jose et al., 2013). Consistently, endocytic mutants in which corralling is perturbed display widened, unfocused exocytic poles and an unstable polarity axis (Hervas-Aguilar and Penalva, 2010; Jose et al., 2013).
Basic questions regarding the relationship of endocytosis and exocytosis require clarification. What are the key molecular players required for the spatial organization of endocytosis and exocytosis? Are the endocytic and exocytic pathways obligatorily coupled during polarity establishment and maintenance, or can one pathway be affected while leaving the other intact? The amenability of budding yeast to genetic analyses and sensitive imaging techniques provides the opportunity to address these questions.
RESULTS
An imaging-based screen identifies mutants affecting the polarization of endocytic and exocytic trafficking domains independently
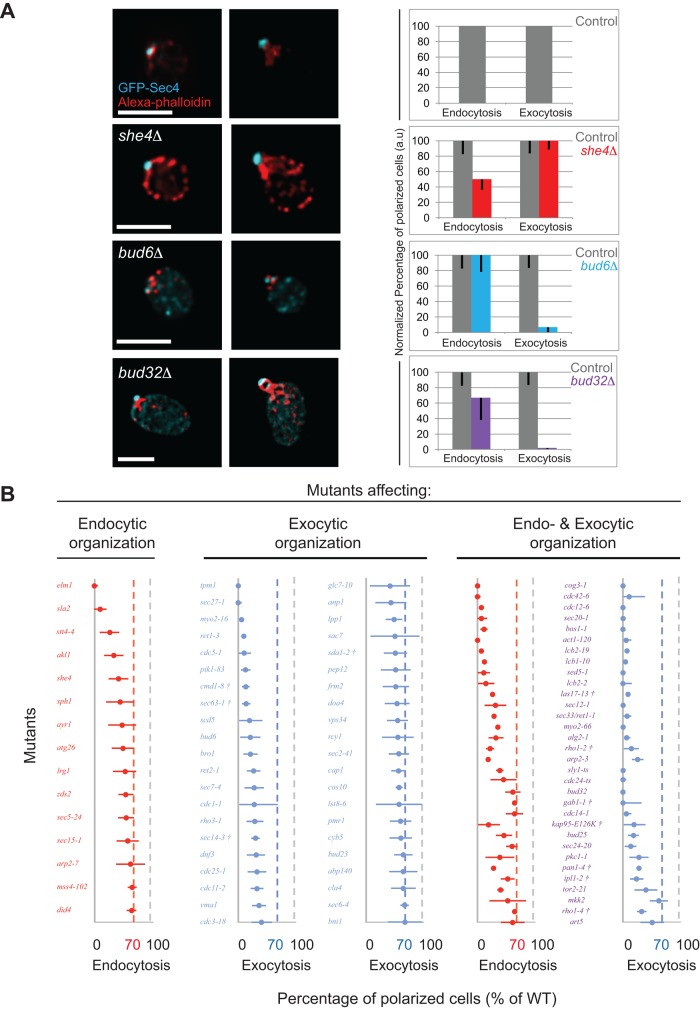
To identify proteins required for the spatial organization of endocytic and exocytic trafficking domains and test whether the two pathways are obligatorily coupled during polarized growth, we performed an imaging-based screen of 360 candidate mutants that were chosen based on previously identified roles in endocytosis, exocytosis, or cell polarity (Supplemental Table S1). Exocytic vesicles were marked with a green fluorescent protein (GFP)–tagged version of the Rab GTPase Sec4 (Calero et al., 2003), whereas endocytosis-associated actin patches were labeled with phalloidin, and the cell wall was defined by concanavalin A staining. The results of the screen were based on the localization of these reporters in mutant versus control cells. We selected 216 mutants displaying defects in domain organization after initial qualitative analysis for subsequent semiautomated quantitative analysis to identify mutants displaying altered distributions of endocytic and exocytic vesicles compared with control cells. Analysis was restricted to small-budded cells, in which endocytosis and exocytosis are polarized in control cells (McCusker et al., 2012). The analysis process is schematically represented in Supplemental Figure S1, A and B, and detailed in Materials and Methods. We quantified how much an individual cell is polarized by calculating the fluorescence ratio (FR) of the bud to the mother cell (Supplemental Figure S1). Statistical analysis of the FR for endocytosis and exocytosis identified three classes of mutants: those exhibiting depolarized endocytosis, depolarized exocytosis, or both depolarized (Supplemental Tables S1 and S2). These three classes are represented by she4Δ, bud6Δ, and bud32Δ mutants, respectively (Figure 1A).
FIGURE 1:
The identification of mutants affecting the spatial organization of the endocytic and/or exocytic pathways. (A) Steady-state images of fixed cells at different cell cycle stages. Scale bars, 5 μm. Bar charts show the percentage of polarized cells normalized to control levels. (B) Mutants predominantly affecting endocytosis, exocytosis, or both pathways. Percentage of cells with polarized endocytic and exocytosis (normalized to the control level) is shown as red and blue dots, respectively, with error bars showing the uncertainty in the fraction of (de)polarized cells (see Materials and Methods). The vertical dotted lines indicate the polarization level of control cells (gray) and the threshold (70% of control level) for endocytosis (red) and exocytosis (blue). †Strains shifted to 37ºC instead of 34°C before fixation.
Deletion of SHE4, a myosin regulator, resulted in depolarized endocytosis, consistent with previous studies (Figure 1A; Wendland et al., 1996; Wesche et al., 2003). However, polarized exocytosis appeared to be largely unaffected (Figure 1A). Of interest, whereas other strong endocytic mutants such as sla2Δ cells displayed dense endocytic clusters (Jose et al., 2013), she4Δ cells exhibited an increase in the number of brighter depolarized endocytic vesicles labeled by phalloidin or marked by Abp1–red fluorescent protein (RFP) in the mother cell compared with control cells (Supplemental Figure S2).
In contrast, deletion of BUD6, an actin- and formin-interacting protein, resulted in strongly depolarized exocytosis with few depolarized endocytic patches (Figure 1A). Quantitative analysis indicated that although exocytosis was affected, the endocytic fluorescence ratio of bud6Δ cells was not significantly different from that for control cells. Actin cables, which mediate polarized exocytic vesicle delivery, were weakly stained in bud6Δ mutants, as observed by others (Amberg et al., 1997; Graziano et al., 2011). Conversely, we observed that actin patches appeared brighter in bud6Δ cells than in control cells, consistent with the idea of competition between endocytosis and exocytosis for the available monomeric actin pool (Supplemental Figure S2, A and C; Burke et al., 2014).
Deletion of BUD32, a kinase component of the conserved KEOPS complex that is required for guiding polarity axis establishment (Ni and Snyder, 2001; Downey et al., 2006), exhibited depolarized endocytic and exocytic domains, although not to the same extent, and an enlarged average cell size (Figure 1, A and B). In contrast to control cells, in which actin cables were oriented along the mother–daughter axis, bud32Δ mutants displayed misoriented cables (Supplemental Figure S2A). Although secretion was previously reported to be defective in bud32Δ mutants (Bonangelino et al., 2002; Banuelos et al., 2010), the role of this protein in the organization of both endocytic and exocytic domains was not previously demonstrated. In contrast to the she4Δ mutant, for which a large number of cells exhibited depolarized endocytosis, as illustrated by the broad distribution of endocytic signal along the bud–mother axis (Supplemental Figure S3), bud32Δ mutants exhibited a sharper, although weaker than wild-type, endocytic signal in the bud, indicative of a large number of entirely depolarized cells and a few well-polarized cells (Supplemental Figure S3).
In addition to known trafficking regulators, the screen identified proteins required for the spatial organization of endocytic and exocytic trafficking domains (Figure 1B). Moreover, the identification of mutants that affect the spatial organization of one but not both pathways in cells that had polarized and formed a bud, suggested that the spatial coupling of endocytosis and exocytosis might not be obligatory during polarity maintenance. It is noteworthy that the mutants identified by the screen affected the distribution of endocytic and exocytic fluorescence distribution along the mother–bud axis in different ways. For simplification, cells were scored as having polarized or nonpolarized trafficking domains, but it is evident from the fluorescence distributions that many interesting intermediate phenotypes were also detected (Supplemental Figure S3).
In vivo imaging reveals the coupling of endocytic and exocytic domains during polarity establishment
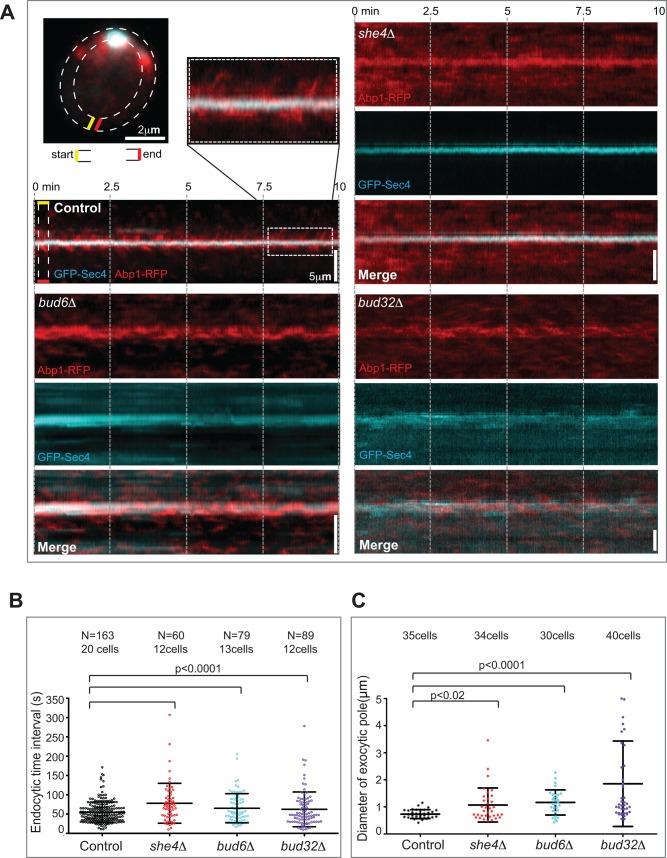
The relatively independent distribution of endocytic and exocytic pathways in polarized small-budded cells prompted us to address whether the spatial coupling of these domains is important during polarity establishment. We monitored the dynamics of the trafficking domains in unbudded polarizing control and mutant cells by simultaneous dual-color evanescent-field imaging (near–total internal reflection fluorescence microscopy [TIRFM]). Abp1-RFP and GFP-Sec4 were used as endocytic and exocytic markers. The endocytic and exocytic kymographs generated from the cell cortex of unbudded control cells displayed a focused exocytic pole corralled by endocytic vesicles (Figure 2A). However, bud6Δ and she4Δ mutants exhibited defects in the polarization of exocytic and endocytic domains over time, respectively, whereas both trafficking domains were affected in bud32Δ cells (Figure 2A), consistent with their steady-state phenotypes in our screen.
FIGURE 2:
The endocytic and exocytic pathways are coupled during polarity establishment. (A) The kymographs were generated from a region selected around the cell cortex of unbudded, polarizing cells (dashed lines). The magnified image illustrates endocytic corralling of the exocytic pole. (B) Scatter dot plot showing the time intervals between consecutive endocytic events during polarity establishment. Control cells (black) and she4Δ (red), bud6Δ (cyan), and bud32Δ (purple) mutants. The black bars indicate the mean and SD over N events. (C) The distributions of exocytic pole size for polarized control cells (black) compared with she4Δ (red), bud32Δ (purple), and bud6Δ (cyan) mutants.
Of interest, although these mutants displayed a fraction of nonpolarized trafficking domains, there was also a significant fraction of endocytic and exocytic vesicles that remained polarized in the mutants during polarity establishment (Figure 2A). These results indicate that even in mutants that specifically disrupt the spatial organization of either endocytic or exocytic trafficking domains during polarity maintenance, coupling of the domains is evident during polarity establishment.
Consistent with this, the mutants representative of all three classes displayed altered endocytic dynamics during the formation of an exocytic pole (Figure 2B). Previously we showed that regular, periodic endocytic events constitute a signature of normal polarity establishment in wild-type cells (Jose et al., 2013). In contrast to control cells, for which endocytic events were indeed periodic (time interval of 54 s between events, with a low SD of 26 s), endocytic events were more temporally random in the mutants, resulting in a larger spread of endocytic time intervals and a higher SD. The endocytic signature was perturbed irrespective of whether endocytosis (she4Δ, 78 s, SD = 52), exocytosis (bud6Δ, 65 s, SD = 38), or both pathways (bud32Δ, 62 s, SD = 45) were predominantly affected (Figure 2B). Moreover, delayed internalization of the lipophilic dye FM4-64 revealed defective endocytosis in the she4Δ mutant, whereas the intracellular accumulation of the dye in bright puncta in bud32Δ mutants (Supplemental Figure S4) indicated that the spatial disorganization of endocytic vesicles in these mutants reflects underlying functional defects.
Previously we found that mutants defective in endocytic corralling often display widened, less focused exocytic poles (Jose et al., 2013). The she4Δ, bud6Δ, and bud32Δ mutants also displayed significant widening of the exocytic pole compared with control cells (Figure 2C). These in vivo studies reveal the spatial and functional coupling of endocytic and exocytic compartments during polarity establishment. Together with the steady-state observations, our results suggest that the coupling of these trafficking pathways is essential during polarity establishment but dispensable during polarity maintenance.
Endoytic and exocytic mutants display opposite defects in exocytic vesicle dynamics
To understand whether spatial disorganization of the trafficking pathways reflects underlying defects in vesicle transport dynamics, we compared exocytic vesicle dynamics in mutants and control cells. However, monitoring post-Golgi vesicle dynamics presents a formidable technical challenge in yeast, since these vesicles are highly dynamic and localize to the bud at high density, making single-vesicle tracking difficult. To monitor single vesicles, we therefore tagged the exocytic marker Sec4 with a photoswitchable fluorophore, mEos, enabling stochastic switching of the fluorophore and the simultaneous visualization of only a subset of vesicles. The tagged protein was verified to be functional (Supplemental Figure S5). Single-vesicle tracking was compared between control cells and the endocytic mutant she4Δ, the exocytic mutant bud6Δ, and the bud32Δ mutant affecting both endocytic and exocytic vesicle organization.
Post-Golgi vesicles were imaged in small-budded cells and then localized and tracked in time to obtain trajectories of vesicle transport. We calculated the mean square displacement (MSD) and then derived the instantaneous diffusion coefficient (D) of each trajectory to identify different vesicle subpopulations (Zajac et al., 2013; Sibarita, 2014), allowing us to discern whether their kinetics were altered in the endocytic and exocytic mutants.
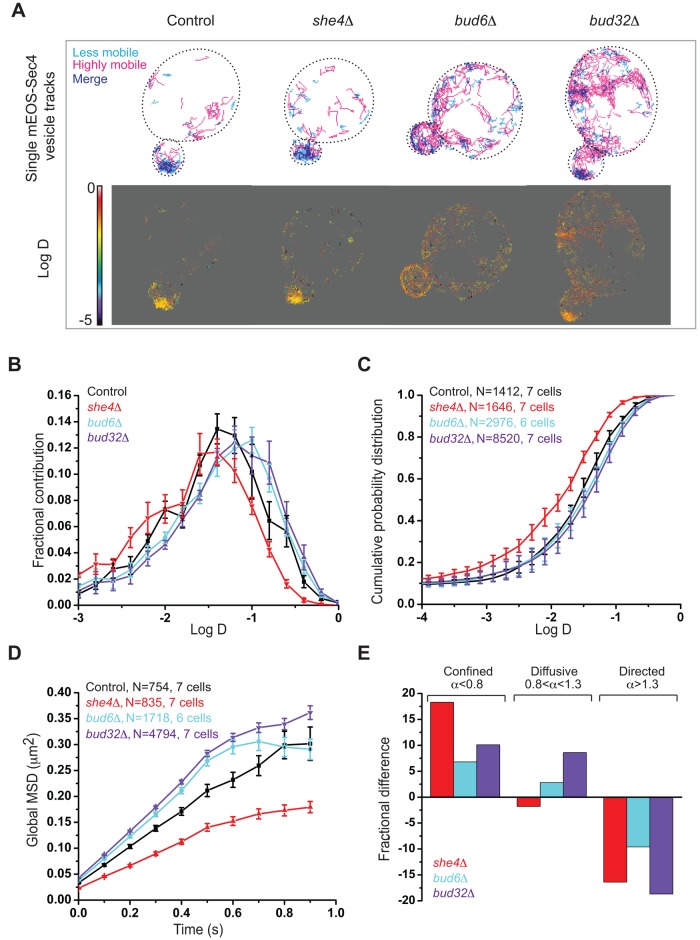
We initially compared two windows of trajectories: a less mobile pool (Figure 3A, top, cyan, D = 0.01–0.03 μm2/s) and a highly mobile cohort (magenta, D > 0.1 μm2/s), in control and mutant cells. Control and she4Δ cells displayed a major pool of the less mobile trajectories in the buds, whereas this pool was reduced in bud6Δ and bud32Δ mutants (Figure 3A, top). Consistently, the heat map of D including all vesicle trajectories in control and she4Δ cells displayed lower mobility in the bud (Figure 3A, bottom), contrary to buds of bud6Δ and bud32Δ mutants.
FIGURE 3:
Endocytic and exocytic mutants display opposite effects on exocytic vesicle dynamics. (A) Top, single-vesicle trajectories of mEos-Sec4 in small-budded she4Δ, bud6Δ, and bud32Δ cells vs. control cells. Less mobile (cyan, D = 0.01–0.03 μm2/s) and highly mobile trajectories (magenta, D > 0.1 μm2/s). Bottom, heat maps of D of exocytic vesicles for control cells and mutants. Scale bar, log D values on the heat map. (B) Normalized histograms of log D of exocytic vesicles in control cells (black) and she4Δ (red), bud6Δ (cyan), and bud32Δ (purple) mutants. (C) The cumulative probability distributions of log D of exocytic vesicles in control cells (black), she4Δ (red), bud6Δ (cyan), and bud32Δ (purple) mutants. (D) The average MSD curves for control cells (black) and she4Δ (red), bud6Δ (cyan). and bud32Δ (purple) mutants. (E) The change in the fraction of mEOS-Sec4 vesicles displaying confined, diffusive, or directed movement in mutants compared with control. Error bars indicate SE in B–D, and N is the number of trajectories.
To quantify these differences in mEOS-Sec4 tagged vesicle dynamics, we analyzed the distribution of raw D and the cumulative probability distribution of D for the entire range of diffusion for all of the trajectories in control and mutant cells (Figure 3, B and C). In addition, we analyzed the MSD averaged over all trajectories for each strain (Figure 3D). Finally, to determine whether overall differences in vesicle mobility reflects varying populations of differentially moving vesicle cohorts, we also analyzed the MSD of individual trajectories using a general fitting function whose scaling exponent α can be used to identify subpopulations of vesicles with confined (α < 0.8), diffusive (0.8 < α < 1.3), or directed (α > 1.3) motions (Figure 3E; Materials and Methods; Sibarita, 2014). The histograms and probability distributions were normalized to the total number of trajectories for each sample (>1000). In contrast to she4Δ cells, for which the histogram was shifted to lower D values (less-mobile trajectories) compared with the control, histograms of bud6Δ and bud32Δ mutants were shifted to higher D values (more-mobile trajectories). The median D obtained by pooling all of the trajectories was significantly lower in she4Δ (0.02 μm2/s ± 0.001) than in control cells (0.036 μm2/s ± 0.002), whereas it was higher in bud6Δ (0.043 μm2/s ± 0.002) and bud32Δ (0.047 μm2/s ± 0.001) cells (Supplemental Table S3). Consistently, the average MSD curves, whose initial slope reflects the instantaneous mobility of vesicles, displayed lower mobility in she4Δ cells compared to the control, in contrast to bud6Δ and bud32Δ cells (Figure 3D).
Relative to control cells, all mutants showed an increase in the confined population of exocytic vesicle trajectories, whereas the fraction of exocytic vesicles undergoing directed movement was reduced in all mutants. The fraction of vesicles undergoing diffusive movement increased in bud6Δ and bud32Δ cells compared with control cells, but slightly decreased in she4Δ cells (Figure 3E).
Next we analyzed the MSD curves of these different vesicle subpopulations to study how their mobility was altered and understand how this contributed to the global changes in the exocytic vesicle dynamics of the control and mutants. The MSD curves of the confined vesicle fraction showed lower mobility in she4Δ cells, in contrast to bud6Δ and bud32Δ cells, which displayed similar mobility to that of control cells (Supplemental Figure S6A). Conversely, the MSD curves of the diffusive fraction displayed increased mobility in bud6Δ and bud32Δ cells compared with control cells (Supplemental Figure S6B). Thus the increase in the confined fraction of lower D in she4Δ cells contributed to a reduction in the global mobility of exocytic vesicles, whereas the increase in the diffusive fraction of exocytic vesicles in bud6Δ and bud32Δ cells displaying higher D increased the global exocytic vesicle mobility in these mutants.
In summary, the perturbation of She4 or Bud6, whose activity is required for the spatial organization of endocytic and exocytic trafficking domains, respectively, alters post-Golgi vesicle dynamics in vivo but in opposite ways.
The screen identifies the exocyst complex as a network hub connecting the endocytic and exocytic pathways
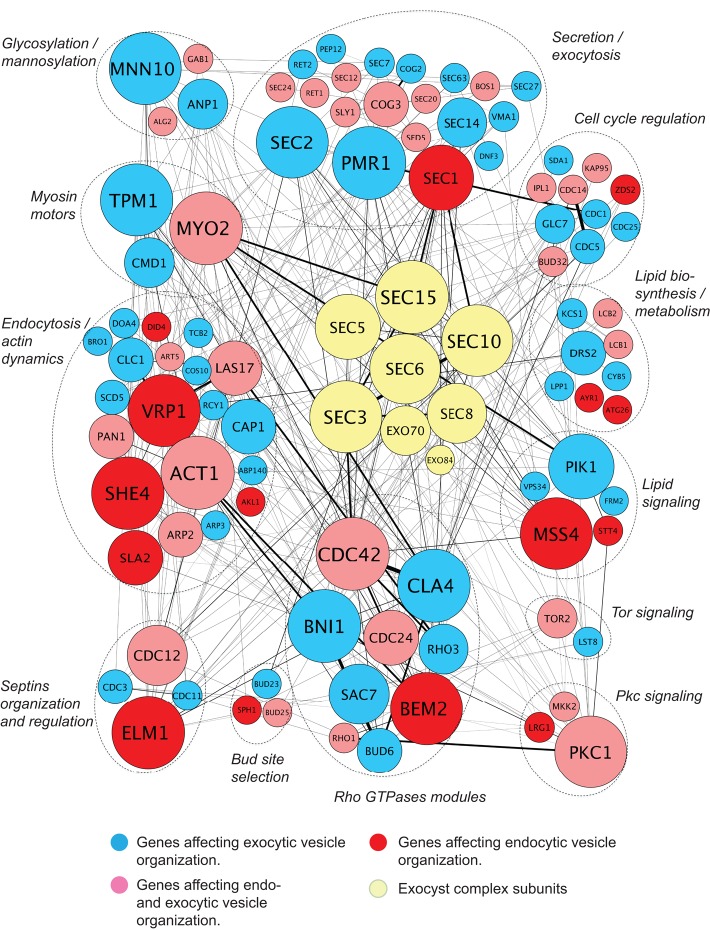
We next integrated the mutants identified by our imaging screen with functional data from genetic interactions to visualize the network of components required for the spatial organization of endocytic and exocytic domains. By identifying the players displaying the most functional interactions with the network, our goal was to identify pivotal components that organize endocytic and exocytic domains. Key polarity proteins such as Cdc42, the p21-activated kinase (PAK) Cla4, and actin-associated proteins, including Vrp1 and Myo2, displayed the most connections to other genes in the network (Figure 4). Whereas the average number of interactions displayed by a component of the network was 7, the exocyst vesicle-tethering complex, an essential octameric complex required for exocytosis in yeast (TerBush and Novick, 1995), showed a significantly higher average of 13 interactions for each subunit (p < 0.001). Because the exocyst is an octameric complex whose subunits display genetic interactions with each other, we also analyzed the number of unique genetic interactions between the exocyst complex and nonexocyst genes to avoid introducing bias due to the number of subunits in the exocyst complex. The exocyst interacted with 26 other components of our network, ∼25% of the entire network, compared with an average of seven interactions for nonexocyst genes. The exocyst complex thus emerged as a network hub connecting key components required for the spatial organization of the endocytic and exocytic pathways.
FIGURE 4:
Systems analysis identifies the exocyst complex as a hub linking the endocytic and exocytic pathways. In the genetic interaction map, the mutants identified by the screen are classified according to their function and connected by lines representing genetic interactions with other network components. Each gene is represented by a circle whose size increases with the number of interactions with other genes. Note that the size of the exocyst genes is not proportional to the number of interactions, as the gene names would be too large for presentation. On average, genes encoding exocyst complex subunits are more connected with genes affecting the spatial organization of trafficking pathways than are those genes with each other.
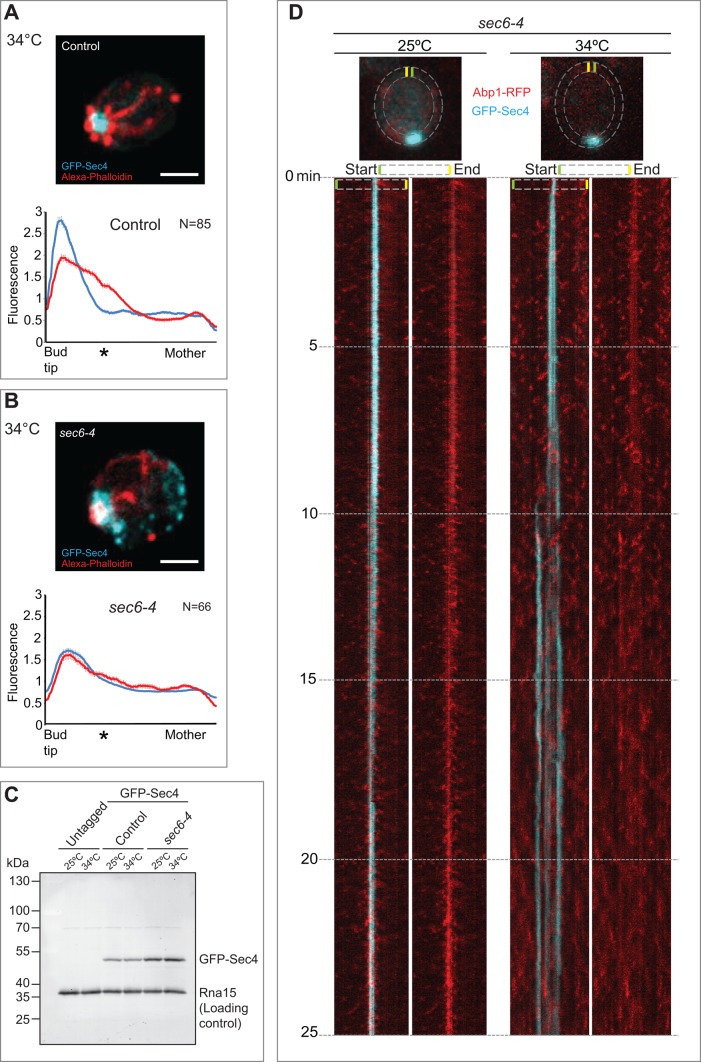
In addition to its role in tethering exocytic vesicles at the plasma membrane in yeast, the exocyst complex colocalizes with endocytosis-associated clathrin and its adapters in fly and human cells (Folsch et al., 2003; Prigent et al., 2003; Langevin et al., 2005; Sommer et al., 2005). The exocyst has been ascribed functions during endocytosis at a step between the internalization and recycling of diverse cargoes, including Cadherin and the vitellogenin receptor in flies (Langevin et al., 2005; Sommer et al., 2005) and GLUT4, hTAC, and DAF-4 basolateral recycling in worms (Chen et al., 2014). In yeast, exocyst localization at the plasma membrane also requires clathrin (Shen et al., 2013), and the exocyst has been shown to play a role in the endocytic recycling of Ste3, the mating factor receptor (Sommer et al., 2005). Indeed, some of the earliest pioneering work on the secretory pathway in budding yeast demonstrated a role for the exocyst in endocytosis (Riezman, 1985). Thus evidence in diverse systems suggests a link between exocyst function and endocytosis/exocytosis. However, the role of the exocyst in the spatial organization of endocytic and exocytic trafficking domains is not fully understood. To investigate this role, we monitored endocytic and exocytic vesicle organization in the exocyst mutant sec6-4 fixed after shifting to the restrictive temperature of 34ºC for 1 h. Rather than the usual bull's-eye configuration of endocytic and exocytic vesicles, sec6-4 cells displayed endocytic sites interspersed with the exocytic pole (Figure 5, A and B). This phenotype was observed in many of the exocyst mutants examined, with varying severity, as illustrated by the superimposed endocytic and exocytic fluorescence distribution peaks derived by averaging fluorescence along the bud–mother axis in multiple cells (Figures 5B and 6E and Supplemental Figure S3). The interspersal of endocytic and exocytic markers was evident in exocyst mutants but not in another late-acting sec mutant, sec1-1, or in the myo2-16 mutant (Supplemental Figure S3). Western blotting with anti-GFP antibodies did not reveal major differences in expression of GFP-Sec4 between cells grown at 25 and 34°C (Figure 5C).
FIGURE 5:
Endocytic and exocytic vesicles become interspersed in exocyst mutants. (A) Steady-state image of a fixed control cell. The maximum intensity projection of a deconvolved 3D image stack. Scale bar, 2 μm. Bottom, fluorescence distribution (y-axis) of the endocytic (red) and exocytic (cyan) compartments along the bud–mother axis (x-axis, from left to right) in small-budded cells. Error bars denote SD. An asterisk indicates the position of the average bud neck. (B) As in A, but showing a sec6-4 mutant after shift to the restrictive temperature. (C) Immunoblot analysis of the levels of GFP-Sec4 at 25 and 34°C in untagged cells, sec6-4 cells, and an isogenic wild-type control. The protein Rna15 was detected as a loading control. (D) Simultaneous, dual-color near-TIRFM to examine in vivo dynamics of endocytic (red) and exocytic (cyan) vesicles in the sec6-4 exocyst mutant at 25 and 34°C. Cells were grown at 25ºC and then imaged at the indicated temperature. The dashed lines represent the region selected for generating the kymograph. The kymograph on the left shows the merge of the endocytic and exocytic channels; the right kymograph shows the endocytic channel alone.
FIGURE 6:
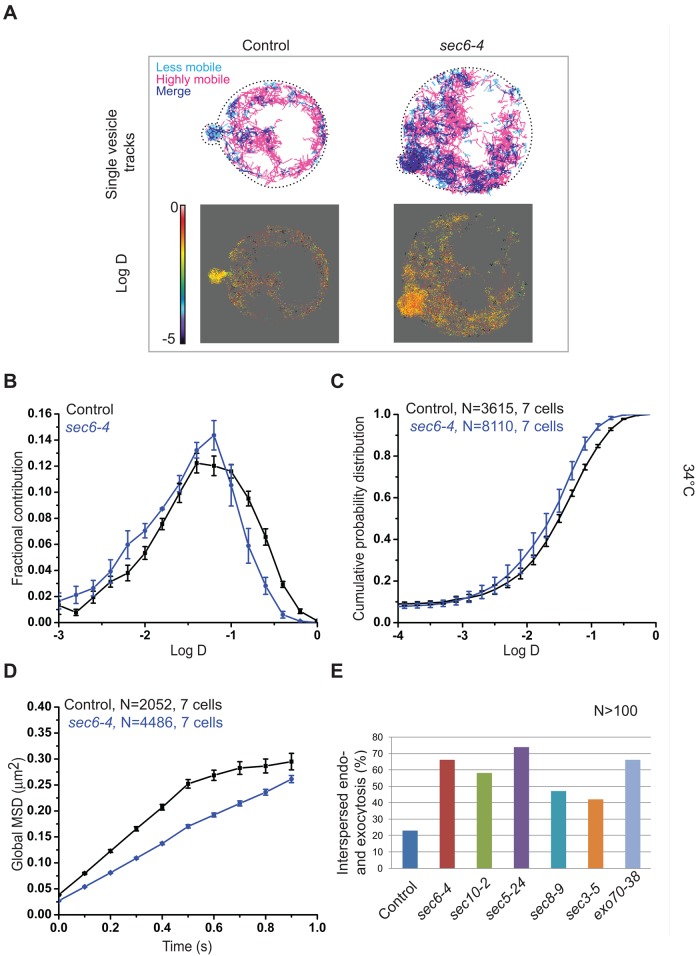
Vesicle tracking in exocyst mutants reveals exocytic dynamics similar to that of endocytic mutants. (A) Top, vesicle trajectories in small-budded control and sec6-4 cells expressing mEos-Sec4. Lower (D = 0.01–0.03 μm2/s) and higher (D > 0.1 μm2/s) mobile vesicle trajectories are shown in cyan and magenta, respectively. Bottom, heat maps of D of exocytic vesicles in control and sec6-4 cells. Scale bar, log D values on the heat map. (B) Normalized histograms of log D of exocytic vesicles in control cells (black) and sec6-4 (blue) mutants. (C) Cumulative probability distributions of log D of exocytic vesicles in control cells (black) and sec6-4 (blue) mutants. (D) Average MSD curves for control cells (black) and sec6-4 mutants (blue). (E) Percentage of cells displaying interspersed endocytic and exocytic trafficking domains in different exocyst mutants. More than 100 small-budded cells were used for the control and each mutant in the statistics. The error bars indicate SE in B–D, and N is the number of trajectories.
In vivo imaging of endocytic and exocytic dynamics by simultaneous, dual-color, evanescent-field imaging in sec6-4 cells revealed striking disruption of the exocytic pole after only 6 min at the restrictive temperature of 34ºC (Figure 5D, right kymograph). Initially, the exocytic pole widened before splitting, at which point endocytic and exocytic events became interspersed (Figure 5D and Supplemental Movie S1). Endocytosis became rapidly depolarized at the restrictive temperature, in contrast to endocytic activity at the permissive temperature, which remained discrete and polarized (Figure 5D, left kymograph, and Supplemental Movie S2). Consistently, at the restrictive temperature, endocytic events in sec6-4 cells were less periodic (82 s, SD = 62) compared to control cells (52 s, SD = 30; Supplemental Figure S7A). Consistent with a functional relationship between the exocyst and the endocytic apparatus, we identified a genetic interaction between the endocytic machinery and sec6-4. The sec6-4 mutant displayed a cold-sensitive phenotype at 15°C, where its growth rate was reduced compared with control cells, and the organization of endocytic and exocytic domains was disrupted. However, when sec6-4 was combined with the endocytic mutant rvs167Δ, the double mutant formed colonies at a faster rate than the sec6-4 mutant alone at 15°C (Supplemental Figure S7B), and the trafficking domains displayed an organization that was indistinguishable from that of control cells (Supplemental Figure S7C).
Having seen the perturbation of endocytic and exocytic dynamics upon exocyst inactivation and a genetic interaction between the exocyst and the endocytic machinery, we next investigated how exocytic vesicle mobility was affected in the exocyst mutant sec6-4. Quantitative analysis of exocytic vesicle dynamics at 34°C revealed a reduction in the mobility of mEOS-Sec4 vesicles in sec6-4 compared with an isogenic wild-type control at the same temperature (Figure 6, A–C). The reduced exocytic vesicle mobility in sec6-4 cells resembled that in the endocytosis-defective she4Δ mutant. The median D for all the analyzed trajectories was significantly lower for sec6-4 (D = 0.032 μm2/s ± 0.001) than for the control (D = 0.042 μm2/s ± 0.002; p < 0.001). The confined, diffusive, or directed vesicle subpopulations did not show any prominent differences in their fractional contribution in sec6-4 cells compared with the control (<1%). Of interest, analysis of the MSDs of these different vesicle fractions revealed lower D for all of them compared with the control, irrespective of their mode of movement, explaining the overall lower exocytic vesicular mobility in this mutant (Supplemental Figure S6, C and D). Given that vesicle accumulation, particularly in the buds of sec6-4 mutant, occurs within a limited volume of space because cell surface expansion is blocked (Novick et al., 1980; Zhang et al., 2008), the lower D may reflect the limited mobility of vesicles due to steric hindrance. Although moving with lower mobility, exocytic vesicles in sec6-4 did not display complete confinement at any time during the measurement, in contrast to control cells (Figure 6C and Supplemental Figure S6, C and D). This observation is consistent with a defect in vesicle tethering, since tethered vesicles at the plasma membrane would be expected to exhibit confinement before or concomitant with fusion.
DISCUSSION
The endocytic and exocytic pathways appear to be interdependent during polarity establishment but independent during polarity maintenance
In this study, we used a quantitative imaging-based screen to identify mutants that affect the spatial organization of endocytic and exocytic trafficking compartments in order to understand whether these trafficking domains are obligatorily connected. The phenotypes identified by the screen were based on the localization differences of the endocytic and endocytic reporters, namely GFP-Sec4 and F-actin (labeled by phalloidin), essential components of these vesicular compartments. The screen identified mutants affecting the spatial organization of endocytic or exocytic domains independently in small-budded cells undergoing a growth phase requiring polarity maintenance, consistent with the idea that the two pathways are not obligatorily coupled.
Mutants that specifically affect only one pathway in small-budded cells during polarity maintenance include she4, an interaction partner of all myosins in budding yeast (Toi et al., 2003; Wesche et al., 2003). The endocytosis-associated actin patch localization defects and endocytic defects observed in she4Δ mutants are likely linked to dysfunction of the type 1 myosins Myo3 and Myo5 (Toi et al., 2003; Wesche et al., 2003). She4 also physically interacts with the exocytosis-associated motor Myo2; however, she4Δ cells are not reported to display defects in Myo2 localization, explaining why she4Δ mutants were found to specifically affect endocytic but not exocytic vesicle localization in our screen (Wesche et al., 2003).
The screen also identified mutants, including bud6Δ, that specifically affect exocytic vesicle organization during polarity maintenance. Bud6 feeds the formin Bni1 with actin monomers, functioning as an actin nucleation-promoting factor of actin filaments (Moseley et al., 2004; Moseley and Goode, 2005; Graziano et al., 2011). The filaments nucleated by formins serve as tracks for myosin motor-mediated exocytic vesicle transport (Evangelista et al., 1997; Sagot et al., 2002; Schott et al., 2002). Bud6 is therefore an important regulator of formin-mediated actin assembly, explaining the faint actin cable staining and depolarized exocytosis identified by the screen. Indeed, others reported thinner actin cables in bud6Δ mutants (Graziano et al., 2011). We found that endocytosis-associated actin patches were not significantly depolarized in bud6Δ mutants; however, the observed reduction of actin cable staining was associated with brighter actin patches in the mutant (Supplemental Figure S2). This may reflect a shift in the equilibrium of the monomeric actin pool between endocytic and exocytic associated actin filament assembly mechanisms, as reported in fission yeast for Arp2/3 and formin mutants (Burke et al., 2014).
Whereas mutants such as she4Δ and bud6Δ affected either endocytic or exocytic vesicle organization, respectively, in small-budded cells, both pathways were simultaneously affected in these mutants during polarity establishment. These results suggest that during polarity establishment, the endocytic and exocytic systems may be obligatorily coupled such that perturbation of one pathway inevitably impinges upon the other, likely reflecting a need for the rapid recycling of endocytic and exocytic components to optimize polarized growth.
The spatial disorganization of endocytic and exocytic domains in the mutants identified by the screen reflected underlying defects in these transport pathways. Both she4Δ and bud32Δ cells displayed defects in endocytic uptake of FM4-64 (Supplemental Figure S4). The high-resolution single-vesicle tracking studies in small-budded cells indicated that secretory vesicle dynamics was altered in the endocytic and exocytic mutant she4Δ and bud6Δ cells, but in opposite ways. When compared with control cells, the endocytic mutant she4Δ displayed a decrease in the mobility of Sec4-marked exocytic vesicles. This decrease might reflect defects in the endocytic recycling of factors required for releasing Sec4 from the membrane during fusion; candidates being the Sec4 GAPs Msb3/4 or a downstream effector of Sec4. The reduced mobility of exocytic vesicles in the exocyst mutant sec6-4 may emanate from a similar defect, since we observed that exocyst mutants, which are known to exhibit recycling defects (Sommer et al., 2005), are also defective in endocytosis. This idea is consistent with a report that the recycling of the myosin motor Myo2 is dependent on Sec4 GTPase activity and the exocyst; when the GTPase activity of Sec4 is perturbed or the exocyst is inactivated, post-Golgi vesicles accumulate in the bud (Donovan and Bretscher, 2012).
Conversely, bud6Δ and bud32Δ mutants displayed depolarized exocytosis, which may contribute to the increase in the diffusive fraction of vesicles displaying higher mobility, as identified in our high-density vesicle tracking experiments.
The exocyst is a network hub connecting endocytosis and exocytosis
Three lines of evidence support the idea that the exocyst complex is a network hub connecting endocytosis and exocytosis. First, exocyst mutants display defects in both endocytosis and exocytosis, as would be expected of an important regulator of these pathways. Second, exocyst mutants display a striking interspersal of endocytic and exocytic domains, sharing the cortical region predominantly occupied by exocytic vesicles in wild-type cells. This phenotype, which is seen in many exocyst mutants to a varying degree, is distinct from that seen in the late-acting sec mutant sec1-1 or myo2-16, indicating that exocyst activity is critical for the normal spatial organization of endocytic and exocytic domains. During polarity establishment in wild-type cells, the endocytic and exocytic zones tend to be spatially segregated, forming a bull's-eye pattern in which the exocytic pole is surrounded by endocytosis-associated actin patches (Jose et al., 2013). In contrast, the endocytic and exocytic zones display significant overlap at the restrictive temperature in exocyst mutants, associated with an ensuing block in polarized growth. These observations illustrate the importance of the spatial segregation of the trafficking compartments to optimize polarized growth.
The third line of evidence supporting a key role for the exocyst in endocytosis/exocytosis was derived by studying the repertoire of genetic interactions displayed between the mutants identified by our imaging screen. By integrating the data from our imaging-based screen with large-scale genetic screens (Schuldiner et al., 2005; Costanzo et al., 2010), we built a network of functionally connected components required for the spatial organization of endocytic and exocytic domains. This analysis revealed the exocyst complex as being more highly connected to regulators of endocytosis, exocytosis, and polarity than other components of the network. This is consistent with previous work in diverse systems, including yeast, flies, and humans, demonstrating a role for the exocyst in exocytosis and endocytic-based recycling (Riezman, 1985; Langevin et al., 2005; Sommer et al., 2005). Moreover, it endorses the utility of combining orthogonal quantitative approaches in the identification of important regulators of membrane trafficking (Schuldiner et al., 2005; Costanzo et al., 2010; Liberali et al., 2014).
Collectively the use of dual-color, evanescent-field imaging, high-resolution single-vesicle tracking, and systems-level and genetic analyses points to the role of the exocyst as a network hub connecting the endocytic and exocytic pathways to optimize polarized growth. Understanding the signals that determine endocytic and exocytic fluxes via the exocyst will shed light on how these trafficking pathways control cellular polarity and tune polarized membrane growth to generate appropriate cell shape.
MATERIALS AND METHODS
Strains and fixation
The yeast strains used for live imaging are listed in Table 1, and those used for candidate screen are listed in Supplemental Table S1. For the imaging-based screen, cells were transformed with a CEN LEU2 GFP-Sec4 plasmid (pDM18, a gift from R. Collins, Cornell University), grown to log phase, and fixed with 3.7% formaldehyde, 10 mM sodium azide, and 10 mM sodium fluoride for 10 min (Calero et al., 2003). Cells were centrifuged and resuspended in 1× phosphate-buffered saline (PBS) containing 3.7% formaldehyde, 10 mM sodium azide, and 10 mM sodium fluoride for additional 50 min. The fixed cells were labeled with Alexa 546–phalloidin (1000 U/ml) and Alexa 356–concanavalin A (1.5 mg/ml) before imaging in 1× PBS. For live-cell imaging, PCR-based homologous recombination at the endogenous gene locus was used to tag open reading frames (ORFs) at the C-terminus.
TABLE 1:
Yeast strains used for live imaging.
| Name | Genotype | Reference |
|---|---|---|
| DMY 1129 | MATa, SSD1-V, Δbar1::kanMX6, his3^1, leu2^0, met15^0, ura3^0 | Haploid deletion library |
| DMY 1145 | MATa, SSD1-V, Δbud6::kanMX6, his3^1, leu2^0, met15^0, ura3^0 | Haploid deletion library |
| DMY 1185 | MATa, SSD1-V, Δshe4::kanMX6, his3^1, leu2^0, met15^0, ura3^0 | Haploid deletion library |
| DMY 1426 | MATa, SSD1-V, Δbud32::kanMX6, his3^1, leu2^0, met15^0, ura3^0 | Haploid deletion library |
| DMY 1698 | MATa, SSD1-V, Δbar1::kanMX6, ABP1-RFP::HIS3, his3^1, leu2^0, met15^0, ura3^0 | This study |
| DMY 1892 | MATa, SSD1-V, Δbud6::kanMX6, ABP1-RFP::HIS3, GFP-SEC4::URA3, his3^1, leu2^0, met15^0, ura3^0 | This study |
| DMY 1692 | MATa, SSD1-V, Δshe4::kanMX6, ABP1-RFP::HIS3, his3^1, leu2^0, met15^0, ura3^0 | This study |
| DMY 1706 | MATa, SSD1-V, Δbud32::kanMX6, ABP1-RFP::HIS3, his3^1, leu2^0, met15^0, ura3^0 | This study |
| DMY 349 | MATa, bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 350 | MATα, bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 937 | MATa, sec6−4::HIS3, bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 979 | MATa, sec6−4::kanMX6, bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 1798 | MATa, ABP1-RFP::HIS3, GFP-SEC4::URA3 , bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 1894 | MATa, sec6−4::kanMX6, ABP1-RFP::HIS3, GFP-SEC4::URA3 , bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 1787 | MATα, Δrvs167::natNT2, bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 1919 | MATa, sec6−4::kanMX6, Δrvs167::natNT2, bar1, his3-11,15, leu2-3112, trp1-1, ura3-1, ade2-1, can1-100, GAL+ | This study |
| DMY 1563 | MATa, SSD1-V, sec4-8::kanMX6, his3^1, leu2^0, met15^0, ura3^0 |
DMY1698, 1892, 1692, and 1706 were transformed with a CEN LEU2 GFP-Sec4 plasmid (pDM18) for in vivo imaging of endocytosis/exocytosis. DMY1129, 1145, 1426, 1185, 350, and 979 were transformed with a CEN LEU2 mEos-Sec4 plasmid (pDM415) for single-vesicle tracking.
Steady-state imaging and image processing
The cells were imaged in 1× PBS using a wide-field inverted microscope (Axiovert 200M) equipped with a 100× objective (oil, numerical aperture [NA] 1.4, plan Apo), mercury lamp (HBO10; Carl Zeiss, Marly le Roi, France), and an electron-multiplying charge-coupled device (EMCCD) camera (Evolve; Photometrics, Tuscon, AZ). MetaMorph 7.7 software (Molecular Devices, Sunnyvale, CA) was used for image acquisition and analysis. Filter sets LF488-B-000 (FFO2-482/18, FFO1-525/45, Di01-R488 [exciter, emitter, dichroic]), LF561-A-000 (FFO2-561/14, FFO1-609/54, Di01-R561), and LF405-A-000 (FFO1-390/40, FFO1-452/45, Di01-R405; Semrock, Rochester, NY) were used to sequentially image cells expressing GFP-Sec4 and labeled with Alexa 546–phalloidin and Alexa 356–concanavalin A, respectively.
For steady-state imaging, 3-μm Z-stacks of 0.15-μm step size were acquired. The multicolor three-dimensional (3D) image stacks were deconvolved using a point spread function (PSF)–based iterative constraint deconvolution algorithm with eight iterations (Sibarita, 2005) and displayed as a maximum intensity projection (Figure 1A). The experimental PSF of each wavelength was extracted using diffraction-limited beads (Molecular Probes, Thermo Fisher, Waltham, MA). Deconvolution was performed separately for the GFP and monomeric RFP channels using their respective PSF. The software package for 3D deconvolution was custom written as a plug-in running within the MetaMorph software environment.
Qualitative analysis
The candidate screen was conducted in two parts—an initial qualitative screen, followed by semiautomated quantification. For the qualitative analysis, the steady-state organization of endocytic and exocytic domains in mutant strains was compared with control cells. The analysis was conducted on the untreated image stacks. We restricted the analysis to small-budded cells for which the endocytic and exocytic compartments display a highly polarized organization in control cells. Mutants in which polarization is strongly affected would be expected to display weaker fluorescence in the bud and depolarized vesicles in the mother cell (Supplemental Figure S1B and Figure 1A). More than 100 small-budded cells were analyzed for each strain. Supplemental Table S1 shows the mutants that exhibited phenotypes compared with control cells. These mutants were selected for further quantitative analysis, as described in the next section.
Semiautomated quantitative analysis
Analysis method.
Quantitative analysis of the steady-state multicolor image stacks was performed using Matlab and its Image Processing Toolbox (MathWorks, Natick, MA).
Quantification of fluorescence ratios for individual cells.
Concanavalin A staining of the cell wall was used for cell segmentation. This facilitated the distinction of mother cells from buds and the estimation of their surface area from a projection of the in-focus planes of the image stack. Cells for which the bud size (area) was <30% of the mother cell size were selected for further analysis. This method excluded larger-budded, nonpolarized cells. Subsequently the total endocytic and exocytic fluorescence intensities in the bud and the mother cell were calculated. The bud-to-mother FR was computed separately for the endocytic and exocytic channels, yielding one endocytic FR and one exocytic FR for each cell.
Statistical analysis of FRs.
Image analysis was performed on 20–35 cells/strain that satisfied the stringent selection criterion by which the bud size was <30% of the mother size. Distributions of FRs at a population (strain) level were analyzed statistically in Excel. The FR is a meaningful measure of how well the endocytic or exocytic domain is polarized. However, fluorophores carried by individual vesicles produced a discrete rather than continuous signal (Figure 1A). Therefore small fluctuations in vesicle numbers generated significant biological noise and, owing to the broad range of FR values, resulted in wide distributions, even for control cells. This observation complicated the definition of “polarized” and “depolarized” domains. We found that our qualitative visual observations were best reproduced by quantitative analysis when a “thresholding” method was incorporated, as follows.
A reference FR below which a cell can be considered as depolarized and above which it can be considered as polarized was set at the median of the FR distribution for the control strain (96 cells pooled from four different experiments). Therefore, by definition, 50% of control cells are counted as “depolarized.” This defined a very stringent criterion for polarity, enabling the identification of subtle phenotypes. To estimate the inaccuracy in the determination of this threshold, the medians of FR distributions were calculated independently for the four different experiments on the control strain, and the standard error (SE) of the median (M) was extracted. The polarity state (polarized or depolarized) of mutant cells for which the FR was between M − SE and M + SE might be incorrectly determined owing to the uncertainty in threshold determination. This fraction defines the error bars displayed in Supplemental Figure S1 and Figure 1.
Strains for which the percentage of cells with polarized endocytosis, exocytosis, or both were <70% of control cells were classified into three phenotypes: depolarized endocytosis, depolarized exocytosis, or both depolarized.
Control of analysis results using Student's t tests.
Results obtained using the thresholding method were verified using a direct statistical analysis of the distribution of FR. The mean and SD of the endocytic and exocytic FRs were computed for control and mutant strains. Each mutant condition was compared with the control strain using a two-sample Student's t test. The mutants showing a significantly lower mean for endocytic and/or exocytic FR are listed in Supplemental Table S2, together with the corresponding p values. Student's t tests confirmed the phenotypes obtained with the thresholding method and revealed five additional mutants with a significantly lower polarization level for endocytosis (vrp1Δ) or exocytosis (clc1Δ, drs2Δ, kcs1Δ, and tcb2Δ). These phenotypes were confirmed after visual inspection of the microscopy images.
Imaging of actin cables and patches
Cells were grown to early log phase (OD of ∼0.2) and fixed with 3.7% formaldehyde, 10 mM sodium azide, and 10 mM sodium fluoride for 10 min. After centrifugation, cells were resuspended in 1× PBS containing 3.7% formaldehyde, 10 mM sodium azide, and 10 mM sodium fluoride for an additional 50 min. The fixed cells were labeled with Alexa 546–phalloidin at a final concentration of 1000 U/ml. The cells were imaged on a spinning disk confocal microscope (Observer Z1; Carl Zeiss) equipped with a Yokogawa spinning disk unit (CSU X1-A1N), a 100× differential interference contrast objective (oil, NA 1.4, plan Apo), and an xyz piezo stage by exciting with a 561-nm laser (Jive; Cobolt, Solna, Sweden). Fluorescence was acquired using an Evolve EMCCD camera after passing through a quad-band mirror (405, 491, 561, 642 nm). The confocal microscope was driven using MetaMorph 7.7, and the maximum intensity projections of the image stacks were generated as displayed in Supplemental Figure S2.
Analysis of the number of actin patches and Abp1-marked endocytic vesicles
Maximum intensity projections of 10 plane confocal stacks (2 μm) were generated for control and mutants. Visually resolvable actin patches/vesicles in the mother cell were manually counted in polarized unbudded and small-budded cells. The box-and-whisker plots of the number of patches/vesicles in control cells and mutants were created using GraphPad Prism 5 (Supplemental Figure S2B). A nonparametric two-tailed Mann–Whitney test was performed to determine whether the median number of patches per cell was significantly different between the control and mutants.
Analysis of the brightness of actin patches and Abp1-marked endocytic vesicles
The average intensity of the actin patches was calculated using the Integrated Morphometry Analysis module in MetaMorph as follows. Maximum intensity projections of 10 plane confocal stacks (2 μm) were generated for control cells and mutants. The images were autothresholded to mask all the cells in a frame. The mean average intensity and SD in the intensity were calculated for all the cells. Average intensity + 2×SD was taken as the manual threshold for the untreated intensity projection images of the same cells to calculate the average brightness per pixel corresponding to the patches. This thresholding procedure averaged out any statistical fluctuations or illumination differences between frames. As an additional control, only images with the average fluorescence intensity of the area covered by the cells within 1000-Gy values from that of the control were included for all mutants for this statistics to further overrule any imaging differences. Size filtering was done to select only individual actin patches and avoid clusters. The box-and-whisker plots of the brightness of the patches were compared between the control and the mutants (GraphPad Prism 5; Supplemental Figure S2C). A nonparametric two-tailed Mann–Whitney test was performed to determine whether the median brightness of patches was significantly different between the control and mutants.
FM4-64 labeling and imaging
Cells were grown to log phase. Endocytosis was blocked by incubating the cells at 4°C for 5 min, and FM4-64 (Molecular Probes) was added to a final concentration of 32 μM for 15 min. Cells were washed once with ice cold yeast extract/peptone/dextrose (YPD) and imaged either immediately in YPD or after 40 min at room temperature using a spinning disk confocal microscope, as described in Imaging of actin cables and patches.
Construction of the genetic interaction map
Interactions among genes selected from the imaging-based screen were analyzed using the open-source software Cytoscape (www.cytoscape.org, version 2.7.0; Cline et al., 2007). Genetic interactions identified from previously published studies were loaded onto Cytoscape from the Saccharomyces Genome Database (Cherry et al., 1998; Tong et al., 2004; Schuldiner et al., 2005). The interaction map displays genes affecting endocytosis, exocytosis, or both as circles (Figure 4). The number of interactions that each gene displayed with other genes of the map was quantified directly from Cytoscape. We also computed the number of interactions displayed by the exocyst complex as a whole. To do this, we compiled all the interactions displayed by all exocyst subunits, removed duplicate interactions, and removed interactions with other exocyst genes.
Endocytic and exocytic imaging of deletion mutants
GFP-Sec4 and Abp1-mRFP were used as exocytic and endocytic markers to study trafficking dynamics in vivo. Simultaneous acquisition of exocytic and endocytic vesicles in control cells and mutants expressing these markers were performed using the same microscope system as described in Steady-state imaging and image processing, with the addition of a 100× TIRFM objective (oil, NA 1.46, plan Apo), a motorized TIRFM slider, and an optic fiber–connected laser bench (491, 561 nm; Cobolt). A dual-view module was connected to the microscope for splitting the GFP/mRFP fluorescence and viewing them simultaneously. The module was calibrated using 100-nm beads (Molecular Probes) to align the channels. An additional 1.6× Optovar magnification lens provided sampling at 100 nm/pixel. Filter set 76 HE (excitation 406/493/561 nm, beam splitter 427/503/578 nm, emission 460/525/608 nm; Zeiss) was used in the microscope, and a dichroic 565dcxr and two single band pass filters, 510/30m and 630/75m, were used in the dual-view module for monitoring the GFP and mRFP channels simultaneously, without significant bleedthrough between the channels. Live imaging was done in near-TIRFM mode in all cases. For in vivo imaging of deletion mutants, cells were grown to log phase in selective minimal yeast medium and imaged at room temperature (25°C).
Live-cell imaging of temperature-sensitive mutants
Control and sec6-4 cells were grown to log phase in selective minimal yeast medium at 25°C and shifted to the restrictive temperature (34°C) either immediately or 1 h before imaging at 34°C. An inverted motorized microscope (Nikon Ti) equipped with a 100× objective (oil, NA 1.49, plan Apo), an optic fiber–connected laser bench (491, 561 nm; Cobolt), a perfect focus system, and a temperature-controlled chamber set to 34°C (Ludin chamber; Life Imaging Services, Basel, Switzerland) was used for imaging the cells after the temperature shift. An additional magnification of 1.5× was used in the detection path to have sampling at 107 nm/pixel. Fluorescence was collected by a quad-band dichroic filter (Di01-R405/488/561/635; Semrock) and split by a Dual Cam module (565dcxr dichroic) connected to the microscope and detected by two Evolve EMCCD cameras for simultaneous acquisition of the GFP and mRFP channels. The module was calibrated using 100-nm beads (Molecular Probes) to optimize camera alignment.
Analysis of endocytic frequency
Endocytic dynamics were analyzed from kymographs. The images of the endocytic and exocytic channels were median filtered (filter size 3 × 3), and a region of interest (10 pixels/1 μm) was selected around the cortex. The fluorescence intensity in this region was projected over time to generate the respective kymographs. To analyze the frequency of endocytic events, a line scan (5 pixels wide) of the endocytic kymograph was done along the time axis to monitor the endocytic fluorescence fluctuations over time. These intensity fluctuations were differentiated and smoothened to obtain the individual endocytic peaks over time. A threshold of 50% of the maximum amplitude was set for each sequence of endocytic events, and only the peaks above this threshold were selected. The time interval between the consecutive selected peaks was displayed as a scatter plot with the mean and SD (GraphPad Prism 5; GraphPad Software, San Diego, CA). An unpaired two-tailed test with Welch's correction was used to calculate whether the variance of the distribution of the endocytic intervals of the mutants was significantly different from the control (Figure 2B and Supplemental Figure S7A). This analysis procedure was described previously (Jose et al., 2013). The position of the line scan was chosen adjacent to the center of the exocytic pole, where consecutive endocytic events were observed.
Analysis of exocytic pole size
A fluorescence intensity line scan (5 pixels wide/0.5 μm) around the cortex of the cell was generated from GFP-Sec4 images of fixed cells. This fluorescence distribution along the cortex was fitted with a Gaussian and the full-width at half-maximum calculated (Origin Pro7, Northampton, MA). A nonparametric two-tailed Mann–Whitney test was used to determine whether the pole size of the mutants was significantly different from that of the control (Figure 2C).
Construction of a CEN LEU2 mEos-Sec4 plasmid
A plasmid expressing mEos-Sec4 under the control of the SEC4 promoter was generated using a 600–base pair fragment of the SEC4 promoter and cloned into a BamHI-XbaI–digested pRS315 plasmid, introducing an NdeI site at the 3′ end of the promoter sequence. The SEC4 ORF sequence was then inserted between NdeI-SmaI sites in this plasmid, including 650 base pairs of 3′ regulatory sequence after the SEC4 stop codon. In a final step, mEOS was cloned into the NdeI site, introducing a flexible GAGAGAGA peptide linker between mEOS and the Sec4 protein. The plasmid rescued the growth of a sec4-8 mutant strain at restrictive temperature, consistent with the mEOS-Sec4 fusion being functional (Supplemental Figure S5).
Single-vesicle tracking of mEos-Sec4
Control and mutant cells were transformed with a CEN LEU2 mEOS-SEC4 plasmid. Cells expressing mEos-Sec4 were grown at 25°C to log phase. The cells were imaged at 30°/34°C using the same microscope setup as described in Live-cell imaging of temperature-sensitive mutants. Control cells and sec6-4 mutants were shifted to the restrictive temperature of 34°C for 1 h before imaging at 34°C. Imaging was performed in an oblique illumination mode. mEos-Sec4-containing vesicles were imaged in small-budded cells using a 561-nm laser (Cobolt) with additional continuous photoactivation using a 405-nm laser (Omicron, Dudenhofen, Germany). Laser power was adjusted to optimize single-vesicle imaging. The imaging was performed at similar frequencies to those reported previously for the observation of active transport (Frost et al., 2010; Rossier et al., 2012). The fluorescence was collected on an Evolve EMCCD camera after passing through a combination of dichroic and emission filters (D101-R561 and F39-617 respectively; Chroma, Bellows Falls, VT). Images were acquired in a streaming mode at 10 Hz (100-ms exposure time) using a 200 × 200 pixel region of interest.
Analysis of vesicle trajectories
Two thousand images were generated for each single-vesicle tracking experiment, which were analyzed to extract single-vesicle localization and dynamics. The single vesicles were localized in each image frame and tracked over time using a combination of wavelet segmentation (Izeddin et al., 2012) and simulated annealing algorithms. The software package used to derive quantitative data on protein localization and dynamics was custom written as a plug-in running within the MetaMorph software environment.
Analysis of exocytic vesicle mobility
We observed that all MSDs show a quasilinear dependence at short times, enabling us to calculate an instantaneous diffusion coefficient (D) to characterize mobility. To analyze the global mobility, the software computed the instantaneous diffusion coefficient from the linear regression on the first three points of all trajectories longer than six consecutive frames. The frequency distribution of log D was calculated with a bin width of 0.2 in GraphPad Prism 5 and plotted in Origin Pro7 to generate the histogram of diffusion coefficients and the cumulative probability distributions (Figures 3, B and C, and 6, B and C).
To study the behavior of vesicle trajectories, MSDs for each trajectory greater than six consecutive frames (and <50 frames to avoid clusters) were fitted with a general model MSD = 〈r2〉 = tα + k, where D is the diffusion coefficient, t is the time lag, and α is the scaling exponent (Saxton, 1993, 1995; Sibarita, 2014). The MSD values were imported into Origin Pro7 to calculate the mean values and standard errors corresponding to each time point (Figures 3D and 6D). The trajectories were sorted according to the value of the scaling exponent α into confined, diffusive, and directed fractions and their respective MSDs plotted (Figures 3E and Supplemental Figure S6).
Genetic studies
Exocyst-endocytic mutants for genetic interaction analysis were generated using classical genetics. Endocytic deletion strains of mating phenotype Mat α were crossed with sec6-4 Mat a. The diploids were sporulated in low-nutrient medium (1% KAc, pH 7.0) for 3–5 d, and tetrads were dissected. Progeny were genotyped and selected for genetic analysis. The strains were grown to log phase. We serially diluted 0.1 OD of cells and spotted them on YPD plates to compare their growth at 15°C.
Supplementary Material
Acknowledgments
We thank David Drubin, Charlie Boone, Lionel Minvielle-Sebastia, and Ruth Collins for yeast strains, anti-Rna15 sera, and plasmids. Work in D.M.'s lab is generously funded by FP7 Marie Curie Grant IRG249298, Agence Nationale de la Recherche Grant 2010 JCJC 1210 01, Fondation pour la Recherche Medicale Grant INE20100518678, Agence pour la Recherche sur le Cancer Grant PDF20120605172, and Centre National de la Recherche Scientifique, Université Bordeaux Segalen, and Conseil Régional d'Aquitaine Volet Recherche Grants 20091301015 and 2012 13 01 012. M.J. and S.T. were funded by the Fondation pour la Recherche Medicale, Agence Nationale de la Recherche, and Université Bordeaux Segalen. Work at CNS, IISC was funded by the Indian Institute of Science, Bangalore, and the Department of Biotechnology, India. A. R. was funded by CNRS and Conseil régional d'Aquitaine grant 2011 130 4008.
Abbreviations used:
- FR
fluorescence ratio
- MSD
mean square displacement
- PSF
point spread function
- TIRFM
total internal reflection fluorescence microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1527) on May 6, 2015.
REFERENCES
- Amberg DC. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol Biol Cell. 1998;9:3259–3262. doi: 10.1091/mbc.9.12.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Zahner JE, Mulholland JW, Pringle JR, Botstein D. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol Biol Cell. 1997;8:729–753. doi: 10.1091/mbc.8.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuelos MG, Moreno DE, Olson DK, Nguyen Q, Ricarte F, Aguilera-Sandoval CR, Gharakhanian E. Genomic analysis of severe hypersensitivity to hygromycin B reveals linkage to vacuolar defects and new vacuolar gene functions in Saccharomyces cerevisiae. Curr Genet. 2010;56:121–137. doi: 10.1007/s00294-009-0285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calero M, Chen CZ, Zhu W, Winand N, Havas KA, Gilbert PM, Burd CG, Collins RN. Dual prenylation is required for Rab protein localization and function. Mol Biol Cell. 2003;14:1852–1867. doi: 10.1091/mbc.E02-11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli B, Hurlbut WP, Mauro A. Turnover of transmitter and synaptic vesicles at the frog neuromuscular junction. J Cell Biol. 1973;57:499–524. doi: 10.1083/jcb.57.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Li L, Li J, Liu B, Zhu X, Zheng L, Zhang R, Xu T. SEC-10 and RAB-10 coordinate basolateral recycling of clathrin-independent cargo through endosomal tubules in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2014;111:15432–15437. doi: 10.1073/pnas.1408327111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesneau L, Dupre S, Burdina A, Roger J, Le Panse S, Jacquet M, Cuif MH. Gyp5p and Gyl1p are involved in the control of polarized exocytosis in budding yeast. J Cell Sci. 2004;117:4757–4767. doi: 10.1242/jcs.01349. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan KW, Bretscher A. Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Dev Cell. 2012;23:769–781. doi: 10.1016/j.devcel.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- Echauri-Espinosa RO, Callejas-Negrete OA, Roberson RW, Bartnicki-Garcia S, Mourino-Perez RR. Coronin is a component of the endocytic collar of hyphae of Neurospora crassa and is necessary for normal growth and morphogenesis. PLoS One. 2012;7:e38237. doi: 10.1371/journal.pone.0038237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Folsch H, Pypaert M, Maday S, Pelletier L, Mellman I. The AP-1A and AP-1B clathrin adaptor complexes define biochemically and functionally distinct membrane domains. J Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost NA, Shroff H, Kong H, Betzig E, Blanpied TA. Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron. 2010;67:86–99. doi: 10.1016/j.neuron.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y, Hyams JS. Endocytosis in fission yeast is spatially associated with the actin cytoskeleton during polarised cell growth and cytokinesis. J Cell Sci. 2005;118:4231–4242. doi: 10.1242/jcs.02530. [DOI] [PubMed] [Google Scholar]
- Graziano BR, DuPage AG, Michelot A, Breitsprecher D, Moseley JB, Sagot I, Blanchoin L, Goode BL. Mechanism and cellular function of Bud6 as an actin nucleation-promoting factor. Mol Biol Cell. 2011;22:4016–4028. doi: 10.1091/mbc.E11-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger ED, Kessels MM, Qualmann B. Temporal and spatial coordination of exocytosis and endocytosis. Nat Rev Mol Cell Biol. 2003;4:127–139. doi: 10.1038/nrm1016. [DOI] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S. A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol. 2010;8:e1000282. doi: 10.1371/journal.pbio.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas-Aguilar A, Penalva MA. Endocytic machinery protein SlaB is dispensable for polarity establishment but necessary for polarity maintenance in hyphal tip cells of Aspergillus nidulans. Eukaryot Cell. 2010;9:1504–1518. doi: 10.1128/EC.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE, Reese TS. Evidence for recycling of synaptic vesicle membrane during transmitter release at the frog neuromuscular junction. J Cell Biol. 1973;57:315–344. doi: 10.1083/jcb.57.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Boulanger J, Racine V, Specht CG, Kechkar A, Nair D, Triller A, Choquet D, Dahan M, Sibarita JB. Wavelet analysis for single molecule localization microscopy. Opt Express. 2012;20:2081–2095. doi: 10.1364/OE.20.002081. [DOI] [PubMed] [Google Scholar]
- Jose M, Tollis S, Nair D, Sibarita JB, McCusker D. Robust polarity establishment occurs via an endocytosis-based cortical corralling mechanism. J Cell Biol. 2013;200:407–418. doi: 10.1083/jcb.201206081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Adams AE. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J Cell Biol. 1984;98:922–933. doi: 10.1083/jcb.98.3.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Wabnik K, Martiniere A, Langowski L, Willig K, Naramoto S, Leitner J, Tanaka H, Jakobs S, Robert S, et al. Recycling, clustering, and endocytosis jointly maintain PIN auxin carrier polarity at the plasma membrane. Mol Syst Biol. 2011;7:540. doi: 10.1038/msb.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli M, Galati V, Boudier K, Roberson RW, Philippsen P. Growth-speed-correlated localization of exocyst and polarisome components in growth zones of Ashbya gossypii hyphal tips. J Cell Sci. 2008;121:3878–3889. doi: 10.1242/jcs.033852. [DOI] [PubMed] [Google Scholar]
- Langevin J, Morgan MJ, Sibarita JB, Aresta S, Murthy M, Schwarz T, Camonis J, Bellaiche Y. Drosophila exocyst components Sec5, Sec6, and Sec15 regulate DE-Cadherin trafficking from recycling endosomes to the plasma membrane. Dev Cell. 2005;9:365–376. doi: 10.1016/j.devcel.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberali P, Snijder B, Pelkmans L. A hierarchical map of regulatory genetic interactions in membrane trafficking. Cell. 2014;157:1473–1487. doi: 10.1016/j.cell.2014.04.029. [DOI] [PubMed] [Google Scholar]
- McCusker D, Royou A, Velours C, Kellogg D. Cdk1-dependent control of membrane-trafficking dynamics. Mol Biol Cell. 2012;23:3336–3347. doi: 10.1091/mbc.E11-10-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Sagot I, Manning AL, Xu Y, Eck MJ, Pellman D, Goode BL. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol Biol Cell. 2004;15:896–907. doi: 10.1091/mbc.E03-08-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostov K, Su T, ter Beest M. Polarized epithelial membrane traffic: conservation and plasticity. Nat Cell Biol. 2003;5:287–293. doi: 10.1038/ncb0403-287. [DOI] [PubMed] [Google Scholar]
- Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Prigent M, Dubois T, Raposo G, Derrien V, Tenza D, Rosse C, Camonis J, Chavrier P. ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J Cell Biol. 2003;163:1111–1121. doi: 10.1083/jcb.200305029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riezman H. Endocytosis in yeast: several of the yeast secretory mutants are defective in endocytosis. Cell. 1985;40:1001–1009. doi: 10.1016/0092-8674(85)90360-5. [DOI] [PubMed] [Google Scholar]
- Riquelme M, Bredeweg EL, Callejas-Negrete O, Roberson RW, Ludwig S, Beltran-Aguilar A, Seiler S, Novick P, Freitag M. The Neurospora crassa exocyst complex tethers Spitzenkorper vesicles to the apical plasma membrane during polarized growth. Mol Biol Cell. 2014;25:1312–1326. doi: 10.1091/mbc.E13-06-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos J, Kelly RB. The endocytic machinery in nerve terminals surrounds sites of exocytosis. Curr Biol. 1999;9:1411–1414. doi: 10.1016/s0960-9822(00)80087-1. [DOI] [PubMed] [Google Scholar]
- Rossier O, Octeau V, Sibarita JB, Leduc C, Tessier B, Nair D, Gatterdam V, Destaing O, Albiges-Rizo C, Tampe R, et al. Integrins beta1 and beta3 exhibit distinct dynamic nanoscale organizations inside focal adhesions. Nat Cell Biol. 2012;14:1057–1067. doi: 10.1038/ncb2588. [DOI] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Saxton MJ. Lateral diffusion in an archipelago. Single-particle diffusion. Biophys J. 1993;64:1766–1780. doi: 10.1016/S0006-3495(93)81548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ. Single-particle tracking: effects of corrals. Biophys J. 1995;69:389–398. doi: 10.1016/S0006-3495(95)79911-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott DH, Collins RN, Bretscher A. Secretory vesicle transport velocity in living cells depends on the myosin-V lever arm length. J Cell Biol. 2002;156:35–39. doi: 10.1083/jcb.200110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Shen D, Yuan H, Hutagalung A, Verma A, Kummel D, Wu X, Reinisch K, McNew JA, Novick P. The synaptobrevin homologue Snc2p recruits the exocyst to secretory vesicles by binding to Sec6p. J Cell Biol. 2013;202:509–526. doi: 10.1083/jcb.201211148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibarita JB. Deconvolution microscopy. Adv Biochem Eng Biotechnol. 2005;95:201–243. doi: 10.1007/b102215. [DOI] [PubMed] [Google Scholar]
- Sibarita JB. High-density single-particle tracking: quantifying molecule organization and dynamics at the nanoscale. Histochem Cell Biol. 2014;141:587–595. doi: 10.1007/s00418-014-1214-1. [DOI] [PubMed] [Google Scholar]
- Slaughter BD, Unruh JR, Das A, Smith SE, Rubinstein B, Li R. Non-uniform membrane diffusion enables steady-state cell polarization via vesicular trafficking. Nat Commun. 2013;4:1380. doi: 10.1038/ncomms2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B, Oprins A, Rabouille C, Munro S. The exocyst component Sec5 is present on endocytic vesicles in the oocyte of Drosophila melanogaster. J Cell Biol. 2005;169:953–963. doi: 10.1083/jcb.200411053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Talesh N, Horio T, Araujo-Bazan L, Dou X, Espeso EA, Penalva MA, Osmani SA, Oakley BR. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toi H, Fujimura-Kamada K, Irie K, Takai Y, Todo S, Tanaka K. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesche S, Arnold M, Jansen RP. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr Biol. 2003;13:715–724. doi: 10.1016/s0960-9822(03)00264-1. [DOI] [PubMed] [Google Scholar]
- Whalley T, Terasaki M, Cho MS, Vogel SS. Direct membrane retrieval into large vesicles after exocytosis in sea urchin eggs. J Cell Biol. 1995;131:1183–1192. doi: 10.1083/jcb.131.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac AL, Goldman YE, Holzbaur EL, Ostap EM. Local cytoskeletal and organelle interactions impact molecular-motor-driven early endosomal trafficking. Curr Biol. 2013;23:1173–1180. doi: 10.1016/j.cub.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Orlando K, He B, Xi F, Zhang J, Zajac A, Guo W. Membrane association and functional regulation of Sec3 by phospholipids and Cdc42. J Cell Biol. 2008;180:145–158. doi: 10.1083/jcb.200704128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T. Vesicle trafficking dynamics and visualization of zones of exocytosis and endocytosis in tobacco pollen tubes. J Exp Bot. 2008;59:861–873. doi: 10.1093/jxb/ern007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.